Abstract
Short RNA expression in several distinct stages of T-lymphocyte development was comprehensively profiled. The total number of microRNAs (miRNAs) expressed per cell at different stages of development varies over nearly an order of magnitude in parallel with changes in total cellular RNA content, suggesting that global miRNA levels are coregulated with the translational capacity of the cell. However, individual miRNAs were dynamically regulated during T-cell development, with at least one miRNA or miRNA family overrepresented at each developmental stage. miRNA regulation in this developmental pathway is characterized by analog rather than switch-like behavior, with temporal enrichments at distinct stages of development observed against a background of constant, basal expression of the miRNA. Enrichments of these miRNAs are temporally correlated with depletions of the transcript levels of targets containing seed matches to the specific miRNAs, and may have specific functional consequences. miR-181a, which is specifically enriched at the CD4+CD8+ (DP) stage of thymocyte development, can repress the expression of Bcl-2, CD69, and the T-cell receptor, all of which are coordinately involved in positive selection.
Keywords: CD69, development, genomics, microRNAs, microarray, thymocyte
Short RNAs are known to control gene expression at several different levels in organisms ranging from yeast to plants to mammals (Kloosterman and Plasterk 2006). In mammals, the best-characterized class of short RNA species is the microRNA (miRNA) class. These 22-nucleotide (nt) RNA species repress gene expression at the level of translation by binding to the 3′ untranslated region (UTR) of target mRNAs. The precise mechanisms of this repression are probably multifold (Bagga et al. 2005; Pillai et al. 2005; Giraldez et al. 2006; Leung et al. 2006; Petersen et al. 2006).
Both bioinformatic and in vitro experimental data indicate that miRNA:mRNA recognition is mediated predominantly by an interaction between the 5′ end of a miRNA and a complementary sequence in the mRNA target (Doench and Sharp 2004; Brennecke et al. 2005; Lim et al. 2005). Bases 2–7 or 2–8 of the miRNA are primary contributors to target specificity and are referred to as the miRNA “seed” region (Doench and Sharp 2004; Brennecke et al. 2005). Genomic experiments examining the effects of transfection of individual miRNAs in HeLa cells have demonstrated that a high proportion of the transcripts down-regulated in response to a transfection of a miRNA contain sequences complementary to the seed region (Lim et al. 2005). Based on these observations, it is thought that miRNA family members that share common seed sequences have similar target specificity. These observations have been extended to predict mRNA targets of miRNAs (Lewis et al. 2003, 2005) and to demonstrate a general reduction in the transcript levels of these targets in tissues where these miRNAs are expressed (Farh et al. 2005; Krutzfeldt et al. 2005).
The number of described mammalian miRNAs continues to increase through direct cloning efforts and bioinformatic prediction. Expression of several individual miRNAs such as miR-142 (lymphoid), miR-223 (myeloid), miR-1 (muscle), and miR-9 (neuronal) is largely restricted to single tissues or organ systems, consistent with these miRNAs having a role in the developmental specification of these cells (Lagos-Quintana et al. 2002; Chen et al. 2004). However, a large number of miRNAs exhibit a more wide-ranging pattern of expression, consistent with these regulators being involved in events such as growth and homeostasis. A systematic understanding of the roles of miRNAs in such events is incomplete, as few direct studies of changes in mRNA and miRNA expression over the course of a single developmental pathway have been conducted.
Specific expression of individual miRNAs in several compartments of the mammalian hematopoietic system has been well described (Chen et al. 2004; Monticelli et al. 2005). However, a close examination of the dynamic regulation of miRNAs and the consequences of this regulation on global gene expression during sequential stages of development of a single cell lineage has not been performed. T-lymphocyte development in the thymus has been extensively characterized (Starr et al. 2003). The DN1 stage of development is a mix of lymphoid and myeloid progenitor cells (Porritt et al. 2004). T-lineage commitment is fixed upon rearrangement of the genetic locus encoding the β chain of the antigen receptor at the DN3 stage; cells that successfully rearrange this locus proceed to the highly proliferative DN4 stage. Following the DN4 stage, expression of the CD4 and CD8 coreceptors defines the DP stage of thymocyte development, at which point the thymocytes rearrange the α chain of the T-cell receptor (TCR) and undergo positive and negative selection. Those cells that are positively selected mature to CD4 SP (helper) or CD8 SP (killer) T lymphocytes and egress the thymus to the periphery. Importantly, individual cell populations along this developmental series are enriched for cells undergoing fundamental cellular processes such as proliferation, differentiation, and apoptosis. Dynamic regulation of individual miRNAs within these stages might be expected to influence these processes without perhaps being critical for development of the T lineage per se.
Here we describe discovery-compatible profiling and quantitative measurement of short RNA expression from several ordered stages of T-lymphocyte development. The pool of miRNAs at a given developmental stage correlates closely with the cytoplasmic volume as indicated by amount of total RNA. Although nearly all miRNAs that we observe in this process are constitutively expressed, specific miRNAs are enriched at distinct stages of thymocyte development. Specific enrichment of miRNAs can be correlated with the depletion of transcripts harboring seed matches to these miRNAs. In addition, miRNA enrichment bears functional consequences; in reporter assays, the DP-enriched miR-181a confers repression through the 3′ UTR elements of three predicted targets (CD69, Bcl-2, and the TCR) known to be regulated at this stage of development.
Results
Short RNA profiling using low nanogram amounts of total RNA
We directly cloned short RNAs from developing thymocytes to enable profiling of known miRNAs while allowing for the discovery of additional small RNA species of potential interest. This involved developing a variation of existing protocols (Lagos-Quintana et al. 2001; Lau et al. 2001) to allow cloning of short RNA species from low nanogram amounts of total RNA (Supplementary Online Material 1 [SOM1]).
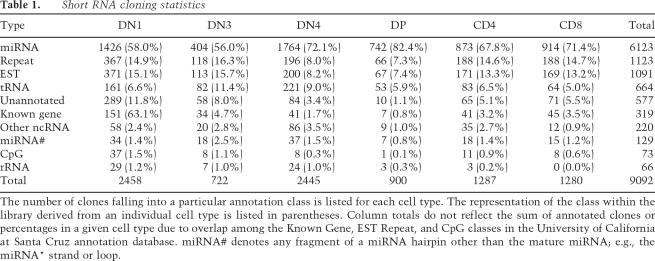
We collected and analyzed 10,533 short RNA clones comprised of 3445 nonredundant sequences from six stages of T-lymphocyte development, each sorted to >95% purity (Table 1; SOM2–4). The CD25+CD44+ DN2 population, which is a transient intermediate between the DN1 and DN3 populations, was omitted because of the rarity of cells representing this stage. The cloned sequences ranged in length from 16 to 33 nt. Each sequence in the library was assigned a cloning frequency from each cell type. Nine-hundred-fifty-seven of the sequences were observed only once, suggesting that the library is not close to saturation. The library was searched using the BLAST algorithm against full-length miRNA hairpins in mirBase (release 8.1), tRNA, and NONCODE databases (Liu et al. 2005) and the mouse genome (mm8). While 67% of the clones matched precisely to sequences in the above databases, an additional 19% of the distinct sequences could be annotated if we allowed a single internal mismatch or up to three mismatches at the end of the sequence. We also noticed that >80% of the clones annotated as mismatches using the above criteria fell into the category of 3′ end mismatches with a bias for adenosine or uracil in these positions. This pattern could reflect biases in the cloning procedures or might reflect nucleotide modification or addition in vivo following processing by Drosha or Dicer. Indeed, similar observations have been reported in cloning efforts by other groups (Aravin and Tuschl 2005). Because of this, we elected to include these sequences in our analysis and were thus able to annotate 9092 (86.3%) of the clones using the above criteria.
Table 1.
Short RNA cloning statistics
The number of clones falling into a particular annotation class is listed for each cell type. The representation of the class within the library derived from an individual cell type is listed in parentheses. Column totals do not reflect the sum of annotated clones or percentages in a given cell type due to overlap among the Known Gene, EST Repeat, and CpG classes in the University of California at Santa Cruz annotation database. miRNA# denotes any fragment of a miRNA hairpin other than the mature miRNA; e.g., the miRNA* strand or loop.
