Abstract
Sulfation is an important Phase II conjugation reaction involved in the synthesis and metabolism of steroids in humans. Two different isoforms (2B1a and 2B1b) are encoded by the sulfotransferase (SULT) 2B1 gene utilizing different start sites of transcription resulting in the incorporation of different first exons. SULT2B1a and SULT2B1b are 350 and 365 amino acids in length, respectively, and the last 342 aa are identical. Message for both SULT2B1 isoforms is present in human tissues although SULT2B1b message is generally more abundant. However, to date only SULT2B1b protein has been detected in human tissues or cell lines. SULT2B1b is localized in the cytosol and/or nuclei of human cells. A unique 3′-extension of SULT2B1b is required for nuclear localization in human BeWo placental choriocarcinoma cells. Nuclear localization is stimulated by forskolin treatment in BeWo cells and serine phosphorylation has been identified in the 3′-extension. SULT2B1b is selective for the sulfation of 3β-hydroxysteroids such as dehydroepiandrosterone and pregnenolone, and may also have a role in cholesterol sulfation in human skin. The substrate specificity, nuclear localization, and tissue localization of SULT2B1b suggest a role in regulating the responsiveness of cells to adrenal androgens via their direct inactivation or by preventing their conversion to more potent androgens and estrogens.
Keywords: Sulfotransferase, sulfation, Phase II metabolism, dehydroepiandrosterone
INTRODUCTION
Conjugation with a sulfonate group is a major pathway involved with the synthesis, inactivation and excretion of steroid hormones in human tissues. The family of cytosolic sulfotransferases (SULTs) catalyzes the transfer of the sulfonate group from 3′-phosphoadenosine 5′-phosphosulfate (PAPS) to a wide variety of compounds containing hydroxyl or amine groups to form sulfates or sulfamates, respectively. One of the major SULT catalyzed reactions involves the sulfonation of hydroxysteroids such as dehydroepiandrosterone (DHEA) or estrogens to form sulfate esters (Figure 1). The sulfonate group is charged at physiological pHs rendering the steroid sulfates incapable of binding and activating their receptors (1, 2).
Figure 1.
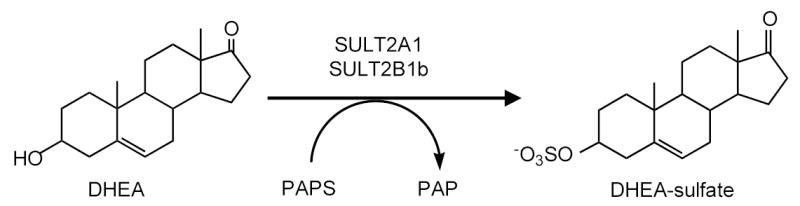
Sulfonation of dehydroepiandrosterone (DHEA) catalyzed by either SULT2A1 or SULT2B1b. The SULTs catalyze the transfer of the sulfonate group from 3′-phosphoadenosine 5′-phosphosulfate to the 3-hydroxyl group of DHEA to form a sulfate ester.
Steroid sulfation in humans and higher primates is unique from that in laboratory animals due primarily to the synthesis and secretion of large amounts of adrenal androgens. High levels of adrenal androgens are secreted by the fetal adrenal during development as well as by the reticular layer of the adult adrenal after adrenarche (3, 4). By far, the most abundant adrenal androgen secreted is DHEA-sulfate (5, 6). DHEA-sulfate is present at micromolar quantities in the plasma of young adults and decreases slowly with age until the 7th or 8th decade of life (7). DHEA is important as a source of precursor for the synthesis of androgens and estrogens in peripheral tissues and after the removal of the sulfate moiety by sulfatase activity can be converted into testosterone in the prostate or estrogens in breast tissue. After menopause in women all estrogen synthesis uses DHEA as a precursor (5) (8). Therefore, the desulfation and conversion of DHEA to either potent androgens or estrogens in hormonally responsive tissues is important in regulation of these tissues. Sulfation of DHEA as well as other androgens and estrogens in these tissues then becomes an important mechanism for controlling the interconversion and activity of these steroids.
CYTOSOLIC SULT FAMILY
The human SULT gene family consists of at least twelve genes encoding thirteen different isoforms (Table 1). Five members of the SULT1 or phenol SULT family (SULT1A1, 1A3, 1E1, 1C2, 1C4) sulfate steroids, however these isoforms primarily conjugate the phenolic groups of estrogens and related compounds. SULT1E1 is responsible for the sulfation of β-estradiol at picomolar and nanomolar concentrations and in contrast to the high affinity for β-estradiol conjugates DHEA at relatively high micromolar concentrations (9, 10). The SULT2 family contains two genes encoding three proteins that are primarily involved with the conjugation of hydroxysteroids. SULT2A1 is expressed in the reticular layer of the adrenal where it is involved with the synthesis and secretion of DHEA-sulfate (3). SULT2A1 is also highly expressed in the liver and has a role in the conjugation of xenobiotics, therapeutic drugs and bile acids (11–15). SULT2A1 has a broad substrate reactivity and is capable of sulfating both 3α- and 3β-hydroxysteroids as well as the 3-phenolic hydroxyl of estrogens and the 17-hydroxyl of testosterone (11, 16). SULT2A1 also conjugates steroid-like therapeutic compounds including budesonide (15) and tibolone (14), and has a protective role in the conjugation of cytotoxic bile acids when the levels increase due to liver disease or cholestasis (17). SULT2A1 has been purified from human liver (11) and the cDNA has been cloned and expressed from human liver (18, 19) and adrenal RNA (20).
Table 1.
