Abstract
The p16INK4A and p14ARF proteins, encoded by the INK4A-ARF locus, are key regulators of cellular senescence, yet the mechanisms triggering their up-regulation are not well understood. Here, we show that the ability of the oncogene BMI1 to repress the INK4A-ARF locus requires its direct association and is dependent on the continued presence of the EZH2-containing Polycomb-Repressive Complex 2 (PRC2) complex. Significantly, EZH2 is down-regulated in stressed and senescing populations of cells, coinciding with decreased levels of associated H3K27me3, displacement of BMI1, and activation of transcription. These results provide a model for how the INK4A-ARF locus is activated and how Polycombs contribute to cancer.
Keywords: Senescence, cancer, INK4B, INK4A, ARF, Polycomb
Cellular senescence is an irreversible growth arrest triggered by several types of stress, including DNA damage, telomere shortening, and oncogene activation (Dimri 2005). Recently, its relevance as a bona fide tumor-suppressive mechanism in vivo has been highlighted (for review, see Narita and Lowe 2005). The Polycomb group (PcG) proteins BMI1, CBX7, and CBX8 are capable of delaying the onset of senescence in mouse and human embryonic fibroblasts (MEFs and HEFs) (Jacobs et al. 1999; Gil et al. 2004; Dietrich et al. 2007). This has been shown to correlate with a decrease in the levels of p16INK4A and, in some cases, p14ARF (p19Arf in mice). Both of these proteins are encoded by the INK4A-ARF locus and are tumor suppressors that act upstream of the pRB and p53 pathways, respectively (Lowe and Sherr 2003).
The BMI1-containing Polycomb-Repressive Complex 1 (PRC1), of which many variants are thought to exist, also contains the CBX (CBX2, CBX4, CBX6, CBX7, and CBX8), PHC1-3, RNF1-2, and SCML1-2 proteins (Levine et al. 2004). A second complex, PRC2, contains the histone methyltransferase EZH2, which together with EED and SUZ12 trimethylates histone H3 on Lys 27 (H3K27me3) (Cao and Zhang 2004; Pasini et al. 2004b). The ability of PRC1 to bind to chromatin is dependent on PRC2 function, and it has been proposed that this is primarily achieved via binding to the H3K27me3 mark (Rastelli et al. 1993; Hernandez-Munoz et al. 2005).
In this study, we address several outstanding questions concerning the regulation of the INK4A-ARF locus by BMI1. We establish that BMI1 together with other PcGs and the associated H3K27me3 mark “blanket” the locus both in vivo and in vitro (tissue culture) in both mouse and human cells. We show that the repression of the locus by BMI1 is dependent on the continued association of the EZH2-containing PRC2 complex and that the levels of EZH2 are down-regulated in stressed and senescent cells. This down-regulation leads to the loss of H3K27me3, displacement of BMI1, and activation of INK4A transcription, resulting in senescence. Taken together, our results provide a model for how the INK4A-ARF locus is regulated in response to multiple cellular signals and how increased expression of the PcGs contributes to cancer.
Results and Discussion
PcGs and associated H3K27me3 ‘blanket’ the INK4A-ARF locus both in vitro and in vivo
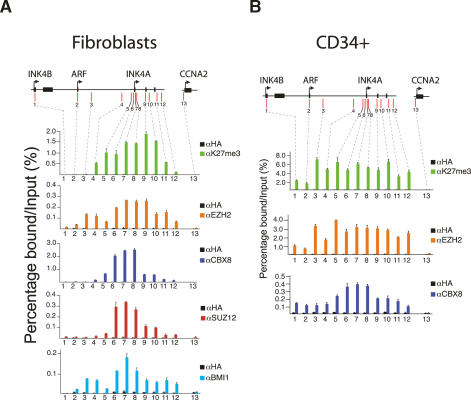
To determine how PcGs bind to the human INK4A-ARF locus we probed tiled arrays representing the entire INK4B and INK4A-ARF loci for the presence of PRC1 (CBX8), PRC2 (SUZ12), and the H3K27me3 mark in TIG3-TERT HEFs. This revealed very strong enrichments of SUZ12, CBX8, and H3K27me3 beginning upstream of the INK4A promoter and extending downstream beyond the end of the gene, a stretch of ∼10 kb (Supplementary Fig. S1). Similar “blanket” type enrichments have recently been observed on several PcG regulated genes in mammalian embryonic cells (Boyer et al. 2006; Bracken et al. 2006; Lee et al. 2006; Squazzo et al. 2006). This chromatin immunoprecipitation coupled with microarray analysis (ChIP-on-chip) data was validated by performing real-time quantitative PCR (qPCR) of immunoprecipitated DNA from an independent ChIP experiment (Fig. 1A). Using this approach we also established that BMI1 and EZH2 bind to the INK4A-ARF locus with similar, but interestingly not entirely identical, profiles to CBX8 and SUZ12 (Fig. 1A). No significant PcG or H3K27me3 enrichments were observed on the ARF promoter. Therefore, even though the genetic evidence for the regulation of ARF by PcGs is very strong in mice (Jacobs et al. 1999; Bruggeman et al. 2005; Molofsky et al. 2005), our data may suggest that it is not a direct PcG target, at least in human cells. However, this does not appear to be the case, because EZH2, CBX8, and H3K27me3 are significantly enriched throughout the INK4B-ARF–INK4A locus in primary human CD34+ bone marrow (BM) cells (Fig. 1B). Interesting, these CD34+ cells express high levels of EZH2 and do not express detectable levels of INK4A, INK4B, or ARF (Supplementary Fig. S3). A similar experiment performed with mouse lin− BM cells revealed similar EZH2, CBX8, and H3K27me3 enrichments throughout the mouse Ink4a-Arf locus (Supplementary Fig. S2). Taken together, our data suggest that in addition to INK4A, both ARF and INK4B are directly regulated by Polycomb proteins in both human and mouse cells. This would be consistent with the recent hypothesis that the all three genes are coordinately regulated (Gil and Peters 2006; Kim and Sharpless 2006).
Figure 1.
