Abstract
The ubiquitin proteasome system has been implicated in both cardiac physiology and pathophysiologies. Research in this area has been hampered by the lack of a simple, reproducible method to assess 26S-proteasome peptidase activities. The current report demonstrates that one reason for lack of reproducibility is the myriad of ATP concentrations, many of them excessive, that have been used to stimulate peptidase activity. The chymotrypsin-like or caspase-like activities of 26S-proteasome in cardiac tissue isolates was determined using Suc-LLVY-AMC or Z-LLE-AMC, respectively, over a range of ATP concentrations up to 2 mmol/L. The optimal ATP concentration to assess both peptidase activities was found to be in the low micromolar range (from 6 to 100 μmol/L) depending on the cardiac tissue isolate protein (10 to 90 μg protein) contained in the reaction. Increasing ATP beyond the optimal range was inhibitory. In general, chymotrypsin-like and caspase-like activities could be stimulated 2 to 2.5-fold and 1.4 to 1.8-fold, respectively, over basal (ATP, 0 μmol/L), and could be effectively inhibited with lactacystin or Z-Pro-Nle-Asp-CHO, respectively. Based on these observations, an optimized method is presented for ex vivo determination of cardiac 26S-proteasome peptidase activities which was used to confirm inactivation of this complex by myocardial ischemia and reperfusion.
Keywords: 26S-proteasome, activity, method, ATP, heart tissue, ischemia, reperfusion
Introduction
The ubiquitin-proteasome system (UPS) is the major nonlysosomal system for turnover of intracellular proteins and has been implicated in regulation of numerous cellular processes, including the cell cycle, apoptosis, and protein quality control [1]. Dysfunction of the UPS has been implicated in several cardiac pathophysiologies, including myocardial ischemia [2] and several forms of cardiomyopathy [3]. Given the importance of the UPS in cardiac function and dysfunction (reviewed in [4]), the growing number of investigators in this field have need for an accurate and reproducible method to assess proteasome function.
Currently, there are two methods available for this determination. The less common method utilizes transgenic animals expressing a Green Fluorescent Protein (GFP)-CL1 degron fusion protein whose degradation can be monitored in situ using fluorescence microscopy [5]. The more common method follows cleavage of proteasome-specific peptides linked to a fluorophore, usually amido-4-methylcoumarin (AMC) or β-napthylamide (βNA), and is applied to ex vivo systems containing either a purified proteasome preparation or a tissue isolate. Many studies have simply added the substrate to the tissue isolate and have assumed that the observed activity is representative of 26S-proteasome activity. As pointed out by Kisselev and Goldberg [6, 7] this is inappropriate for a variety of reasons, the most relevant being a requirement for ATP. Other studies have added 2 mmol/L ATP or higher citing one of two studies [8, 9] although these were not determining proteasome activity in tissue isolates but in purified or semi-purified preparations.
In our initial study [2] assessing 26S-proteasome in ischemic myocardium, up to 1 mmol/L ATP was added to the assay mixture, with large variations in results and often lack of proteasome peptidase activity. Because of these variable results, in subsequent studies [10, 11] ATP concentration was decreased to less than 100 μmol/L, which decreased variability but was still not optimal. In this report, we determine 26S-proteasome peptidase activities in cardiac tissue isolates over a range of ATP concentrations and show that the optimal concentration is actually in the low micromolar range depending on the protein concentration and that higher concentrations may be inhibitory.
Methods
Chemicals and Reagents
Suc-LLVY-AMC, Z-LLE-AMC, lactacystin (Lac), and Z-Pro-Nle-Asp-CHO (ZPNAC) were obtained from Biomol Research Lab (Plymouth Meeting, PA). Adenosine 5′ -triphosphate disodium salt (ATP), ultrapure grade, was obtained from Sigma – Aldrich (St. Louis, MO). All other reagents were obtained from reputable sources.
Animals
Male Sprague Dawley rats (250–300 g) were obtained from Taconic Farms (Germantown, NY). All protocols were approved by the Institutional Animal Care and Utilization Committee and were in compliance with the NIH Guide for the Care and Use of Laboratory Animals (revised 1996).
