Abstract
mRNA surveillance system represses the expression of nonstop mRNA by rapid mRNA degradation and translation repression. Here we show that the level of protein product of nonstop mRNA containing a poly(A) tail was reduced 100-fold, and this reduction was due to rapid mRNA degradation, translation repression, and protein destabilization, at least in part, by the proteasome. Insertion of a poly(A) tract upstream of a termination codon resulted in translation repression and protein destabilization, but not rapid mRNA decay. We propose that translation of the poly(A) tail plays crucial roles in nonstop mRNA surveillance via translation repression and protein destabilization.
Keywords: Nonstop mRNA, polylysine, translational repression, poly(A), proteasome
Poly(A) tail is a common modification of eukaryotic mRNA and plays many fundamental roles in mRNA biogenesis, translation, and mRNA stability (Mangus et al. 2003; Coller and Parker 2004). It is well established that the cap structure at the 5′ end and the poly(A) tail at the 3′ end of mRNA stimulate translation initiation synergistically (Tarun and Sachs 1995; Preiss and Hentze 1998; Kahvejian et al. 2001). It has been shown that interaction of poly(A)-binding protein (PABP) with eIF4G–4E complex stimulates the recruitment of 43S ribosome complex to the 5′ end of mRNA (Kahvejian et al. 2001, 2005). PABP also stimulates the 60S ribosomal subunit-joining step after the formation of the 48S preinitiation complex (Searfoss et al. 2001; Kahvejian et al. 2005).
It has been shown that an aberrant mRNA lacking a termination codon (nonstop mRNA) is rapidly degraded by a nonstop mRNA decay pathway (NSD) in eukaryotes (Frischmeyer et al. 2002; van Hoof et al. 2002). In yeast, Ski7p is proposed to recognize a stalled 80S ribosome at the 3′ end of a nonstop mRNA and recruit the exosome and the Ski complex, resulting in rapid degradation of nonstop mRNA. We recently showed that translation of nonstop mRNA is repressed after initiation, probably because ribosome translating nonstop mRNA is stalled at the 3′ end of mRNA and represses the further round of translation by the roadblock of ribosome immediately following the leading one (Inada and Aiba 2005). Recently it was shown that nonstop mRNA lacking the poly(A) tail was translated efficiently in yeast (Meaux and Van Hoof 2006), and we speculated that translation of the poly(A) tail and its product may play crucial role(s) in repressing the expression of nonstop mRNA.
In this study, we found that the expression of a nonstop reporter gene was reduced 100-fold, and this strong reduction was due to translation repression and protein destabilization, as well as rapid mRNA degradation. The products of nonstop mRNA were destabilized, at least in part, by the proteasome. Insertion of the poly(A) sequences just upstream of a termination codon of the reporter gene strongly reduced the expression due to protein destabilization by the proteasome and translation repression, but not rapid mRNA decay, and this down-regulation occurred when the poly(A) sequences can be translated into polylysine and by a cotranslational mechanism. We propose that translation of the poly(A) tail plays novel roles in quality control of gene expression in eukaryotes.
Results and Discussion
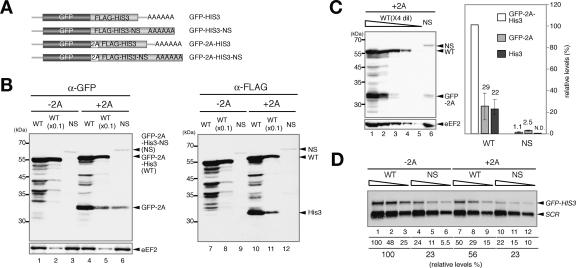
Recently, we showed that the product of nonstop mRNA was hardly detectable, and translation of nonstop mRNA was repressed after initiation (Inada and Aiba 2005). To know the mechanism of the quality-control system for nonstop mRNA, we tried to detect the nonstop mRNA product. Four promoters different in strength were used to express nonstopHIS3 mRNA products. The product of nonstopHIS3 mRNA was successfully detected with GPD1 promoter, and the level of nonstop gene product was ∼1% of the control product (data not shown). We suspected that this strong repression might be due to translation repression and/or protein destabilization. To evaluate the contributions of these regulations, we constructed GFP-HIS3 fusion genes and inserted 2A sequence of FMDV between two genes (Fig. 1A). Since a FMDV 2A sequence induces cotranslational release of nascent peptide from tRNA followed by the synthesis of the downstream translation products using the same ribosomes (Donnelly et al. 2001), we expected that both truncated products might be detected. Indeed, truncated products were detected with anti-GFP and anti-Flag antibody, respectively, in addition to an intact GFP-2A-His3 product (Fig. 1B, lanes 4,5,10,11). The quantification of the products revealed that the relative amounts of truncated products, GFP-2A or His3, to GFP-2A-His3 were almost the same (Fig. 1B, lanes 5,11). We also confirmed that the insertion of the FMDV 2A sequence did not affect the species of reporter mRNAs (data not shown). These results are consistent with an idea that the nascent peptide-containing 2A sequence (GFP-2A) is cotranslationally released from ribosome and downstream His3 protein is produced by the same ribosome. Therefore, we assume that the level of GFP-2A might reflect translation efficiency regardless of the stability of the full-length protein products. The level of GFP-2A in cells harboring pGFP-2A-HIS3-NS was 9% of that in cells harboring pGFP-2A-HIS3 (Fig. 1C). Northern blot analysis revealed that the level of GFP-2A-HIS3-NS mRNA was 40% of the GFP-2A-HIS3 (Fig. 1D), and this reduction in nonstop mRNA might be too moderate to explain the down-regulation of the GFP-2A level. These results are consistent with the proposal that translation repression contributes to the reduction of nonstop mRNA expression, in addition to rapid mRNA decay (Inada and Aiba 2005).