PcG proteins bind throughout the INK4A-ARF locus. (A) ChIP analysis of the INK4B and INK4A-ARF loci in TIG3-TERT HEFs. The precipitated DNA was amplified by real-time qPCR using primers specific for the regions indicated by red bars in the top of the panel. Enrichments are presented as percentages of total input. (B) ChIP analysis of the INK4B and INK4A-ARF loci in CD34-positive BM cells.
Ectopically expressed BMI1 represses the INK4A-ARF locus by direct association
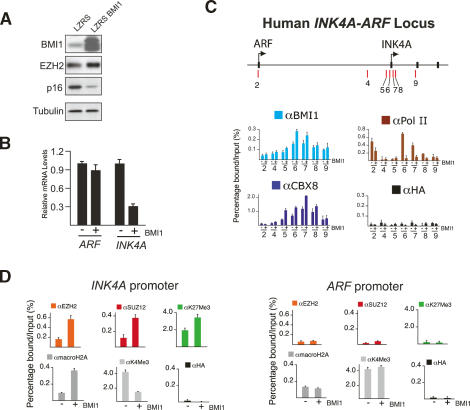
To address how BMI1 can prevent stress-induced senescence and thereby contribute to tumorigenesis, we ectopically expressed the protein in both MEFs and HEFs (Fig. 2; Supplementary Fig. S4). As previously reported, BMI1 overexpression leads to transcriptional repression of INK4A but not ARF in HEFs (Fig. 2A,B), whereas both Ink4a and, to a lesser extent, Arf expression are repressed in MEFs (Supplementary Fig. S4). Concomitant with the repression of INK4A, BMI1 binding increased approximately twofold principally around the first exon (exon 1α) of INK4A in HEFs (Fig. 2C). The lack of BMI1 association with the ARF promoter and consequent lack of repression may be due to the loss of H3K27me3 upon transfer of cells to tissue culture (see Supplementary Fig. S5). Interestingly, the increase in BMI1 association with the INK4A promoter coincided with an increase in association of the PRC1 component CBX8, suggesting that ectopic BMI1 associates as part of an intact PRC1 complex. Similar results were obtained in MEFs albeit with a slight but significant increase in CBX8 enrichment on the ARF promoter (Supplementary Fig. S4).
Figure 2.
Ectopically expressed BMI1 is recruited to the INK4A-ARF locus. (A) Western blot analysis of LZRS- and LZRSBMI1-infected TIG3-TERT HEFs. (B) qPCR analysis of the mRNA levels of ARF and INK4A in cells from A. (C) ChIP analysis of BMI1, CBX8, and RNA polymerase II enrichments in LZRS- and LZRSBMI1-infected TIG3-TERT HEFs. (D) ChIP analysis at the human “peak” PcG-binding region (primer set 7 in C) and the ARF promoter in LZRS- and LZRSBMI1-infected TIG3-TERT HEFs.
Next we wished to determine if the chromatin state around the INK4A-ARF locus is altered in cells ectopically expressing BMI1. In human cells we observed an increase in deposition of the histone variant macroH2A on the INK4A promoter, but not on the ARF promoter (Fig. 2D). This histone variant is known to block transcriptional initiation by limiting nucleosomal remodeling and histone acetylation and has previously been linked to PRC1-mediated repression (Henikoff et al. 2004; Hernandez-Munoz et al. 2005). In addition, BMI1 overexpression led to a decrease in the levels of H3K4me3, a mark associated with an open, active/poised transcriptional state. We also observed greater levels of the PRC2 complex and H3K27me3 in BMI1-overexpressing cells. This may be the consequence of increased EZH2 mRNA and protein levels in these cells (Fig. 2A; data not shown). Interestingly, Trojer and Reinberg (2006) speculated recently that a dimer of PRC1 may function during replication to tether the PRC2 complex to the daughter strand in order to propagate the H3K27me3 mark. Supporting this speculation is the observation of a transient interaction between PRC1 and PRC2 components during Drosophila development (Poux et al. 2001). It will be interesting to perform double ChIPs on single nucleosomes to determine if both PRC2 and PRC1 complexes coexist on target genes such as INK4A. In any case, our results suggest that BMI1, as part of an intact PRC1 complex, represses INK4A transcription by direct binding to its promoter and downstream coding region, leading to chromatin alterations and displacement of RNA polymerase II.
EZH2 down-regulation in late passage fibroblasts correlates with displacement of BMI1 from the INK4A-ARF locus
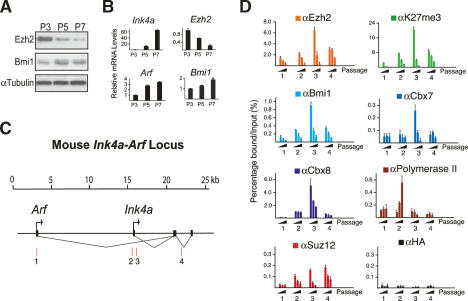
We next wished to determine if the known increase in Ink4A and Arf expression in MEFs approaching cellular senescence could be a result of decreased PcG binding to the Ink4a-Arf locus. As shown in Figure 3B, Ink4a and to a lesser extent, Arf levels increase in MEFs undergoing senescence. Consistent with previous observations (Kamminga et al. 2006), we observed a strong decrease in Ezh2, but not Bmi1, protein levels in MEFs undergoing senescence (Fig. 3A). This decrease correlated with the disappearance of Ezh2, Suz12, and the H3K27me3 mark along the Ink4a-Arf locus in senescing cells and the progressive displacement of the PRC1 components Bmi1, Cbx7, and Cbx8 (Fig. 3D). The low or background levels of Suz12, Cbx7, and Cbx8 observed on the Arf promoter are due to the fact that most PcGs and H3K27me3 are already displaced between Passage 0 and Passage 3 (Supplementary Fig. S5). Consistent with the fact that ectopic expression of BMI1 decreases RNA polymerase II association and transcription, the dissociation of the PcGs during cellular senescence has the opposite effect of increasing RNA polymerase II association and transcription (Fig. 3D; Supplementary Fig. S5).
Figure 3.