As expected, the majority of clones were miRNAs. The average percentage of clones representing miRNA sequences from each cell type was 68%, ranging from a low of 56% in DN3 cells to a high of 82% in DP thymocytes (Table 1). The relative representation of individual sequence classes within each cell type varied similarly. Sequences corresponding to genomic regions annotated as repeats or to which ESTs have been mapped were generally the second most abundant classes, observed with an average cloning frequency of 12%. The third tier of abundance consisted of sequences associated with tRNAs or unannotated regions of the genome, with average cloning frequencies of 6.6%. An exception was noted in the highly proliferative DN4 population, where tRNA-associated sequences were exceeded in abundance only by miRNAs. Sequences corresponding to ribosomal RNA, “other” non-tRNA and nonribosomal ncRNA classes, CpG islands, and the RefSeq “Known Gene” category constituted the fourth tier of abundance, with an average cloning frequency of 1.9%. The vast majority of clones associated with ncRNA loci corresponded to the sense strand of the specific ncRNAs. Interestingly, although 319 clones overlapped RefSeq “Known” genes, only 36% of these clones mapped to these annotations in the sense orientation. Fully half of the clones overlapping annotated genes mapped antisense to the protein-coding region of the gene. The remaining 14%, while associated with RefSeq genes, mapped to more than one region of the genome, so the precise location from which these sequences originated, and the strand polarity associated with these sequences, could not be determined.
Quantitative validation of clone representation
We profiled short RNA cloning frequency from two independent preparations of DN4 thymocytes and observed a high degree of similarity (Pearson correlation coefficient = 0.925) (SOM6). This high degree of similarity demonstrated that the short RNA profiling method was reproducible. To determine whether the profile of short RNAs accurately reflected their relative abundance in the cell, a quantitative solution–hybridization-based Trilogy assay (Neely et al. 2006) was used to directly measure the abundance of 17 individual miRNAs in the DP thymocyte population (SOM 7–9). Reassuringly, there was a high degree of correlation between the calculated number of copies per cell and relative cloning frequency of each of these miRNAs (Pearson coefficient = 0.974). Surprisingly, this quantification revealed that miRNAs are expressed at much lower levels in DP thymocytes than in other mammalian cells in which miRNA expression has been directly quantitated. For example, miR-181a, the miRNA cloned with the highest frequency in this cell type, was expressed at ∼810 copies per cell as compared with the up to 50,000 copies per cell reported for miRNAs in HeLa cells (Lim et al. 2003).
To determine that the changes in cloning frequency of individual miRNAs throughout thymocyte development reflected the relative abundance of these miRNAs at each stage, we used the Trilogy assay to calculate the copy number of each of three to five miRNAs independently for each cell stage except DN1 (the scarcity of these cells did not permit isolation of sufficient RNA to perform the Trilogy assay). The copy number for each miRNA in each of the cell types was independently determined and plotted against the cloning frequency of the miRNA (SOM9). We observed generally high correlations between miRNA copy number measured by the Trilogy assay and cloning frequency in each cell type, allowing the copy number of each miRNA species (and of the overall miRNA pool) to be estimated for each cell.
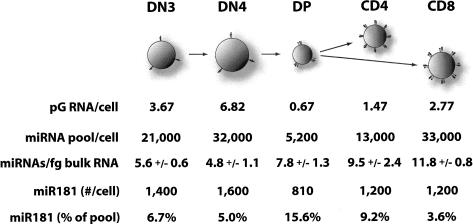
The amount of total RNA per cell at various stages of thymocyte development varied widely, ranging from a maximum of 6.8 pg/cell in highly proliferative DN4 thymocytes to a minimum of 0.7 pg/cell in DP thymocytes undergoing selection. The calculated miRNA pool was also highly dynamic throughout T-lymphocyte development, ranging from ∼5000 copies per cell in DP thymocytes to 33,000 copies per cell in DN4 thymocytes. However, the estimated size of the miRNA pools covaried closely with changes in total RNA level at the various stages of T-cell development (Fig. 1; SOM9). This covariation is reflected in a remarkably constant value for the calculated number of miRNAs per femtogram of cellular RNA. Indeed, although the per-cell RNA and miRNA pool vary by 10-fold and sevenfold, respectively, the calculated miRNAs:total RNA ratio slowly and steadily increased by a factor of two between the immature DN3 and mature SP stages. In sum, these results strongly suggest that the total pool of miRNAs is tightly regulated relative to the levels of ribosomal and messenger RNA.
Figure 1.
Measurements of total RNA content per cell, miRNAs per cell, and miRNAs per total RNA content. Values were derived from cell counts, total RNA quantification, and measurement of the concentration of individual miRNAs in each cell type normalized to a recovery control. The calculated miRNA pool represents an estimate based on the average of constants relating miRNA copy number to cloning frequency. Additional data is in SOM7–9. We were not able to obtain enough DN1 cells to perform miRNA quantitation analysis.
Dynamic regulation of miRNA species during thymocyte development
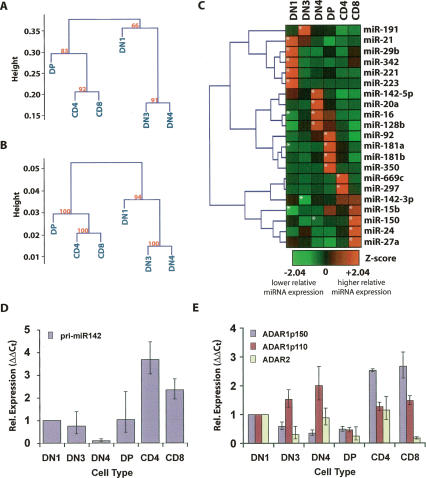
The possibility that each stage of T-cell development might have a signature miRNA pattern was next investigated. We tested whether the ontogeny relationships of the developing thymocytes to one another could be predicted with the miRNA signature of these cells. We performed hierarchical clustering of the cloning frequency of miRNAs to infer cell lineage relationships. A ranked correlation coefficient was calculated for all pairwise cell type combinations (SOM10) and was used as a similarity matrix to generate a rooted tree (Fig. 2A). A second rooted tree was generated using mRNA array data from Hoffman et al. (2003), who clustered the same cell types (Fig. 2B). A multiscale bootstrapping method was used to analyze the clustering in each case (Suzuki and Shimodaira 2006). The two dendrograms revealed strikingly similar clustering patterns. The immature DN3 and DN4 cells clustered together, the mature CD4 and CD8 SP cells clustered together, and DP thymocytes fell at an intermediate position. The DN1 population (which contains several different cell types) clustered in the same location in both dendrograms. These data suggest that the miRNA profile reflects the developmental relationships between individual cells to a similar degree as the mRNA profile.
Figure 2.
miRNA expression is dynamically regulated during thymocyte development. Rooted trees were generated from miRNA and mRNA expression data derived from each thymocyte population. (A) Relationship of cells determined by Spearman correlation of miRNA profiles. (B) Relationship of cells determined by Spearman correlation of mRNA profiles (Hoffmann et al. 2003). Values at branch points in A and B denote multiscale bootstrapping significance values. (C) Heat map representing relative expression of the 21 miRNAs that were identified as enriched or depleted during thymocyte development by the χ2 test. (Asterisks) Statistically significant enrichment or depletion. (D) qRT–PCR analysis of pri-miR-142 in thymocyte populations. (E) qRT–PCR analysis of ADAR family members in thymocyte populations.
