Abstract
Chromatin remodeling complexes (remodelers) are large, multisubunit macromolecular assemblies that use ATP hydrolysis to alter the structure and positioning of nucleosomes. The mechanisms proposed for remodeler action on nucleosomes are diverse, and require structural evaluation and insights. Previous reconstructions of remodelers using electron microscopy revealed interesting features, but also significant discrepancies, prompting new approaches. Here, we use the orthogonal tilt reconstruction method, which is well suited for heterogeneous samples, to provide a reconstruction of the yeast RSC (remodel the structure of chromatin) complex. Two interesting features are revealed: first, we observe a deep central cavity within RSC, displaying a remarkable surface complementarity for the nucleosome. Second, we are able to visualize two distinct RSC conformers, revealing a major conformational change in a large protein “arm,” which may shift to further envelop the nucleosome. We present a model of the RSC-nucleosome complex that rationalizes the single molecule results obtained by using optical tweezers and also discuss the mechanistic implications of our structures.
Keywords: chromatin remodeling, single particle electron microscopy
Cells have evolved a variety of mechanisms to overcome the physical barrier imposed by the packaging of DNA into chromatin. Chromatin structure affects all DNA transactions and a large collection of cellular factors are involved in the process of regulating it and its basic repeating unit, the nucleosome. Noncovalent modification of nucleosomes is performed by the ATP-dependent chromatin remodeling complexes, large macromolecular assemblies (termed “remodelers”) that often surpass 1 MDa in molecular mass (1, 2). Although the different activities catalyzed by these complexes have been extensively characterized, the actual mechanism by which they are accomplished is still a source of controversy. Critical to elucidating this mechanism will be a detailed structural analysis of the remodeling complexes and their interactions with the nucleosomal substrate.
Cryo-electron microscopy (cryo-EM) has emerged as a powerful technique for the structural characterization of large macromolecular assemblies that are either refractory to crystallization or difficult to overexpress and/or purify in the quantities required for x-ray studies. Furthermore, cryo-EM has the potential to describe conformational flexibility, which is likely to play an important role in the activity of large complexes, with a level of detail not amenable to other techniques (3).
A critical step in obtaining a 3D reconstruction by EM is the generation of the initial model. This step is particularly challenging in the absence of symmetry and when heterogeneity (biochemical and/or conformational) is present in the sample. The most robust approach to obtain an initial model under these circumstances is arguably the random conical tilt method (RCT) (4). In this technique, images are collected first with the sample tilted to a high angle and then untilted. The individual untilted molecular images are aligned and classified into distinct views and a 3D reconstruction of each view is obtained by using the corresponding tilted images. Because the initial sorting of views can separate different species present in the sample, this reconstruction method does not rely on the assumption of homogeneity that underlies the angular reconstitution method (5).
One of the difficulties faced with the RCT method arises from the limitation in the extent to which the sample can be tilted in the microscope. This restriction prevents certain views from being obtained and results in an incomplete sampling of the particle in Fourier space (“missing cone”) that affects the final reconstruction. A further complication arises from the fact that initial reconstructions are typically obtained from negatively stained samples (where contrast is higher). One of the most severe potential artifacts introduced by the staining procedure is sample flattening resulting from drying the sample after staining.
We recently proposed a method, the orthogonal tilt reconstruction (OTR) method (6), that eliminates the missing cone and its resulting artifacts and has as its only requirement that the sample adopt a large number of orientations on the grid. Data are collected at −45° and +45° such that each half of the data constitutes a set of views orthogonal to the other half.
To understand how remodelers function, EM reconstructions of three remodelers have been performed (7–9). All three complexes belong to the SWI/SNF family. The first two structures solved were those of the yeast RSC (8) and SWI/SNF (7) complexes. These were followed by our own publication of the structure of the human PBAF complex, the homolog of yeast RSC (9). These reconstructions were obtained from negatively stained samples using either an angular reconstitution approach (7) or the RCT method (8, 9). Despite the significant degree of conservation among these complexes (10–12), the reconstructions were remarkably different. In particular, the yeast complexes, which share the highest level of similarity, resulted in seemingly unrelated structures. The yeast RSC (8) and the human PBAF (9) complexes, though different, have an overall similar general architecture. However, both reconstructions were affected by the missing cone problem (8, 9). It was also evident from these two reconstructions that these complexes were flexible and potentially biochemically heterogeneous.