Human Cytosolic Sulfotransferases (SULTs). The major SULT families and the isoform names suggested by Blanchard et al. (39) are listed as well as some of the common names used to describe these isoforms.
| SULT1 or Phenol SULT family | ||
| SULT1A1: | Phenol-sulfating PST, P-PST-1, thermal-stable (TS)-PST | |
| SULT1A2: | P-PST-2 | |
| SULT1A3: | Monoamine-sulfating PST, M-PST, thermal-labile (TL)-PST | |
| SULT1B1: | Thyroid hormone-sulfating ST, ST1B2 | |
| SULT1C2: | ||
| SULT1C4: | ||
| SULT1E1: | Estrogen-ST, EST | |
| SULT2 or Hydroxysteroid SULT Family | ||
| SULT2A1: | Dehydroepiandrosterone-ST, DHEA-ST | |
| SULT2B1a: | ||
| SULT2B1b: | ||
| Other SULTs | ||
| SULT4A1: | Brain-specific-ST | |
| SULT6A1: | ||
More recently, a second SULT2 gene was identified during sequence analysis of the human genome (21). The SULT2B1 gene encodes two SULT isoforms, SULT2B1a and SULT2B1b, that were initially identified by cloning of their messages. The two separate messages are generated via the use of different start sites of transcription resulting in the incorporation of different first exons (21). The expressed SULT2B1 isoforms show selectivity for the sulfation of 3β-hydroxysteroids including DHEA and pregnenolone, and demonstrate little activity with 3α-hydroxysteroids and estrogens (22). Expressed SULT2B1a has been reported to possess more pregnenolone sulfation activity than SULT2B1b whereas expressed SULT2B1b has more cholesterol sulfation activity (23). SULT2B1b is also capable of conjugating several drugs and xenobiotics including genistein (22), tibolone (14) and raloxifene (13).
CLONING AND STRUCTURE OF SULT2B1 ISOFORMS
The SULT2B1b gene is located at the same chromosomal loci as the SULT2A1 gene suggesting that even though they are not highly homologous they may have been derived by a gene duplication event (24). The two isoforms of SULT2B1 are generated from two different transcriptional products of the SULT2B1 gene. Her et al. (21) initially identified the SULT2B1 cDNAs by screening a human placental expressed sequence tag database with a highly conserved amino acid sequence motif “RKGxxGDWKNxFT” present in the SULTs. Two related cDNA sequences transcribed from the same gene were identified. These transcripts utilized different transcriptional start sites to incorporate different first exons. SULT2B1a is 350 aa in length whereas SULT2B1b is 365 aa in length with the additional 15 aa being at the amino-terminal end of the protein (Figure 2). The final 344 aa of both sequences are identical since they are derived from the same exons. Compared to the other human SULTs, the SULT2B1 isoforms possess a unique 3′ sequence consisting of 50 aa. This unique 3′-sequence in the SULT2B1 isoforms is enriched in proline residues suggesting a fairly rigid structure. Homology searches of protein databases does not identify a similar sequence with significant homology. Although The SULT2A and 2B genes are at the same chromosomal loci, Figure 2 shows that the amino acid sequences of the enzymes display only 48% homology.
Figure 2.

Sequence alignment of human SULT2B1a, SULT2B1b and SULT2A1. The sequences were aligned using the Clustal program in the MacVector Sequence Analysis software. Gaps denoted as dashes (-) were inserted to optimize the alignments. Amino acids that are different from the sequence of SULT2B1b are shown. Also only the 5′-end of SULT2B1a is shown since the last 344 aa are identical. The asterisk denotes the stop codon in the SULT2A1 sequence. The arrow denotes the start of the 3′ sequence. The underlined sequence represents the phosphorylated peptide identified by mass spectroscopy and the number signs (#) denote the putative phosphorylated serine residues.
EXPRESSION OF SULT2B1 ISOFORMS
SULT2B1a and SULT2B1b message expression occurs in a number of human tissues including prostate, placenta, respiratory system and skin (21, 25–27). Analysis of the expression of the two specific SULT2B1 messages in different human tissues using RT-PCR shows variability especially in SULT2B1a expression (23, 28). The levels of SULT2B1b specific message are generally several-fold greater than those for SULT2B1a and SULT2B1b message is detectable in more tissue types. Immunoblot analysis of the expressed native forms of SULT2B1a and SULT2B1b has shown that these proteins can be easily distinguished by their different molecular masses that result from the differences in their amino terminal sequences (22). In contrast to message levels of the SULT2B1 isoforms, immunoblot analysis of several human tissues detects only the presence of SULT2B1b protein (25). SULT2B1b has been identified by immunoblot in prostate, placenta, breast, lung, skin and platelets (25–27, 29). To date, immunoblot analysis of cytosol prepared from both commercial and autopsy samples of human brain has not detected the presence of either SULT2B1b isoform (unpublished observation). These results suggest that SULT2B1a and SULT2B1b expression in human tissues is controlled at both the transcriptional and translational levels and that the SULT2B1a message may not be efficiently translated in human tissues. For example, the SULT2B1 cDNAs were originally isolated from a placental cDNA library (21) and both messages have been amplified from placental RNA (23); however, only SULT2B1b has been immunologically detected in placental tissue (25). The mechanistic basis for the lack of translation of the SULT2B1a message in human tissues remains to be elucidated. Possibly the SULT2B1a message is translated in specific tissues or at specific developmental times.