PcGs and H3K27me3 are lost from the Ink4a-Arf locus in cells undergoing senescence. (A) Western blots of cell lysates prepared from MEFs at increasing passage numbers were probed with the indicated antibodies. (B) qPCR analysis of the indicated genes on mRNA prepared from the cells shown in A. (C) Representation of the mouse Ink4a-Arf gene locus. Amplified regions in D are indicated as red bars. (D) ChIP analysis using the indicated antibodies and the cells shown in A on the Ink4a-Arf locus.
Similar results were also observed in human TIG3 HEFs passaged to senescence (Supplementary Fig. S6). Consistent with previous reports using human fibroblasts, INK4A was moderately up-regulated between mid- and late passages, whereas ARF levels were unchanged (Alcorta et al. 1996; Wei et al. 2001). The BMI1 mRNA and protein levels were slightly down-regulated in late passage HEFs, as described previously (Itahana et al. 2003). However, EZH2 mRNA and protein levels were significantly more strongly down-regulated. This correlated with a decreased binding of EZH2 along the INK4A-ARF locus and loss of the associated H3K27me3 mark, displacement of BMI1, and an increase in RNA polymerase II association in late passage TIG3 cells.
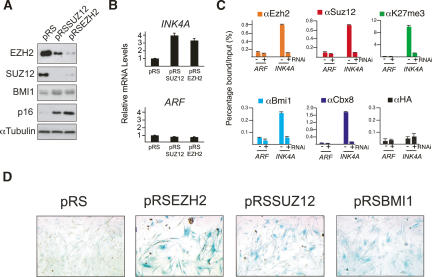
Interestingly, we also observed a strong decrease in the mRNA levels of EZH2, but not of other PcG genes, in cells exposed to multiple types of cellular stress, including UV, IR, and bleomycin-induced DNA damage (Supplementary Fig. S7; data not shown). In agreement with this, the expression of EZH2 was recently shown to be repressed by the p53 pathway (Tang et al. 2004). Indeed, the decrease in EZH2 mRNA levels in DNA damaged TIG3-TERT cells is dependent on the presence of p53 (data not shown). This decrease in EZH2 mRNA levels during cellular stress and the loss of PcGs from the INK4A promoter (Supplementary Fig. S8) led us to speculate that it may be causally linked to the gradual activation of the INK4A-ARF locus in senescing populations of cells. Therefore, to formally establish that a decreasing pool of the PRC2 complex directly leads to senescence and displacement of BMI1, we targeted the expression of EZH2 and SUZ12 by short hairpin interference (Fig. 4A). The loss of either protein results in the disruption of the PRC2 complex (Pasini et al. 2004b) and leads to the decrease of H3K27me3 and displacement of both BMI1 and CBX8 from the INK4A promoter (Fig. 4C). Significantly, cells depleted of EZH2 or SUZ12, similar to cells depleted of BMI1, activate INK4A transcription and become senescent (Fig. 4; Supplementary Fig. S9).
Figure 4.
Targeted depletion of PRC2 displaces BMI1 from the INK4A-ARF locus, increases the expression of INK4A, and leads to senescence. (A) Western blot analysis of cell lysates prepared from TIG3-TERT HEFs infected with pRS, pRSSUZ12, or pRSEZH2, 7 d after selection with puromycin. (B) qPCR analysis of INK4A and ARF mRNA levels from cells from A. (C) ChIP analysis of PcGs and H3K27me3 enrichments on the INK4A and ARF promoters in pRS- and pRSSUZ12-infected cells. (D) β-galactosidase staining to detect senescent cells in TIG3-TERT cells infected with pRS, pRSSUZ12, pRSEZH2, or pRSBMI1 constructs.
At present it is unclear how the H3K27me3 mark is lost in senescing cells. Two apparent possibilities exist, which include histone replacement or the action of an as yet unidentified H3K27me3 demethylase. Considering our demonstration that the histone variant macroH2A increases on the locus in BMI1-overexpressing cells this suggests that future studies should be directed at evaluating if there are alterations in the distribution of histone H3 variants levels along the INK4A-ARF locus during entry into senescence. Such studies may also implicate the involvement of the various chaperone complexes required for deposition of histone variants into chromatin in the senescence process. Furthermore, it is very likely that systematic approaches to identify chromatin modifiers of the locus will uncover several novel oncogenes and tumor suppressors. An example of this is the demonstration that the tumor suppressor and chromatin remodeling factor hSNF5 binds to and activates transcription from the INK4A promoter (Oruetxebarria et al. 2004).
Implications for our understanding of the role of PcGs in cancer, stem cell biology, and cellular stress responses
Bmi1 is essential for the maintenance of both adult (hematopoetic and neural) and cancer stem cell populations (Lessard and Sauvageau 2003; Molofsky et al. 2003; Park et al. 2003; Leung et al. 2004), and this has primarily been attributed to its ability to repress the Ink4a-Arf locus (Bruggeman et al. 2005; Molofsky et al. 2005). Recently, Ink4a and Arf have been shown to accumulate in stem cells of old mice; however, the levels of Bmi1 are unchanged (Janzen et al. 2006; Krishnamurthy et al. 2006; Molofsky et al. 2006). Consistent with these results, we did not find any change in Bmi1 levels in late passage MEFs despite the accumulation of Ink4a and Arf. Taken together with the demonstration that EZH2 levels are decreased in senescent cells and that the continuous presence of the PRC2 complex is required for Bmi1 function, this suggests that Ezh2 levels may decrease in old stem cells, leading to the activation of the Ink4-Arf locus and subsequent decrease in stem cell self-renewal. To test this, it will be very interesting to determine if Ezh2 levels decrease and if PcGs are displaced from the Ink4a-Arf locus in stem cells of old mice compared with those from young mice.