Given the discrepancies observed among the reconstructions and their potential heterogeneity, it is unclear at this point to which degree they represent the true underlying structure, which is necessary to fully understand nucleosome interaction and remodeling. Therefore, we set out to obtain a reconstruction of the yeast RSC complex using the OTR method, which should be particularly well suited to address this type of sample. To our knowledge, this work constitutes the first application of the method. Our reconstruction suggests that both the original RSC structure (8) and our own PBAF reconstruction (9) were affected by significant sample flattening in addition to the anisotropy of the merged data. Whereas the former structure of RSC suggests that the nucleosome resides between two “pincer-like” lobes, our new structure reveals instead a large central cavity with a remarkable surface complementarity to the nucleosome, suggesting that the complex envelops its substrate. Furthermore, we identify conformational variability in the complex reminiscent of that observed with the human PBAF remodeler (9). We present the structures of two conformers and suggest how these conformers may contribute to nucleosome interaction and remodeling. Finally, we also examine the remodeling of mononucleosomes by RSC in single molecule experiments under the optical tweezers and discuss our results in the context of our structural model.
Results
Negatively Stained RSC Prepared by the Sandwich Method Is Amenable to the OTR Method.
The main prerequisite for the application of the OTR method (6) is that the sample must adopt a large number of orientations on the grid. A general concern when obtaining initial reconstructions with negatively stained samples is that the staining procedure often results in a significant degree of flattening, complicating the interpretation of the final structure. We sought to address both the orientation requirement and the flattening concern before attempting a reconstruction. Our rationale is as follows: if we perform reference-free alignment and classification on sets of projections obtained at 0° and 45°, the presence of a number of characteristic views in both sets would indicate that the sample is neither adopting a small number of preferred orientations, nor significantly flattened. This conclusion is justified because, within a class representing a given view, the actual particles adopt random rotations around an axis perpendicular to the projection plane. This axis is that of the electron beam. However, although the projection plane is parallel to the support in an untilted sample, there is no single physical plane that corresponds to the projection plane for a tilted sample. Thus, each projection image within a given class in the tilted sample corresponds to a particle adopting a different orientation on the grid, and a number of similar views in both the tilted and untilted images indicate the presence of multiple orientations in the sample. A similar reasoning applies to the presence of flattening. Assuming that flattening manifests itself mainly along a direction normal to the support, particles that only differ in their in-plane rotation will be classified together in an untilted sample, regardless of their flattening. However, because images in a class from a tilted sample come from particles adopting different orientations on the grid, they will no longer be similar once affected by flattening. The presence of a number of similar views from both tilted and untilted data sets indicates that a sample is not severely flattened provided that the images in the classes from the tilted data show random distributions of in-plane rotations.
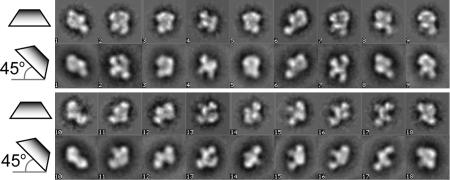
To address the issues outlined above, we performed reference-free alignment and classification for two data sets collected from negatively stained RSC: one consisting of 12,241 particles collected at 0° and the other consisting of 39,960 particles collected at ±45° [see supporting information (SI) Fig. 5A]. A comparison of class averages obtained with each sample indicates that (i) the sample is not significantly flattened and (ii) it is amenable to reconstruction by the OTR method (Fig. 1).
Fig. 1.
Analysis of RSC's amenability to the orthogonal tilt reconstruction method. Comparison of class averages obtained through reference-free alignment and classification of images collected at either 0° (horizontal rhomboid) or ±45° tilt (tilted rhomboid) showing the presence of similar characteristic views in both samples.
OTR of RSC.
The RSC samples used lacked ATP, DNA, divalent cations, and other cofactors that might influence conformational states. We obtained a data set composed of 19,980 pairs of RSC projections collected at +45° and −45° (6) (SI Fig. 5A). In a departure from our original description of the methodology (6), we combined the particles obtained at the two tilts into a single data set. The reasoning was that, whereas a +45° projection corresponds to the +90° tilted mate of a −45° projection, this −45° projection is the −90° tilted mate of the +45° projection. Although the projections coming from the second exposure are less desirable, the double exposure is a relatively small concern when working with negatively stained samples and aiming for an initial, low-resolution model. On the other hand, there is a significant advantage to doubling the size of the data set.