Examination of the libraries indicated that the vast majority of the 136 known miRNAs that were cloned in this analysis were present at every stage of thymocyte development. However, there were a number of cases in which an individual miRNA’s expression was significantly higher in one cell type than in others by χ2 analysis of the frequency data set (SOM11). The relative abundance of 21 individual miRNAs varied among the six cell populations in a statistically significant fashion (Fig. 2C; SOM11). Statistically significant changes were heavily biased toward overrepresentation, with 20 events in which miRNAs were significantly enriched in a specific cell type compared with five events in which miRNAs were significantly depleted. Three of the five depletions observed (miR-15b, miR-16, and miR-181a) were at the DN1 stage, suggesting up-regulation of these miRNAs upon commitment to the T lineage. The other two depletions were striking in that they occurred at intermediate stages of T-lymphocyte development. Most striking was miR-142-3p, which was down-regulated over sixfold at the DN3 stage relative to the temporally adjacent DN1 and DN4 stages. Mir-142-5p was concomitantly depleted at the same stage. The uniqueness of these events in regards to global miRNA expression patterns led us to examine them more closely.
Expression of the miR-142 primary transcript (pri-miR-142) was first examined. Surprisingly, quantitative RT–PCR (qRT–PCR) analysis of this transcript revealed that there was no significant reduction in expression of pri-miR-142 relative to GAPDH between the DN1 and DN3 stages (Fig. 2D). Editing of the pri-miR-142 transcript by the p110 isoform of adenosine deaminase 1 (ADAR1) has been shown to inhibit processing of this transcript to the pre-miRNA hairpin by the Drosha enzyme (Yang et al. 2006). Since ADAR activity has been demonstrated in rat thymus (Paul and Bass 1998; Yang et al. 2003), we wondered whether we could correlate the specific depletion of miR-142-3p and miR-142-5p at the DN3 stage to an increase in the expression of ADAR1p110. qRT–PCR analysis demonstrated an increase in expression of the message encoding the ADAR1p110 isoform between the DN1 and DN3 stages of development, concomitant with a decrease in expression of the messages encoding ADAR1p150 and ADAR2 (Fig. 2E). Strikingly, although pri-miR-142 expression was reduced at the DN4 stage relative to GAPDH, we noticed an increase in miR-142 expression at this stage concomitant with a relative increase of the ADAR2 transcript at this stage. Later increases in miR-142 expression correlated with an increase in pri-miR-142 expression and/or a higher relative expression of ADAR2 or ADAR1p150 relative to the ADAR1p110 isoform.
In summary, the global picture of miRNA expression dynamics during T-lymphocyte development suggests a mode of expression in which miRNAs that are consistently expressed at a basal level throughout this process exhibit specific enrichment at distinct maturational stages. In contrast, stage-specific depletion is exceedingly rare. In the case examined, this depletion is correlated with an increase in transcript levels encoding a known post-transcriptional regulator of the miR-142 gene product.
Genomic signatures of miRNA families at individual stages of thymocyte development
Several studies have reported that mRNAs containing seed matches to tissue-specific miRNAs tend to be expressed at lower levels in the tissue(s) where the miRNA is expressed than in other tissues (Brennecke et al. 2005; Farh et al. 2005; Krutzfeldt et al. 2005; Stark et al. 2005). This depletion is thought to reflect a selective pressure against the presence of potential miRNA target sites in the bulk of transcripts coexpressed with a given miRNA. We analyzed the relationship between stage-specific enrichment of miRNAs during T-lymphocyte development and mRNA expression in these stages using the microarray data of Hoffman et al. (2003).
The mRNA array data were normalized to reflect the level of relative expression of a transcript in a particular stage as compared with the mean expression of all six stages examined. In this representation, one might expect that genes whose mRNA levels changed most between stages would be enriched for large-scale differences controlled at the level of transcription, obscuring the more subtle effects that have been observed for miRNAs in a global analysis. We therefore eliminated the 5% of genes exhibiting the greatest degree of enrichment and the 5% of genes exhibiting the greatest degree of depletion at each stage, examining the cumulative distribution of the remaining 90% of transcripts at each stage using a nonparametric Kolmogorov-Smirnov (KS) statistic. The hypothesis that relative expression of genes with UTR elements harboring seed matches was increased or decreased in each cell type was tested.
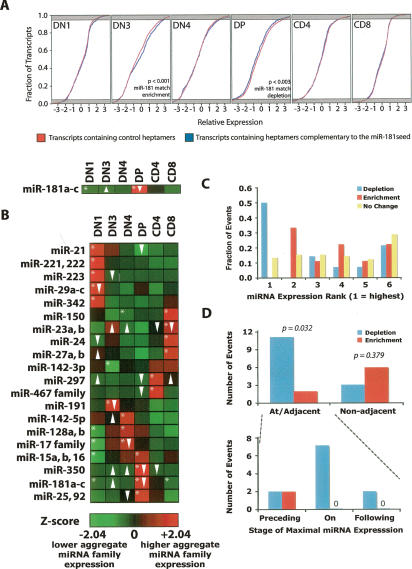
The largest enrichment in cloning frequency observed was that of the miR-181 miRNA family at the DP stage of thymocyte maturation. This family was chosen for a preliminary analysis. The most significant depletion of transcripts containing seed matches to the miR-181 family at the DP stage relative to control sets was observed when the experimental set was defined as all transcripts containing a sequence complementary to positions 2–8 of the miR-181 family seed region, and the control sets were defined as sets of transcripts containing heptameric sequences of similar abundance (p = 0.003) (Fig. 3A). Interestingly, there was also a significant enrichment of transcripts containing miR-181 family seed matches at the DN3 stage, with no enrichment or depletion occurring at the DN1, DN4, CD4 SP, or CD8 SP stages.
Figure 3.
Genomic signatures in response to changes in miRNA family member dxpression. (A) Cumulative distribution plots of relative expression of transcripts with 3′ UTR elements harboring matches to position 2–8 of the seed region of the miR-181 family versus sets of control heptamers at each stage of thymocyte development. (B) χ2 analysis of aggregate expression of miRNA family members. (Arrows) Significant (p < 0.05) enrichment (up) or depletion (down) of transcripts containing seed-match heptamers relative to a cohort of sets of transcripts containing control heptamers with similar (±10) occurrence; (asterisks) statistically significant enrichment or depletion of the indicated miRNA family. (C) Enrichment and depletion of transcripts containing heptamer seed matches relative to control sequences in relation to ranked relative miRNA expression in the six cell populations examined. Results for the 20 significantly changed miRNA families shown in B are presented. (D) Temporal analysis of enrichments and depletions during thymocyte development relative to stage of maximum miRNA family aggregate expression. Significance of these enrichments when compared with 1000 randomizations of the data set is shown.
The analysis was extended to other miRNA families that were statistically enriched or depleted at various stages of thymocyte development, using the same definitions and controls outlined above for the broader analysis. Again, the aggregate expression of all miRNA family members in a given cell type (e.g., summing the expression of miR-15a, miR-15b, and miR-16, all of which share a common seed) was considered rather than the abundance of individual statistically enriched or depleted miRNAs. Of 91 seed families tested, 20 were demonstrated to vary significantly over the course of thymocyte development (Fig. 3B).
When requiring a p value <0.05 in the KS test, 24 enrichments or depletions were observed in a set of 120 comparisons (20 miRNA family aggregates in six stages). These enrichment and depletion events were identified for 14 of 20 significantly changed miRNA families, with depletion of seed match-containing transcripts for at least one significantly enriched miRNA family in each cell type (Fig. 3B). Relative miRNA expression was ranked in each cell stage, and enrichment and depletion events were examined in regards to these levels. Fully 50% of transcript depletion events were observed at the stage at which a miRNA family was most highly expressed, while no enrichment events were observed at this stage (Fig. 3C). Surprisingly, the majority of enrichment events for transcripts containing seed matches were observed at the stage in which the miRNA was expressed at the second highest level. We took advantage of the temporal arrangement of the various thymocyte developmental stages to address this observation.
The general pattern of expression of miRNA families mirrored that of individual miRNAs, characterized by constitutive expression with significant enrichments at a single stage of thymocyte development. Examination of the enrichment and depletion of transcripts containing seed matches revealed an overrepresentation of depletion events prior to, at, or following the cell stages where the highest aggregate miRNA expression was observed (Fig. 3D). Closer examination of these adjacent stages revealed that enrichment occurred only prior to, but never at or following, the cell stage with the highest level of miRNA expression (Fig. 3E).
