Abstract
Epigenetic maintenance of the expression state of the genome is critical for development. Drosophila GAGA factor interacts with FACT and modulates chromatin structure for the maintenance of gene expression. Here we show that the GAGA factor–FACT complex and its binding site just downstream from the white gene are crucial for position effect variegation. Interestingly there is a dip of histone H3 Lys 9 methylation and a peak of H3 Lys 4 methylation at this site. The GAGA factor and FACT direct replacement of histone H3 by H3.3 through association of HIRA at this site, and maintain white expression under the heterochromatin environment. Based on these findings we propose that the GAGA factor and FACT-dependent replacement of Lys 9-methylated histone H3 by H3.3 counteracts the spreading of silent chromatin.
Keywords: GAGA factor, FACT, histone H3.3, position effect variegation, heterochromatin
Chromatin can transmit genetic information without changes in primary DNA sequence from one cell generation to the next. The method of transmission, termed epigenetic gene expression, depends on post-translational modifications of four core histones. Recent studies have demonstrated that the degree or the pattern of the selected histone modification on the nucleosome is tightly correlated with the regulation of gene expression (Jenuwein and Allis 2001). Two types of epigenetic gene expression have been studied extensively in Drosophila. One is maintenance of Hox gene expression governed by Polycomb and trithorax group genes (Simon and Tamkun 2002; Ringrose and Paro 2004). The other is position effect variegation (PEV) (Reuter and Spierer 1992; Grewal and Elgin 2002). Silencing and counteracting maintenance of the active state are involved in the phenomena. The silencing is achieved through methylation of histone H3 at K27 and/or K9 followed by binding of Polycomb group or heterochromatin proteins recognizing these silent marks (Rea et al. 2000; Czermin et al. 2002; Grewal and Elgin 2002; Müller et al. 2002; Ringrose and Paro 2004). To elucidate the mechanism for the maintenance of the active state, we focused on the Trithorax-like (Trl) gene encoding the GAGA factor. Trl is a member of trithorax group, and Trl mutation is an enhancer of PEV (Farkas et al. 1994), suggesting a common mechanism underlying the maintenance of Hox gene expression and PEV. Previous study has revealed that the GAGA factor interacts with a Drosophila counterpart of human FACT (Orphanides et al. 1998, 1999) and that the GAGA factor–FACT complex facilitates chromatin remodeling and contributes to the maintenance of Hox gene expression (Shimojima et al. 2003). However, it remained elusive how the GAGA factor-triggered chromatin alterations lead to the maintenance of gene expression. Here we addressed the issue by focusing on PEV. Our data demonstrate a critical role for the GAGA factor and FACT in the replacement of K9-methylated histone H3 by H3.3 to counteract the spreading of silent chromatin.
Results
GAGA factor–FACT complex is involved in PEV
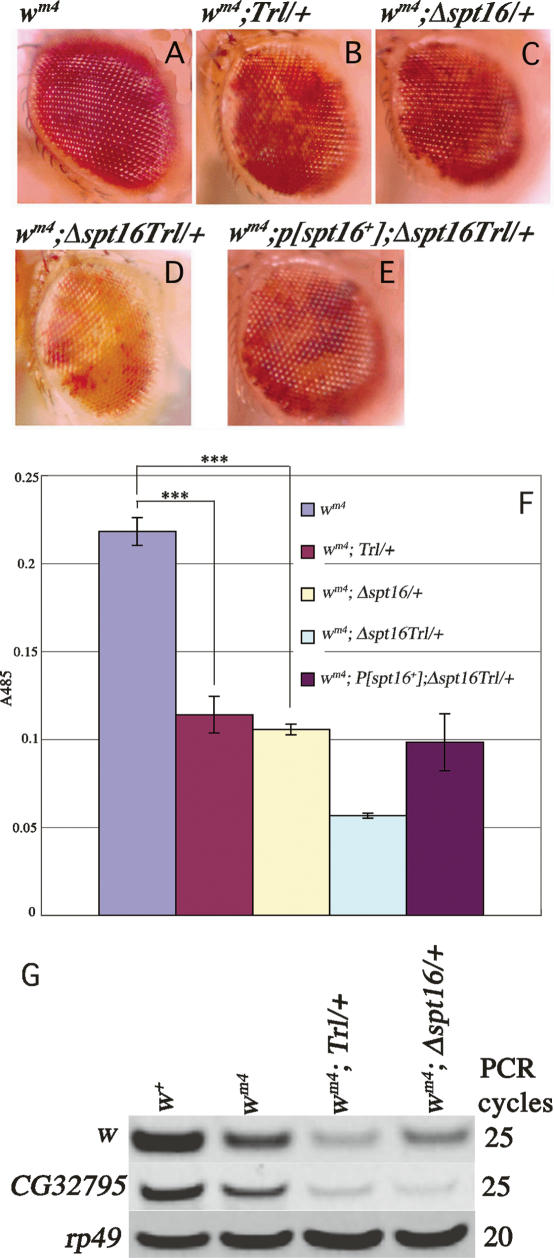
When an actively transcribed white (w) gene is juxtaposed with centromeric heterochromatin by chromosome inversion such as In (1) wm4 (Fig. 2A, below), its expression is subject to variable but heritable silencing, which gives rise to variegated eye color. This phenomenon, termed PEV, provides evidence for a critical role for chromatin structure in gene expression (Muller 1930; Tartof et al. 1984). In our genetic background, In (1) wm4 showed almost red eye with small white spots (Fig. 1A). The variegated eye color phenotype of wm4 was enhanced in a Trl13C/+ line as revealed by expanded white regions (Fig. 1, A vs. B, F; P < 0.001). This confirms the previous conclusion that Trl is an enhancer of PEV (Farkas et al. 1994). Similarly, the PEV was enhanced when a single dose of spt16 was removed in the wm4 background (Fig. 1, A vs. C, F; P < 0.001). The PEV was further enhanced in flies doubly heterozygous for Trl13C and Δspt16 than in each single heterozygote (Fig. 1, D vs. B,C, F ). This effect was reversed by expression of dSPT16 from a transgene (Fig. 1, E vs. D, F ). As a negative control, reduction of a single dose of scf (Kobayashi et al. 1998) did not enhance the PEV of wm4 (data not shown). These results indicate that the GAGA factor–FACT complex plays a role in PEV.
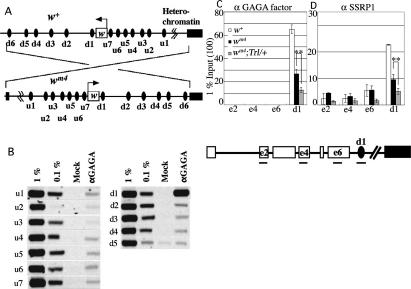
Figure 2.