Figure 1.
Multiple steps are involved in the repression of nonstop gene expression. (A) Schematic drawing of the construction of fusion genes. The boxes indicate the ORFs and the stretch of A indicates a poly(A) tail on reporter mRNAs. (B) Translation repression and post-translational regulation are involved in the reduced expression of nonstop mRNA. W303 cells were transformed with pGPDp-GFP-HIS3 (WT, −2A) or pGPDp-GFP-HIS3-NS (NS, −2A) or pGPDp-GFP-2A-HIS3 (WT, +2A) or pGPDp-GFP-2A-HIS3-NS (NS, +2A). Cells were grown in SC-Ura, and samples were prepared as described previously (Inada and Aiba 2005). The levels of proteins were analyzed by Western blotting with anti-GFP antibody (left panel) or anti-Flag antibody (right panel). The samples were applied after 10-fold dilution when indicated. (C) The quantification of GFP-2A and GFP-2A-His3 products derived from pGPDp-GFP-2A-HIS3 (WT) or pGPDp-GFP-2A-HIS3-NS (NS) genes. (Left) Protein samples prepared from cells harboring indicated plasmids were diluted fourfold and analyzed by Western blotting with anti-GFP antibody. (Right) The relative levels of each product normalized to the level of GFP-2A-His3 as 100% are shown as the mean values of three independent experiments with standard deviations. (D) The insertion of 2A sequence has little effect on the nonstop mRNA level. RNA samples prepared from cells shown in B were diluted twofold (5–1.25 μg) and analyzed by Northern blotting with GFP and SCR probes. (Lane 1) Numbers immediately below the figure represent the level of mRNA normalized to the level of GFP-HIS3 mRNA as 100%. The numbers are shown as the mean values of three independent experiments.
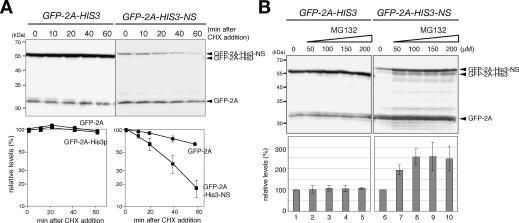
The level of GFP-2A-His3-NS was 100-fold lower than that of GFP-2A-His3, and the down-regulation of the full-length product was significantly stronger than that of the GFP-2A level (Fig. 1C). Based on the assumption that the level of GFP-2A may reflect translation repression and rapid mRNA decay, we suspected that protein destabilization might also contribute to the quality control for nonstop mRNA. To address this possibility, the stability of protein was examined by the determination of the levels of remaining proteins after the inhibition of translation. We found that GFP-2A-His3-NS (t1/2 = 25 min) was more unstable than GFP-2A-His3 or GFP-2A (t1/2 > 60 min) (Fig. 2A). To address the involvement of proteasome in a rapid degradation of GFP-2A-His3-NS, we examined the effect of a proteasome-specific inhibitor on the level of GFP-2A-His3-NS. For this purpose, the erg6Δ mutant was used, since the mutant shows the increased uptake of MG132, a specific inhibitor of proteasome (Lee and Goldberg 1996). We found that the level of GFP-2A-His3-NS in the W303erg6Δ mutant was increased 2.5-fold 2 h after the addition of MG132 (Fig. 2B, right panels). We observed little effects of the addition of MG132, if any, on the level of GFP-2A-His3 or GFP-2A (Fig. 2B, left panels). This is the first evidence to show that the products derived from aberrant mRNA are destabilized, at least in part, by the proteasome in vivo. It remains to be determined whether the degradation of the nonstop product by proteasome depends on ubiquitination or not.
Figure 2.
Protein product of nonstop mRNA is destabilized by the proteasome. (A) The product of nonstop mRNA is unstable. (Top panels) Samples of W303 cells harboring an indicated plasmid were prepared at indicated times after the addition of cycloheximide (0.1 mg/mL). The levels of the remaining proteins were determined by Western blotting. The samples of cells harboring pGPDp-GFP-2A-HIS3 (WT) were applied after 10-fold dilution. (Bottom panels) The relative levels of proteins were quantified and shown as a function of time after the inhibition of translation by cycloheximide (CHX). Relative amounts are shown as the mean values of three independent experiments with standard deviations. (B) MG132, a proteasome-specific inhibitor, stabilizes the products of nonstop mRNA. (Top panels) W303erg6Δ cells harboring an indicated plasmid were cultured and samples were prepared 2 h after the addition of MG132 at the indicated concentration. The samples of cells harboring pGPDp-GFP-2A-HIS3 (WT) were applied after 10-fold dilution. (Bottom panels) The qualification of GFP-2A-His3 and GFP-2A-His3-NS levels. The relative numbers are shown as the mean values of three independent experiments with standard deviations.