The observed patterns of miRNA expression are consistent with three modes of activity. In the first case (miR-181 family), depletion of transcripts containing seed matches is observed at the stage where the miRNA is acutely enriched. In the second (miR-25,92 family), a more gradual up-regulation of the miRNA family results in depletion of transcripts containing seed matches prior to the maximal expression of the miRNA family. In the final case, enrichment of miR-142-5p at the DN1 and DN4 stages appears to hold transcripts containing seed matches at a background level, with depletion of this miRNA at DN3 allowing enrichment of transcripts containing seed matches at this stage. Importantly, all three of these potential modes of action are consistent with negative effects on transcript expression at stages of high miRNA expression. We wondered whether these global effects could be correlated with the regulation of genes known to be involved in thymocyte maturation at specific stages.
miR-181a represses expression of genes with roles in thymocyte maturation through their 3′ UTR elements
We focused again on the miR-181 family; this family of miRNAs is strikingly up-regulated at the DP stage of thymocyte development, and might thus be expected to most dramatically impact gene expression at this stage. miR-181 has been reported to be highly expressed in thymocytes (Chen et al. 2004); however, the developmental specificity of this expression has not been described. The Targetscan 3.0 server (Lewis et al. 2005) was used to identify candidate targets of the miR-181 family that might play a role in the processes of positive or negative selection characteristic of this stage of T-cell development. Notably, CD69 and Bcl-2 are predicted targets of the miR-181a family. Bcl-2 is known to be selectively down-regulated at the DP stage of thymocyte development (Gratiot-Deans et al. 1993), but expression levels of this gene are restored following positive selection to the CD4 or CD8 SP stage. CD69 expression is increased on DP cells that have undergone positive or negative selection; this marker has been shown to appear on the surface of stimulated T cells <30 min following stimulation (Hara et al. 1986). Interestingly, surface expression of the antigen receptor is also increased in DP thymocytes following positive selection; this is known to occur post-transcriptionally (Bonifacino et al. 1990; Maguire et al. 1990). Although neither the α nor the β chain of the TCR is a predicted miRNA target on the Targetscan server, we noticed a seed match to the miR-181 family in the 3′ UTR of the TCRα transcript. The precise location of this seed match is conserved to rat, and the human TCRα UTR features a miR-181 seed match nearby. We wondered whether the miR-181 family might control expression of Bcl-2, CD69, the TCRα chain, and other genes with known or putative roles in positive selection through interactions with the 3′ UTR elements of their transcripts
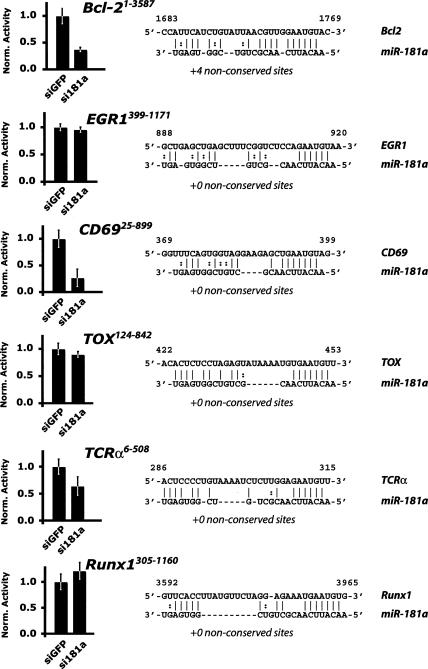
The 3′ UTR elements of several predicted targets of miR-181a were fused to a luciferase reporter, and we examined the ability of a synthetic miR-181a small interfering RNA (siRNA) duplex to repress the expression of these constructs in HeLa cells. Relative to transfection of a control siRNA, miR-181a significantly down-regulated reporters fused to the 3′ UTRs of the TCRα chain, CD69, and Bcl2 (Fig. 4). The down-regulation of Bcl-2 was marginally enhanced but not dependent upon the presence of the AU-rich stability element (ARE) in the 3′ UTR of this gene (data not shown). Expression of reporters fused to the predicted miR-181 targets TOX, Runx1, EGR1, Bcl2AF1, and FoxP1 were not significantly down-regulated by transfection of synthetic miR-181a (Fig. 4; data not shown), even though each of these genes is a predicted target of the miR-181 family and contain similar seed matches. We concluded that miR-181a is able to selectively repress the expression of a reporter gene fused to the 3′ UTR elements of the Bcl-2, TCRα, or CD69 genes.
Figure 4.
Repression of predicted miR-181a target UTRs in a reporter assay. The 3′ UTR elements of several genes identified by the Targetscan 3.0 server (Lewis et al. 2005) as potential miR-181a targets were tested in a dual-luciferase assay. An alignment of the conserved seed matches is indicated for each predicted target. Graphs indicate expression of the construct in HeLa cells transfected with a control siRNA duplex versus a miR-181a duplex normalized to the effect of each siRNA on a control UTR in a dual luciferase assay. UTR fragment length and miR-181a target sites are indicated. Results are representative of a minimum of three independent experiments performed in triplicate. Error bars represent one SD.
Dysregulation of miR-181a targets in Dicer-deficient DP thymocytes
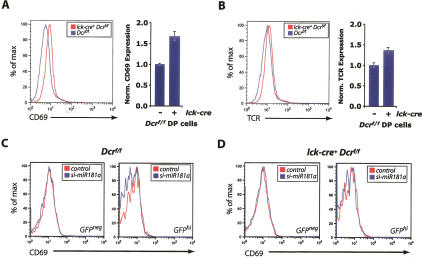
The finding that miR-181a was able to directly repress the expression of a reporter gene fused to the 3′ UTR elements of CD69, Bcl-2, or the TCRα chain suggested that the expression of these genes would increase if miR-181a levels were decreased in DP thymocytes. Thus, expression of these genes in Dicer-deficient DP thymocytes was examined. An lck-cre transgene (Hennet et al. 1995) was crossed into the floxed Dicer background (Harfe et al. 2005) to effect recombination of Dcrf alleles at the DN3 stage of thymocyte development.
The overall pattern of CD69 expression was conserved in Dicer-deficient DP thymocytes; however, these cells exhibited a reproducibly higher median fluorescence intensity when compared with sex-matched littermate controls harboring a functional allele of Dicer (Fig. 5A; SOM15). Similarly, the TCR was expressed at a higher level in Dicer-deficient cells (Fig. 5B; SOM15). Interestingly, the relative increases in expression of CD69 and the TCR in Dicer-deficient DP thymocytes observed in vivo correlated with the relative repression observed in the above 3′ UTR assays.
Figure 5.
Dysregulation of CD69 and the TCR in Dicer-deficient DP thymocytes. CD69 (A) and TCR (B) staining on electronically gated DP thymocytes from lck-cre+ and control Dicerf/f mice. Derepression was normalized via median fluorescence intensity (bar graphs). (C) CD69 levels on GFPhi and GFPneg Dicerf/f DP thymocytes transfected with a miR-181a or control siRNA. (D) As in C, examining lck-cre+ Dicerf/f DP thymocytes. Median fluorescence intensities for each parameter are provided in SOM16. Results are representative of a minimum of three individual experiments.
Finally, Bcl-2 expression in Dicer-deficient DP thymocytes was examined. Dicer-deficient DP thymocytes expressed levels of Bcl-2 protein markedly lower than those observed in control cells (data not shown). Consistent with a previous study (Cobb et al. 2005), Dicer-deficient thymocytes were more apoptotic than control cells. We suggest that loss of Dicer function induces a transcriptional program in DP thymocytes that obscures any specific effect of miR-181a on levels of Bcl-2 protein expression.