GAGA factor–FACT complex is present on d1. (A) Schematic presentation of chromosomal inversion in wm4. Arrows indicate the direction of w transcription. u1–u7 and d1–d5 represent potential GAGA factor-binding sequences around w. d6 is a site abutting the proximal break point of wm4 inversion. (B) ChIP assay for the presence of GAGA factor around the w locus in the w+ context. (C,D) Semiquantitative ChIP assays for occupancies of GAGA factor (C) and FACT subunit dSSRP1 (D) on w exons, d1 and d6 in w+, wm4, and wm4;Trl13C/+. e2, e4, and e6 represent w exons 2, 4, and 6, respectively. Asterisks indicate statistical significance: (**) P < 0.005.
Figure 1.
GAGA factor–FACT complex is involved in PEV. (A–F) Functional interaction between Trl and Spt16 as revealed by enhancement of PEV. Male eyes of wm4 (A), wm4;Trl13C/+ (B), wm4;Δspt16/+ (C), wm4;Δspt16 Trl13C/+ (D), and wm4;P[spt16+];Δspt16 Trl13C/+ (E). (F) Eye pigmentation in the indicated lines. Error bars represent SD throughout the paper. Asterisks indicate statistical significance: (***) P < 0.001. (G) RT–PCR for relative abundance of w, CG32795, and rp49 mRNAs.
To confirm the involvement of the GAGA factor–FACT complex in PEV, we compared w expression in embryos 10–22 h after egg laying (AEL) among w+, wm4, wm4;Trl13C/+ and wm4;Δspt16/+ lines. At this stage, w is expressed mainly in the Malpigian tubules (Fjose et al. 1984), and its expression is subject to PEV (Schultz 1956). Actually, the Trl13C/+ or Δspt16/+ population consisted of embryos heterozygous for the mutation (50%), homozygous for the mutation (25%), and homozygous for a balancer (25%), but we call the population simply Trl13C/+ or Δspt16/+ here. As expected, expression of w or its neighboring gene CG32795 measured by RT–PCR was lower in wm4 than in w+, and its expression was further decreased in wm4;Trl13C/+ and wm4; Δspt16/+, while expression of a control gene rp49 was almost the same among the four lines (Fig. 1G). By contrast, reduction of a single dose of either Trl or spt16 or both did not affect w or CG32795 expression when these genes are located on their normal chromosomal positions (Supplementary Fig. S1). These results support the involvement of the GAGA factor–FACT complex in PEV.
GAGA factor–FACT complex on a site just downstream from w plays a role in PEV
If the GAGA factor–FACT complex directly participates in the maintenance of w expression, these proteins should be present in the w locus. Using chromatin immunoprecipitation (ChIP) assays, we analyzed the presence of the GAGA factor on 12 potential GAGA factor-binding sequences around the w locus (Fig. 2A, u1–u7 and d1–d5) in embryos 10–22 h AEL. Significant amounts of the GAGA factor were detected only in a site termed d1 (Fig. 2B). This site seems to be important because it is present between w and heterochromatin in the wm4 context and just downstream from w. w transcription appears to terminate before d1 because no transcripts were detectable on d1 by RT–PCR (Supplementary Fig. S2).
We then compared occupancies of the GAGA factor and FACT on w exons 2, 4, 6, and d1 among w+, wm4, and wm4;Trl13C/+ lines. The GAGA factor was detectable on d1 but not on w exons. Compared with w+, the level of the GAGA factor on d1 reduced in wm4 and further reduced in wm4;Trl13C/+ (Fig. 2C). Consistent with the previous observation that virtually all GAGA factors are associated with FACT in embryonic nuclei (Shimojima et al. 2003), FACT was also present on d1, and its level reduced in a similar manner as the GAGA factor (Fig. 2D). The reduced level of FACT is not due to diminished expression because Western analyses showed that expression of the FACT subunits is normal in wm4 and wm4; Trl13C/+ (Supplementary Fig. S3). Taken together, the results presented thus far suggest that the GAGA factor–FACT complex on d1 plays a role in the maintenance of w expression under the heterochromatin environment.
Importance of d1 in PEV
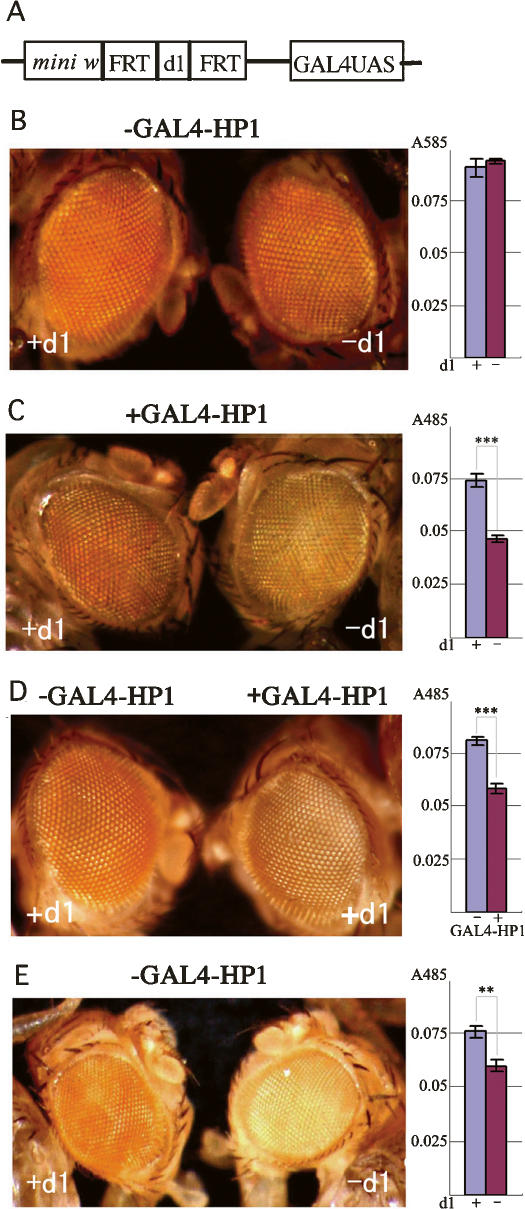
To assess the importance of d1 in the protection of gene expression from the heterochromatin spreading, we made transgenic fly lines harboring the mini w gene with the d1 sequence encompassed by FRTs, a 201-base-pair (bp) spacer DNA, and GAL4-UAS (Fig. 3A). Note that the conventional mini w terminates just before d1 and lacks it). ChIP assays showed that the GAGA factor and FACT are present at d1 on the transgene (Supplementary Fig. S4). Most of the lines showed normal red eye color before or after flip out of d1 (Fig. 3B). However, when GAL4-HP1 fusion proteins were expressed, we observed variegated eye color in the lines with d1 (Fig. 3C, left). The variegation was further enhanced in the lines without d1 (Fig. 3C, right). These results suggest an importance of d1 in PEV, but the effect could be due to the diminished distance between mini w and GAL4-UAS after excision of d1. To exclude the possibility, we made similar transgenic fly lines in which the GAGA factor-binding site was destroyed by base substitutions keeping the distance between the functional units. These lines also showed the enhanced variegation in the presence of GAL4-HP1, while the same lines showed normal red eye color in the absence of GAL4-HP1 (Fig. 3D). The enhanced levels of variegation in the presence of GAL4-HP1 were almost the same as those without d1 (Fig. 3, cf. D and C, right panel). These data reveal a critical role for the GAGA factor-binding sequence at d1 to prevent the heterochromatin spreading. One of the lines with the wild-type GAGA factor-binding sequence showed variegation even without expression of GAL4-HP1 due to the transgene insertion into a region neighboring centromeric heterochromatin on the second chromosome in the order of mini w–d1–heterochromatin (Fig. 3E, left). This variegation was also enhanced upon excision of d1 (Fig. 3E, right), demonstrating that d1 is functional at a place many kb away from the centromeric heterochromatin as in the wm4 context. These observations indicate that the d1 sequence is necessary and sufficient to counteract the heterochromatin spreading.