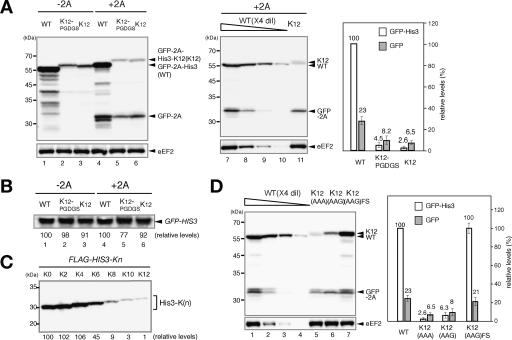
The poly(A) tail is a common modification for eukaryotic mRNA, but should not be translated. Translation of nonstop mRNA containing the poly(A) tail results in translation of the poly(A) sequences, and we suspected that the aberrant translation may have a role in the repression of nonstop gene expression. To examine the effect of translation of the poly(A) sequences, we inserted annealed oligonucleotides encoding 12 consecutive lysine residues (K12). Since it was shown that translation termination is regulated by nascent peptide in prokaryotes (Gong and Yanofsky 2002), we also examined the position effect of a polylysine by the insertion of 12 lysine residues plus five amino acid residues (K12-PGDGS) just upstream of a termination codon of the GFP-2A-HIS3 reporter. The insertion of the K12 sequence resulted in a drastic reduction of the levels of GFP-2A-His3 and GFP-2A (Fig. 3A) without affecting the level of GFP-2A-HIS3 mRNA (Fig. 3B). Since an additional five residues (PGDGS) after a polylysine did not affect the repression, we suspect that K12 may not repress translation termination. The expression of the reporter genes was significantly reduced when more than eight consecutive lysine residues (K8) were added to the C terminus of Flag-His3p, and 10 consecutive lysine residues (K10) showed the same severe effect on the reporter expression as K12 (Fig. 3C). To see whether an amino acid sequence was a determinant for the reduction, we examined the protein levels derived from two reporter genes, pGPDp-GFP-2A-HIS3-K12(AAG) and pGPDp-GFP-2A-HIS3-K12(AAG)FS. pGPDp-GFP-2A-HIS3-K12(AAG) reporter contains 12 lysine codons made up of four repeats of the sequence AAGAAGAAA. pGPDp-GFP-2A-HIS3-K12(AAG)FS construct is identical to the pGPDp-GFP-2A-HIS3-K12(AAG) construct, but contains an additional nucleotide just before the four repeats of AAGAAGAAA to put it out of frame and two more nucleotides following the sequence to restore the +0 reading frame just before the stop codon. The levels of the GFP-2A-His3 and GFP-2A product derived from pGPDp-GFP-2A-HIS3-K12(AAG) were reduced (Fig. 3D, lane 6), and this reduction was eliminated by introduction of frameshift mutations (Fig. 3D, lane 7), indicating that the amino acid but not nucleotide sequence is a determinant for the repression. These results suggest that the level of protein bearing a polylysine may be reduced by protein destabilization as well as translation repression, and a tract of >10 consecutive lysine residues is sufficient for the repression.
Figure 3.
The insertion of a poly(A) tract upstream of a termination codon reduced the expression. (A) Translation repression and protein destabilization by the insertion of a poly(A) tract upstream of a termination codon. (Left panel) The levels of proteins in W303 cells harboring indicated plasmid were analyzed as described in Figure 1B. (Lane 1) pGPDp-GFP-HIS. (Lane 2) pGPDp-GFP-HIS3-K12PGDGS. (Lane 3) pGPDp-GFP-HIS3-K12. (Lane 4) pGPDp-GFP-2A-HIS3. (Lane 5) pGPDp-GFP-2A-HIS3-K12PG DGS. (Lane 6) pGPDp-GFP-2A-HIS3-K12. (Middle panel) Protein samples prepared from cells harboring pGPDp-GFP-2A-HIS3 (WT) or pGPDp-GFP-2A-HIS3-K12 (K12) were diluted fourfold and analyzed by Western blotting with anti-GFP antibody. (Right panel) The relative levels of each product normalized to the level of GFP-2A-His3 as 100% are shown as the mean values of three independent experiments with standard deviations. (B) The insertion of the poly(A) tract does not affect the mRNA level. W303 cells described in A were cultured, and the levels of reporter mRNAs were determined by Northern blotting with GFP probe. (C) Effect of the number of inserted lysine residues at C terminus on the expression. W303 cells were transformed with a series of reporter genes (pGPDp-Flag-HIS3-Kn) encoding proteins bearing an indicated number of lysine residues at C terminus. The levels of protein products were determined with Western blotting, and numbers immediately below the figure represent the level of protein normalized to the level of His3p as 100%. (D) An amino acid sequence is a determinant to reduce the level of protein produced from mRNA containing the poly(A) tract in a coding region. (Left) The levels of proteins in W303 cells harboring pGPDp-GFP-2A-HIS3 (WT), pGPDp-GFP-2A-HIS3-K12 [K12(AAA)], pGPDp-GFP-2A-HIS3-K12(AAG) [K12(AAG)], or pGPDp-GFP-2A-HIS3-K12(AAG)FS [K12(AAG)FS] plasmid were analyzed as described in Figure 1C. (Right) The relative levels of each product normalized to the level of GFP-2A-His3 as 100% are shown as the mean values of three independent experiments with standard deviations.