Modulation of potential miR-181a targets by transfection of nucleic acids into Dicer-deficient DP thymocytes
The increased expression of CD69 and the TCR in Dicer-deficient DP thymocytes is consistent with the regulation of these gene products by miR-181a and miR-181b, both of which are specifically up-regulated at the DP stage. However, Dicer deletion would be expected to reduce the levels of all miRNAs. Thus, the increase in expression of CD69 and the TCR in DP thymocytes might be due to reasons other than the reduction in levels of the miR-181a family. To demonstrate direct regulation of CD69 and the TCR in DP thymocytes by the miR-181 family, Dicer-deficient and control DP thymocytes were isolated from mice and cotransfected with miR-181a or control siRNAs in addition to a GFP marker plasmid. There was a reproducible reduction in the surface levels of CD69 in GFPhi thymocytes when compared with GFPhi thymocytes that had been transfected with a control siRNA duplex (Fig. 5C,D; SOM16). A similar repression of the TCR complex was not observed within the time frame of the experiment (data not shown), potentially due to the fact that the fully assembled TCR has an exceptionally long half-life (Minami et al. 1987). We concluded that in Dicer-deficient DP cells, specific introduction of miR-181a is able to repress the observed increase in CD69 expression. The reporter assays demonstrate that miR-181a is able to effect this repression directly through the 3′ UTR elements of these genes. Thus, it is likely that miR-181 family members directly regulate the levels of CD69 and the TCR through their 3′ UTR elements at the DP stage of thymocyte development.
Discussion
We quantitatively profiled short RNA species from sequential stages of development of a single mammalian cell lineage, documenting dynamic regulation of individual miRNAs and miRNA families during this process. This regulation is correlated to changes in gene expression at the genomic level in the processes of lineage commitment, stepwise maturation, and the developmental decision to mature to one of two related but highly different cell types. The functional significance of these dynamics is demonstrated in the context of miR-181a and its predicted targets.
The degree of variation in the total miRNA pool and total cellular RNA content across the T-lymphocyte developmental progression is striking. The miRNA pool varies per cell from a minimum of 5000 molecules per cell in DP thymocytes to 33,000 molecules per cell in DN4 cells. At the same time, total RNA content ranges from a low of 0.7 pg of RNA per cell in DP thymocytes to a maximum of 6.8 pg of RNA per cell in DN4 thymocytes. The major changes that we observe in per-cell miRNA pool and total RNA content stand in stark contrast to the constant, progressive increase in the ratio of the total miRNA pool to the total RNA pool. This progressive increase is consistent with previous studies in which overall miRNA levels are correlated to the level of differentiation of a cell (Lu et al. 2005), but our data also suggest that global miRNA levels are tightly coupled to cytoplasmic volume and/or ribosomal content. Cells that are rapidly proliferating (e.g., DN4 cells) are likely to be more transcriptionally and translationally active. The corresponding increase in the number of transcripts and level of translation would necessitate an increase in miRNA levels to maintain the same degree of control of the transcriptome. Although the absolute copy number of miR-181a is lower in DP cells than at any other developmental stage, it represents a higher fraction of the miRNA pool. It is only in the context of relative concentration that miR-181a mediates its biological effect at the DP stage of development. Thus, the concentration of an individual miRNA in relation to its targets is likely to be more important than its absolute copy number.
In relative terms, the vast majority of short RNAs were expressed at constant levels throughout thymocyte development. Of 136 miRNAs that were observed in the study, only 21 were observed to vary by cell type. Consideration of the aggregate expression of miRNA family members yielded largely consistent observations in regards to miRNA expression dynamics. However, there were instances in which the aggregate miRNA family revealed additional enrichment (miR-23) or changed the stage at which significance was observed (miR-15 family). The significantly changed miRNAs and miRNA families were continually expressed throughout thymocyte development, with transitory enrichments at specific cell stages. Curiously, while a high level of enrichment for at least one miRNA or miRNA family was observed at each distinct stage of thymocyte development, the reverse pattern of specific depletion at a single stage was much more rare. A major depletion in the cloning frequency of both products of the miR-142 gene at the DN3 stage is one exception (Figs. 2, 3). The relative expression of various ADAR family members at and adjacent to this developmental stage coupled with the lack of transcriptional down-regulation of pri-miR-142 at the DN3 stage suggest the possibility that ADAR2 and/or ADAR1p150 compete with ADAR1p110 to inhibit editing of pri-miR-142, ensuring proper maturation of the mature miR-142 miRNAs. Indeed, the perturbed pri-miR-142 editing patterns described in ADAR null mutants are consistent with this model (Yang et al. 2006). This may partially explain why this miRNA is an exception to the rule of basal expression with stage-specific enrichment observed for other miRNA species.
Statistically significant changes in the relative levels of expression of predicted mRNA targets were observed for 14 of 20 dynamically regulated miRNA families and for at least one miRNA family in each developmental stage examined. Consistent with a role in directly repressing the levels of proteins encoded by target mRNAs or reinforcing transcriptional down-regulations, in most cases underrepresentation of predicted targets was observed around local maxima in miRNA family expression. This strong correlation indicates that regulation by miRNAs is a characteristic of many intermediate developmental stages, and not solely of mature, differentiated cells or “one-way” developmental specification.
The dispensability of the RNA interference (RNAi) pathway for thymocyte maturation has been suggested by conditional deletion of Dicer in the T lineage (Cobb et al. 2005; Muljo et al. 2005). However, it is unclear from these studies whether individual cells had completely lost Dicer activity and miRNA function at the point that positive selection and lineage choice occurred. Indeed, even using the early-acting lck-cre transgene, DP cells exhibit detectable levels of mature miRNAs (Cobb et al. 2005). The 10-fold reduction in thymocyte number in these mice at stages immediately following deletion of the floxed locus (Cobb et al. 2005; data not shown), coupled with a marked increase in apoptosis of Dicer-deficient thymocytes both in vivo and ex vivo (Cobb et al. 2005; data not shown), leads us to believe that elimination of Dicer is quite deleterious, manifesting after Dicer protein and the miRNA pool have been depleted. Indeed, peripheral T cells in lck-cre Dicerf/f mice are enriched for nonrecombined alleles of Dicerf (data not shown).
In any case, it is clear that even individual miRNAs can influence hematopoietic development. Mice reconstituted with cells transduced with a miR-181a overexpression vector exhibit a paucity of T lymphocytes (Chen et al. 2004). While this was interpreted as a facilitated commitment to the B lineage, the results of this study provide an alternative explanation. Disruption of CD69 signaling has been shown to negatively impact the egress of lymphocytes from lymphoid organs, including the thymus (Nakayama et al. 2002; Alfonso et al. 2006; Shiow et al. 2006). Forced expression of miR-181a past the DP stage of thymocyte development would be expected to decrease CD69 levels on positively selected thymocytes, resulting in retention of these cells in the thymus and an apparent decrease in peripheral T cells. Our data in regard to miR-181a’s effect on the expression of Bcl-2 and the TCR in vivo are less complete. However, the effects of forced expression of miR-181a on these gene products would further exacerbate the phenotype resulting from lowered CD69 expression. Reduced expression of the TCR would be expected to shift the threshold for positive and negative selection, while an inability to increase Bcl2 expression upon positive selection would result in cell death. The degree to which miR-181a or other miRNAs can influence expression of these genes during T-cell maturation will not be known until genetic inactivation of this miRNA is described.
Returning to the more global observations of this study, the lower relative degree of enrichment or depletion of predicted targets and subtle effects on gene expression in this study, (particularly in regard to similar computational studies) at first seem unremarkable. However, other studies have compared gene expression profiles in tissues specifically expressing a given miRNA or from cell lines in which an ectopic miRNA is introduced. This analysis stands in stark contrast for the reason that nearly every miRNA that we observe is expressed throughout T-lymphocyte development, with graded increases or decreases at specific times during this process. This pattern of constant expression with transient enrichment is not consistent with the “switch-like” function that has been attributed to miRNAs with known developmental roles (Reinhart et al. 2000; Fazi et al. 2005; Giraldez et al. 2006) and suggests two things: First, any individual miRNA would be expected to exert some level of post-transcriptional control at all times, with more or less robust effects during times of significant enrichment or depletion, respectively. It follows that these miRNAs would not be expected to dramatically effect gene activation and repression, instead exerting most of their effects to modulate gene expression at an intermediate level. Second, the role of individual miRNAs and miRNA families extends beyond the temporal window of their first expression, continuing to play an active role in modulation of gene expression in subsequent cell stages.