We performed reference-free alignment and classification for this combined data set, generating 75 final classes. For each class we plotted the distribution of in-plane rotation angles for the projections comprising it. Whenever a plot showed a nonrandom distribution, manifested as the presence of significant wedges where no particles where present (see SI Fig. 5B), it was taken as an indication of the presence of flattened particles and the corresponding class was discarded. About a quarter of the classes were discarded by using this criterion.
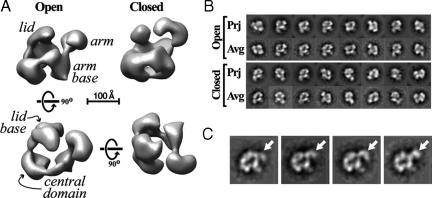
We generated initial volumes for each of the remaining classes and subjected them to 3D alignment and classification to identify the main characteristic features present in the data set. A subset (eight) of the best single-class volumes based on general appearance and the similarity between their reprojections and the experimental reference-free class averages were selected for single- and multireference projection-matching refinement against 0° data. Although most class volumes shared some common features, two were particularly stable in these refinements. Fig. 2A shows the refined versions of these two reconstructions. Their resolution is 37 Å according to the 0.5 Fourier shell correlation criterion (13) and plots of the distribution of angles assigned to the 0° experimental images after projection-matching refinement shows uneven but complete coverage of Euler space (see SI Fig. 5C).
Fig. 2.
RSC conformational variability. (A) Reconstructions of two conformers of RSC. The initial models were obtained by using the Orthogonal Tilt Reconstruction method and then refined by projection-matching against 0° data. The names of the main features of the reconstructions are indicated in italics. The “open” and “closed” conformations differ mainly in the position of the arm (at the right of the structures shown in the top row). The bottom row shows the “top” and “back” views of the open conformer. A scale bar corresponding to 100 Å is shown. (B) Comparison of reprojections (“Prj”) of the structures shown in A with the best-matching reference-free class averages (“Avg”). (C) Four selected experimental class averages for RSC showing a variety of positions occupied by the arm (arrows).
RSC Bears a Large Central Cavity, and Shows Conformational Flexibility.
The two RSC reconstructions show a common architecture: a large ring of density encircling a central cavity and two protrusions on one side of this ring, one longer than the other. This central ring of density was characteristic of the vast majority of single-class volumes obtained. Interestingly, the two reconstructions shown differ dramatically in the actual position of the longer protrusion, which we refer to as the “arm”: the “open” conformation (Fig. 2A Left) shows this arm pointing away from the central cavity, whereas the “closed” conformation (Fig. 2A Right) has it pointing inwards and capping the cavity. The open conformation, which is more abundant (data not shown) and was relatively more stable during projection-matching refinement, shows an open central ring composed of three large densities linked by thinner connections (see Fig. 2A). In the “front” view (Fig. 2A Upper Left) the rightmost density in the open ring serves also as the base of the arm and is termed the “arm base.” It is connected to another density at the front of the complex (the “central base” domain), in turn connected to a large density at the back, from which stems the shorter protrusion, termed “lid,” with its base being the “lid base.” The ring is interrupted at this point. This gap is not present in the closed conformation and, in general, the two reconstructions differ most in this region, which also includes the lid and its base.
The conformational flexibility in the arm can also be observed in the experimental class averages (see Fig. 2C).
The RSC Reconstructions Can Accommodate a Nucleosome.
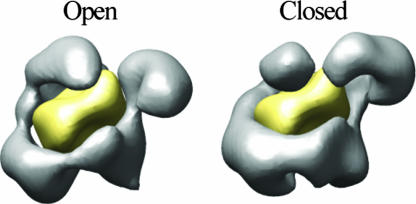
The central cavity and surrounding densities observed in the RSC reconstructions strongly suggest a possible binding site for the nucleosome. To test whether the central cavity could indeed accommodate a nucleosome and whether this putative binding site agreed with experimental observations, we manually placed a low-pass filtered (37 Å) version of the x-ray crystal structure of the nucleosome core particle (14) [Protein Data Bank (PDB) ID code 1AOI] in this potential binding pocket. Fig. 3 shows the result of this docking for the open and closed conformations of RSC; the fit is excellent and no major clashes can be observed in either conformation (see SI Movie 1 of the open conformation with the high resolution nucleosome modeled within its cavity). Although the closed conformation of RSC brings the arm right on top of and in very close proximity to the modeled bound nucleosome, it does not affect the goodness of the fit (Fig. 3). This model for RSC/nucleosome binding is in good agreement with experimental observations and suggests a potential role in the remodeling mechanism for the conformationally variable arm (see Discussion and Fig. 4).