Figure 3.
Importance of d1 for counteracting the heterochromatin spreading. (A) Transgene construct. (B,C) Male eye and eye pigmentation of a typical transgenic line with or without d1 before (B) or after (C) expression of GAL4-HP1. (D) Male eye and eye pigmentation of a typical transgenic line carrying base substitutions in the GAGA factor-binding sequence in the presence (right panel) or absence (left panel) of GAL4-HP1. (E) Male eye and eye pigmentation of another line (B2) carrying the wild-type GAGA factor-binding sequence with or without d1. Asterisks indicate statistical significance: (**) P < 0.005; (***) P < 0.001.
d1 is a DNase-hypersensitive site
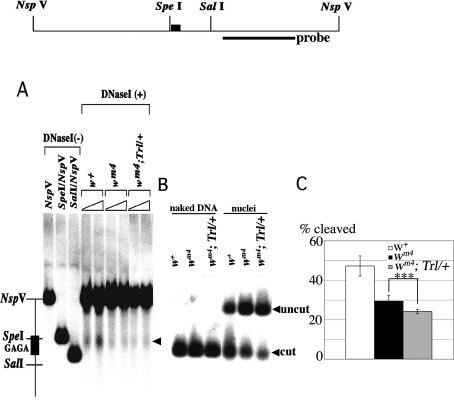
What is the mechanism underlying the protection from the heterochromatin spreading? To address the issue we analyzed the chromatin states around the w locus. As the GAGA factor–FACT complex stimulates chromatin remodeling around the GAGA factor-binding site (Shimojima et al. 2003), some alterations of chromatin structure would also occur at d1. To test the possibility, we carried out nuclease sensitivity assays using nuclei isolated from embryos 10–22 h AEL. DNase I preferentially cleaved at w+ d1 within a 1.6-kb region encompassing two NspV sites (Fig. 4A). The DNase I hypersensitivity at d1 became less prominent in wm4 and wm4;Trl13C/+ than in w+. To make quantitative comparison of the nuclease sensitivity at d1 among the three lines, we performed restriction enzyme accessibility assays. Figure 4B shows that an Spe I site neighboring the GAGAG cluster in d1 was highly accessible in w+. The accessibility reduced in wm4 and further reduced in wm4;Trl13C/+ (Fig. 4C; P < 0.001). By contrast, the accessibilities of Spe I at other chromosomal regions were low, and no significant changes were detectable among the three lines (data not shown). These results clearly show that d1 is a DNase-hypersensitive site, and suggest that the GAGA factor promotes chromatin remodeling at d1.
Figure 4.
GAGA factor induces DNase-hypersensitive site at d1. (A) DNase I hypersensitivity assay. Thick line and filled box in the restriction map represent the probe for hybridization and the GAGAG cluster in d1, respectively. The DNase I-cut products are marked by an arrowhead. (B,C) Restriction enzyme accessibility assay. A typical example of the assay (B) and a summary of the results (C). Asterisks indicate statistical significance: (**) P < 0.005; (***) P < 0.001.
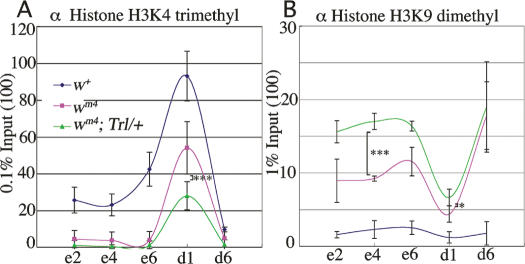
Histone H3 K4 methylation peaks and H3 K9 methylation dips at d1
Next we compared distribution of K4-methylated histone H3 and K9-methylated histone H3 around the w locus among w+, wm4 and wm4;Trl13C/+ lines. The euchromatin marker K4-methylated histone H3 was present on w+ exons and d1 and its level decreased in wm4 and further decreased in wm4;Trl13C/+ (Fig. 5A). The observed decrease in the level of K4 methylation in the Trl13C/+ line could be due to decrease in the absolute number of K4-methylated histone H3 or due to more pronounced masking of the K4-methyl epitope through the spreading of heterochromatin. Whatever the reason may be, the data are consistent with the idea that the GAGA factor counteracts the spreading of heterochromatin. By contrast, the heterochromatin marker K9-methylated histone H3 was barely detectable on w+ exons and d1, but its level increased in wm4 and further increased in wm4; Trl13C/+ (Fig. 5B). Site d6 is very close to the centromeric heterochromatin in the wm4 context (Fig. 2A) and was heavily loaded with the K9-methylation signal in wm4 and wm4; Trl13C/+ but not in w+. The level of K9 methylation on w exons in wm4;Trl13C/+ was as high as that on d6 (Fig. 5B; P < 0.001). These results strongly support the importance of the GAGA factor on d1 for protection from the heterochromatin spreading. The results also highlight interesting properties of d1: There are a dip of K9 methylation (Fig. 5B) and a peak of K4 methylation (Fig. 5A) at d1.
Figure 5.
Histone modifications around w as revealed by semiquantitative ChIP assays. Distribution of K4-methylated histone H3 (A) or K9-methylated histone H3 (B) around w in w+, wm4, and wm4;Trl13C/+. Asterisks indicate statistical significance: (*) P < 0.05; (***) P < 0.001.