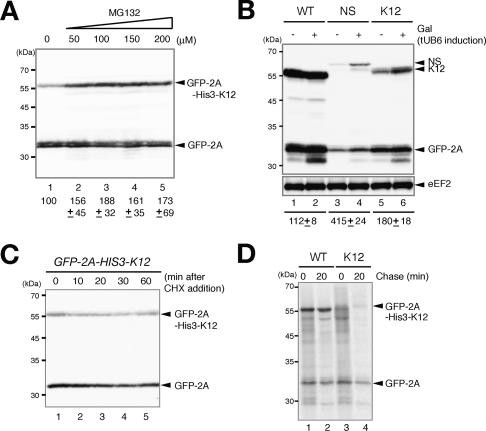
To examine the degradation of protein bearing a polylysine by proteasome, we determined the levels of the proteins after the inhibition of proteasome activity by MG132. The level of GFP-2A-His3-K12 in the W303erg6Δ strain was increased almost twofold 2 h after the addition of MG132 (Fig. 4A). We observed little effects of the addition of MG132, if any, on the level of GFP-2A (Fig. 4A). In addition, the levels of GFP-2A-His3-NS and GFP-2A-His3-K12 were significantly increased when the proteasome activity was inhibited by the overproduction of tandem ubiquitins, tUb6 (Saeki et al. 2005), although the levels of GFP-2A derived from GFP-2A-HIS3-NS was increased to some extent (Fig. 4B). To confirm the destabilization of proteins bearing a polylysine, the levels of remaining proteins were determined after the inhibition of translation. We found that the levels of GFP-2A-His3-K12 protein were not drastically reduced, even up to 60 min after the inhibition of translation (Fig. 4C). Therefore, we examined the protein stability by pulse-chase analysis. The half-life of GFP-2A-His3-K12 was almost the same as that of GFP-2A by >40-min chase time after pulse-labeling for 7 min (data not shown), indicating that the full-length GFP-2A-His-K12 may not be rapidly degraded. We suspected that the nascent peptide produced from aberrant mRNA containing the poly(A) sequence may be degraded cotranslationally, and the complete polypeptide released from ribosome may not be subjected to the rapid degradation. To address this, cells expressing GFP-2A-His3-K12 were radiolabeled for 1 min, a predicted time required for complete synthesis, and chased with amino acids to see the stability of nascent peptides. The full-length GFP-2A-His3-K12 product was hardly detected in a 1-min pulse-label, and several products smaller than GFP-2A-His3-K12 were detected (Fig. 4D, lane 3). These incomplete products disappeared over a 20-min chase, and only small amounts of the intact products were detected (Fig. 4D, lane 4). In contrast, the full-length GFP-2A-His3 protein was synthesized in a 1-min pulse-label, and the level was slightly increased after a 20-min chase (Fig. 4D, lanes 1,2). These results suggest that translation elongation may be arrested when the poly(A) sequence is translated and incomplete nascent peptides are rapidly degraded cotranslationally.
Figure 4.
Translation of the poly(A) sequence results in the destabilization of products by the proteasome. (A) MG132 increased the level of the protein bearing a polylysine. Samples of W303erg6Δ cells harboring pGPDp-GFP-2A-HIS3-K12 were prepared 2 h after the addition of MG132 at the indicated concentration. The relative levels of GFP-2A-His3-K12 are shown as the mean values of three independent experiments with standard deviations. (B) The induction of tUb6 increased the levels of GFP-2A-His3-NS and GFP-2A-His3-K12 products. W303 cells harboring pKK43 (p424GAL1p-tUB6) were transformed with pGPDp-GFP-2A-HIS3 (WT) pGPDp-GFP-2A-HIS3-NS (NS), or pGPDp-GFP-2A-HIS3-K12 (K12). Cells were grown in SC medium containing 2% raffinose until OD600 = 0.8, and samples were prepared for Western blotting. When indicated (+), galactose was added to induce the expression of tUb6 that inhibits the proteasome activity. Numbers immediately below the figure represent the ratio of the full-length protein levels under tUb6 induction condition to noninduction condition. The level of GFP-2A derived from GFP-2A-HIS3-NS was increased 1.5-fold by tUb6 overproduction. The numbers are shown as the mean values of three independent experiments. (C) Stability of GFP-2A-His3-K12 protein measured by Western blotting after the inhibition of translation. Samples of W303 cells harboring a pGPDp-GFP-2A-HIS3-K12 plasmid were prepared at indicated times after the addition of cycloheximide (0.1 mg/mL). The levels of the remaining proteins were determined by Western blotting. (D) Stability of translation products of GFP-2A-HIS3-K12 measured by pulse-chase analysis. W303 cells harboring pGPDp-GFP-2A-HIS3 (WT) or pGPDp-GFP-2A-HIS3-K12 (K12) were labeled with [35S]methionine and cysteine (25 μCi/mL) for 1 min and chase for 20 min. Cells were harvested and cell extracts were prepared for immunoprecipitation with anti-GFP antibody. Samples were subjected to SDS-PAGE, and the radioactivity was measured using Typhoon9400 (GE).