Fig. 3.
Fitting of a nucleosome into the open and closed conformations of RSC. The crystal structure of the nucleosome (23) (PDB ID code 1AOI) was filtered to the resolution of the two reconstructions (37 Å) and manually fitted into each of the two reconstructed conformations. The filtered nucleosome is shown in yellow, and the RSC reconstructions are shown in gray.
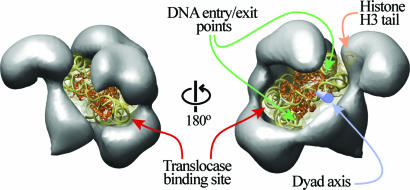
Fig. 4.
Model of nucleosome binding by RSC. The x-ray crystal structure of the nucleosome (23) (PDB ID code 1AOI) was manually fitted into the central cavity of RSC. The nucleosome is shown as a ribbon diagram within a translucent surface representation filtered to 10 Å. The DNA is represented in gold, and the protein is represented in orange. Back (Left) and front (Right) views of the complex are shown. The entry/exit points of the nucleosomal DNA are indicated with green arrows, the dyad axis (blue cylinder) is indicated with a blue arrow, the histone H3 tail visible in the crystal structure is indicated with an orange arrow, and the binding site for the translocase domain is shown on the DNA with maroon arrows.
RSC Binds Mono- and Dinucleosomes with Similar Affinity.
The striking complementarity between a nucleosome and the central cavity in our RSC reconstructions suggests that a mononucleosome is the natural substrate of this remodeler. To test this possibility, we performed electrophoretic mobility shift assays with RSC and mono- or dinucleosomes in the absence of ATP (as was the case for the samples used for our reconstructions). We find that the affinity of RSC for mono- or dinucleosome species was very similar (SI Fig. 6), suggesting that dinucleosomes are not a preferred substrate.
ATP-Dependent Translocation and Loop Formation by RSC on Single Nucleosomal Substrates.
Because our structure and biochemical work support the binding of RSC to a single nucleosome, we tested the DNA translocation parameters of RSC on a single nucleosome, rather than nucleosome arrays (15). Briefly, a single nucleosomal template attached to two beads was stretched between a pipette and an optical trap. DNA end-to-end distance was monitored in real time under constant tension as a function of single molecule remodeler activity (15). To eliminate signal arising from the interaction between RSC and bare DNA, the measurements were performed under a constant 4 pN tension, i.e., above the limit at which those interactions are observed (15, 16). As we have shown previously, this reaction depends not only on a nucleosome substrate under these conditions but also on the presence of ATP (15, 16).
RSC can target a nucleosome and translocate DNA relative to it generating a large DNA loop (SI Fig. 7). RSC appears to be able to translocate several hundreds of base pairs at an average velocity of 12 bp/s with a step size just below our detection limit of 10 bp. The appearance of discontinuous jumps of the tether length back to the base line indicates cooperative loop dissipation which may occur either by release of the DNA loop on the same side from which it formed, or on the opposite side, leading to the net displacement of the nucleosome (SI Fig. 7).
Discussion
We present a reconstruction of RSC using the OTR method. A former structure of RSC by others (8) shares with our structure a region for nucleosome binding. However, our RSC structure suggests that the nucleosome binding site resembles a deep cavity rather than a pincer/cleft, which impacts our understanding of the remodeling mechanism, as detailed below. Furthermore, from a population of multiple conformers we were able to resolve two different, specific conformations of the complex's long arm, suggesting a potential role in either substrate binding or the remodeling mechanism.
We have also presented a relatively simple way of gauging whether a given sample adopts the large number of orientations required for the application of the OTR method. This test only requires collecting a data set at 0° in addition to the +/− 45° data needed for the reconstruction method itself. Given that 0° data would typically be collected at the beginning of any project for an initial characterization of the sample, this approach does not involve a significant additional effort. Furthermore, the untilted data can be used for an initial refinement of the reconstructions.