GAGA factor and FACT direct histone H3 replacement by H3.3 at d1
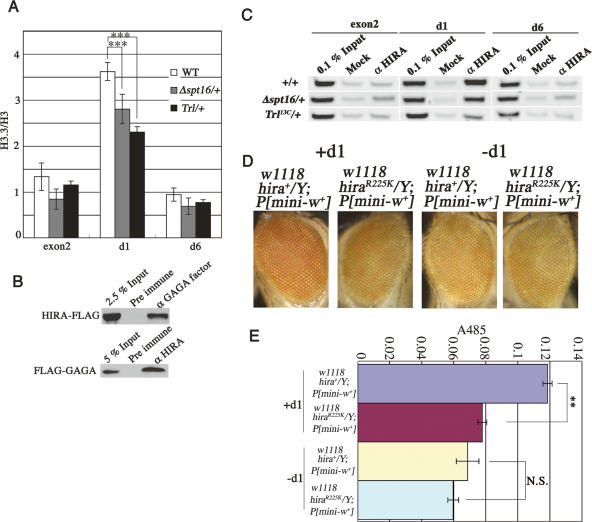
The K9 methylation could be removed at d1 by histone replacement or demethylation. We considered replication-independent replacement of histone H3 by its variant H3.3 (Ahmad and Henikoff 2002) rather than demethylation because histone H3.3 is a preferred target of K4 but not K9 methylation (McKittrick et al. 2004), while a demethylated histone H3 could be a target of K9 methylation again. To test the possibility, we made transgenic fly lines expressing Flag-tagged histone H3 or Flag-tagged histone H3.3 and analyzed occupancies of these histones on d1 and other control sites. The expression level of the tagged H3 or H3.3 was ∼1% the level of endogenous histone H3 (Supplementary Fig. S5). ChIP assays using an anti-Flag antibody showed that the histone H3.3 to H3 ratio was high at d1 compared with w exon 2 or d6 in the wild type (Fig. 6A). This is due to an increase in the H3.3 level and a decrease in the H3 level at d1 (data not shown). However, in Trl13C/+ or Δspt16/+, the ratio reduced significantly at d1 (P < 0.001) but not at exon 2 or d6 (Fig. 6A). These results suggest that the GAGA factor and FACT direct replacement of histone H3 by H3.3 at d1. As expected, reChIP assays showed that histone H3.3 but not histone H3 received K4 methylation at d1 (Supplementary Fig. S6).
Figure 6.
GAGA factor and FACT direct replacement of histone H3 by H3.3 at d1. (A) Occupancies of Flag-histone H3 or Flag-histone H3.3 at d1, exon2, and d6 as measured by semiquantitative ChIP assays using an anti-Flag antibody in wild type, Trl13C/+, or Δspt16/+. H3.3 abundance is shown by ratio of Flag-H3.3 to Flag-H3. Asterisks indicate statistical significance: (***) P < 0.001. (B) Physical interaction between GAGA factor and HIRA. (Top panel) Proteins in a nuclear extract from a transgenic line expressing Flag-HIRA from the cognate promoter (Loppin et al. 2005) were immunoprecipitated with preimmune serum or anti-GAGA factor antiserum, and detected with the anti-Flag antibody. (Bottom panel) Proteins in a nuclear extract from a transgenic line expressing Flag-GAGA factor (Shimojima et al. 2003) were immunoprecipitated with preimmune serum or anti-HIRA antiserum (Loppin et al. 2005), and detected with the anti-Flag antibody. (C) GAGA factor and FACT participate in association of HIRA to d1. ChIP assays using anti-HIRA antibodies (Loppin et al. 2005) in wild type, Trl13C/+, or Δspt16/+. (D,E) PEV is enhanced by a hira mutation in a d1-dependent manner. (D) Male eyes of the mini-w transgenic line B2 (see Fig. 3E) with the indicated genotypes with or without d1. (E) Eye pigmentation in the indicated lines. Asterisks indicate statistical significance: (**) P < 0.005. (N.S.) Not significant.
HIRA is a chaperone specific for H3.3 deposition (Tagami et al. 2004). If the GAGA factor and FACT critically participate in the replacement of histone H3 by H3.3, these factors may physically interact with HIRA. To test the possibility, we carried out immunoprecipitation experiments and ChIP assays. We found that HIRA was coimmunoprecipitated with anti-GAGA factor antibodies (Fig. 6B, top panel; Supplementary Fig. S8). In a reciprocal test, anti-HIRA antibodies immunoprecipitated the GAGA factor (Fig. 6B, bottom panel; Supplementary Fig. S8). We also observed occupancy of HIRA at d1 but not at exon 2 or d6 (Fig. 6C). The amounts of HIRA at d1 decreased to 70% and 40% of the wild-type level in Δspt16/+ and Trl13C/+, respectively. The reduced level of HIRA at d1 in Trl13C/+ or Δspt16/+ is not due to decreased expression of HIRA in these mutants (Supplementary Fig. S7). Furthermore, a mutation in the Drosophila hira gene (Loppin et al. 2005) enhanced PEV (Fig. 6D [left], E; P < 0.005). Importantly, no difference in the variegation was observed between wild type and the mutant when the GAGA factor cannot access to the w region due to excision of d1 (Fig. 6D [right], E). Taken together, these findings indicate that the GAGA factor and FACT participate association of HIRA to d1 for the histone H3.3 replacement, and prevent the spreading of heterochromatin.
GAGA factor and FACT-dependent histone H3 replacement by H3.3 in Fab-7 boundary of Abdominal-B
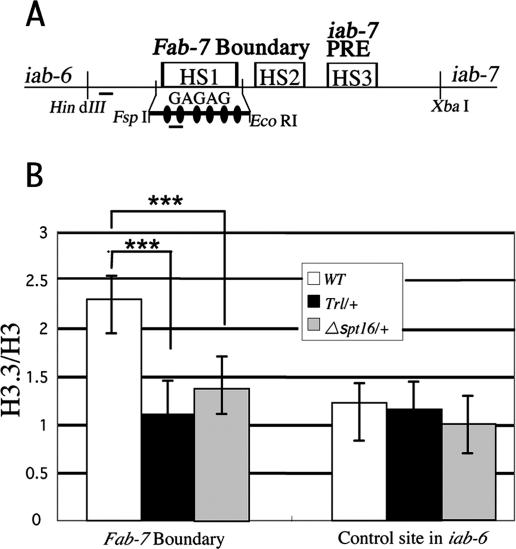
Is the GAGA factor and FACT-dependent replacement of histone H3 by H3.3 specific to d1 or a general mechanism? To address the issue, we examined the histone H3.3 to H3 ratio at the DNase-hypersensitive site (HS) 1 in the Fab-7 boundary of Abdominal-B (Abd-B). We chose the HS1 because there exists a nice cluster of six GAGAG sequences within the site (Fig. 7A). As shown in Figure 7B, the histone H3.3 to H3 ratio was high at the HS1 in the wild type. However, the ratio reduced significantly in Trl13C/+ or Δspt16/+ (P < 0.001), while these mutations did not affect the ratio at a control site lacking the GAGA factor-binding sequence. These results suggest that the GAGA factor and FACT-directed replacement of histone H3 by H3.3 also occurs at the DNase HS1 in the Fab-7 boundary of Abd-B.
Figure 7.
GAGA factor and FACT-dependent replacement of histone H3 by H3.3 in the Fab-7 boundary of Abd-B. (A) Schematic presentation of the Fab-7 region. Small horizontal bars indicate the PCR regions in the ChIP assays. Each filled oval represents the GAGAG sequence. (B) Occupancy of Flag-histone H3 or Flag-histone H3.3 at the DNase HS1 in the Fab-7 boundary and at a control site in iab-6 lacking the GAGA factor-binding sequence was measured as in Figure 6A. Asterisks indicate statistical significance: (***) P < 0.001.