In cancer the levels of EZH2 or BMI1 are frequently found to be up-regulated, and speculations have been made that this may contribute to cancer development (Pasini et al. 2004a; Raaphorst 2005). One possibility is that it may allow cancer stem cells to avoid stress/age-induced senescence, thereby increasing the chance of acquiring additional mutations on the road to becoming full-blown tumors. However, surprisingly, unlike BMI1, ectopically expressed EZH2 is not recruited to the INK4A-ARF locus and therefore is incapable of conferring repression in TIG3 cells (data not shown). This could be because EZH2, a gene whose expression is very tightly associated with proliferation status, is not limiting in unstressed, asynchronously growing embryonic fibroblasts. Interestingly, in a previous study we found that ectopically expressed EZH2 did not confer a proliferative advantage to asynchronously growing MEFs, whereas it did when cells were plated under stressful conditions (Bracken et al. 2003). Mechanistically, it is possible that another component of the PRC2 complex, an associated factor, or a transcription factor capable of recruiting PRC2 to the locus is also limiting. Future studies will be required to determine the capability of ectopically expressed EZH2 to associate and repress the INK4A-ARF locus in different cellular contexts such as in adult stem cells or cells of epithelial origins. Despite this, the very presence of EZH2 on the INK4A promoter, as reported here, raises the exciting possibility that it contributes directly to its epigenetic silencing by DNA methylation (Esteller et al. 2001). However, since the INK4A promoter is not methylated in normal cells, there must be a trigger independent of EZH2 to induce methylation during cancer progression.
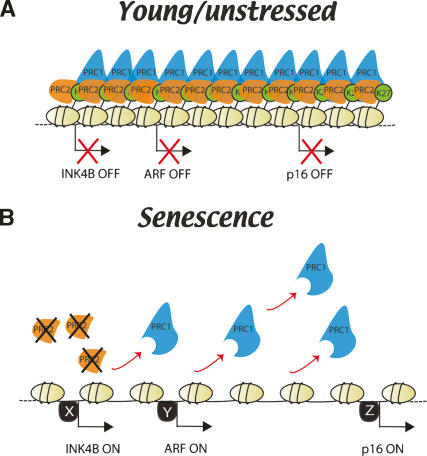
Finally, our results should provide a valuable model for future studies of the activation of the INK4B, INK4A, and ARF genes in response to various cellular signals. One can envision that, although the promoters of the three genes may respond to different signals and different promoter-specific transcription factors, the PcGs may act as master regulators coordinately determining the relative access of transcription factors (Fig. 5). Furthermore, we predict that the level of H3K27me3 along the INK4B and INK4A-ARF loci in vivo acts as an epigenetic index or memory of prior aging and stress.
Figure 5.
Model for how PcG proteins regulate cellular senescence in vivo. (A) In young proliferating normal diploid cells, the PRC2 complex is highly abundant and maintains the levels of H3K27me3 along the INK4B and INK4A-ARF loci. This in turn ensures the continued association of the BMI1-containing PRC1 complex and repression of the INK4A, INK4B, and ARF genes. (B) In older or stressed cells the levels of EZH2 are down-regulated, leading to the disruption of the PRC2 complex and loss of H3K27me3 along the INK4B and INK4A-ARF loci. Without H3K27me3, the BMI1-containing PRC1 complex is displaced. We propose a model in which cumulative stress leads to the progressive alteration of the chromatin state around the INK4B and INK4A-ARF loci, thereby conferring accessibility to specific transcriptional activators (X, Y and Z).
Materials and methods
Tissue culture
The human diploid lung embryonic fibroblast cell line TIG3 was grown in DMEM supplemented with 10% (v/v) FCS. Primary MEFs were prepared from C57BL6 embryos taken at embryonic day 13.5 during embryogenesis. SA-β-gal assays were performed as previously described (Narita et al. 2003).
Generation of antibodies
Mouse monoclonal antibodies for BMI1 were generated against full-length human BMI1 fused to the maltose-binding protein (MBP). Clone DC9 was used for immunoblotting. To attain an antibody capable of performing BMI1 ChIPs, we screened 15 monoclonal antibodies first for their ability to function in normal immunoprecipitation and subsequently for their ability to efficiently coimmunoprecipitate histone H3 in ChIP conditions. Clone AF27 was selected as the only antibody capable of immunoprecipitating both BMI1 and H3 in ChIP conditions. Two polyclonal CBX7 antibodies, “GAPEL” and “RELF,” were produced in rabbits using synthetic peptides corresponding to amino acids 121–144 (GAPELVDKGPLVPTLPFPLRKPRK[K]) and 154–178 (KKFPPRGPNL ESHSHRRELFLQEPP) of human CBX7, respectively. These were coupled to Keyhole Limpet Hemocyanin (KLH) through the C- or N-terminal lysine residues, respectively, and injected subcutanously into rabbits according to standard procedures (DAKO). Specific antibodies from positive sera were affinity-purified on the respective peptide antigens according to standard procedures. For ChIP assays, 5 μg of a pool of both affinity-purified antibodies were used per immunoprecipitation.
Western blot analysis
Immunoblotting was performed with the following antibodies: rabbit anti-Suz12 (Upstate Biotechnology), mouse anti-EZH2 (BD43) (Pasini et al. 2004b), mouse anti-EED (Bracken et al. 2003), mouse anti-BMI1 (DC9, described above), mouse anti-p16 (DCS50), and rabbit anti-p19 (Abcam).
ChIP assays
ChIPs were performed and analyzed essentially as described (Bracken et al. 2003). The antibodies used were rabbit anti-H3K27me3 (07-449; Upstate Biotechnology), anti-SUZ12 (07-379; Upstate Biotechnology), anti-PolII (sc-899; Santa Cruz Biotechnology), anti-HA (sc-805; Santa Cruz Biotechnology), anti-macroH2A (07-219; Upstate Biotechnology), anti-H3K4Me3 (ab8580; Abcam), an equal mix of anti-CBX8 “LAST” and “GALD” (Bracken et al. 2006), an equal mix of anti-CBX7 “GAPEL” and “RELF” described above, and mouse monoclonals specific for EZH2 (AC22) (Pasini et al. 2004b) and BMI1 (AF27) described above. For normal ChIPs, the immunoprecipitated DNA was quantified by real-time qPCR. The sequences of the PCR primers are available on request. Instead, for the ChIP-on-chip analysis, the immunoprecipitated DNA was amplified by LMPCR as described previously (Bracken et al. 2006) and hybridized to NimbleGen (http://www.nimblegen.com) tiled arrays spanning 50 kb of DNA spanning positions 21,953,000 to 22,003,000 on chromosome 9 according to the University of California at Santa Cruz Genome Browser (hg17) at http://genome.ucsc.edu/cgi-bin/hgGateway. High-resolution mapping was achieved by representing 690 probes of 50 base pairs (bp) with an average spacing of 22 bp between probes.