Our reconstruction joins three existing ones for ATP-dependent chromatin remodeling complexes (7–9), all obtained from negatively stained samples. The differences and similarities between this reconstruction and the previously published one for RSC (8) as well as our own reconstruction of its human homolog PBAF (9) have revealed limitations in the methods used previously (discussed below). When all three structures are compared from the same viewpoint (Fig. 2 in this article as well as in refs. 8 and 9), normal to the sample support in refs. 8 and 9, their similar outlines are most evident. This is particularly the case for the two RSC structures. However, these similarities disappear when the structures are compared in a direction parallel to the support plane (compare bottom-left corner of Fig. 2 in this article with bottom of figure 3 in ref. 8 and bottom of figure 2 in ref. 9); our reconstruction shows significantly increased thickness and distinct features not restricted to a single plane. Furthermore, this increased thickness is not simply due to a larger mass but rather to the appearance of the central cavity. Two observations suggest that the previous structures suffer from significant flattening: (i) all features of the structures lie on a single plane and (ii) the dimensions on the reconstructions are significantly smaller in the direction perpendicular to the support, where flattening should have its largest effect. It is also likely that our PBAF reconstruction suffered from incomplete stain penetration; whereas the central mass of that complex (which we termed the “platform”) is of considerable thickness, it consists of a single, uninterrupted density. We suggest that a combination of flattening and incomplete staining could have altered the geometry of a RSC-like cavity to a region of continuous density.
A striking parallel between our reconstructions of RSC and PBAF is the conformational variability in one of their protrusions (termed “knobs” in the case of PBAF). Although we cannot make an unambiguous assignment of equivalent features in the two structures, this protrusion is, in both cases, the one attached to the thinner part of the complex and the one exhibiting a more noticeable constriction at its base (compare Fig. 2 in this paper with figure 2 in ref. 9). This similarity between these two homologous complexes suggests that conformational flexibility of this protrusion might be shared by SWI/SNF family remodelers. The far more dramatic differences observed between the yeast SWI/SNF structure (7) and the other three reconstructions are harder to rationalize. The SWI/SNF structure was solved by using an angular reconstitution approach, which assumes that the characteristic views present in the sample correspond to different views of the same object. Application of this approach to heterogeneous samples (conformers), where this assumption does not hold, could lead to an incorrect final structure.
We identified a variety of positions for the arm in the experimental class averages (see Fig. 2C), indicating that the conformational variability is not restricted to the two conformations we were able to reconstruct. The variability seen in the back of the complexes (where the open and closed conformations differ most) might also be the result of a failure to fully resolve heterogeneity with our current refinements. Preliminary tests using a 3D maximum likelihood (ML) approach (17) have confirmed the presence of multiple conformers (data not shown) that explain the differences seen between the open and closed conformations outside the arm itself.
The RSC reconstructions immediately suggest a potential binding site for the nucleosome in the central cavity. We tested whether this was sterically possible by manually fitting a filtered version of the atomic structure of the nucleosome (14) into the RSC reconstructions (Fig. 3). The contour of the central cavity in RSC is remarkably complementary to that of the nucleosome structure for both the open and closed conformations (Fig. 3). Additionally, despite the extensive wrapping of the nucleosome by the remodeler, no clashes can be seen in our model.
Given the quality of the fit shown in Fig. 3, we generated a model of a RSC-nucleosome complex using an unfiltered nucleosome. In principle, our model accommodates any rotational positioning of the nucleosome (orientation of the DNA entry/exit points relative to RSC). However, we chose to place the entry/exit sites pointing toward the largest opening of the cavity at the front of the complex (Fig. 4 Right) as this arrangement best accommodates DNA emitting from the nucleosome. Furthermore, this model is in overall agreement with experimental observations and has potential mechanistic implications as outlined below.
SWI/SNF family remodelers can generate dinucleosome-like products (18–20), raising the possibility that a nucleosome dimer might be the preferred substrate. However, gel shift analyses with RSC along with ATPase studies strongly favored a RSC-mononucleosome complex as the standard unit of remodeling. The results presented here show that dinucleosomes display nearly identical affinity for RSC as mononucleosomes (SI Fig. 6). Taken together, the striking complementarity between the RSC central pocket and a nucleosome (Figs. 3 and 4 and Movie 1), the similar affinity of RSC for mono- and dinucleosomes (SI Fig. 6), and the fact that the translocation observed with mononucleosomal substrates in single molecule assays (SI Fig. 7) is identical to that observed on nucleosomal arrays (15), strongly suggest that remodeling by RSC only requires the binding of a single nucleosome. We note, however, that these observations do not rule out a possible influence of a neighboring nucleosome on RSC activity in vivo; it simply suggests that the standard unit of remodeling is a RSC–mononucleosome complex.