Discussion
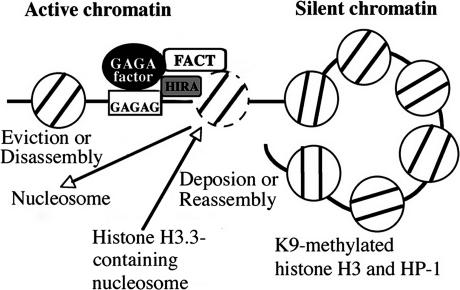
In this study, we show that the GAGA factor–FACT complex is present on d1, a site just downstream from w, and participates in PEV. d1 appears to be a peculiar site where histone H3 K4 methylation peaks and H3 K9 methylation dips, and necessary and sufficient to counteract the heterochromatin spreading. The GAGA factor and FACT contribute to replacement of histone H3 by H3.3 through association of HIRA to d1, and maintains w expression under the heterochromatin environment. Based on these data, we propose the following model for the maintenance of the active state against the spreading of silent chromatin (Fig. 8). Heterochromatin is marked by K9-methylated histone H3 and its binding protein HP1, and has a tendency to spread into neighboring regions (Rea et al. 2000; Grewal and Elgin 2002). Histone H3.3 replacement is thought to be achieved through either eviction of a nucleosome and deposition of a H3.3-containing nucleosome or stepwise disassembly–reassembly without eviction of a nucleosome. As the GAGA factor–FACT complex facilitates chromatin remodeling (Shimojima et al. 2003) and the GAGA factor is known to generate a nucleosome-free region around its binding site (Lu et al. 1993), it is most likely that eviction or disassembly of a nucleosome occurs at the DNase-hypersensitive site of d1. The GAGA factor and FACT participate association of HIRA to d1 and the histone replacement would be accomplished by subsequent deposition or reassembly of a H3.3-containing nucleosome. This process would be repeated constantly to eliminate K9-methylated histone H3 at d1 and counteract the spreading of silent chromatin.
Figure 8.
Model for GAGA factor and FACT-dependent histone replacement to counteract the heterochromatin spreading. We propose that the replacement is achieved through the GAGA factor–FACT complex-dependent eviction or disassembly of a nucleosome, the GAGA factor, and FACT-dependent association of HIRA, and the subsequent HIRA-mediated deposition or reassembly of a H3.3-containing nucleosome. See the text for details. It remains unknown whether the GAGA factor interacts with HIRA directly or through other GAGA factor-associated proteins.
Schwartz and Ahmad (2005) have reported histone H3.3 replacement triggered by transcription elongation. However, genome-wide profiling has shown histone H3.3 replacement from upstream of to downstream from transcription units (Mito et al. 2005). Although some of the replacement may be explained by elongation during intergenic transcription, the histone H3.3 replacement at d1 appears to occur independent of transcription elongation. Thus, the present study indicates a distinct pathway for histone H3.3 replacement.
In FlyBase, transcription of the w adjacent gene CG32795 has been reported to start immediately after the GAGA factor-binding sequence of d1, suggesting that d1 is a part of the promoter region of CG32795. Therefore, we examined the effect of Trl and spt16 mutations on expression of CG32795. The reduction of a single dose of Trl or spt16 affected the CG32795 expression in the wm4 context (Fig. 1) but not in the w+ context (Supplementary Fig. S1). These data are consistent with the idea that d1 is a functional promoter element of CG32795 in the w+ context, although Trl and spt16 become haplo-insufficient only when the accessibility of the GAGA factor–FACT complex to d1 decreased under the heterochromatin environment. This raises the possibility that the protection from heterochromatin spreading by the GAGA factor and FACT at d1 is a consequence of their function within the CG32795 promoter. However, conventional promoters do not have a barrier function against heterochromatin silencing. For example, the presence of GAL4 (or E2F) on a promoter carrying GAL4 (or E2F)-binding site did not modify PEV of the attached reporter gene (Seum et al. 2000). Genome-wide profiling of H3.3 replacement in Drosophila has revealed the clear dip of H3.3-containing nucleosomes at immediately upstream of the transcription start sites of active genes (Mito et al. 2005). This is in sharp contrast with the case of d1, where we observed peaks of both the H3.3/H3 ratio and the actual H3.3 level, and illuminates the difference between d1 and ordinary promoters. Furthermore, the GAGA factor-dependent histone H3.3 replacement was detected also at the DNase HS1 in the Fab-7 boundary of Abd-B (Fig. 7), where no promoter activity has been demonstrated. These findings indicate that the GAGA factor and FACT-dependent histone H3.3 replacement can occur without promoter functions. Nevertheless, the barrier function could be assisted by the putative promoter activity of d1 such as formation of a transcription initiation complex.
The GAGA factor-binding sequence at d1 consists of (GA)8. As the GAGA factor forms an oligomer through its BTB domain, the factor can make a cooperative and stable binding to closely spaced GAGAG elements (Katsani et al. 1999). This is presumably the reason why d1 gave a prominent signal among the GAGAG sequences around w in our ChIP assay (Fig. 2B). Because the GAGA factor occupies many closely spaced GAGAG sequences within the Drosophila genome (Lu et al. 1993; van Steensel et al. 2003) including the Polycomb/trithorax response elements of Hox genes, the proposed mechanism (Fig. 8) may operate not only in loci juxtaposed with heterochromatin but also in other loci such as the regulatory regions of Hox genes. Indeed we observed the GAGA factor and FACT-dependent histone H3.3 replacement in the Fab-7 boundary of Abd-B. High levels of histone H3.3 have been also reported at the locus control region of the chicken folate receptor gene (Jin and Felsenfeld 2006), suggesting that the barrier function against the chromatin silencing via histone H3.3 replacement may be evolutionarily conserved up to vertebrates.
Materials and methods
Fly stocks
To obtain transgenic fly lines expressing Flag-histone H3 or H3.3, we inserted an entire ORF of histone H3 or H3.3 with a six-amino-acid spacer (SRPVAT) and a Flag peptide at its C terminus into a pCaSpeR-hsp83-Act5C-EGFP vector in which the mini w marker of pCaSpeR-hsp83 (Horabin and Schedl 1993) was replaced with the Act5C-EGFP marker and used it for germline transformation. Transgenic fly lines carrying the 212-bp d1 sequence (FlyBase, Chr. X: 2646116–2645905) were made by inserting a DNA fragment FRT-d1-FRT-a spacer DNA-GAL4UAS after mini w of the pCaSpeR-hsp83 vector. We also made transgenic lines as above in which the GAGA factor-binding sequence at d1, (TC)8, was replaced with ACACAGCCG GCACACA. Excision of d1 was carried out by crossing with a line P[β2-Tubulin FLP] expressing the flippase. Trl13C is a P-element insertion line (Farkas et al. 1994). The Δdspt16 and Δdspt16 Trl13C lines have been described (Shimojima et al. 2003). P[Act5C-EGFP, spt16+] line was prepared by inserting an entire ORF of spt16+ into pCaSpeR-hsp83-Act5C-EGFP and the chromosome containing the P-element was introduced into the Δspt16 Trl13C line to yield a rescue line P[Act5C-EGFP, spt16+]; Δspt16 Trl13C. hiraR225K is ssm185b described by Loppin et al. (2005).