The selective reactivity of SULT2B1b with 3β-hydroxysteroids overlaps the broader substrate reactivity of SULT2A1 (11, 22). This is reflected in the ability of both SULTs to efficiently sulfate DHEA and pregnenolone. The initial studies on the tissue expression of the two isoforms suggest they may not be highly expressed in the same tissues. SULT2A1 is highly expressed in liver and adrenal whereas SULT2B1b protein is not detectable in liver cytosol (11, 22, 30). In contrast, SULT2B1b is present in lung, skin, breast and prostate tissues where SULT2A1 expression is not detectable (25, 26). Analyzing the physiological functions of the two SULT2 enzymes in human tissues will need to involve their overlapping but distinct substrate specificities as well as apparently different tissue distributions.
Expression of both of the SULT2B1 cDNAs in Cos-1 cells using the pCR3.1 vector generated low levels of the catalytically competent proteins (21). Subsequent expression studies in E. coli demonstrated that enzymatically active SULT2B1a was readily expressed whereas the native form of expressed SULT2B1b was not catalytically active (15). However, bacterial expression of SULT2B1b with an amino-terminal histidine (His)-tag allowed recovery of active enzyme suggesting that the His-tag stabilized the active form of the enzyme or permitted more efficient folding of the enzyme in E. coli. The presence of the amino-terminal His-tag does alter the kinetics of both expressed SULT2B1a and SULT2B1b for DHEA and pregnenolone (15). SULT2B1b can be expressed in E. coli as a GST-fusion protein (31) or with a His-tag (22) and after affinity purification the active enzyme can be recovered following enzymatic removal of the tag sequences.
The difficulties in expressing SULT2B1b in bacterial and mammalian systems is reflected in the inability to efficiently isolate active SULT2B1b from human tissues or cultured cells. One reason for the apparent delay in the identification of human SULT2B1b is that the enzyme has not been isolated in an active form from human tissues or cells. The loss of DHEA and pregnenolone sulfation activity during the preparation of cytosol apparently hindered the detection and characterization of SULT2B1b. DHEA sulfation activity has been reported to low or absent in cytosol prepared from human breast cancer cell lines (32). Figure 3 shows that even though cytosol prepared from MCF-7 cells does not sulfate DHEA, intact MCF-7 cells readily sulfate DHEA added to the medium. These cells express SULT2B1b and not SULT2A1 (32). SULT2A1 is responsible for the sulfation of DHEA in human adrenal and liver. He et al. (25) have reported a similar loss of SULT2B1b activity during lysis of human LNCaP prostate adenocarcinoma cells. Apparently, disruption of the cell structure in the preparation of lysate or cytosol is sufficient to inactivate SULT2B1b at least with 3β-hydroxysteroids as substrates. He et al. (25) have also reported that intact nuclei isolated from human placenta sulfate DHEA and possess immunoreactive SULT2B1b whereas disruption of the nuclei results in the loss of DHEA sulfation activity. Apparently, protein-protein interactions or the environment of the intact cell or isolated nuclei is sufficient to maintain the enzyme in an active configuration.
Figure 3.
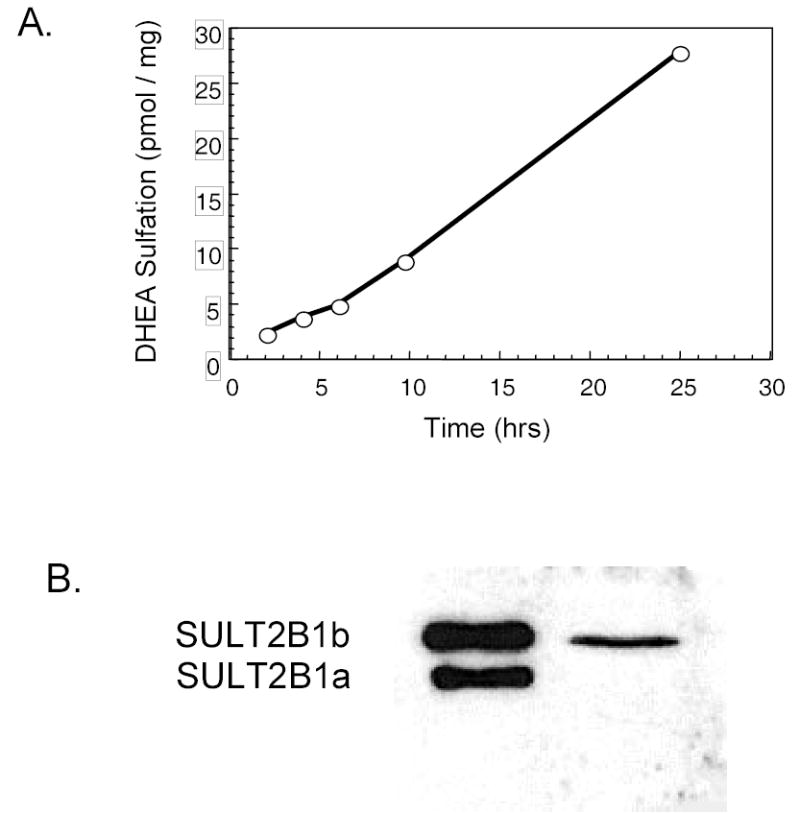
Loss of SULT2B1b activity during cytosol preparation of human MCF-7 breast cancer cells. Panel A shows the formation of 3H-DHEA-sulfate when intact MCF-7 cells are cultured with 2 μM 3H-DHEA. Aliquots of the culture medium were extracted with chloroform to separate DHEA and DHEA-sulfate (9, 11) then DHEA-sulfate synthesis was quantified by scintillation spectroscopy. Panel B shows the immunoblot analysis of cytosol prepared from the MCF-7 cells using a rabbit anti-SULT2B1 polyclonal antibody (22). SULT2B1a and 2B1b are readily distinguishable due to their different molecular masses. Only SULT2B1b is detected in MCF-7 cytosol. No DHEA sulfation activity was observed in the cytosol prepared from the cells in Panel A. Additionally SULT2A1 is not expressed in MCF-7 cells (32).