Quantification of mRNA levels by qPCR
cDNA was generated by RT–PCR using the PE Applied Biosystems TaqMan Reverse Transcription Reagents. Relative mRNA levels were determined using the SyBR Green I detection chemistry system (Applied Biosystems), using an ABI Prism 7300 Sequence Detection System. Ubiquitin was used as a control gene for normalization. The sequences of the primers used are listed in Supplementary Table 1.
Acknowledgments
We thank Eros Lazzerini Denchi for critical reading of the manuscript and Anders Lund and members of the Helin laboratory for very helpful discussions. We thank Michael Lees for producing some of the MEFs used in this study. This work was supported by grants from the Association for International Cancer Research, the Danish Cancer Society, the Novo Nordisk Foundation, the Danish Medical Research Council, the Danish Natural Science Research Council, and the EU-FP6 programs “INTACT” and “DIAMONDS.”
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.415507
References
- Alcorta D.A., Xiong Y., Phelps D., Hannon G., Beach D., Barrett J.C., Xiong Y., Phelps D., Hannon G., Beach D., Barrett J.C., Phelps D., Hannon G., Beach D., Barrett J.C., Hannon G., Beach D., Barrett J.C., Beach D., Barrett J.C., Barrett J.C. Involvement of the cyclin-dependent kinase inhibitor p16 (INK4a) in replicative senescence of normal human fibroblasts. Proc. Natl. Acad. Sci. 1996;93:13742–13747. doi: 10.1073/pnas.93.24.13742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer L.A., Plath K., Zeitlinger J., Brambrink T., Medeiros L.A., Lee T.I., Levine S.S., Wernig M., Tajonar A., Ray M.K., Plath K., Zeitlinger J., Brambrink T., Medeiros L.A., Lee T.I., Levine S.S., Wernig M., Tajonar A., Ray M.K., Zeitlinger J., Brambrink T., Medeiros L.A., Lee T.I., Levine S.S., Wernig M., Tajonar A., Ray M.K., Brambrink T., Medeiros L.A., Lee T.I., Levine S.S., Wernig M., Tajonar A., Ray M.K., Medeiros L.A., Lee T.I., Levine S.S., Wernig M., Tajonar A., Ray M.K., Lee T.I., Levine S.S., Wernig M., Tajonar A., Ray M.K., Levine S.S., Wernig M., Tajonar A., Ray M.K., Wernig M., Tajonar A., Ray M.K., Tajonar A., Ray M.K., Ray M.K., et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- Bracken A.P., Pasini D., Capra M., Prosperini E., Colli E., Helin K., Pasini D., Capra M., Prosperini E., Colli E., Helin K., Capra M., Prosperini E., Colli E., Helin K., Prosperini E., Colli E., Helin K., Colli E., Helin K., Helin K. EZH2 is downstream of the pRB–E2F pathway, essential for proliferation and amplified in cancer. EMBO J. 2003;22:5323–5335. doi: 10.1093/emboj/cdg542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracken A.P., Dietrich N., Pasini D., Hansen K.H., Helin K., Dietrich N., Pasini D., Hansen K.H., Helin K., Pasini D., Hansen K.H., Helin K., Hansen K.H., Helin K., Helin K. Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes & Dev. 2006;20:1123–1136. doi: 10.1101/gad.381706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruggeman S.W., Valk-Lingbeek M.E., van der Stoop P.P., Jacobs J.J., Kieboom K., Tanger E., Hulsman D., Leung C., Arsenijevic Y., Marino S., Valk-Lingbeek M.E., van der Stoop P.P., Jacobs J.J., Kieboom K., Tanger E., Hulsman D., Leung C., Arsenijevic Y., Marino S., van der Stoop P.P., Jacobs J.J., Kieboom K., Tanger E., Hulsman D., Leung C., Arsenijevic Y., Marino S., Jacobs J.J., Kieboom K., Tanger E., Hulsman D., Leung C., Arsenijevic Y., Marino S., Kieboom K., Tanger E., Hulsman D., Leung C., Arsenijevic Y., Marino S., Tanger E., Hulsman D., Leung C., Arsenijevic Y., Marino S., Hulsman D., Leung C., Arsenijevic Y., Marino S., Leung C., Arsenijevic Y., Marino S., Arsenijevic Y., Marino S., Marino S., et al. Ink4a and Arf differentially affect cell proliferation and neural stem cell self-renewal in Bmi1-deficient mice. Genes & Dev. 2005;19:1438–1443. doi: 10.1101/gad.1299305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao R., Zhang Y., Zhang Y. The functions of E(Z)/EZH2-mediated methylation of lysine 27 in histone H3. Curr. Opin. Genet. Dev. 2004;14:155–164. doi: 10.1016/j.gde.2004.02.001. [DOI] [PubMed] [Google Scholar]
- Dietrich N., Bracken A.P., Trinh E., Schjerling C.K., Koscki H., Rappsilber J., Helin K., Hansen K.H., Bracken A.P., Trinh E., Schjerling C.K., Koscki H., Rappsilber J., Helin K., Hansen K.H., Trinh E., Schjerling C.K., Koscki H., Rappsilber J., Helin K., Hansen K.H., Schjerling C.K., Koscki H., Rappsilber J., Helin K., Hansen K.H., Koscki H., Rappsilber J., Helin K., Hansen K.H., Rappsilber J., Helin K., Hansen K.H., Helin K., Hansen K.H., Hansen K.H. Bypass of senescence by the Polycomb group protein CBX8 through direct binding to the INK4A-ARF locus. EMBO J. 2007 doi: 10.1038/sj.emboj.7601632. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimri G.P. What has senescence got to do with cancer? Cancer Cell. 2005;7:505–512. doi: 10.1016/j.ccr.2005.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteller M., Corn P.G., Baylin S.B., Herman J.G., Corn P.G., Baylin S.B., Herman J.G., Baylin S.B., Herman J.G., Herman J.G. A gene hypermethylation profile of human cancer. Cancer Res. 2001;61:3225–3229. [PubMed] [Google Scholar]