PEV analysis
To observe PEV, wm4 females were crossed to males of yw; +/TM3y+, yw; Δspt16/TM3y+, yw; Trl13C/TM3y+, yw; Δspt16 Trl13C/TM3y+, or yw;P[Act5C-EGFP, spt16+]; Δspt16 Trl13C/TM3y+. After raising at 18°C, the eye colors of male offsprings were observed within 6 h after eclosion. To examine the effect of d1 on w variegation, the flies in which d1 had been flipped out or not were crossed with GHP1–6 (Seum et al. 2000), then the fusion protein GAL4-HP1 was expressed by heat shock at day 7 AEL for 30 min in a 37°C water bath, and the eye colors were observed after raising at 25°C. To measure eye pigmentation, 10 (wm4 lines in Fig. 1F) or 30 (transgenic lines in Figs. 3, 6E) male adult heads within 24 h after eclosion were homogenized in 20 μL of 0.1% NH4OH. The homogenates were extracted once with chloroform. Absorbance of the supernatants was measured at 485 nm.
ChIP assays
Embryos from each line were collected 10–22 h AEL and dechorionated. ChIP assays were performed as described (Orlando et al. 1998; Andrulis et al. 2000) without the CsCl centrifugation step. After incubation with chromatin fragments derived from 50-mg embryos, antibody-loaded protein A agarose beads were washed seven times with RIPA buffer (10 mM Tris-HCl at pH 8.0, 1 mM EDTA at pH 8.0, 0.5 mM EGTA at pH 8.0, 140 mM NaCl, 0.1% SDS, 0.1% sodium deoxycholate, 1% Triton X-100), once with RIPA high-salt buffer (RIPA containing 500 mM NaCl), once with LiCl wash buffer (250 mM LiCl, 10 mM Tris-HCl at pH 8.0, 1 mM EDTA at pH 8.0, 0.5% sodium deoxycholate, 0.5% NP-40), and then once with TE (10 mM Tris-HCl at pH 8.0, 1 mM EDTA at pH 8.0). After washing, the beads were resuspended in 300 μL of reverse cross-link buffer (300 mM NaCl, 0.5% SDS, and 300 μg/mL Proteinase K in TE) and incubated overnight at 65°C. The beads were spun down and the supernatant was saved. The beads were washed with 150 μL of reverse cross-link buffer and spun down, and the supernatant was saved. DNA was recovered from the combined supernatant by phenol-chloroform treatment followed by ethanol precipitation and resuspended in 50 μL of TE, and a 1-μL portion was used for PCR. The PCR products were resolved by 10% polyacrylamide gel electrophoresis, visualized with SybrGreen I (Molecular Probe), and quantitated using LuminoImager LAS 1000 (FujiFilm). Primers used were 5′-AGATGTCAGACAGG GACAGC-3′ and 5′-GCAGTTCGTGTTTGCTATCTG-3′ for u1, 5′-CAGGCGACTGGGCTGCAATT-3′ and 5′-GGCCGT GGGATTTCCTTCTG-3′ for u2, 5′-CACCAACAACCAACAA CGAAAC-3′ and 5′-CTGTGAAATACTTACGAAGAAAC-3′ for u3, 5′-CATAATGTCAAATGTTGTGGCAC-3′ and 5′-AGCCAAGGGAGCGAGATCC-3′ for u4, 5′-CTTAGAAATA ATTCCCGAATTGG-3′ and 5′-TGCGCATTTCGAAAAGTT GTT-3′ for u5, 5′-TCGCGCACATACACTGCAAC-3′ and 5′-CTCATCTGTGCAACAGTTGCG-3′ for u6, 5′-CTAACTAT GTGACCCGAACCC-3′ and 5′-GAGTTTTCCGGGGGCCAA GG-3′ for u7, 5′-GCTTGGATTTCACTGGAACTAG-3′ and 5′-AAAAACACACTGCCCACACGC-3′ for d1, 5′-GCAGAA GAGGACAACAATGCG-3′ and 5′-GATTGCTCGATTTAA TTTGCC-3′ for d2, 5′-CCATATGTCACACTCACACATG-3′ and 5′-AACACGAAAAATTGAAACGATGC-3′ for d3, 5′-CT AAAGAGATTTAATAGCAAG-3′ and 5′-AATGAAAATGAA AATGAAAGACAGAC-3′ for d4, 5′-GCTTATGCTGATTTC GTATC-3′and 5′TTTTTTTTAGTGCACAAATAGC-3′ for d5, 5′-CGTCGAGTGTAAACGCTGTG-3′ and 5′-CGGCAGGAC TTTTGCGGAAT-3′ for d6, 5′-GCCAACTAGCCGAGAA CCTC-3′ and 5′-TAGTCCGCGTGTCCGGTTGA-3′ for e2, 5′-ACAGGTGTTGGCCGTTGTGC-3′ and 5′-CATATCCCGGG CTACTTTGC-3′ for e4, 5′-AGCCGGGCGAAATTAGCTGC-3′ and 5′-TGGCCAGACCCACGTAGTCC-3′ for e6, 5′-CTAA CAAGATTTCAAGCTGTGTG-3′ and 5′-GCGCGTTGATAT GCCCCAATG-3′ for HS1 in Fab-7 boundary, and 5′-CGAGC CGTGTCTTTGCCACTG-3′ and 5′-CGAGGACGAGCAACG GACGTC-3′ for a control site in iab-6. To generate antiserum against GAGA factor, a histidine-tagged factor lacking N-terminal five amino acids was expressed in Escherichia coli BL21 (DE3) pLys S, purified by Ni-NTA chromatography followed by Mono S column chromatography, and then used for immunization of a rabbit. Polyclonal antibodies against histone H3 K4 di- or trimethyl were raised by using each H3 peptide carrying di- or trimethylated K4 and affinity-purified. Other antibodies were anti-dSSRP1 and anti-dSPT16 (Shimojima et al. 2003), anti-HIRA (Loppin et al. 2005) (a gift of B. Loppin, Université Claude Bernard, Villeurbanne, France), antihistone H3 K9 dimethyl (Upstate Biotechnology), and anti-Flag antibody M2 (Sigma). In Figures 2, C and D; 5, A and B; 6A; and 7B, averages and SDs (standard deviations) from at least five independent experiments are shown.
Immunoprecipitation
Nuclear extracts were prepared as described (Shimojima et al. 2003) from yw, yw; P[w+, Flag-HIRA]/TM6B (Loppin et al. 2005), or yw;P[w+, Flag-GAGA] embryos 10–22 h AEL and dialyzed against binding buffer (20 mM HEPES at pH 8.0, 150 mM NaCl, 3 mM MgCl2, 0.1 mM EGTA, 5% glycerol, 0.05% NP-40, 0.5 mM DTT, protease cocktail [Sigma]). Portions of nuclear extract containing 500–600 μg of proteins were incubated with anti-GAGA, anti-dSSRP1, or anti-HIRA antibody-loaded Protein G agarose (Upstate Biotechnology) for 2 h at 4°C. Then the beads were collected and washed with binding buffer containing 200 μg/mL BSA.