Isolated Heart Preparation
Rats were heparinized (500 U ip.) and anesthetized (pentobarbital sodium, 70 mg/kg ip.). The hearts were rapidly removed and perfused in the Langendorff mode [12] as previously described [13] at a constant pressure equivalent to 90 cm H2O. The perfusate was a modified Krebs-Henseleit buffer containing (mmol/L): NaCl 118, KCl 4.9, MgSO4 1.2, KH2PO4 1.2, NaHCO3 25, glucose 11, CaCl2 2.5, and oxygenated with 95% O2/5% CO2, at 37°C. Systolic pressure was monitored by means of a fluid-filled latex balloon (0.1 ml) connected to a pressure transducer that was inserted into the left ventricle and expanded to a physiologic end diastolic pressure of 5 mm Hg. Hearts were excluded from the study if they developed persistent arrhythmias, had systolic pressure lower than 70 mm Hg, or had a heart rate below 220 beats/minute during the equilibration period.
Preparation of tissue isolate and 26S Proteasome activity
Discussed in detail at end of Results and Discussion section.
Results and Discussion
Optimal conditions for determination of cardiac 26S-proteasome activity in tissue isolates
The 26S proteasome is a macromolecular structure consisting of the 20S proteasome, containing the catalytic sites for proteolysis, which is capped on one or both ends by 19S-regulatory particle(s). The general function of the regulatory component is to recognize substrates; remove ubiquitin-like proteins from substrates; and then present the substrate to the 20S proteasome in such a manner so to gain access to the catalytic chamber. The binding of the regulatory component usually activates the 20S proteasome resulting in a change in configuration of the NH2-termini of the α-subunits opening up the access pore [1, 14]. These functions require hydrolysis of ATP by constituent ATPase subunits [15].
Determination of 26S proteasome peptidase activities with fluorogenic peptide substrates absolutely requires addition of ATP in order to assess function of the 19S-regulatory particle. Chymotrypsin-like and caspase-like activities were assessed by following degradation of the fluorogenic substrates, Suc-LLVY-AMC and Z-LLE-AMC, respectively. Figure 1A illustrates the effect of increasing concentrations of ATP on cardiac tissue isolate 26S-proteasome peptidase activities. When 30 μg tissue isolate protein was included in the assay, the optimal ATP concentration was 28 μmol/L for the chymotrypsin-like activity and 14 μmol/L for the caspase-like activity and higher concentrations were inhibitory. In general, a 2 to 2.5-fold activation of chymotrypsin-like activity over basal (0 μmol/L ATP) was observed while stimulation of caspase-like activity by ATP is much lower (1.4 to 1.8-fold increase) (Figure 1A inset). The optimal concentration of ATP was dependent on the amount of isolate protein included in the reaction mixture (Figure 1B). Stimulation of chymotrypsin-like activity was observed with as little as 10 μg protein where maximal stimulation was at 6 μmol/L ATP. Addition of up to 90 μg protein increased basal activity but also increased the optimal ATP concentration accordingly (Figure 1B). Addition of 2 mmol/L ATP to the 30 and 90 μg assays was quite inhibitory resulting in activities below basal. Heat-inactivated protein had no basal or ATP-stimulated peptidase activity (not shown). Similar protein effects were observed for the caspase-like activity (Figure 1B). These observations illustrate the importance of determining optimal assay conditions for 26S-proteasome determination whether the reaction mixture contains purified proteasome or simple tissue isolate protein to avoid under-estimation.
Figure 1. Optimal conditions for determination of cardiac 26S-proteasome activity. Panel A: The effect of ATP concentration.