We showed that the levels of GFP-2A derived from GFP-2A-HIS3 were decreased threefold when the poly(A) sequence was inserted upstream of a termination codon (Fig. 3A, lanes 4–6). This may suggest that translation of the poly(A) sequence induces elongation arrest that leads to queuing and arrest of ribosome all the way back to the GFP coding sequences. Translation regulation by the interaction of nascent peptide with ribosome tunnel has been extensively examined in prokaryotes (Nakatogawa and Ito 2001, 2002; Gong and Yanofsky 2002) and eukaryotes (Gaba et al. 2005; Onouchi et al. 2005). Putative translation arrest products were detected by pulse-chase analysis (Fig. 4D), and the level of the protein bearing a polylysine was increased after the inhibition of the proteasome activity (Fig. 4A,B). These results let us propose that the insertion of consecutive lysine codons just upstream of the stop codon induces translation arrest when it is translated into polylysine, and destabilizes the protein products only by a cotranslational mechanism. We also propose that translation elongation of nonstop mRNA is arrested due to translation of the poly(A) tail, and nonstop products are produced only when translation elongation can be completed. The nonstop products may remain as peptidyl-tRNA on stalled ribosomes at the 3′ end of mRNA, and degrade by the proteasome cotranslationally. Further experiments are needed to make clear the mechanism of translation repression and protein degradation caused by translation of the poly(A) sequence.
In this study, we showed several results to support the model that translation of the poly(A) tail plays a crucial role in the quality control for nonstop mRNA with the poly(A) tail. First, translation of the poly(A) sequence results in the partial but significant repression of further rounds of translation (Figs. 1, 3). Second, the aberrant protein derived from nonstop mRNA is destabilized, at least in part, by proteasome (Figs. 2, 4). Third, the insertion of the poly(A) sequence upstream of a termination codon is sufficient for the down-regulation via translation repression and protein degradation by proteasome (Figs. 3, 4). The poly(A) tail is a common modification in eukaryotes and plays many important roles in gene expression, but it should not be translated. Therefore, we propose that translation of the poly(A) tail is recognized as aberrant by ribosome and results in translation repression and destabilization of aberrant products.
Materials and methods
Strains and general methods
Escherichia coli DH5α was used for DNA manipulations. Standard procedures were followed for yeast manipulations (Kaiser et al. 1994). To construct W303 erg6Δ strains, a DNA fragment containing the erg6Δ∷LEU2 was amplified from the genomic DNA of KNY43 (Nakatsukasa et al. 2004) by PCR using primers 5′-GATGCAACAGGGTAAGATCAGCAA-3′ and 5′-ACATGCCGTTTCACCACAGGAT-3′, and introduced into W303-1a cells along with YCplac22 (Gietz and Sugino 1988) and one of the reporter plasmids. Recombinant DNA procedures were carried out as described previously (Sambrook and Russell 2001). Northern blotting and Western blotting were performed as described (Inada and Aiba 2005). To detect GFP proteins, anti-GFP antibody (sc-9996, Santa Cruz Biotechnology) was used in 1000-fold dilution. Pulse-chase analysis was performed as described previously (Inada and Aiba 2005) and anti-GFP antibody was used for immunoprecipitation.