Nuclease accessibility assay
Restriction enzyme or DNase I accessibility assays were carried out as described (Thomas and Elgin 1988). PCR primers 5′-GG TAGCGGTCCGGTTGTTTTC-3′ and 5′-CACTCGTTTTTCA GCGAATCG-3′ were used to prepare a probe for Southern hybridization.
RT–PCR
First strand cDNAs were prepared by using random primers on total cellular RNA from embryos. PCR primers used were 5′-AACGGAGCCATCTTCCTCTTCC-3′ and 5′-GGGCCTCCC TCATAAAAACTGGC-3′ for w, 5′-TACTCAACGTAATTG GACTGATAATG-3′ and 5′-GGTCAGGGTGCAGTAGTAC CAG-3′ for CG32795, and 5′-CCACCAGTCGGATCGATA TG-3′ and 5′-CACGTTGTGCACCAGGAACT-3′ for rp49.
Acknowledgments
We thank J. Workman, S. Elgin, S. Henikoff, K. Ahmad, V. Pirrotta, and R. Paro for discussions, and C. Wu, P. Spierer, B. Loppin, and the Bloomington Stock Center for reagents or fly lines. This work was supported by Grants in Aid for Scientific Research from MEXT Japan and Takeda Science Foundation. T.N. and T.S. were supported by a Center of Excellence Program of Japan and the Japan Society for the Promotion of Science, respectively.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1503407
References
- Ahmad K., Henikoff S., Henikoff S. The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol. Cell. 2002;9:1191–1200. doi: 10.1016/s1097-2765(02)00542-7. [DOI] [PubMed] [Google Scholar]
- Andrulis E.D., Guzman E., Doring P., Werner J., Lis J.T., Guzman E., Doring P., Werner J., Lis J.T., Doring P., Werner J., Lis J.T., Werner J., Lis J.T., Lis J.T. High-resolution localization of Drosophila Spt5 and Spt6 at heat shock genes in vivo: Roles in promoter proximal pausing and transcription elongation. Genes & Dev. 2000;14:2635–2649. doi: 10.1101/gad.844200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czermin B., Melfi R., McCabe D., Seitz V., Imhof A., Pirrotta V., Melfi R., McCabe D., Seitz V., Imhof A., Pirrotta V., McCabe D., Seitz V., Imhof A., Pirrotta V., Seitz V., Imhof A., Pirrotta V., Imhof A., Pirrotta V., Pirrotta V. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell. 2002;111:185–196. doi: 10.1016/s0092-8674(02)00975-3. [DOI] [PubMed] [Google Scholar]
- Farkas G., Gausz J., Galloni M., Reuter G., Gyurkovics H., Karch F., Gausz J., Galloni M., Reuter G., Gyurkovics H., Karch F., Galloni M., Reuter G., Gyurkovics H., Karch F., Reuter G., Gyurkovics H., Karch F., Gyurkovics H., Karch F., Karch F. The Trithorax-like gene encodes the Drosophila GAGA factor. Nature. 1994;371:806–808. doi: 10.1038/371806a0. [DOI] [PubMed] [Google Scholar]
- Fjose A., Polito L.C., Weber U., Gehring W., Polito L.C., Weber U., Gehring W., Weber U., Gehring W., Gehring W. Developmental expression of the white locus of Drosophila melanogaster. EMBO J. 1984;3:2087–2094. doi: 10.1002/j.1460-2075.1984.tb02095.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewal S.I., Elgin S.C.R., Elgin S.C.R. Heterochromatin: New possibilities for the inheritance of structure. Curr. Opin. Genet. Dev. 2002;12:178–187. doi: 10.1016/s0959-437x(02)00284-8. [DOI] [PubMed] [Google Scholar]
- Horabin J.I., Schedl P., Schedl P. Sex-lethal antoregulation requires multiple cis-acting elements upstream and downstream of the male exon and appears to depend largely on controlling the use of the male exon 5′ splice site. Mol. Cell. Biol. 1993;13:7734–7746. doi: 10.1128/mcb.13.12.7734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenuwein T., Allis C.D., Allis C.D. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- Jin C., Felsenfeld G., Felsenfeld G. Distribution of histone H3.3 in hematopoietic cell lineages. Proc. Natl. Acad. Sci. 2006;103:574–579. doi: 10.1073/pnas.0509974103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsani K.R., Hajibagueri M.A.N., Verrijzer C.P., Hajibagueri M.A.N., Verrijzer C.P., Verrijzer C.P. Cooperative DNA binding by GAGA transcription factor requires the conserved BTB/POZ domain and reorganizes promoter topology. EMBO J. 1999;18:698–708. doi: 10.1093/emboj/18.3.698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi M., Aita N., Hayashi S., Okada K., Ohta T., Hirose S., Aita N., Hayashi S., Okada K., Ohta T., Hirose S., Hayashi S., Okada K., Ohta T., Hirose S., Okada K., Ohta T., Hirose S., Ohta T., Hirose S., Hirose S. DNA supercoiling factor localizes to puffs on polytene chromosomes in Drosophila melanogaster. Mol. Cell. Biol. 1998;18:6737–6744. doi: 10.1128/mcb.18.11.6737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loppin B., Bonnefoy E., Anselme C., Laurencon A., Karr T.L., Couble P., Bonnefoy E., Anselme C., Laurencon A., Karr T.L., Couble P., Anselme C., Laurencon A., Karr T.L., Couble P., Laurencon A., Karr T.L., Couble P., Karr T.L., Couble P., Couble P. The histone H3.3 chaperone HIRA is essential for chromatin assembly in the male pronucleus. Nature. 2005;437:1386–1390. doi: 10.1038/nature04059. [DOI] [PubMed] [Google Scholar]
- Lu Q., Wallrath L.L., Granok H., Elgin S.C.R., Wallrath L.L., Granok H., Elgin S.C.R., Granok H., Elgin S.C.R., Elgin S.C.R. (CT)n · (GA)n repeats and heat shock elements have distinct roles in chromatin structure and transcriptional activation of the Drosophila hsp26 gene. Mol. Cell. Biol. 1993;13:2802–2814. doi: 10.1128/mcb.13.5.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKittrick E., Gafken P.R., Ahmad K., Henikoff S., Gafken P.R., Ahmad K., Henikoff S., Ahmad K., Henikoff S., Henikoff S. Histone H3.3 is enriched in covalent modifications associated with active chromatin. Proc. Natl. Acad. Sci. 2004;101:1525–1530. doi: 10.1073/pnas.0308092100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mito Y., Henikoff J.G., Henikoff S., Henikoff J.G., Henikoff S., Henikoff S. Genome-scale profiling of histone H3.3 replacement patterns. Nat. Genet. 2005;37:1090–1097. doi: 10.1038/ng1637. [DOI] [PubMed] [Google Scholar]
- Muller H. Types of visible variations induced by X-rays in Drosophila. J. Genet. 1930;22:299–334. [Google Scholar]