All remodelers bear a similar ATPase subunit (21, 22) related to known DNA translocases, leading to the idea that ATP-dependent DNA translocation is central to the remodeling mechanism (1, 16, 23, 24). Recent studies have mapped the binding site for Sth1, the translocase in RSC, to ≈50 bp inside the nucleosome (23, 25). One of the two possible translocase-binding sites in the nucleosome (which is 2-fold symmetric) in our model is adjacent to the central base domain (see Fig. 4). This part of the complex shows the most extensive area of close proximity between RSC and the nucleosomal DNA. The other potential binding site for the translocase is fully exposed in our structure. We note, however, that the affinity of RSC for the nucleosome is considerably higher in the presence of ATP than in its absence (18). Thus, it remains possible that a conformational change helps the translocase engage at this alternative position. Because our samples lack ATP, this conformation may not be adopted in our current structures.
Current models for remodeling involve the formation of small DNA loops/waves on the surface of the nucleosome that propagate around the nucleosome to provide sliding, and/or the generation of a large DNA loop that accumulates on the nucleosome surface, providing factor access to the nucleosome surface (15, 23). Our model provides both support and challenges for these models. First, the shape of the deep cavity and its remarkable fit to the nucleosome would appear to set limits on the size of the DNA waves/loop that can be propagated along the entire length of the nucleosome surface, as large loops would provide a steric clash. Although conformational changes could occur to accommodate larger loops, the current structure suggests that very small loops are likely the primary intermediate for the sliding reaction. Our structural model also reveals several areas where the nucleosomal DNA is accessible to solvent, suggesting that the large DNA loops observed in our optical trap experiments (SI Fig. 7) could be generated and maintained at one or more of these positions on the nucleosome surface (Fig. 4). We suggest that both mechanistic models are consistent with our structures when additional features of the mechanism are considered. According to one model of remodeling, DNA is pumped toward the dyad from the site of DNA translocation, located ≈50 bp inside the nucleosome. The pumped DNA can have one of two fates: (i) it continues to propagate in the form of a small wave/loop through the dyad and around the nucleosome (inside the central cavity) to finally exit out the other side, or (ii) it remains at the exposed dyad, with subsequent pumping/translocation events adding to the size of the loop present at the dyad. This “building up” of DNA at the dyad may underlie the observation of RSC forming large loops on the nucleosome in single-molecule studies described here and previously (15).
Our structure reveals several additional points of close proximity with nucleosomal DNA, including regions near the DNA entry/exit sites and around the dyad axis (Fig. 4). The RSC features involved include the lid and its base, as well as the central base at the front of the complex (Fig. 4). These features might mediate the much higher affinity for the nucleosome observed with RSC complex compared with its isolated ATPase subunit (Sth 1) alone (15, 23). The presence of more than a single DNA-binding site in RSC had already been suggested by studies showing the formation of loops by the remodeler on bare DNA (16). In addition, portions of the lid and base face histone components. This proximity could allow regulated recognition of histones and their modifications and might contribute to the overall binding affinity. Future work will determine which particular RSC proteins and domains constitute these features. Along these lines, the ATPase subunit of all SWI/SNF family remodelers contains a bromodomain, a protein motif involved in the binding of acetylated histone tails (1). RSC includes an additional four subunits containing these motifs (11, 26, 27). Among them, Rsc4 (72 kDa) contains tandem bromodomains and was shown to bind to a histone H3 tail peptide acetylated at Lys-14 (27). Mapping the location of bromodomain-containing subunits within RSC will further help us understand how the complex engages its substrate as well as any potential role the observed conformational flexibility might play in the recognition of modified histone tails.
This work can be extended to vitrified samples, using our initial reconstructions and a maximum likelihood approach (among other tools) to visualize and characterize the conformations present in fully hydrated complexes. This extension, in conjunction with a RSC/nucleosome structure and mapping of key RSC components will allow us to start making detailed testable hypotheses regarding the interactions between RSC and the nucleosome and the remodeling mechanism.
Materials and Methods
Sample Preparation and Electron Microscopy.
RSC was purified as reported (24). Negatively stained samples were prepared following the sandwich method using uranyl formate (28) (see SI Text). Data were collected on a Tecnai 12 microscope equipped with a LaB6 source and operated at 120 keV and a nominal magnification of ×49,000. Data were collected either at 0° (for sample characterization and refinement) or at +45° (first) and −45° (second) (for reconstruction) under low-dose conditions on Kodak SO-163 film.