Plasmids
A HIS3 SpeI–BamHI fragment containing an entire ORF except the initiation and termination codons was amplified by PCR using the primers 5′-GGACTAGTGGACCTGGTGACTACAAGGACGACGATGACAAG GGTCCTGGTATGACAGAGCAGAAAGCCC-3′ and 5′-CGGGATCC CATAAGAACACCTTTGGTGGAGG-3′ and was inserted into p416CYC vector (Mumberg et al. 1995) to generate pSA134. A GFP XbaI–SpeI fragment was amplified using the primers 5′-GCTCTAGAATGAGTAAAG GAGAAGAACTTTTCAC-3′ and 5′-GGACTAGTTTTGTATAGTTC ATCCATGCCA-3′ and was inserted into the corresponding sites of p416CYC (Mumberg et al. 1995) to generate pSA135. BamHI–EcoRI fragments of pIT798 (HIS3) or pIT799 (HIS3-NS) containing a 3′ untranslated region (UTR) (Inada and Aiba 2005) were inserted into pSA134 to generate pSA136 (pFlag-HIS3) and pSA137 (pFlag-HIS3-NS), respectively. A XbaI–SpeI GFP fragment of pSA135 was inserted into the corresponding sites of pSA136 and pSA137 to generate pSA138 (pCYCp-GFP-Flag-HIS3-CYC1ter) and pSA139 (pCYCp-GFP-Flag-HIS3-NS-CYC1ter), respectively. To insert a FMDV 2A sequence, two oligonucleotides (5′-CTAGC CAGCTGTTGAATTTTGACCTTCTTAAGCTTGCGGGAGACGTCG AGTCCAACCCTGGGCCCA-3′ and 5′-CTAGTGGGCCCAGGGTTG GACTCGACGTCTCCCGCAAGCTTAAGAAGGTAAAATTCAACAG CTGG-3′) were annealed and ligated to SpeI-digested pSA138 or pSA139 to generate pSA142 (pCYCp-GFP-2A-Flag-HIS3-CYC1ter) and pSA143 (pCYCp-GFP-2A-Flag-HIS3-NS-CYC1ter), respectively. In order to express reporter genes under the GPD promoter, XbaI–EcoRI fragments of pSA138, pSA139, pSA142, or pSA143 were inserted into the corresponding sites of p416GPD vector (Mumberg et al. 1995). Resulting plasmids were designated as pSA144 (pGPDp-GFP-Flag-HIS3-CYC1ter), pSA145 (pGPDp-GFP-Flag-HIS3-NS-CYC1ter), pSA146 (pGPDp-GFP-2A-Flag-HIS3-CYC1ter), and pSA147 (pGPDp-GFP-2A-Flag-HIS3-NS-CYC1ter), respectively. Then, SacI–EcoRI fragments of pSA144, pSA145, pSA146, or pSA147 were ligated to the corresponding sites of YCplac33 (Gietz and Sugino 1988) to eliminate transcriptional terminator sequence (CYC1ter) and construct pGPDp-GFP-2A-Flag-HIS3 and pGPDp-GFP-2A-Flag-HIS3-NS. Resulting plasmids were designated as pSA156 (pGPDp-GFP-Flag-HIS3), pSA157 (pGPDp-GFP-Flag-HIS3-NS), pSA158 (pGPDp-GFP-2A-Flag-HIS3), and pSA159 (pGPDp-GFP-2A-Flag-HIS3-NS), respectively. In order to generate a pKK2 plasmid encoding HIS3-K12, two oligonucleotides (5′-GATCCGGCGCGCGGAAAAAAAAAAAAAAA AAAAAAAAAAAAAAAAAAAAATAACCCGGGCGCG-3′ and 5′-GA TCCGCGCCCGGGTTATTTTTTTTTTTTTTTTTTTTTTTTTTTTTT TTTTTTCCGCGCGCCG-3′) were annealed and ligated into the BamHI site of pSA136. BsiWI–EcoRI fragments of pKK2 were inserted into the corresponding sites of pSA156 and pSA158 to generate pKK6 (pGPDp-GFP-Flag-HIS3-K12) and pKK7 (pGPDp-GFP-2A-Flag-HIS3-K12), respectively. For pKK1 encoding HIS3-K12PGDGS, two oligonucleotides (5′-GATCCGGCGCGCGGAAAAAAAAAAAAAAAAAAAAAAAAAAAA AAAAAAAACCCGGGCGCG-3′ and 5′-GATCCGCGCCCGGGTTTT TTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTCCGCGCGCCG-3′) were annealed and ligated into the BamHI site of pSA136. BsiWI–EcoRI fragments of pKK1 were inserted into the corresponding sites of pSA156 and pSA158 to generate pKK4 (pGPDp-GFP-Flag-HIS3-K12PGDGS) and pKK5 (pGPDp-GFP-2A-Flag-HIS3-K12PGDGS), respectively. pKK32 [pGPDp-GFP-2A-HIS3-K12(AAG)] was constructed as pKK7, but two oligonucleotides (5′-GATCGCGCGCGAAGAAGAAAAAGAAGAAAAAGA AGAAAAAGAAGAAAGGAGGGG-3′ and 5′-GATCCCCCTCCTTTC TTCTTTTTCTTCTTTTTCTTCTTTTTCTTCTTCGCGCGC-3′) were annealed and inserted instead of the oligonucleotides used to construct pKK7. pKK41 [pGPDp-GFP-2A-HIS3-K12(AAG)FS] was also constructed as pKK7 with two oligonucleotides (5′-GATCGCGCGCCCAAGA AGAAAAAGAAGAAAAAGAAGAAAAAGAAGAAAGAGGGGGGG-3′ and 5′-GATCCCCCCCCTCCTTTCTTCTTTTTCTTCTTTTTCTTCT TTTTCTTCTTGGGCGCGC-3′). To construct pGPDp-Flag-HIS3-Kn plasmdis, oligonucleotides 5′-GATCCGGCGCGCGG(A)nTAACCCG GGCGCG-3 ′ and 5′-GATCCGCGCCCGGGTTA(T)nCCGCGCGCCG-3′ were annealed and inserted into the BamHI site of pIT912 (pGPDp-Flag-HIS3-CYC1ter). pKK43 (p424GAL1p-tUB6) was constructed by the insertion of an EcoRI–XhoI fragment of pEK206 (Saeki et al. 2005) containing tUB6 into the corresponding sites of p424 GAL1 (Mumberg et al. 1994).