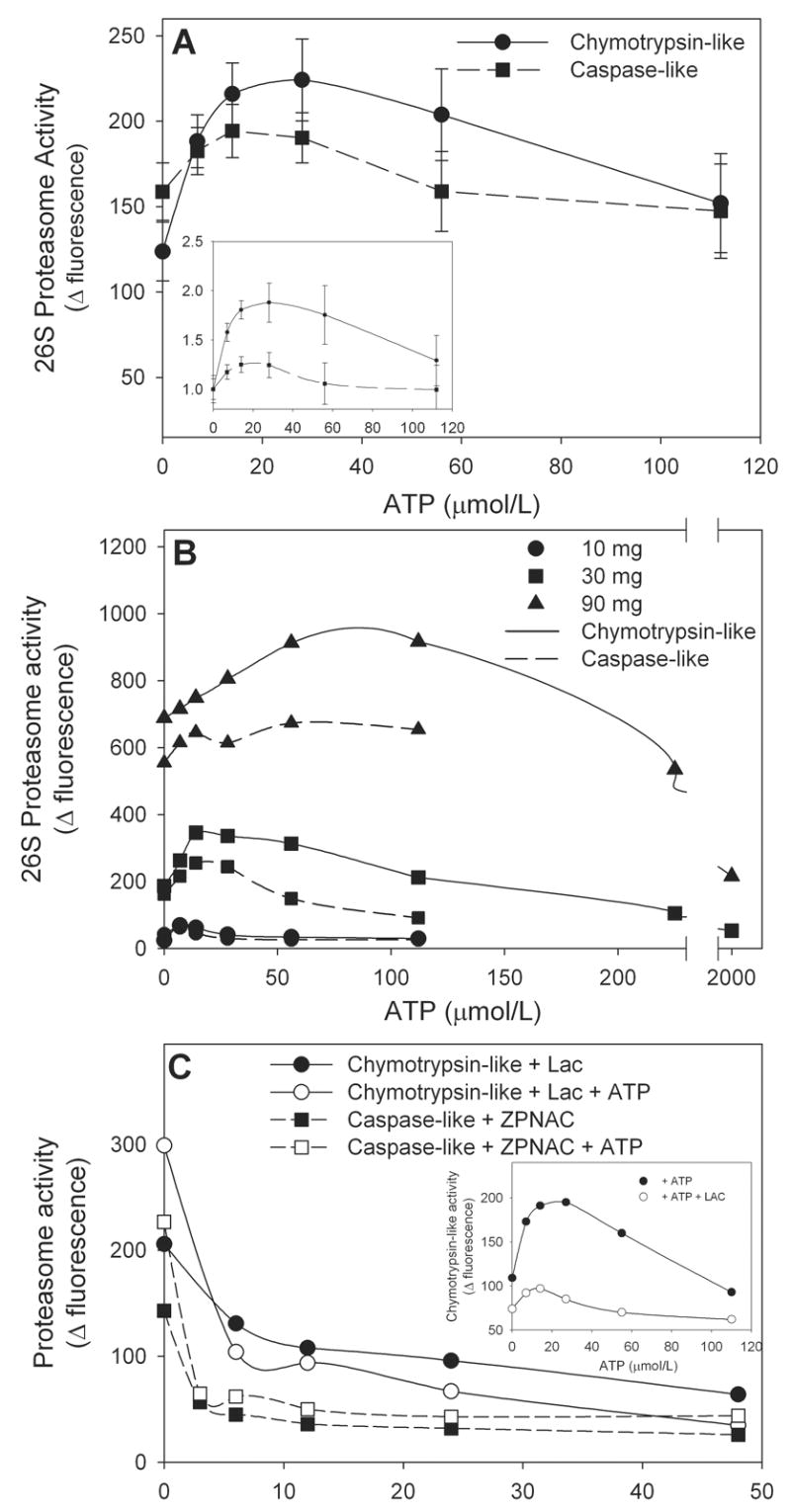
Aliquots (30 μg protein) of rat heart tissue cytosol was assayed for chymotrypsin-like and caspase-like activities in the presence of increasing concentrations of ATP. The inset shows the data normalized to the 0 μmol/L ATP or basal activity to determine magnitude of activation. The values represent the mean ± SEM of 5 to 7 determinations. Panel B: The effect of increasing protein concentrations. Aliquots of rat heart tissue cytosol containing up to 90 μg protein were assayed for chymotrypsin-like and caspase-like activities in the presence of increasing concentrations of ATP. The values represent the mean of 2 to 3 determinations. Panel C: The effect of proteasome inhibitors. Aliquots (30 μg protein) of rat heart tissue cytosol was assayed for chymotrypsin-like and caspase-like activities at the optimal ATP concentration of 28 μmol/L and in the presence of increasing concentrations of the proteasome inhibitors, lactacystin (Lac) and Z-Pro-Nle-Asp-CHO (ZPNAC). The inset shows the effect of increasing ATP concentrations on chymotrypsin-like activity in the absence and presence of Lac, 10 μmol/L. The values represent the mean of 2 to 3 determinations.