SUBCELLULAR LOCALIZATION
The subcellular localization of SULT2B1b is unique among the human SULT isoforms. SULT2B1b has been localized to the cytosol and nuclei of both human cells and tissues (25). In human term placenta, SULT2B1b is almost totally localized in the nuclei of synchiotrophoblast cells (25). SULT2B1b activity can be assayed in intact purified placenta nuclei and SULT2B1b protein detected by immunoblotting. In contrast, the localization of SULT2B1b varies in other tissues. SULT2B1b is expressed only in the cytosol of human prostate epithelial cells, prostate benign prostatic hyperplasia, prostate adenocarcinoma and LNCaP prostate adenocarcinoma cells (25). Intact nuclei from LNCaP cells do not sulfate DHEA added to the medium nor do the nuclei contain readily detectable immunoreactive SULT2B1b.
In human T47D and MCF-7 breast cancer cells, SULT2B1b is present in cytosol as well as intact nuclei (figure 4). Low levels of the enzyme are consistently detected in microsomes. Disruption of either T47D or MCF-7 breast cancer cells results in the loss of DHEA sulfation activity. In normal human breast tissue SULT2B1b is detectable in breast epithelial cells (figure 5). The staining is in general heterogeneous with staining in both cytosol and nuclei although the cytosolic staining is apparently dominant. In preliminary studies of human breast cancer tissues, figure 6 shows that SULT2B1b is primarily localized in cytosol of cancer cells. No nuclear staining has been observed in Caucasian breast cancer patients although nuclear localization has been detected in African American breast cancer samples. The levels of immunoreactive SULT2B1b are apparently greater in breast cancer tissue as compared to normal breast tissue which is consistent with the increase in SULT2B1b message reported by Bieche et al. (33) in human breast cancer tissues. More detailed studies of SULT2B1b expressed in Caucasian and African American breast cancer as well as the role of SULT2B1b in the synthesis of estrogens and progression or regulation of breast cancer are underway.
Figure 4.
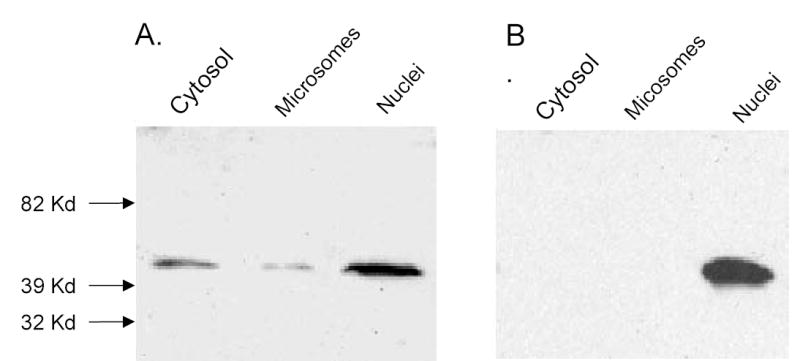
Immunoblot analysis of subcellular fractions of MCF-7 cell. Nuclei were prepared from MCF-7 cells using differential centrifugation and Opti-prep gradient centrifugation (25). Microsomes and cytosol were then isolated from the supernatant of the initial low speed centrifugation to obtain the nuclear pellet. Aliquots (100 μg) of the isolated fractions were resolved by SDS-polyacrylamide gel electrophoresis, transferred to a nitrocellulose membrane and reacted with a rabbit anti-SULT2B1b polyclonal antibody. The secondary was goat anti-rabbit HRP-conjugated IgG (Southern Biotechology) and the immunoconjugates were visualized with the West-Pico system (Pierce).
Figure 5.
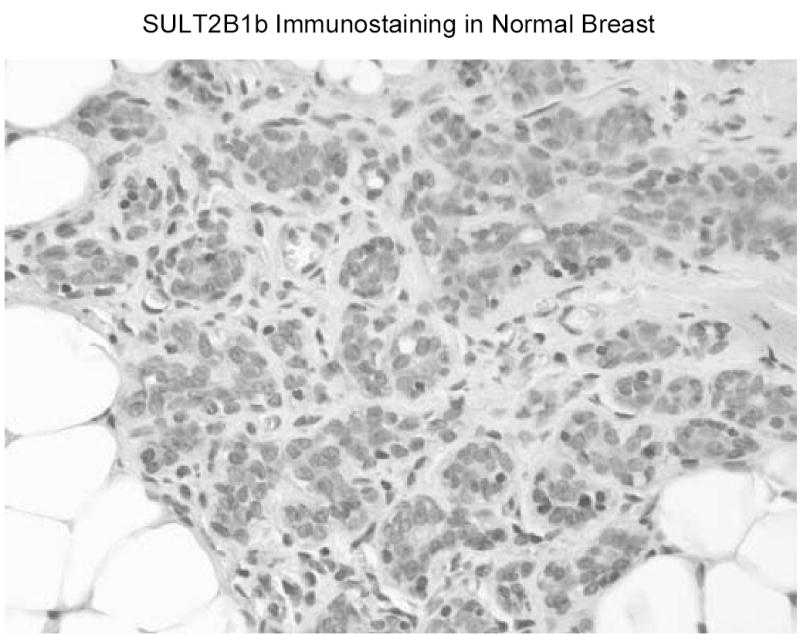
SULT2B1b immunostaining in normal human breast tissue. Samples of normal breast tissue were obtained from the Tissue Procurement Service of the UAB Comprehensive Cancer Center. Sections of paraffin embedded tissue were immunostained with a rabbit anti-SULT2B1b polyclonal antibody following antigen retrieval (25).