Acknowledgments
We thank Dr. Toshiya Endo and Dr. Shuh-ichi Nishikawa for erg6Δ strain, and Dr. Akio Toh-e for plasmids and yeast strains. We also thank Dr. Hiroji Aiba, Dr. Tohru Yoshihisa, Dr. Yasuko Murakami, Dr. Takumi Kamura, and Dr. Koichi Ito for helpful discussion and critical reading of manuscript. This work was supported by Grants-in-Aid from the Ministry of Education, Culture, Sports, Science, and Technology of Japan, and Naito foundation (to T.I).
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1490207
References
- Coller J., Parker R., Parker R. Eukaryotic mRNA decapping. Annu. Rev. Biochem. 2004;73:861–890. doi: 10.1146/annurev.biochem.73.011303.074032. [DOI] [PubMed] [Google Scholar]
- Donnelly M.L., Luke G., Mehrotra A., Li X., Hughes L.E., Gani D., Ryan M.D., Luke G., Mehrotra A., Li X., Hughes L.E., Gani D., Ryan M.D., Mehrotra A., Li X., Hughes L.E., Gani D., Ryan M.D., Li X., Hughes L.E., Gani D., Ryan M.D., Hughes L.E., Gani D., Ryan M.D., Gani D., Ryan M.D., Ryan M.D. Analysis of the aphthovirus 2A/2B polyprotein ‘cleavage’ mechanism indicates not a proteolytic reaction, but a novel translational effect: A putative ribosomal ‘skip.’. J. Gen. Virol. 2001;82:1013–1025. doi: 10.1099/0022-1317-82-5-1013. [DOI] [PubMed] [Google Scholar]
- Frischmeyer P.A., van Hoof A., O’Donnell K., Guerrerio A.L., Parker R., Dietz H.C., van Hoof A., O’Donnell K., Guerrerio A.L., Parker R., Dietz H.C., O’Donnell K., Guerrerio A.L., Parker R., Dietz H.C., Guerrerio A.L., Parker R., Dietz H.C., Parker R., Dietz H.C., Dietz H.C. An mRNA surveillance mechanism that eliminates transcripts lacking termination codons. Science. 2002;295:2258–2261. doi: 10.1126/science.1067338. [DOI] [PubMed] [Google Scholar]
- Gaba A., Jacobson A., Sachs M.S., Jacobson A., Sachs M.S., Sachs M.S. Ribosome occupancy of the yeast CPA1 upstream open reading frame termination codon modulates nonsense-mediated mRNA decay. Mol. Cell. 2005;20:449–460. doi: 10.1016/j.molcel.2005.09.019. [DOI] [PubMed] [Google Scholar]
- Gietz R.D., Sugino A., Sugino A. New yeast–Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- Gong F., Yanofsky C., Yanofsky C. Instruction of translating ribosome by nascent peptide. Science. 2002;297:1864–1867. doi: 10.1126/science.1073997. [DOI] [PubMed] [Google Scholar]
- Inada T., Aiba H., Aiba H. Translation of aberrant mRNAs lacking a termination codon or with a shortened 3′-UTR is repressed after initiation in yeast. EMBO J. 2005;24:1584–1595. doi: 10.1038/sj.emboj.7600636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahvejian A., Roy G., Sonenberg N., Roy G., Sonenberg N., Sonenberg N. The mRNA closed-loop model: The function of PABP and PABP-interacting proteins in mRNA translation. Cold Spring Harb. Symp. Quant. Biol. 2001;66:293–300. doi: 10.1101/sqb.2001.66.293. [DOI] [PubMed] [Google Scholar]
- Kahvejian A., Svitkin Y.V., Sukarieh R., M’Boutchou M.N., Sonenberg N., Svitkin Y.V., Sukarieh R., M’Boutchou M.N., Sonenberg N., Sukarieh R., M’Boutchou M.N., Sonenberg N., M’Boutchou M.N., Sonenberg N., Sonenberg N. Mammalian poly(A)-binding protein is a eukaryotic translation initiation factor, which acts via multiple mechanisms. Genes & Dev. 2005;19:104–113. doi: 10.1101/gad.1262905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser C.A., Adams A., Gottschling D.E., Adams A., Gottschling D.E., Gottschling D.E. Methods in yeast genetics. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1994. [Google Scholar]
- Lee D.H., Goldberg A.L., Goldberg A.L. Selective inhibitors of the proteasome-dependent and vacuolar pathways of protein degradation in Saccharomyces cerevisiae. J. Biol. Chem. 1996;271:27280–27284. doi: 10.1074/jbc.271.44.27280. [DOI] [PubMed] [Google Scholar]