Materials and methods
Mice
C57/BL6 mice were from Taconic or Jackson laboratories. The Dicerf/f mouse has been described (Harfe et al. 2005). The lck-cre transgenic mouse was from the Jackson laboratories. All mice were housed, handled, and euthanized in accordance with federal and institutional guidelines.
Thymocyte and RNA isolation
Thymocytes were isolated from female mice aged 5–10 wk and stained with antibodies from BD-Pharmingen. Populations were designated as follows: CD4SP, CD4+CD8+TCRhi; CD8SP, CD8+CD4−TCRhi; DP, CD4+CD8+TCRmed/lo; DN, CD4−CD8− CD19−GR1−γδ−TER-119−CD11b−DX5−. DN cells were further divided as follows: DN1, CD44+CD25−; DN3, CD44−CD25+; DN4, CD25−CD44−. Cells were sorted to >95% purity and processed using Ambion’s miRVana kit per the manufacturer’s instructions to isolate both short and long fractions of RNA. For preparation of bulk DP cell RNA in the direct quantification experiments, a one-step positive selection with biotinylated anti-CD8 and anti-biotin magnetic beads was used (Miltenyi). This routinely resulted in purities of >95%.
Short RNA cloning
Short RNAs were cloned essentially as described (Lagos-Quintana et al. 2001; Lau et al. 2001). We cloned directly from the short RNA fraction eluted from miRvana columns. There was no gel isolation between the 5′ and 3′ adapter ligation steps. Following RT (SuperScript III, Invitrogen) and 10 cycles of PCR amplification with AmpliTaq Gold (Perkin Elmer), amplifications were digested with the appropriate enzyme (StuI or PvuII, both from New England Biolabs) and gel isolated from a denaturing polyacrylamide gel using a 10-base-pair (bp) ladder (Invitrogen) and SyBR Gold (Molecular Probes). After the first round of amplification, Pfu Turbo was used, iterating amplification, digestion, and gel isolation until cloneable product could be identified. For this study, various libraries were amplified anywhere from 30 to 38 cycles in total. Bioinformatic extraction of individual short RNA clones from sequence reads was as described (Houbaviy et al. 2003). 3′linkers: Stu, 5′-CCTGTATC TGTGTATGGddC-3′; Pvu, 5′-CTGG-TATCTGTGTATGG ddC-3′. 5′ linkers: Stu, 5′-ACCACAGAGAAACCGrArGrG-3′; Pvu, 5′-ACCACAGAGAAACCGrCrArG-3′. 3′RTprime/PCR oligos: Stu, 5′-GACTAGCTTGGTGCC-ATACACAGATACA GG-3′; Pvu, 5′-GACTAGCTTGGTGCCATACACAGATAC CAG-3′. 5′ PCR oligos: Stu, 5′-GAGCCAACAGGCACCA CAGAGAAACCGAGG-3′; Pvu, 5′-GAGCCAACAGGCACCA CAGAGAAACCGCAG-3′. A detailed protocol is available on request.
Direct miRNA quantitation and calculation of miRNA copy number per cell
Thymocytes were sorted to >95% purity, directly counted in a hemacytometer, and lysed in Trizol. Five femtomoles of miR-196a was spiked into each lysis and later detected as a recovery control. Initial experiments in DP thymocytes were performed without a recovery control. In later experiments, the average recovery for four independent isolations of DP thymocytes was calculated, and data from earlier DP quantitations were normalized to this value. RNA was processed per the manufacturer’s instructions, quantitated on a Nanodrop spectrophotometer, and assayed using the Trilogy Assay (U.S. Genomics) essentially as described (Neely et al. 2006). The molarity of a given miRNA in each sample was determined by fitting the coincident events in the RNA sample to a curve generated with a synthetic miRNA template in a complex background.
Phylogenetic analysis
The complete linkage method of hierarchical clustering in the R software package was used to generate rooted trees from miRNA and mRNA expression data. To eliminate noise, we omitted miRNAs cloned less than five times overall and set mRNA array values less than log2(20) to log2(20). Significance of clustering was analyzed with the Pvclust package (Suzuki and Shimodaira 2006).
qRT–PCR analysis of ADAR expression
RNA was extracted with Trizol, DNase-I-digested, and reverse-transcribed using the SuperScript III kit. Quantitative real-tme PCR was performed using a SyBR-Green kit (Applied Biosystems) on an ABI 7500 instrument using the following oligos: A1LF, 5′-GACTACGCGTTGGGACTAGC-3′; A1LR, 5′-TGC TGAAGCTGGAAACTCCT-3′; A1SF, 5′-CTTGCCGGCACTA TGTCTC-3′; A1SR, 5′-TGCTGAAGCTGGAAACTCCT-3′; AB1F, 5′-CCAGTCAA-GAAGCCCTCAAA-3′; AB1R, 5′-GCG GTACTTGGAGTGACCAT-3′. Changes in relative expression of ADARp110 and ADARp150 were similar whether GAPDH or Tubulin was used as a control. The pri-miR-142 oligos have been described (Yang et al. 2006).
Luciferase assays
HeLa cells were transfected at 80% confluency in 24-well plates with 100 nM siRNA, 100 ng of pGL3, and 700 ng of Renilla/UTR reporter. Cells were split 1:3 4–6 h after transfection and assayed at 48 h in a Dual Luciferase Assay (Promega). All results were normalized to the effect of siRNAs on pRL-TK-RenCX6X (Doench et al. 2003). Si181a is the predicted post-Dicer-processing product of the miR-181a pre-miRNA hairpin. UTRs were amplified with the following oligos and subcloned into the NotI and XhoI sites of pRL-TK-CX6x: ToxF, 5′-AGCTAGCTCGAG CATGTGAGCTTGTGGGTCAC-3′; ToxR, 5′-ATGCATGCG GCCGCAGCACTTAGCTAGCGCGTTC-3′; RunxF, 5′-AGC TAGCTCGAGTGCATCTGGGTGGTCATTTA-3′; RunxR, 5′ATGCATGCGGCCGCTTGGATCTTTGGGGTACAGC-3′; CD69F, 5′-AGCTAGCTCGAGACTGTGCCATAGCACCACA G-3′; CD69R, 5′-ATGCATGCGGCCGCACAGCTTAAACTT TATAGTGGGTTTT-3′; EGRF, 5′-AGCTAGCTCGAGCATC TGTGCCATGGATTTTG-3′; EGRR, 5′-ATGCATGCGGCC GCTATCCCATGGGCAATAGAGC-3′; Foxp1F, 5′-AGCTA GCTCGAGAGACCGAAGATTGGGGAAAA-3′; Foxp1R, 5′-ATGCATGCGGCCGCTGAGGTCAGAACTTAAAATG-3′; Bcl2af1F, 5′-AGCTAGCTCGAGGCAAACATAAGGAGGA CAGCTT-3′; Bcl2af1R, 5′-ATGCATGCGGCCGCAGGG GAGCATCATGCAATAC-3′; TcrbF, 5′-AGCTAGCTCGAG TATGCATCCTGAGCCGTTCT-3′; TcrbR, 5′-ATGCATGCG GCCGCCTCCATGTTTTTATTGATTTAGTCTG-3′; TcraF2, 5′-AGCTAGCTCGAGGCAAGACTGACAGAGCCTGA-3′; TcraR2, 5′-ATGCATGCGGCCGCGAATCACCTTTAATGAT GTCATGG-3′.
Transfection of DP thymocytes
Whole thymocytes were isolated and nucleofected using the Primary T-cell Nucleofection Kit (Amaxa) and program X-001. Cells (5 × 106) were nucleofected with 1.5–3 μg of siRNA duplexes and 1 μg of pMaxGFP. Levels of surface markers were monitored at 12–16 h post-nucleofection.