- Müller J., Hart C.M., Francis N.J., Vargas M.L., Sengupta A., Wild B., Miller E.L., O’Connor M.B., Kingston R.E., Simon J.A., Hart C.M., Francis N.J., Vargas M.L., Sengupta A., Wild B., Miller E.L., O’Connor M.B., Kingston R.E., Simon J.A., Francis N.J., Vargas M.L., Sengupta A., Wild B., Miller E.L., O’Connor M.B., Kingston R.E., Simon J.A., Vargas M.L., Sengupta A., Wild B., Miller E.L., O’Connor M.B., Kingston R.E., Simon J.A., Sengupta A., Wild B., Miller E.L., O’Connor M.B., Kingston R.E., Simon J.A., Wild B., Miller E.L., O’Connor M.B., Kingston R.E., Simon J.A., Miller E.L., O’Connor M.B., Kingston R.E., Simon J.A., O’Connor M.B., Kingston R.E., Simon J.A., Kingston R.E., Simon J.A., Simon J.A. Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell. 2002;111:197–208. doi: 10.1016/s0092-8674(02)00976-5. [DOI] [PubMed] [Google Scholar]
- Orlando V., Jane E.P., Chinwalla V., Harte P.J., Paro R., Jane E.P., Chinwalla V., Harte P.J., Paro R., Chinwalla V., Harte P.J., Paro R., Harte P.J., Paro R., Paro R. Binding of Trithorax and Polycomb proteins to bithorax complex: Dynamic changes during early Drosophila embryogenesis. EMBO J. 1998;17:5141–5150. doi: 10.1093/emboj/17.17.5141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orphanides G., LeRoy G., Chang C.H., Luse D.S., Reinberg D., LeRoy G., Chang C.H., Luse D.S., Reinberg D., Chang C.H., Luse D.S., Reinberg D., Luse D.S., Reinberg D., Reinberg D. FACT, a factor that facilitates transcription elongation through nucleosomes. Cell. 1998;92:5141–5150. doi: 10.1016/s0092-8674(00)80903-4. [DOI] [PubMed] [Google Scholar]
- Orphanides G., Wu W.H., Lane W.S., Hampsey M., Reinberg D., Wu W.H., Lane W.S., Hampsey M., Reinberg D., Lane W.S., Hampsey M., Reinberg D., Hampsey M., Reinberg D., Reinberg D. The chromatin-specific transcription elongation factor FACT comprises human SPT16 and SSRP1 proteins. Nature. 1999;400:284–288. doi: 10.1038/22350. [DOI] [PubMed] [Google Scholar]
- Rea S., Eisenhaber F., O’Carroll D., Strahl B.D., Sun Z.W., Schmid M., Opravil S., Mechtler K., Ponting C.P., Allis C.D., Eisenhaber F., O’Carroll D., Strahl B.D., Sun Z.W., Schmid M., Opravil S., Mechtler K., Ponting C.P., Allis C.D., O’Carroll D., Strahl B.D., Sun Z.W., Schmid M., Opravil S., Mechtler K., Ponting C.P., Allis C.D., Strahl B.D., Sun Z.W., Schmid M., Opravil S., Mechtler K., Ponting C.P., Allis C.D., Sun Z.W., Schmid M., Opravil S., Mechtler K., Ponting C.P., Allis C.D., Schmid M., Opravil S., Mechtler K., Ponting C.P., Allis C.D., Opravil S., Mechtler K., Ponting C.P., Allis C.D., Mechtler K., Ponting C.P., Allis C.D., Ponting C.P., Allis C.D., Allis C.D., et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406:593–599. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- Reuter G., Spierer P., Spierer P. Position effect variegation and chromatin proteins. Bioessays. 1992;14:604–612. doi: 10.1002/bies.950140907. [DOI] [PubMed] [Google Scholar]
- Ringrose L., Paro R., Paro R. Epigenetic regulation of cellular memory by the Polycomb and Trithorax gene proteins. Annu. Rev. Genet. 2004;38:413–443. doi: 10.1146/annurev.genet.38.072902.091907. [DOI] [PubMed] [Google Scholar]
- Schultz J. The relation of the heterochromatic chromosome regions to the nucleic acids of the cell. Cold Spring Harb. Symp. Quant. Biol. 1956;21:307–328. doi: 10.1101/sqb.1956.021.01.025. [DOI] [PubMed] [Google Scholar]
- Schwartz B.E., Ahmad K., Ahmad K. Transcriptional activation triggers deposition and removal of the histone H3.3. Genes & Dev. 2005;19:804–814. doi: 10.1101/gad.1259805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seum C., Spierer A., Delattre M., Pauli D., Spierer P.A., Spierer A., Delattre M., Pauli D., Spierer P.A., Delattre M., Pauli D., Spierer P.A., Pauli D., Spierer P.A., Spierer P.A. A GAL4-HP-1 fusion protein targeted near heterochromatin promotes gene silencing. Chromosoma. 2000;109:453–459. doi: 10.1007/s004120000101. [DOI] [PubMed] [Google Scholar]
- Shimojima T., Okada M., Nakayama T., Ueda H., Okawa K., Iwamatsu A., Handa H., Hirose S., Okada M., Nakayama T., Ueda H., Okawa K., Iwamatsu A., Handa H., Hirose S., Nakayama T., Ueda H., Okawa K., Iwamatsu A., Handa H., Hirose S., Ueda H., Okawa K., Iwamatsu A., Handa H., Hirose S., Okawa K., Iwamatsu A., Handa H., Hirose S., Iwamatsu A., Handa H., Hirose S., Handa H., Hirose S., Hirose S. Drosophila FACT contributes to Hox gene expression through physical and functional interactions with GAGA factor. Genes & Dev. 2003;17:1605–1616. doi: 10.1101/gad.1086803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon J.A., Tamkun J.W., Tamkun J.W. Programming off and on states in chromatin: Mechanisms of Polycomb and trithorax group complexes. Curr. Opin. Genet. Dev. 2002;12:210–218. doi: 10.1016/s0959-437x(02)00288-5. [DOI] [PubMed] [Google Scholar]
- Tagami H., Ray-Gallet D., Almouzni G., Nakatani Y., Ray-Gallet D., Almouzni G., Nakatani Y., Almouzni G., Nakatani Y., Nakatani Y. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell. 2004;115:51–61. doi: 10.1016/s0092-8674(03)01064-x. [DOI] [PubMed] [Google Scholar]
- Tartof K.D., Hobbs C., Jones M., Hobbs C., Jones M., Jones M. A structural basis for variegating position effects. Cell. 1984;37:869–878. doi: 10.1016/0092-8674(84)90422-7. [DOI] [PubMed] [Google Scholar]
- Thomas G.H., Elgin S.C.R., Elgin S.C.R. Protein/DNA architecture of the DNase I hypersensitive region of the Drosophila hsp26 promoter. EMBO J. 1988;7:2191–2201. doi: 10.1002/j.1460-2075.1988.tb03058.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Steensel B., Dlrow J., Bussemaker J., Dlrow J., Bussemaker J., Bussemaker J. Genomewide analysis of Drosophila GAGA factor target genes reveals context-dependent DNA binding. Proc. Natl. Acad. Sci. 2003;100:2580–2585. doi: 10.1073/pnas.0438000100. [DOI] [PMC free article] [PubMed] [Google Scholar]