- Mangus D.A., Evans M.C., Jacobson A., Evans M.C., Jacobson A., Jacobson A. Poly(A)-binding proteins: Multifunctional scaffolds for the post-transcriptional control of gene expression. Genome Biol. 2003;4:223. doi: 10.1186/gb-2003-4-7-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meaux S., Van Hoof A., Van Hoof A. Yeast transcripts cleaved by an internal ribozyme provide new insight into the role of the cap and poly(A) tail in translation and mRNA decay. RNA. 2006;12:1323–1337. doi: 10.1261/rna.46306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumberg D., Muller R., Funk M., Muller R., Funk M., Funk M. Regulatable promoters of Saccharomyces cerevisiae: Comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res. 1994;22:5767–5768. doi: 10.1093/nar/22.25.5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumberg D., Muller R., Funk M., Muller R., Funk M., Funk M. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene. 1995;156:119–122. doi: 10.1016/0378-1119(95)00037-7. [DOI] [PubMed] [Google Scholar]
- Nakatogawa H., Ito K., Ito K. Secretion monitor, SecM, undergoes self-translation arrest in the cytosol. Mol. Cell. 2001;7:185–192. doi: 10.1016/s1097-2765(01)00166-6. [DOI] [PubMed] [Google Scholar]
- Nakatogawa H., Ito K., Ito K. The ribosomal exit tunnel functions as a discriminating gate. Cell. 2002;108:629–636. doi: 10.1016/s0092-8674(02)00649-9. [DOI] [PubMed] [Google Scholar]
- Nakatsukasa K., Okada S., Umebayashi K., Fukuda R., Nishikawa S., Endo T., Okada S., Umebayashi K., Fukuda R., Nishikawa S., Endo T., Umebayashi K., Fukuda R., Nishikawa S., Endo T., Fukuda R., Nishikawa S., Endo T., Nishikawa S., Endo T., Endo T. Roles of O-mannosylation of aberrant proteins in reduction of the load for endoplasmic reticulum chaperones in yeast. J. Biol. Chem. 2004;279:49762–49772. doi: 10.1074/jbc.M403234200. [DOI] [PubMed] [Google Scholar]
- Onouchi H., Nagami Y., Haraguchi Y., Nakamoto M., Nishimura Y., Sakurai R., Nagao N., Kawasaki D., Kadokura Y., Naito S., Nagami Y., Haraguchi Y., Nakamoto M., Nishimura Y., Sakurai R., Nagao N., Kawasaki D., Kadokura Y., Naito S., Haraguchi Y., Nakamoto M., Nishimura Y., Sakurai R., Nagao N., Kawasaki D., Kadokura Y., Naito S., Nakamoto M., Nishimura Y., Sakurai R., Nagao N., Kawasaki D., Kadokura Y., Naito S., Nishimura Y., Sakurai R., Nagao N., Kawasaki D., Kadokura Y., Naito S., Sakurai R., Nagao N., Kawasaki D., Kadokura Y., Naito S., Nagao N., Kawasaki D., Kadokura Y., Naito S., Kawasaki D., Kadokura Y., Naito S., Kadokura Y., Naito S., Naito S. Nascent peptide-mediated translation elongation arrest coupled with mRNA degradation in the CGS1 gene of Arabidopsis. Genes & Dev. 2005;19:1799–1810. doi: 10.1101/gad.1317105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preiss T., Hentze M.W., Hentze M.W. Dual function of the messenger RNA cap structure in poly(A)-tail-promoted translation in yeast. Nature. 1998;392:516–520. doi: 10.1038/33192. [DOI] [PubMed] [Google Scholar]
- Saeki Y., Isono E., Shimada M., Kawahara H., Yokosawa H., Toh E.A., Isono E., Shimada M., Kawahara H., Yokosawa H., Toh E.A., Shimada M., Kawahara H., Yokosawa H., Toh E.A., Kawahara H., Yokosawa H., Toh E.A., Yokosawa H., Toh E.A., Toh E.A. Knocking out ubiquitin proteasome system function in vivo and in vitro with genetically encodable tandem ubiquitin. Methods Enzymol. 2005;399:64–74. doi: 10.1016/S0076-6879(05)99005-8. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Russell D.W., Russell D.W. Molecular cloning. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, Cold Spring Harbor, NY: 2001. [Google Scholar]
- Searfoss A., Dever T.E., Wickner R., Dever T.E., Wickner R., Wickner R. Linking the 3′ poly(A) tail to the subunit joining step of translation initiation: Relations of Pab1p, eukaryotic translation initiation factor 5b (Fun12p), and Ski2p–Slh1p. Mol. Cell. Biol. 2001;21:4900–4908. doi: 10.1128/MCB.21.15.4900-4908.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarun S.Z., Jr., Sachs A.B., Sachs A.B. A common function for mRNA 5′ and 3′ ends in translation initiation in yeast. Genes & Dev. 1995;9:2997–3007. doi: 10.1101/gad.9.23.2997. [DOI] [PubMed] [Google Scholar]
- van Hoof A., Frischmeyer P.A., Dietz H.C., Parker R., Frischmeyer P.A., Dietz H.C., Parker R., Dietz H.C., Parker R., Parker R. Exosome-mediated recognition and degradation of mRNAs lacking a termination codon. Science. 2002;295:2262–2264. doi: 10.1126/science.1067272. [DOI] [PubMed] [Google Scholar]