Figure 6.
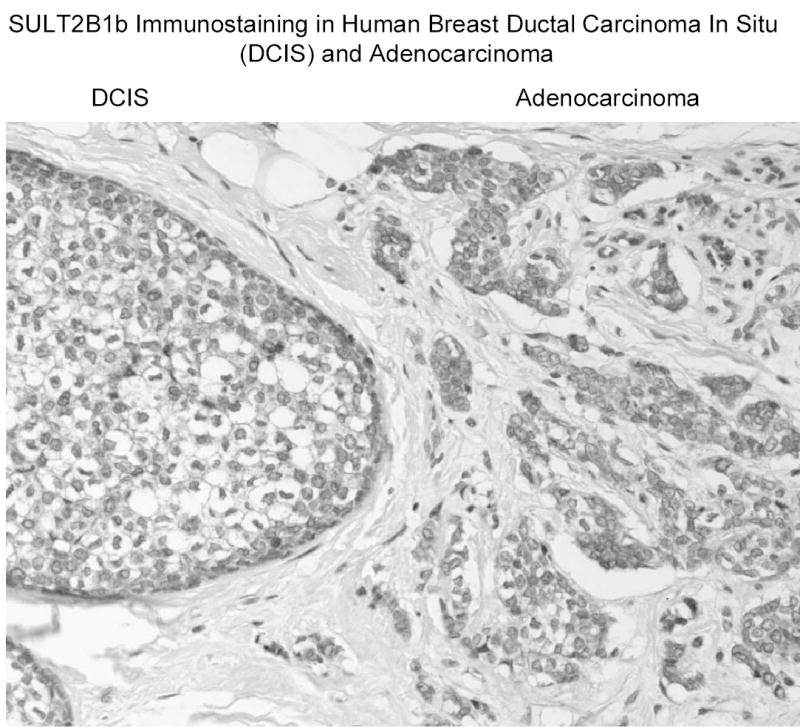
SULT2B1b immunostaining in human breast cancer tissue. Samples of breast cancer tissue were obtained from the Tissue Procurement Service of the UAB Comprehensive Cancer Center. Sections of paraffin embedded tissue were immunostained with a rabbit anti-SULT2B1b polyclonal antibody following antigen retrieval (25).
SULT2B1b has been identified in ciliated cuboidal or columnar cells of terminal bronchae in human lung (26). Immunohistochemical characterization indicates the expression in the terminal bronchial cells is primarily cytosolic. No immunoreactive SULT2B1b was detected in alveolar tissue. Hagashi et al. (27) have reported that SULT2B1b, but not SULT2B1a, is detectable by immunoblotting in human skin. Figure 7 shows staining for SULT2B1b in the outer root sheath of the hair follicle, sebaceous glands and in epidermis of human skin. The presence of SULT2B1b in the epidermis is consistent with its high activity in cholesterol sulfation (23). SULT1A1 has also been localized to the outer root sheath of the hair follicle where it is proposed to have a role in the bioactivation of minoxidil (34).
Figure 7.
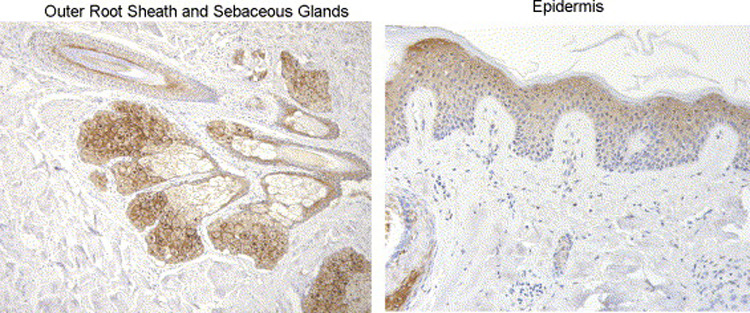
SULT2B1b immunostaining in normal human skin tissue. Samples of normal skin were obtained from the Tissue Procurement Service of the UAB Comprehensive Cancer Center. Sections of paraffin embedded tissue were immunostained with a rabbit anti-SULT2B1b polyclonal antibody following antigen retrieval (25).
The human SULT2B1 isoforms are structurally unique compared to the other human SULTs. Both SULT2B1 isoforms possess a 50 amino acid N-terminal extension that is enriched in serine and proline residues (figure 2). The presence of the 17 proline residues in the sequence of the 3′-extension suggests a fairly rigid structure. Attempts to obtain a crystal structure of SULT2B1b resulted in a structure in which the structure of the amino end or the 3′-terminal extension could not be delineated (35). The 3′- terminal extension has several putative phosphorylation sites as predicted by sequence analysis using the Phosite 1 and Netphos 1 programs. Limited proteolysis and MALDI-TOF mass spectroscopy indicate that a single phosphorylation event is occurring on a 25 aa peptide (figure 2) generated by Glu C digestion from the unique 3′-terminal extension (36). The phosphorylation event may be associated with nuclear localization. Human BeWo choriocarcinoma cells do not express SULT2B1b message or protein. Stable transfection of BeWo cells results in the localization of SULT2B1b in cytosol with lower levels of the enzyme in both the nuclear and microsomal fractions (22). Treatment of BeWo cells with forskolin results in an increase of SULT2B1b in the nuclear fraction (22). Forskolin stimulates cAMP production via an induction of adenyl cyclase activity. Homogenization of the isolated nuclei results in immunoreactive SULT2B1b in the cytosolic fraction suggesting that SULT2B1b is not tightly bound in the nuclei.