Data Processing.
Micrographs were digitized in a robotized Nikon Coolscan 8000 scanner (29) with a pixel size of 12.71 μm (2.59 Å at the sample level). For reconstruction by the OTR method, pairs of particles (at −45° and +45°) were windowed out using Spider's graphic interface WEB (30), binned 2-fold, ramp-corrected, and normalized. The tilt angle and tilt axes positions were obtained by using the program CTFTILT (31) on each individual micrograph. All of the particles (39,960) were combined in a single file and reference-free alignment and classification were performed by using the IMAGIC software suite (32). High spatial frequencies were incorporated gradually throughout the cycles of alignment and classification with an initial low-pass filter of 40 Å and a final low-pass filter of 15 Å. After convergence, a final set of 75 classes were generated. The in-plane rotation angles from the alignment were combined with the angles previously obtained with CTFTILT to build the angular file. All further processing was done with standard Spider operations (30). Class volumes were reconstructed for all classes that showed an even distribution of in-plane rotation angles (psi) (see Results). Alignment and classification of the single-class volumes was performed to characterize the main features present in the reconstructed volumes. A small subset (eight) of the best single-class volumes (selected on the basis of the similarity between their calculated projections and the experimental class averages) were used as references for single-reference and multireference projection-matching refinements against a data set consisting of 12,241 0° particles (prepared identically to those used for OTR reconstruction). The two most stable refined models (Fig. 2A) were used for further analysis. Volumes were contoured to enclose a mass of 1 MDa, the expected molecular weight of RSC and were displayed by using UCSF Chimera (33).
See SI Text for optical tweezers measurements and electrophoretic mobility shifts assays.
Supplementary Material
Acknowledgments
This work was supported by funding from National Institutes of Health/National Institute of General Medical Science (B.R.C., C.B., and E.N.), the Huntsman Cancer Institute (J.W.), the Agouron Foundation (E.N.), and the Office of Biological and Environmental Research of the U.S. Department of Energy through its GTL program (C.B. and E.N.). B.R.C., C.B., and E.N. are Howard Hughes Medical Institute Investigators.
Abbreviations
- RCT
random conical tilt
- cryo-EM
cryo-electron microscopy
- OTR
orthogonal tilt reconstruction.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0700706104/DC1.
References
- 1.Saha A, Wittmeyer J, Cairns BR. Nat Rev Mol Cell Biol. 2006;7:437–447. doi: 10.1038/nrm1945. [DOI] [PubMed] [Google Scholar]
- 2.Workman JL, Kingston RE. Annu Rev Biochem. 1998;67:545–579. doi: 10.1146/annurev.biochem.67.1.545. [DOI] [PubMed] [Google Scholar]
- 3.Leschziner AE, Nogales E. Annu Rev Biophys Biomol Struct. 2007 doi: 10.1146/annurev.biophys.36.040306.132742. [DOI] [PubMed] [Google Scholar]
- 4.Radermacher M, Wagenknecht T, Verschoor A, Frank J. J Microsc. 1987;146:113–136. doi: 10.1111/j.1365-2818.1987.tb01333.x. [DOI] [PubMed] [Google Scholar]
- 5.Van Heel M. Ultramicroscopy. 1987;21:111–123. doi: 10.1016/0304-3991(87)90078-7. [DOI] [PubMed] [Google Scholar]
- 6.Leschziner AE, Nogales E. J Struct Biol. 2006;153:284–299. doi: 10.1016/j.jsb.2005.10.012. [DOI] [PubMed] [Google Scholar]
- 7.Smith CL, Horowitz-Scherer R, Flanagan JF, Woodcock CL, Peterson CL. Nat Struct Biol. 2003;10:141–145. doi: 10.1038/nsb888. [DOI] [PubMed] [Google Scholar]