Used under proper conditions many of the fluorogenic substrates have proven to be highly specific with Suc-LLVY-AMC reported to be 90–95% specific for the chymotrypsin-like activity [7]. Nonetheless, several studies [7, 8, 16] have suggested the use of inhibitors of proteasome peptidases to enhance specificity. Basal and ATP-stimulated chymotrypsin-like and caspase-like activities were inhibited by inclusion of Lac or ZPNAC, respectively (Figure 1C). The inhibition curves for ATP-stimulated chymotrypsin-like and caspase-like activity showed 70% and 78% inhibition by 12 μmol/L Lac and ZPNAC, respectively, when performed at optimal ATP concentrations (28 μmol/L) (Figure 1C inset). Increasing the concentration of the inhibitors to 48 μmol/L yielded little additional inhibition (88% and 80% for Lac and ZPNAC, respectively (Figure 1C)) suggesting that the inhibitors and substrates are not completely specific. Similar profiles were obtained for the non-ATP stimulated peptidase activities with these curves virtually superimposable at the higher inhibitor concentrations. Incubation with ZPNAC had no effect on chymotrypsin-like activity and Lac had no discernable effect on caspase-like activity (data not shown), the latter being of interest since initial studies [17] suggested that all three peptidase activities were targeted. However, more recent studies indicate that Lac is almost 50-fold more specific for the chymotrypsin-like activity [6].
The effect of myocardial ischemia and reperfusion on 26S-proteasome activity – revisited
We [2] and others [18] have previously shown that myocardial ischemia and reperfusion can lead to inactivation of the proteasome. In retrospect, the analysis of proteasome activity in these studies was not performed under optimal conditions, at least as suggested by the results of this study. To reexamine this, isolated rat hearts were subjected to 30 min normothermic global ischemia followed by 60 min aerobic reperfusion and then freeze-clamped and frozen at −80°. The preparation of the tissue isolate and conditions for the assay are described in detail in the next section. Ischemia and reperfusion resulted in an overall 30–35% decrease in chymotrypsin-like activity consistent with our previous results (Figure 2, top left) [2]. These results were reproducible and significant (P<0.05, ANOVA) even with N=3. Lesser effects (20% decrease) were observed for the caspase-like activity (Figure 2, bottom left) suggesting an overall inhibition of 26S proteasome peptidase activities. Normalizing the data to basal activity resulted in similar activation curves suggesting less active proteasome that can still be maximally stimulated (Figure 2, top and bottom right). This result is suggestive of damage to the 19S-regulatory component which would be consistent with previous studies demonstrating that 26S proteasome is more sensitive than 20S-proteasome to oxidative inactivation [8] and a recent study demonstrating selective oxidative injury to the Rpt5 ATPase subunit [19].
Figure 2. Inhibition of 26S-Proteasome peptidase activity by ischemia and reperfusion.
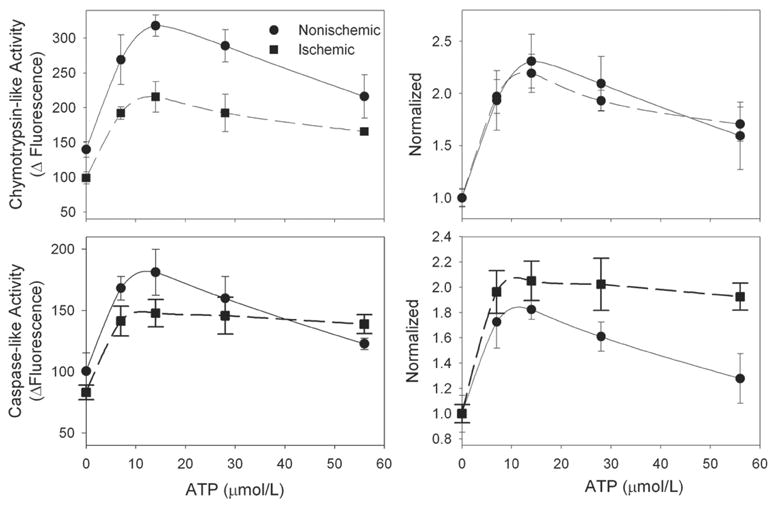
Isolated rat hearts were subjected to 30 min normothermic global ischemia followed by 60 min reperfusion. Hearts were harvested and analyzed for chymotrypsin-like and caspase-like activities. The values represent the mean ± SEM of 3 separate experiments and are expressed as raw data (left panels) and normalized to the 0 μmol/L ATP point (right panels).