Acknowledgments
We are indebted to Hristo Houbaviy, Gene Yeo, and Charlie Whittaker for designing scripts and processes used in the initial analysis of library sequences, and Reinhard Hoffman for providing raw expression data. We are grateful to the MIT CCR shared FACS facility for assisting in cell isolation. We thank U.S. Genomics for use of the Trilogy and Trilogy 2020 instruments as well as miRNA probes and outstanding assistance. We thank C.Z. Chen and M.M. Davis for providing data prior to publication, and for the generous sharing of reagents. Members of the Sharp, Burge, Jacks, and Bartel laboratories all participated in invaluable discussions. In particular, we thank R. Sandberg, B. Friedman, K. Cante-Barrett, and M. Winslow for advice, critical discussion, and review of the manuscript. J.R.N. is funded by the Cancer Research Institute.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1522907
References
- Alfonso C., McHeyzer-Williams M.G., Rosen H., McHeyzer-Williams M.G., Rosen H., Rosen H. CD69 down-modulation and inhibition of thymic egress by short- and long-term selective chemical agonism of sphingosine 1-phosphate receptors. Eur. J. Immunol. 2006;36:149–159. doi: 10.1002/eji.200535127. [DOI] [PubMed] [Google Scholar]
- Aravin A., Tuschl T., Tuschl T. Identification and characterization of small RNAs involved in RNA silencing. FEBS Lett. 2005;579:5830–5840. doi: 10.1016/j.febslet.2005.08.009. [DOI] [PubMed] [Google Scholar]
- Bagga S., Bracht J., Hunter S., Massirer K., Holtz J., Eachus R., Pasquinelli A.E., Bracht J., Hunter S., Massirer K., Holtz J., Eachus R., Pasquinelli A.E., Hunter S., Massirer K., Holtz J., Eachus R., Pasquinelli A.E., Massirer K., Holtz J., Eachus R., Pasquinelli A.E., Holtz J., Eachus R., Pasquinelli A.E., Eachus R., Pasquinelli A.E., Pasquinelli A.E. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell. 2005;122:553–563. doi: 10.1016/j.cell.2005.07.031. [DOI] [PubMed] [Google Scholar]
- Bonifacino J.S., McCarthy S.A., Maguire J.E., Nakayama T., Singer D.S., Klausner R.D., Singer A., McCarthy S.A., Maguire J.E., Nakayama T., Singer D.S., Klausner R.D., Singer A., Maguire J.E., Nakayama T., Singer D.S., Klausner R.D., Singer A., Nakayama T., Singer D.S., Klausner R.D., Singer A., Singer D.S., Klausner R.D., Singer A., Klausner R.D., Singer A., Singer A. Novel post-translational regulation of TCR expression in CD4+CD8+ thymocytes influenced by CD4. Nature. 1990;344:247–251. doi: 10.1038/344247a0. [DOI] [PubMed] [Google Scholar]
- Brennecke J., Stark A., Russell R.B., Cohen S.M., Stark A., Russell R.B., Cohen S.M., Russell R.B., Cohen S.M., Cohen S.M. Principles of microRNA-target recognition. PLoS Biol. 2005;3:e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.Z., Li L., Lodish H.F., Bartel D.P., Li L., Lodish H.F., Bartel D.P., Lodish H.F., Bartel D.P., Bartel D.P. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- Cobb B.S., Nesterova T.B., Thompson E., Hertweck A., O’Connor E., Godwin J., Wilson C.B., Brockdorff N., Fisher A.G., Smale S.T., Nesterova T.B., Thompson E., Hertweck A., O’Connor E., Godwin J., Wilson C.B., Brockdorff N., Fisher A.G., Smale S.T., Thompson E., Hertweck A., O’Connor E., Godwin J., Wilson C.B., Brockdorff N., Fisher A.G., Smale S.T., Hertweck A., O’Connor E., Godwin J., Wilson C.B., Brockdorff N., Fisher A.G., Smale S.T., O’Connor E., Godwin J., Wilson C.B., Brockdorff N., Fisher A.G., Smale S.T., Godwin J., Wilson C.B., Brockdorff N., Fisher A.G., Smale S.T., Wilson C.B., Brockdorff N., Fisher A.G., Smale S.T., Brockdorff N., Fisher A.G., Smale S.T., Fisher A.G., Smale S.T., Smale S.T., et al. T cell lineage choice and differentiation in the absence of the RNase III enzyme Dicer. J. Exp. Med. 2005;201:1367–1373. doi: 10.1084/jem.20050572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doench J.G., Sharp P.A., Sharp P.A. Specificity of microRNA target selection in translational repression. Genes & Dev. 2004;18:504–511. doi: 10.1101/gad.1184404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doench J.G., Petersen C.P., Sharp P.A., Petersen C.P., Sharp P.A., Sharp P.A. siRNAs can function as miRNAs. Genes & Dev. 2003;17:438–442. doi: 10.1101/gad.1064703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farh K.K., Grimson A., Jan C., Lewis B.P., Johnston W.K., Lim L.P., Burge C.B., Bartel D.P., Grimson A., Jan C., Lewis B.P., Johnston W.K., Lim L.P., Burge C.B., Bartel D.P., Jan C., Lewis B.P., Johnston W.K., Lim L.P., Burge C.B., Bartel D.P., Lewis B.P., Johnston W.K., Lim L.P., Burge C.B., Bartel D.P., Johnston W.K., Lim L.P., Burge C.B., Bartel D.P., Lim L.P., Burge C.B., Bartel D.P., Burge C.B., Bartel D.P., Bartel D.P. The widespread impact of mammalian microRNAs on mRNA repression and evolution. Science. 2005;310:1817–1821. doi: 10.1126/science.1121158. [DOI] [PubMed] [Google Scholar]
- Fazi F., Rosa A., Fatica A., Gelmetti V., De Marchis M.L., Nervi C., Bozzoni I., Rosa A., Fatica A., Gelmetti V., De Marchis M.L., Nervi C., Bozzoni I., Fatica A., Gelmetti V., De Marchis M.L., Nervi C., Bozzoni I., Gelmetti V., De Marchis M.L., Nervi C., Bozzoni I., De Marchis M.L., Nervi C., Bozzoni I., Nervi C., Bozzoni I., Bozzoni I. A minicircuitry comprised of microRNA-223 and transcription factors NFI-A and C/EBPα regulates human granulopoiesis. Cell. 2005;123:819–831. doi: 10.1016/j.cell.2005.09.023. [DOI] [PubMed] [Google Scholar]
- Giraldez A.J., Mishima Y., Rihel J., Grocock R.J., Van Dongen S., Inoue K., Enright A.J., Schier A.F., Mishima Y., Rihel J., Grocock R.J., Van Dongen S., Inoue K., Enright A.J., Schier A.F., Rihel J., Grocock R.J., Van Dongen S., Inoue K., Enright A.J., Schier A.F., Grocock R.J., Van Dongen S., Inoue K., Enright A.J., Schier A.F., Van Dongen S., Inoue K., Enright A.J., Schier A.F., Inoue K., Enright A.J., Schier A.F., Enright A.J., Schier A.F., Schier A.F. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science. 2006;312:75–79. doi: 10.1126/science.1122689. [DOI] [PubMed] [Google Scholar]
- Gratiot-Deans J., Ding L., Turka L.A., Nunez G., Ding L., Turka L.A., Nunez G., Turka L.A., Nunez G., Nunez G. bcl-2 proto-oncogene expression during human T cell development. Evidence for biphasic regulation. J. Immunol. 1993;151:83–91. [PubMed] [Google Scholar]