COMPARISION OF HUMAN, RAT AND MURINE SULT2B1
The SULT2B1 cDNAs have also been isolated and characterized from rat (37) and mouse (38) tissues. Both rodent genes have similar exon and intron structures to the human gene. In all three species the promoter for the transcription of the 2B1a message generates a 5′-addition to the sequence that represents the second exon in the 2B1b message resulting in the absence of the first intron observed in the SULT2B1b transcript. The promoter and transcriptional start site for the 2B1b message is encoded by an exon separate from the second exon (38). In humans, both the 2B1a and 2B1b messages have been identified in a number of human tissues although the 2B1b message is more widespread and abundant (21, 23, 28). The tissue distribution and abundance of the SULT2B1 messages differs in rats and mice from that of the human isoforms. High levels of SULT2B1b message are detected in skin and intestine from all three species. In rats, little or no SULT2B1a message is reported in the tissues examined except for brain and testis in which SULT2B1a but not SULT2B1b is expressed (37). In mice, SULT2B1a message is highly expressed in brain and spinal cord relative to intestinal expression (38). In comparison, no SULT2B1 message (21, 23, 28) or protein expression (unpublished observation) is detectable in human brain. To date, immunoblot analysis of SULT2B1 protein expression in brain tissue of either rats or mice has not been reported.
The sequences of the human, rat and mouse SULT2B1b proteins show considerable homology in the core regions that correlate with the functional regions of the other human SULTs (figure 8). The region of amino acids 28–310 of the human SULT2B1b is 84.9% and 83.7% identical to the mouse and rat sequences, respectively. The amino-terminal regions of the proteins that have been suggested to be involved with substrate selectivity are considerably less similar (31). The sequence of the first 28 amino acids of human SULT2B1b is only 39% identical (including a 3-aa gap) to that of the rat and mouse SULT2B1b sequences. Also, the amino-terminal sequences of the rat and mouse SULT2B1b proteins are only 60% identical. In contrast to the human SULT2B1b sequences, the rat and mouse SULT2B1a amino-terminal sequences are 47 and 46 residues longer, respectively, than the predicted human SULT2B1a protein.
Figure 8.

Sequence alignment of human, mouse and rat SULT2B1b sequences. The sequences were aligned using the Clustal program in the MacVector Sequence Analysis software. Gaps denoted as dashes (-) were inserted to optimize the alignments. Amino acids that are different from the sequence of human SULT2B1b are shown. The asterisk denotes the stop codon in the rat and mouse SULT2B1b sequences.
The structure of the 3′-extension of human SULT2B1b is also considerably different from that of the rodent enzymes (Figure 8). The carboxy-terminal ends of the rat and mouse SULT2B1 sequences are 22 and 24 amino acids shorter than the human sequence and show considerable sequence differences. The peptide sequence possessing the phosphorylated serine residue in human SULT2B1b is not present in the rodent proteins. The dissimilarity in the unique amino and carboxyl terminal ends of the human, rat and mouse SULT2B1a and 2B1b proteins suggests that their cellular functions may not be highly conserved. Although the skin localization of SULT2B1b in all three species suggests a role in cholesterol sulfation, the rodent SULT2B1 isoforms appear to have a lower activity in DHEA sulfation (37, 38). In contrast, the specificity for the sulfation of 3β-hydroxysteroids indicates a role for DHEA sulfation as well as cholesterol sulfation in humans. SULT2B1b is expressed in placental, prostate and breast tissues in humans but not in rodents. In humans, DHEA sulfation by SULT2B1b may have a role in regulating the conversion of DHEA to androgens or estrogens in these hormonally responsive tissues.
In summary, SULT2B1b appears to be the major active SULT2B1 isoform in human tissues. The enzyme has a unique carboxy-terminal extension that is rich in prolines and serines and the 3′-extension is not highly conserved between rodents and humans. SULT2B1b is expressed in several human tissues including placenta, breast, prostate, skin and lung. It is the first human SULT to show localization between the cytosolic and nuclear cellular compartments in different cell types. Nuclear localization requires the 3′-carboxy extension and is regulated in part by post-translational modification of this region, at least in placental and breast-derived cell lines. SULT2B1b is selective for the sulfation of 3β-hydroxysteroids and also has activity with cholesterol suggesting it may have different enzymatic functions in different human tissues. The enzyme is suggested to have a role in cholesterol sulfation in skin and in DHEA sulfation in other hormonally responsive tissues.
Acknowledgments
This research was supported in part by PHS grant GM38952 to CNF.