Optimized method for preparation of tissue isolate and assay conditions
The suggested protocols are modifications of the method reported by Reinheckel et al [8] and are scaled for a macro-assay with a final reaction mixture volume of 240 μl which can be scaled down to a micro-assay. Frozen heart tissue is homogenized at low power settings (14,000 RPM; Tissumizer, Tekmar) in 10 volumes Hepes buffer (50 mmol/L) containing: KCl 20 mmol/L, MgCl2 5 mmol/L, DTT 1 mmol/L, pH 7.5, and then centrifuged at 10,000xg for 30 min, all at 4°C. The supernatant is immediately assayed and adjusted for protein content and used for determination of peptidase activities. Studies [8, 16] have suggested the use of 10–20% glycerol or 0.25 mmol/L sucrose presumably to stabilize the complex. We find that if the supernatant is used immediately, addition of these makes little difference, but each investigator should make their own determination. Supernatant should not be frozen and then used as this leads to loss of ATP-dependent activity. To assay for peptidase activity, a 20 μl aliquot of the supernatant is added to 200 μl of same Hepes buffer used for isolation (above). Even though activity was detected with as little as 10 μg, a minimum of 30 μg heart tissue isolate protein is suggested so that differences in peptidase activities can be reproducibly observed. Peptidase activities should be determined over a series of four sequential dilutions of ATP so that the maximal activation is obtained which will depend on the particular sample and protein content. For 30 μg protein, adding sufficient concentrated ATP in 10 μl to yield final concentrations of 0, 7, 14, 28, and 56 μmol/L is suggested. In a separate series of samples, including the 0 μmol/L and two other ATP concentrations around the expected optimal, should be preincubated with a peptidase inhibitor to confirm specificity. For the 30 μg example above, chymotrypsin-like activity is inhibited with LAC, 10–15 μmol/L (final), and caspase-like activity with ZPNAC, 6–10 μmol/L (final). Based on the inhibition curves (Figure 1C), addition of higher concentrations of proteasome inhibitors offers little additional benefit.
The reaction is initiated by the addition of 10 μl concentrated substrate bringing the final volume of the reaction mixture to 240 μl. Chymotrypsin-like and caspase-like activities are assessed with the substrates, Suc-LLVY-AMC or L-ZZE-AMC, at final concentrations of 18 and 45 μmol/L, respectively. Additional substrates are available and Kisselev and Goldberg [7] suggest the substrate, Ac-nLPnLD-AMC, as a possible better choice for caspase-like activity, and Ac-RLR-AMC for trypsin-like activity. We do not routinely determine trypsin-like activity because of high background nonproteasome-mediated cleavage of substrates. The reaction is run at 37°C for 30 min and then quenched by adding 300 μl ice-cold ethanol followed in 10 min by 1 ml H2O. In our experiments, cleavage of substrates was linear over this time period. The samples are read at excitation wavelength, 380 nm, and emission wavelength, 440 nm. ATP-dependent peptidase activities, indicative of 26S-proteasome activity, can then most simply be calculated as the difference in fluorescence between the maximal activity and activity in the presence of the inhibitor. If specific activity is required, the reaction can be run against AMC standards. For some experiments it may be advantageous to present the entire ATP stimulation curve to illustrate shifts in utilization efficiency which may be indicative of ATPase subunit damage.
Acknowledgments
These studies were supported by NIH R01-HL68936 and American Heart Association Heritage Affiliate Grant 0455856T. Saul R. Powell is the Principal Investigator of these two grants.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errorsmaybe discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Glickman MH, Ciechanover A. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol Rev. 2002;82:373–428. doi: 10.1152/physrev.00027.2001. [DOI] [PubMed] [Google Scholar]
- 2.Powell SR, Wang P, Katzeff HL, Shringarpure R, Teoh C, Khaliulin I, et al. Oxidized and ubiquitinated proteins may predict recovery of postischemic cardiac function. Essential role of the proteasome. Antioxid Redox Signal. 2005;7:538–5. doi: 10.1089/ars.2005.7.538. [DOI] [PubMed] [Google Scholar]