- Gil J., Peters G., Peters G. Regulation of the INK4b-ARF–INK4a tumour suppressor locus: All for one or one for all. Nat. Rev. Mol. Cell Biol. 2006;7:667–677. doi: 10.1038/nrm1987. [DOI] [PubMed] [Google Scholar]
- Gil J., Bernard D., Martinez D., Beach D., Bernard D., Martinez D., Beach D., Martinez D., Beach D., Beach D. Polycomb CBX7 has a unifying role in cellular lifespan. Nat. Cell Biol. 2004;6:67–72. doi: 10.1038/ncb1077. [DOI] [PubMed] [Google Scholar]
- Henikoff S., Furuyama T., Ahmad K., Furuyama T., Ahmad K., Ahmad K. Histone variants, nucleosome assembly and epigenetic inheritance. Trends Genet. 2004;20:320–326. doi: 10.1016/j.tig.2004.05.004. [DOI] [PubMed] [Google Scholar]
- Hernandez-Munoz I., Taghavi P., Kuijl C., Neefjes J., van Lohuizen M., Taghavi P., Kuijl C., Neefjes J., van Lohuizen M., Kuijl C., Neefjes J., van Lohuizen M., Neefjes J., van Lohuizen M., van Lohuizen M. Association of BMI1 with polycomb bodies is dynamic and requires PRC2/EZH2 and the maintenance DNA methyltransferase DNMT1. Mol. Cell. Biol. 2005;25:11047–11058. doi: 10.1128/MCB.25.24.11047-11058.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itahana K., Zou Y., Itahana Y., Martinez J.L., Beausejour C., Jacobs J.J., Van Lohuizen M., Band V., Campisi J., Dimri G.P., Zou Y., Itahana Y., Martinez J.L., Beausejour C., Jacobs J.J., Van Lohuizen M., Band V., Campisi J., Dimri G.P., Itahana Y., Martinez J.L., Beausejour C., Jacobs J.J., Van Lohuizen M., Band V., Campisi J., Dimri G.P., Martinez J.L., Beausejour C., Jacobs J.J., Van Lohuizen M., Band V., Campisi J., Dimri G.P., Beausejour C., Jacobs J.J., Van Lohuizen M., Band V., Campisi J., Dimri G.P., Jacobs J.J., Van Lohuizen M., Band V., Campisi J., Dimri G.P., Van Lohuizen M., Band V., Campisi J., Dimri G.P., Band V., Campisi J., Dimri G.P., Campisi J., Dimri G.P., Dimri G.P. Control of the replicative life span of human fibroblasts by p16 and the polycomb protein Bmi-1. Mol. Cell. Biol. 2003;23:389–401. doi: 10.1128/MCB.23.1.389-401.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs J.J., Kieboom K., Marino S., DePinho R.A., van Lohuizen M., Kieboom K., Marino S., DePinho R.A., van Lohuizen M., Marino S., DePinho R.A., van Lohuizen M., DePinho R.A., van Lohuizen M., van Lohuizen M. The oncogene and Polycomb-group gene bmi-1 regulates cell proliferation and senescence through the ink4a locus. Nature. 1999;397:164–168. doi: 10.1038/16476. [DOI] [PubMed] [Google Scholar]
- Janzen V., Forkert R., Fleming H.E., Saito Y., Waring M.T., Dombkowski D.M., Cheng T., DePinho R.A., Sharpless N.E., Scadden D.T., Forkert R., Fleming H.E., Saito Y., Waring M.T., Dombkowski D.M., Cheng T., DePinho R.A., Sharpless N.E., Scadden D.T., Fleming H.E., Saito Y., Waring M.T., Dombkowski D.M., Cheng T., DePinho R.A., Sharpless N.E., Scadden D.T., Saito Y., Waring M.T., Dombkowski D.M., Cheng T., DePinho R.A., Sharpless N.E., Scadden D.T., Waring M.T., Dombkowski D.M., Cheng T., DePinho R.A., Sharpless N.E., Scadden D.T., Dombkowski D.M., Cheng T., DePinho R.A., Sharpless N.E., Scadden D.T., Cheng T., DePinho R.A., Sharpless N.E., Scadden D.T., DePinho R.A., Sharpless N.E., Scadden D.T., Sharpless N.E., Scadden D.T., Scadden D.T. Stem-cell ageing modified by the cyclin-dependent kinase inhibitor p16INK4a. Nature. 2006;443:421–426. doi: 10.1038/nature05159. [DOI] [PubMed] [Google Scholar]
- Kamminga L.M., Bystrykh L.V., de Boer A., Houwer S., Douma J., Weersing E., Dontje B., de Haan G., Bystrykh L.V., de Boer A., Houwer S., Douma J., Weersing E., Dontje B., de Haan G., de Boer A., Houwer S., Douma J., Weersing E., Dontje B., de Haan G., Houwer S., Douma J., Weersing E., Dontje B., de Haan G., Douma J., Weersing E., Dontje B., de Haan G., Weersing E., Dontje B., de Haan G., Dontje B., de Haan G., de Haan G. The Polycomb group gene Ezh2 prevents hematopoietic stem cell exhaustion. Blood. 2006;107:2170–2179. doi: 10.1182/blood-2005-09-3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W.Y., Sharpless N.E., Sharpless N.E. The regulation of INK4/ARF in cancer and aging. Cell. 2006;127:265–275. doi: 10.1016/j.cell.2006.10.003. [DOI] [PubMed] [Google Scholar]