- 8.Asturias FJ, Chung WH, Kornberg RD, Lorch Y. Proc Natl Acad Sci USA. 2002;99:13477–80. doi: 10.1073/pnas.162504299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leschziner AE, Lemon B, Tjian R, Nogales E. Structure (Cambridge, UK) 2005;13:267–275. doi: 10.1016/j.str.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 10.Wang W, Xue Y, Zhou S, Kuo A, Cairns BR, Crabtree GR. Genes Dev. 1996;10:2117–2130. doi: 10.1101/gad.10.17.2117. [DOI] [PubMed] [Google Scholar]
- 11.Cairns BR, Lorch Y, Li Y, Zhang M, Lacomis L, Erdjument-Bromage H, Tempst P, Du J, Laurent B, Kornberg RD. Cell. 1996;87:1249–1260. doi: 10.1016/s0092-8674(00)81820-6. [DOI] [PubMed] [Google Scholar]
- 12.Cairns BR, Erdjument-Bromage H, Tempst P, Winston F, Kornberg RD. Mol Cell. 1998;2:639–651. doi: 10.1016/s1097-2765(00)80162-8. [DOI] [PubMed] [Google Scholar]
- 13.Bottcher B, Wynne SA, Crowther RA. Nature. 1997;386:88–91. doi: 10.1038/386088a0. [DOI] [PubMed] [Google Scholar]
- 14.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 15.Zhang Y, Smith CL, Saha A, Grill SW, Mihardja S, Smith SB, Cairns B, Peterson CL, Bustamante C. Mol Cell. 2006;24:559–568. doi: 10.1016/j.molcel.2006.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lia G, Praly E, Ferreira H, Stockdale C, Tse-Dinh YC, Dunlap D, Croquette V, Bensimon D, Owen-Hughes T. Mol Cell. 2006;21:417–425. doi: 10.1016/j.molcel.2005.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scheres SH, Gao H, Valle M, Herman GT, Eggermont PPB, Frank J, Carazo JM. Nat Methods. 2007;4:27–29. doi: 10.1038/nmeth992. [DOI] [PubMed] [Google Scholar]
- 18.Lorch Y, Cairns BR, Zhang M, Kornberg RD. Cell. 1998;94:29–34. doi: 10.1016/s0092-8674(00)81218-0. [DOI] [PubMed] [Google Scholar]
- 19.Schnitzler G, Sif S, Kingston RE. Cell. 1998;94:17–27. doi: 10.1016/s0092-8674(00)81217-9. [DOI] [PubMed] [Google Scholar]
- 20.Schnitzler GR, Cheung CL, Hafner JH, Saurin AJ, Kingston RE, Lieber CM. Mol Cell Biol. 2001;21:8504–8511. doi: 10.1128/MCB.21.24.8504-8511.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eisen JA, Sweder KS, Hanawalt PC. Nucleic Acids Res. 1995;23:2715–2723. doi: 10.1093/nar/23.14.2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vignali M, Hassan AH, Neely KE, Workman JL. Mol Cell Biol. 2000;20:1899–1910. doi: 10.1128/mcb.20.6.1899-1910.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saha A, Wittmeyer J, Cairns BR. Nat Struct Mol Biol. 2005;12:732–733. doi: 10.1038/nsmb973. [DOI] [PubMed] [Google Scholar]
- 24.Saha A, Wittmeyer J, Cairns BR. Genes Dev. 2002;16:2120–2134. doi: 10.1101/gad.995002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zofall M, Persinger J, Kassabov SR, Bartholomew B. Nat Struct Mol Biol. 2006;13:339–346. doi: 10.1038/nsmb1071. [DOI] [PubMed] [Google Scholar]
- 26.Cairns BR, Schlichter A, Erdjument-Bromage H, Tempst P, Kornberg RD, Winston F. Mol Cell. 1999;4:715–723. doi: 10.1016/s1097-2765(00)80382-2. [DOI] [PubMed] [Google Scholar]
- 27.Kasten M, Szerlong H, Erdjument-Bromage H, Tempst P, Werner M, Cairns BR. EMBO J. 2004;23:1348–1359. doi: 10.1038/sj.emboj.7600143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ohi M, Li Y, Cheng Y, Walz T. Biol Proced Online. 2004;6:23–34. doi: 10.1251/bpo70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Typke D, Nordmeyer RA, Jones A, Lee J, Avila-Sakar A, Downing KH, Glaeser RM. J Struct Biol. 2005;149:17–29. doi: 10.1016/j.jsb.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 30.Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, Leith A. J Struct Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- 31.Mindell JA, Grigorieff N. J Struct Biol. 2003;142:334–347. doi: 10.1016/s1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
- 32.van Heel M, Harauz G, Orlova EV, Schmidt R, Schatz M. J Struct Biol. 1996;116:17–24. doi: 10.1006/jsbi.1996.0004. [DOI] [PubMed] [Google Scholar]
- 33.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.