- 3.Liu J, Chen Q, Huang W, Horak KM, Zheng H, Mestril R, et al. Impairment of the ubiquitin-proteasome system in desminopathy mouse hearts. FASEB J. 2006;20:362–4. doi: 10.1096/fj.05-4869fje. [DOI] [PubMed] [Google Scholar]
- 4.Powell SR. The ubiquitin proteasome system in cardiac physiology and pathology. Am J Physiol Heart Circ Physiol. 2006;291:1–19. doi: 10.1152/ajpheart.00062.2006. [DOI] [PubMed] [Google Scholar]
- 5.Dong X, Liu J, Zheng H, Glasford JW, Huang W, Chen QH, et al. In situ dynamically monitoring the proteolytic function of the ubiquitin-proteasome system in cultured cardiac myocytes. Am J Physiol Heart Circ Physiol. 2004;287:H1417–H1425. doi: 10.1152/ajpheart.01233.2003. [DOI] [PubMed] [Google Scholar]
- 6.Kisselev AF, Goldberg AL. Proteasome inhibitors: from research tools to drug candidates. Chem Biol. 2001;8:739–58. doi: 10.1016/s1074-5521(01)00056-4. [DOI] [PubMed] [Google Scholar]
- 7.Kisselev AF, Goldberg AL. Monitoring activity and inhibition of 26S proteasomes with fluorogenic peptide substrates. Methods Enzymol. 2005;398:364–78. doi: 10.1016/S0076-6879(05)98030-0. [DOI] [PubMed] [Google Scholar]
- 8.Reinheckel T, Sitte N, Ullrich O, Kuckelkorn U, Davies KJ, Grune T. Comparative resistance of the 20S and 26S proteasome to oxidative stress. Biochem J. 1998;335:637–42. doi: 10.1042/bj3350637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.French SW, Mayer RJ, Bardag-Gorce F, Ingelman-Sundberg M, Rouach H, Neve AE, et al. The ubiquitin-proteasome 26s pathway in liver cell protein turnover: effect of ethanol and drugs. Alcohol Clin Exp Res. 2001;25:225S–9S. doi: 10.1097/00000374-200105051-00036. [DOI] [PubMed] [Google Scholar]
- 10.Divald A, Powell SR. Proteasome mediates removal of proteins oxidized during myocardial ischemia. Free Radic Biol Med. 2006;40:156–64. doi: 10.1016/j.freeradbiomed.2005.09.022. [DOI] [PubMed] [Google Scholar]
- 11.Das S, Powell SR, Wang P, Divald A, Nesaretnam K, Tosaki A, et al. Cardioprotection with palm tocotrienol: antioxidant activity of tocotrienol is linked with its ability to stabilize proteasomes. Am J Physiol Heart Circ Physiol. 2005;289:361–7. doi: 10.1152/ajpheart.01285.2004. [DOI] [PubMed] [Google Scholar]
- 12.Langendorff O. Untersuchengen am uberlebenden Saugetierherzen. Pflugers Arch ges Physiol. 1895;61:291–332. [Google Scholar]
- 13.Powell SR, Hall D, Aiuto L, Wapnir RA, Teichberg S, Tortolani AJ. Zinc improves postischemic recovery of the isolated rat heart through inhibition of oxidative stress. Am J Physiol Heart Circ Physiol. 1994;266:H2497–H2507. doi: 10.1152/ajpheart.1994.266.6.H2497. [DOI] [PubMed] [Google Scholar]
- 14.DeMartino GN. Purification of PA700, the 19S regulatory complex of the 26S proteasome. Methods Enzymol. 2005;398:295–306. doi: 10.1016/S0076-6879(05)98024-5. [DOI] [PubMed] [Google Scholar]
- 15.Benaroudj N, Zwickl P, Seemuller E, Baumeister W, Goldberg AL. ATP hydrolysis by the proteasome regulatory complex PAN serves multiple functions in protein degradation. [see comment] Mol Cell. 2003;11:69–78. doi: 10.1016/s1097-2765(02)00775-x. [DOI] [PubMed] [Google Scholar]
- 16.Rodgers KJ, Dean RT. Assessment of proteasome activity in cell lysates and tissue homogenates using peptide substrates. Int J Biochem Cell Biol. 2003;35:716–27. doi: 10.1016/s1357-2725(02)00391-6. [DOI] [PubMed] [Google Scholar]
- 17.Fenteany G, Standaert RF, Lane WS, Choi S, Corey EJ, Schreiber SL. Inhibition of proteasome activities and subunit-specific amino-terminal threonine modification by lactacystin. Science. 1995;268:726–31. doi: 10.1126/science.7732382. [DOI] [PubMed] [Google Scholar]
- 18.Bulteau AL, Lundberg KC, Humphries KM, Sadek HA, Szweda PA, Friguet B, et al. Oxidative modification and inactivation of the proteasome during coronary occlusion/reperfusion. J Biol Chem. 2001;276:30057–63. doi: 10.1074/jbc.M100142200. [DOI] [PubMed] [Google Scholar]
- 19.Ishii T, Sakurai T, Usami H, Uchida K. Oxidative modification of proteasome: identification of an oxidation-sensitive subunit in 26 s proteasome. Biochemistry. 2005;44:13893–901. doi: 10.1021/bi051336u. [DOI] [PubMed] [Google Scholar]