- Krishnamurthy J., Ramsey M.R., Ligon K.L., Torrice C., Koh A., Bonner-Weir S., Sharpless N.E., Ramsey M.R., Ligon K.L., Torrice C., Koh A., Bonner-Weir S., Sharpless N.E., Ligon K.L., Torrice C., Koh A., Bonner-Weir S., Sharpless N.E., Torrice C., Koh A., Bonner-Weir S., Sharpless N.E., Koh A., Bonner-Weir S., Sharpless N.E., Bonner-Weir S., Sharpless N.E., Sharpless N.E. p16INK4a induces an age-dependent decline in islet regenerative potential. Nature. 2006;443:453–457. doi: 10.1038/nature05092. [DOI] [PubMed] [Google Scholar]
- Lee T.I., Jenner R.G., Boyer L.A., Guenther M.G., Levine S.S., Kumar R.M., Chevalier B., Johnstone S.E., Cole M.F., Isono K., Jenner R.G., Boyer L.A., Guenther M.G., Levine S.S., Kumar R.M., Chevalier B., Johnstone S.E., Cole M.F., Isono K., Boyer L.A., Guenther M.G., Levine S.S., Kumar R.M., Chevalier B., Johnstone S.E., Cole M.F., Isono K., Guenther M.G., Levine S.S., Kumar R.M., Chevalier B., Johnstone S.E., Cole M.F., Isono K., Levine S.S., Kumar R.M., Chevalier B., Johnstone S.E., Cole M.F., Isono K., Kumar R.M., Chevalier B., Johnstone S.E., Cole M.F., Isono K., Chevalier B., Johnstone S.E., Cole M.F., Isono K., Johnstone S.E., Cole M.F., Isono K., Cole M.F., Isono K., Isono K., et al. Control of developmental regulators by polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessard J., Sauvageau G., Sauvageau G. Bmi-1 determines the proliferative capacity of normal and leukaemic stem cells. Nature. 2003;423:255–260. doi: 10.1038/nature01572. [DOI] [PubMed] [Google Scholar]
- Leung C., Lingbeek M., Shakhova O., Liu J., Tanger E., Saremaslani P., Van Lohuizen M., Marino S., Lingbeek M., Shakhova O., Liu J., Tanger E., Saremaslani P., Van Lohuizen M., Marino S., Shakhova O., Liu J., Tanger E., Saremaslani P., Van Lohuizen M., Marino S., Liu J., Tanger E., Saremaslani P., Van Lohuizen M., Marino S., Tanger E., Saremaslani P., Van Lohuizen M., Marino S., Saremaslani P., Van Lohuizen M., Marino S., Van Lohuizen M., Marino S., Marino S. Bmi1 is essential for cerebellar development and is overexpressed in human medulloblastomas. Nature. 2004;428:337–341. doi: 10.1038/nature02385. [DOI] [PubMed] [Google Scholar]
- Levine S.S., King I.F., Kingston R.E., King I.F., Kingston R.E., Kingston R.E. Division of labor in polycomb group repression. Trends Biochem. Sci. 2004;29:478–485. doi: 10.1016/j.tibs.2004.07.007. [DOI] [PubMed] [Google Scholar]
- Lowe S.W., Sherr C.J., Sherr C.J. Tumor suppression by Ink4a-Arf: Progress and puzzles. Curr. Opin. Genet. Dev. 2003;13:77–83. doi: 10.1016/s0959-437x(02)00013-8. [DOI] [PubMed] [Google Scholar]
- Molofsky A.V., Pardal R., Iwashita T., Park I.K., Clarke M.F., Morrison S.J., Pardal R., Iwashita T., Park I.K., Clarke M.F., Morrison S.J., Iwashita T., Park I.K., Clarke M.F., Morrison S.J., Park I.K., Clarke M.F., Morrison S.J., Clarke M.F., Morrison S.J., Morrison S.J. Bmi-1 dependence distinguishes neural stem cell self-renewal from progenitor proliferation. Nature. 2003;425:962–967. doi: 10.1038/nature02060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molofsky A.V., He S., Bydon M., Morrison S.J., Pardal R., He S., Bydon M., Morrison S.J., Pardal R., Bydon M., Morrison S.J., Pardal R., Morrison S.J., Pardal R., Pardal R. Bmi-1 promotes neural stem cell self-renewal and neural development but not mouse growth and survival by repressing the p16Ink4a and p19Arf senescence pathways. Genes & Dev. 2005;19:1432–1437. doi: 10.1101/gad.1299505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molofsky A.V., Slutsky S.G., Joseph N.M., He S., Pardal R., Krishnamurthy J., Sharpless N.E., Morrison S.J., Slutsky S.G., Joseph N.M., He S., Pardal R., Krishnamurthy J., Sharpless N.E., Morrison S.J., Joseph N.M., He S., Pardal R., Krishnamurthy J., Sharpless N.E., Morrison S.J., He S., Pardal R., Krishnamurthy J., Sharpless N.E., Morrison S.J., Pardal R., Krishnamurthy J., Sharpless N.E., Morrison S.J., Krishnamurthy J., Sharpless N.E., Morrison S.J., Sharpless N.E., Morrison S.J., Morrison S.J. Increasing p16INK4a expression decreases forebrain progenitors and neurogenesis during ageing. Nature. 2006;443:448–452. doi: 10.1038/nature05091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narita M., Lowe S.W., Lowe S.W. Senescence comes of age. Nat. Med. 2005;11:920–922. doi: 10.1038/nm0905-920. [DOI] [PubMed] [Google Scholar]
- Narita M., Nunez S., Heard E., Narita M., Lin A.W., Hearn S.A., Spector D.L., Hannon G.J., Lowe S.W., Nunez S., Heard E., Narita M., Lin A.W., Hearn S.A., Spector D.L., Hannon G.J., Lowe S.W., Heard E., Narita M., Lin A.W., Hearn S.A., Spector D.L., Hannon G.J., Lowe S.W., Narita M., Lin A.W., Hearn S.A., Spector D.L., Hannon G.J., Lowe S.W., Lin A.W., Hearn S.A., Spector D.L., Hannon G.J., Lowe S.W., Hearn S.A., Spector D.L., Hannon G.J., Lowe S.W., Spector D.L., Hannon G.J., Lowe S.W., Hannon G.J., Lowe S.W., Lowe S.W. Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell. 2003;113:703–716. doi: 10.1016/s0092-8674(03)00401-x. [DOI] [PubMed] [Google Scholar]