References
- 1.Falany CN. Enzymology of human cytosolic sulfotransferases. FASEB Journal. 1997;11:206–216. doi: 10.1096/fasebj.11.4.9068609. [DOI] [PubMed] [Google Scholar]
- 2.Strott CA. Sulfonation and molecular action. Endocr Rev. 2002;23(5):703–32. doi: 10.1210/er.2001-0040. [DOI] [PubMed] [Google Scholar]
- 3.Falany CN, Comer KA, Dooley TP, Glatt H. Human dehydroepiandrosterone sulfotransferase: purification, molecular cloning, and characterization. Annals of the New York Academy of Sciences. 1995;774:59–72. doi: 10.1111/j.1749-6632.1995.tb17372.x. [DOI] [PubMed] [Google Scholar]
- 4.Parker CR, Jr, Jian M, Conley AJ. The localization of DHEA sulfotransferase in steroidogenic and steroid metabolizing tissues of the adult rhesus macaque monkey. Endocr Res. 2000;26(4):517–22. doi: 10.3109/07435800009048564. [DOI] [PubMed] [Google Scholar]
- 5.Bellino FL, Daynes RA, Hornsby PJ, Lavrin DH, Nestler JE, editors. Dehydroepiandrosterone (DHEA) and aging. New York: New York Academy of Sciences; 1995. [Google Scholar]
- 6.Hornsby PJ. Biosynthesis of DHEAS by the human adrenal cortex and its age-related decline. New York Academy of Sciences. 1995;774:29–46. doi: 10.1111/j.1749-6632.1995.tb17370.x. [DOI] [PubMed] [Google Scholar]
- 7.Dharia S, Parker CR., Jr Adrenal androgens and aging. Semin Reprod Med. 2004;22(4):361–8. doi: 10.1055/s-2004-861552. [DOI] [PubMed] [Google Scholar]
- 8.Longcope C. Metabolism of dehydroepiandrosterone. Annals of the New York Academy of Sciences. 1995;774:143–148. doi: 10.1111/j.1749-6632.1995.tb17378.x. [DOI] [PubMed] [Google Scholar]
- 9.Falany CN, Krasnykh V, Falany JL. Bacterial expression and characterization of a cDNA for human liver estrogen sulfotransferase. Journal of Steroid Biochemistry and Molecular Biology. 1995;52:529–539. doi: 10.1016/0960-0760(95)00015-r. [DOI] [PubMed] [Google Scholar]
- 10.Zhang H, Varmalova O, Vargas FM, Falany CN, Leyh TS. Sulfuryl transfer: the catalytic mechanism of human estrogen sulfotransferase. Journal of Biological Chemistry. 1998;273:10888–10892. doi: 10.1074/jbc.273.18.10888. [DOI] [PubMed] [Google Scholar]
- 11.Falany CN, Vazquez ME, Kalb JM. Purification and characterization of human liver dehydroepiandrosterone sulfotransferase. Archives of Biochemistry and Biophysics. 1989;260:641–646. doi: 10.1042/bj2600641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Radominska A, Comer KA, Ziminak P, Falany J, Iscan M, Falany CN. Human liver steroid sulfotransferase sulphates bile acids. Biochemical Journal. 1991;273:597–604. doi: 10.1042/bj2720597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Falany JL, Pilloff DE, Leyh TS, Falany CN. Sulfation of raloxifene and 4-hydroxytamoxifen by human cytosolic sulfotransferases. Drug Metab Dispos. 2006;34(3):361–8. doi: 10.1124/dmd.105.006551. [DOI] [PubMed] [Google Scholar]
- 14.Falany JL, Macrina N, Falany CN. Sulfation of tibolone and tibolone metabolites by expressed human cytosolic sulfotransferases. J Steroid Biochem Mol Biol. 2004;88(4–5):383–91. doi: 10.1016/j.jsbmb.2004.01.005. [DOI] [PubMed] [Google Scholar]
- 15.Meloche CA, Sharma V, Swedmark S, Andersson P, Falany CN. Sulfation of budesonide by human cytosolic sulfotransferase, dehydroepiandrosterone-sulfotransferase (DHEA-ST) Drug Metab Dispos. 2002;30(5):582–5. doi: 10.1124/dmd.30.5.582. [DOI] [PubMed] [Google Scholar]
- 16.Falany CN, Wheeler J, Oh TS, Falany JL. Steroid sulfation by expressed human cytosolic sulfotransferases. Journal of Steroid Biochemistry and Molecular Biology. 1994;48:369–375. doi: 10.1016/0960-0760(94)90077-9. [DOI] [PubMed] [Google Scholar]
- 17.Hofmann AF. Detoxification of lithocholic acid, a toxic bile acid: relevance to drug hepatotoxicity. Drug Metab Rev. 2004;36(3–4):703–22. doi: 10.1081/dmr-200033475. [DOI] [PubMed] [Google Scholar]
- 18.Otterness DM, Wieben ED, Wood TC, Watson RWG, Madden BJ, McCormick DJ, et al. Human liver dehydroepiandrosterone sulfotransferase: molecular cloning and expression of the cDNA. Molecular Pharmacology. 1992;41:865–872. [PubMed] [Google Scholar]
- 19.Comer KA, Falany JL, Falany CN. Cloning and expression of human liver dehydroepiandrosterone sulfotransferase. Biochemical Journal. 1993;289:233–240. doi: 10.1042/bj2890233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Forbes KJ, Hagen M, Glatt H, Hume R, Coughtrie MW. Human fetal adrenal hydroxysteroid sulphotransferase: cDNA cloning, stable expression in V79 cells and functional characterisation of the expressed enzyme. Mol Cell Endocrinol. 1995;112(1):53–60. doi: 10.1016/0303-7207(95)03585-u. [DOI] [PubMed] [Google Scholar]
- 21.Her C, Wood TC, Eichler EE, Mohrenweiser HW, Ramagli LS, Siciliano MJ, Weinshilboum RM. Human hydroxysteroid sulfotransferase SULT2B1: two enzymes encoded by a single chromosome 19 gene. Genomics. 1998;53:284–295. doi: 10.1006/geno.1998.5518. [DOI] [PubMed] [Google Scholar]