- Oruetxebarria I., Venturini F., Kekarainen T., Houweling A., Zuijderduijn L.M., Mohd-Sarip A., Vries R.G., Hoeben R.C., Verrijzer C.P., Venturini F., Kekarainen T., Houweling A., Zuijderduijn L.M., Mohd-Sarip A., Vries R.G., Hoeben R.C., Verrijzer C.P., Kekarainen T., Houweling A., Zuijderduijn L.M., Mohd-Sarip A., Vries R.G., Hoeben R.C., Verrijzer C.P., Houweling A., Zuijderduijn L.M., Mohd-Sarip A., Vries R.G., Hoeben R.C., Verrijzer C.P., Zuijderduijn L.M., Mohd-Sarip A., Vries R.G., Hoeben R.C., Verrijzer C.P., Mohd-Sarip A., Vries R.G., Hoeben R.C., Verrijzer C.P., Vries R.G., Hoeben R.C., Verrijzer C.P., Hoeben R.C., Verrijzer C.P., Verrijzer C.P. P16INK4a is required for hSNF5 chromatin remodeler-induced cellular senescence in malignant rhabdoid tumor cells. J. Biol. Chem. 2004;279:3807–3816. doi: 10.1074/jbc.M309333200. [DOI] [PubMed] [Google Scholar]
- Park I.K., Qian D., Kiel M., Becker M.W., Pihalja M., Weissman I.L., Morrison S.J., Clarke M.F., Qian D., Kiel M., Becker M.W., Pihalja M., Weissman I.L., Morrison S.J., Clarke M.F., Kiel M., Becker M.W., Pihalja M., Weissman I.L., Morrison S.J., Clarke M.F., Becker M.W., Pihalja M., Weissman I.L., Morrison S.J., Clarke M.F., Pihalja M., Weissman I.L., Morrison S.J., Clarke M.F., Weissman I.L., Morrison S.J., Clarke M.F., Morrison S.J., Clarke M.F., Clarke M.F. Bmi-1 is required for maintenance of adult self-renewing haematopoietic stem cells. Nature. 2003;423:302–305. doi: 10.1038/nature01587. [DOI] [PubMed] [Google Scholar]
- Pasini D., Bracken A.P., Helin K., Bracken A.P., Helin K., Helin K. Polycomb group proteins in cell cycle progression and cancer. Cell Cycle. 2004a;3:396–400. [PubMed] [Google Scholar]
- Pasini D., Bracken A.P., Jensen M.R., Lazzerini Denchi E., Helin K., Bracken A.P., Jensen M.R., Lazzerini Denchi E., Helin K., Jensen M.R., Lazzerini Denchi E., Helin K., Lazzerini Denchi E., Helin K., Helin K. Suz12 is essential for mouse development and for EZH2 histone methyltransferase activity. EMBO J. 2004b;23:4061–4071. doi: 10.1038/sj.emboj.7600402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poux S., Melfi R., Pirrotta V., Melfi R., Pirrotta V., Pirrotta V. Establishment of Polycomb silencing requires a transient interaction between PC and ESC. Genes & Dev. 2001;15:2509–2514. doi: 10.1101/gad.208901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raaphorst F.M. Deregulated expression of Polycomb-group oncogenes in human malignant lymphomas and epithelial tumors. Hum. Mol. Genet. 2005;14:R93–R100. doi: 10.1093/hmg/ddi111. [DOI] [PubMed] [Google Scholar]
- Rastelli L., Chan C.S., Pirrotta V., Chan C.S., Pirrotta V., Pirrotta V. Related chromosome binding sites for zeste, suppressors of zeste and Polycomb group proteins in Drosophila and their dependence on Enhancer of zeste function. EMBO J. 1993;12:1513–1522. doi: 10.1002/j.1460-2075.1993.tb05795.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squazzo S.L., O’Geen H., Komashko V.M., Krig S.R., Jin V.X., Jang S., Margueron R., Reinberg D., Green R., Farnham P.J., O’Geen H., Komashko V.M., Krig S.R., Jin V.X., Jang S., Margueron R., Reinberg D., Green R., Farnham P.J., Komashko V.M., Krig S.R., Jin V.X., Jang S., Margueron R., Reinberg D., Green R., Farnham P.J., Krig S.R., Jin V.X., Jang S., Margueron R., Reinberg D., Green R., Farnham P.J., Jin V.X., Jang S., Margueron R., Reinberg D., Green R., Farnham P.J., Jang S., Margueron R., Reinberg D., Green R., Farnham P.J., Margueron R., Reinberg D., Green R., Farnham P.J., Reinberg D., Green R., Farnham P.J., Green R., Farnham P.J., Farnham P.J. Suz12 binds to silenced regions of the genomein a cell-type-specific manner. Genome Res. 2006;16:890–900. doi: 10.1101/gr.5306606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X., Milyavsky M., Shats I., Erez N., Goldfinger N., Rotter V., Milyavsky M., Shats I., Erez N., Goldfinger N., Rotter V., Shats I., Erez N., Goldfinger N., Rotter V., Erez N., Goldfinger N., Rotter V., Goldfinger N., Rotter V., Rotter V. Activated p53 suppresses the histone methyltransferase EZH2 gene. Oncogene. 2004;23:5759–5769. doi: 10.1038/sj.onc.1207706. [DOI] [PubMed] [Google Scholar]
- Trojer P., Reinberg D., Reinberg D. Histone lysine demethylases and their impact on epigenetics. Cell. 2006;125:213–217. doi: 10.1016/j.cell.2006.04.003. [DOI] [PubMed] [Google Scholar]
- Wei W., Hemmer R.M., Sedivy J.M., Hemmer R.M., Sedivy J.M., Sedivy J.M. Role of p14(ARF) in replicative and induced senescence of human fibroblasts. Mol. Cell. Biol. 2001;21:6748–6757. doi: 10.1128/MCB.21.20.6748-6757.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]