- 22.Meloche CA, Falany CN. Expression and characterization of the human 3 beta-hydroxysteroid sulfotransferases (SULT2B1a and SULT2B1b) J Steroid Biochem Mol Biol. 2001;77(4–5):261–9. doi: 10.1016/s0960-0760(01)00064-4. [DOI] [PubMed] [Google Scholar]
- 23.Javitt NB, Lee YC, Shimizu C, Fuda H, Strott CA. Cholesterol and hydroxycholesterol sulfotransferases: identification, distinction from dehydroepiandrosterone sulfotransferase, and differential tissue expression. Endocrinology. 2001;142(7):2978–84. doi: 10.1210/endo.142.7.8244. [DOI] [PubMed] [Google Scholar]
- 24.Otterness D, Mohrenweiser HW, Brandiff BF, Weinshilboum RM. Dehydroepiandrosterone sulfotransferase gene (STD): Localization to human chromosome 19q13. 3 Cytogenet Cell Genet. 1995;70:45–52. doi: 10.1159/000133988. [DOI] [PubMed] [Google Scholar]
- 25.He D, Meloche CA, Dumas NA, Frost AR, Falany CN. Different subcellular localization of sulphotransferase 2B1b in human placenta and prostate. Biochem J. 2004;379(Pt 3):533–40. doi: 10.1042/BJ20031524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.He D, Frost AR, Falany CN. Identification and immunohistochemical localization of sulfotransferase 2B1b (SULT2B1b) in human lung. Biochim Biophys Acta. 2005;1724(1–2):119–26. doi: 10.1016/j.bbagen.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 27.Higashi Y, Fuda H, Yanai H, Lee Y, Fukushige T, Kanzaki T, et al. Expression of cholesterol sulfotransferase (SULT2B1b) in human skin and primary cultures of human epidermal keratinocytes. J Invest Dermatol. 2004;122(5):1207–13. doi: 10.1111/j.0022-202X.2004.22416.x. [DOI] [PubMed] [Google Scholar]
- 28.Geese WJ, Raftogianis RB. Biochemical characterization and tissue distribution of human SULT2B1. Biochem Biophys Res Commun. 2001;288(1):280–9. doi: 10.1006/bbrc.2001.5746. [DOI] [PubMed] [Google Scholar]
- 29.Yanai H, Javitt NB, Higashi Y, Fuda H, Strott CA. Expression of cholesterol sulfotransferase (SULT2B1b) in human platelets. Circulation. 2004;109(1):92–6. doi: 10.1161/01.CIR.0000108925.95658.8D. [DOI] [PubMed] [Google Scholar]
- 30.Comer KA, Falany CN. Immunological characterization of dehydroepiandrosterone sulfotransferase from human liver and adrenal. Mol Pharmacol. 1992;41(4):645–51. [PubMed] [Google Scholar]
- 31.Fuda H, Lee YC, Shimizu C, Javitt NB, Strott CA. Mutational analysis of human hydroxysteroid sulfotransferase SULT2B1 isoforms reveals that exon 1B of the SULT2B1 gene produces cholesterol sulfotransferase, whereas exon 1A yields pregnenolone sulfotransferase. J Biol Chem. 2002;277(39):36161–6. doi: 10.1074/jbc.M207165200. [DOI] [PubMed] [Google Scholar]
- 32.Falany JL, Falany CN. Expression of cytosolic sulfotransferases in normal mammary epithelial cells and breast cancer cell lines. Cancer Res. 1996;56:1551–1555. [PubMed] [Google Scholar]
- 33.Bieche I, Girault I, Urbain E, Tozlu S, Lidereau R. Relationship between intratumoral expression of genes coding for xenobiotic-metabolizing enzymes and benefit from adjuvant tamoxifen in estrogen receptor alpha-positive postmenopausal breast carcinoma. Breast Cancer Res. 2004;6(3):R252–63. doi: 10.1186/bcr784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dooley TP, Walker CJ, Hirshey SJ, Falany CN, Diani AR. Localization of minoxidil sulfotransferase in rat liver and the outer root sheath of anagen pelage and vibrissa follicles. J Invest Dermatol. 1991;96(1):65–70. doi: 10.1111/1523-1747.ep12515856. [DOI] [PubMed] [Google Scholar]
- 35.Lee KA, Fuda H, Lee YC, Negishi M, Strott CA, Pedersen LC. Crystal structure of human cholesterol sulfotransferase (SULT2B1b) in the presence of pregnenolone and 3′-phosphoadenosine 5′-phosphate. Rationale for specificity differences between prototypical SULT2A1 and the SULT2B1 isoforms. J Biol Chem. 2003;278(45):44593–9. doi: 10.1074/jbc.M308312200. [DOI] [PubMed] [Google Scholar]
- 36.He D, Falany CN. Characterization of the proline-serine rich carboxyl terminus in human sulfotransferase 2B1b: immunogenicity, subcellular localization, kinetic properties and phosphorylation. Drug Metab Disp. 2006 doi: 10.1124/dmd.106.011114. (submitted). [DOI] [PubMed] [Google Scholar]
- 37.Kohjitani A, Fuda H, Hanyu O, Strott CA. Cloning, characterization and tissue expression of rat SULT2B1a and SULT2B1b steroid/sterol sulfotransferase isoforms: divergence of the rat SULT2B1 gene structure from orthologous human and mouse genes. Gene. 2006;367:66–73. doi: 10.1016/j.gene.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 38.Shimizu C, Fuda H, Yanai H, Strott CA. Conservation of the hydroxysteroid sulfotransferase SULT2B1 gene structure in the mouse: pre- and postnatal expression, kinetic analysis of isoforms, and comparison with prototypical SULT2A1. Endocrinology. 2003;144(4):1186–93. doi: 10.1210/en.2002-221011. [DOI] [PubMed] [Google Scholar]
- 39.Blanchard RL, Freimuth RR, Buck J, Weinshilboum RM, Coughtrie MW. A proposed nomenclature system for the cytosolic sulfotransferase (SULT) superfamily. Pharmacogenetics. 2004;14(3):199–211. doi: 10.1097/00008571-200403000-00009. [DOI] [PubMed] [Google Scholar]