- Hara T., Jung L.K., Bjorndahl J.M., Fu S.M., Jung L.K., Bjorndahl J.M., Fu S.M., Bjorndahl J.M., Fu S.M., Fu S.M. Human T cell activation. III. Rapid induction of a phosphorylated 28 kD/32 kD disulfide-linked early activation antigen (EA 1) by 12-o-tetradecanoyl phorbol-13-acetate, mitogens, and antigens. J. Exp. Med. 1986;164:1988–2005. doi: 10.1084/jem.164.6.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harfe B.D., McManus M.T., Mansfield J.H., Hornstein E., Tabin C.J., McManus M.T., Mansfield J.H., Hornstein E., Tabin C.J., Mansfield J.H., Hornstein E., Tabin C.J., Hornstein E., Tabin C.J., Tabin C.J. The RNaseIII enzyme Dicer is required for morphogenesis but not patterning of the vertebrate limb. Proc. Natl. Acad. Sci. 2005;102:10898–10903. doi: 10.1073/pnas.0504834102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennet T., Hagen F.K., Tabak L.A., Marth J.D., Hagen F.K., Tabak L.A., Marth J.D., Tabak L.A., Marth J.D., Marth J.D. T-cell-specific deletion of a polypeptide N-acetylgalactosaminyl-transferase gene by site-directed recombination. Proc. Natl. Acad. Sci. 1995;92:12070–12074. doi: 10.1073/pnas.92.26.12070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann R., Bruno L., Seidl T., Rolink A., Melchers F., Bruno L., Seidl T., Rolink A., Melchers F., Seidl T., Rolink A., Melchers F., Rolink A., Melchers F., Melchers F. Rules for gene usage inferred from a comparison of large-scale gene expression profiles of T and B lymphocyte development. J. Immunol. 2003;170:1339–1353. doi: 10.4049/jimmunol.170.3.1339. [DOI] [PubMed] [Google Scholar]
- Houbaviy H.B., Murray M.F., Sharp P.A., Murray M.F., Sharp P.A., Sharp P.A. Embryonic stem cell-specific MicroRNAs. Dev. Cell. 2003;5:351–358. doi: 10.1016/s1534-5807(03)00227-2. [DOI] [PubMed] [Google Scholar]
- Kloosterman W.P., Plasterk R.H., Plasterk R.H. The diverse functions of microRNAs in animal development and disease. Dev. Cell. 2006;11:441–450. doi: 10.1016/j.devcel.2006.09.009. [DOI] [PubMed] [Google Scholar]
- Krutzfeldt J., Rajewsky N., Braich R., Rajeev K.G., Tuschl T., Manoharan M., Stoffel M., Rajewsky N., Braich R., Rajeev K.G., Tuschl T., Manoharan M., Stoffel M., Braich R., Rajeev K.G., Tuschl T., Manoharan M., Stoffel M., Rajeev K.G., Tuschl T., Manoharan M., Stoffel M., Tuschl T., Manoharan M., Stoffel M., Manoharan M., Stoffel M., Stoffel M. Silencing of microRNAs in vivo with ‘antagomirs.’. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana M., Rauhut R., Lendeckel W., Tuschl T., Rauhut R., Lendeckel W., Tuschl T., Lendeckel W., Tuschl T., Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana M., Rauhut R., Yalcin A., Meyer J., Lendeckel W., Tuschl T., Rauhut R., Yalcin A., Meyer J., Lendeckel W., Tuschl T., Yalcin A., Meyer J., Lendeckel W., Tuschl T., Meyer J., Lendeckel W., Tuschl T., Lendeckel W., Tuschl T., Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr. Biol. 2002;12:735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- Lau N.C., Lim L.P., Weinstein E.G., Bartel D.P., Lim L.P., Weinstein E.G., Bartel D.P., Weinstein E.G., Bartel D.P., Bartel D.P. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- Leung A.K., Calabrese J.M., Sharp P.A., Calabrese J.M., Sharp P.A., Sharp P.A. Quantitative analysis of Argonaute protein reveals microRNA-dependent localization to stress granules. Proc. Natl. Acad. Sci. 2006;103:18125–18130. doi: 10.1073/pnas.0608845103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis B.P., Shih I.H., Jones-Rhoades M.W., Bartel D.P., Burge C.B., Shih I.H., Jones-Rhoades M.W., Bartel D.P., Burge C.B., Jones-Rhoades M.W., Bartel D.P., Burge C.B., Bartel D.P., Burge C.B., Burge C.B. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- Lewis B.P., Burge C.B., Bartel D.P., Burge C.B., Bartel D.P., Bartel D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Lim L.P., Lau N.C., Weinstein E.G., Abdelhakim A., Yekta S., Rhoades M.W., Burge C.B., Bartel D.P., Lau N.C., Weinstein E.G., Abdelhakim A., Yekta S., Rhoades M.W., Burge C.B., Bartel D.P., Weinstein E.G., Abdelhakim A., Yekta S., Rhoades M.W., Burge C.B., Bartel D.P., Abdelhakim A., Yekta S., Rhoades M.W., Burge C.B., Bartel D.P., Yekta S., Rhoades M.W., Burge C.B., Bartel D.P., Rhoades M.W., Burge C.B., Bartel D.P., Burge C.B., Bartel D.P., Bartel D.P. The microRNAs of Caenorhabditis elegans. Genes & Dev. 2003;17:991–1008. doi: 10.1101/gad.1074403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim L.P., Lau N.C., Garrett-Engele P., Grimson A., Schelter J.M., Castle J., Bartel D.P., Linsley P.S., Johnson J.M., Lau N.C., Garrett-Engele P., Grimson A., Schelter J.M., Castle J., Bartel D.P., Linsley P.S., Johnson J.M., Garrett-Engele P., Grimson A., Schelter J.M., Castle J., Bartel D.P., Linsley P.S., Johnson J.M., Grimson A., Schelter J.M., Castle J., Bartel D.P., Linsley P.S., Johnson J.M., Schelter J.M., Castle J., Bartel D.P., Linsley P.S., Johnson J.M., Castle J., Bartel D.P., Linsley P.S., Johnson J.M., Bartel D.P., Linsley P.S., Johnson J.M., Linsley P.S., Johnson J.M., Johnson J.M. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- Liu C., Bai B., Skogerbo G., Cai L., Deng W., Zhang Y., Bu D., Zhao Y., Chen R., Bai B., Skogerbo G., Cai L., Deng W., Zhang Y., Bu D., Zhao Y., Chen R., Skogerbo G., Cai L., Deng W., Zhang Y., Bu D., Zhao Y., Chen R., Cai L., Deng W., Zhang Y., Bu D., Zhao Y., Chen R., Deng W., Zhang Y., Bu D., Zhao Y., Chen R., Zhang Y., Bu D., Zhao Y., Chen R., Bu D., Zhao Y., Chen R., Zhao Y., Chen R., Chen R. NONCODE: An integrated knowledge database of non-coding RNAs. Nucleic Acids Res. 2005;33 (Database issue):D112–D115. doi: 10.1093/nar/gki041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J., Getz G., Miska E.A., Alvarez-Saavedra E., Lamb J., Peck D., Sweet-Cordero A., Ebert B.L., Mak R.H., Ferrando A.A., Getz G., Miska E.A., Alvarez-Saavedra E., Lamb J., Peck D., Sweet-Cordero A., Ebert B.L., Mak R.H., Ferrando A.A., Miska E.A., Alvarez-Saavedra E., Lamb J., Peck D., Sweet-Cordero A., Ebert B.L., Mak R.H., Ferrando A.A., Alvarez-Saavedra E., Lamb J., Peck D., Sweet-Cordero A., Ebert B.L., Mak R.H., Ferrando A.A., Lamb J., Peck D., Sweet-Cordero A., Ebert B.L., Mak R.H., Ferrando A.A., Peck D., Sweet-Cordero A., Ebert B.L., Mak R.H., Ferrando A.A., Sweet-Cordero A., Ebert B.L., Mak R.H., Ferrando A.A., Ebert B.L., Mak R.H., Ferrando A.A., Mak R.H., Ferrando A.A., Ferrando A.A., et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- Maguire J.E., McCarthy S.A., Singer A., Singer D.S., McCarthy S.A., Singer A., Singer D.S., Singer A., Singer D.S., Singer D.S. Inverse correlation between steady-state RNA and cell surface T cell receptor levels. FASEB J. 1990;4:3131–3134. doi: 10.1096/fasebj.4.13.1698681. [DOI] [PubMed] [Google Scholar]
- Minami Y., Weissman A.M., Samelson L.E., Klausner R.D., Weissman A.M., Samelson L.E., Klausner R.D., Samelson L.E., Klausner R.D., Klausner R.D. Building a multichain receptor: Synthesis, degradation, and assembly of the T-cell antigen receptor. Proc. Natl. Acad. Sci. 1987;84:2688–2692. doi: 10.1073/pnas.84.9.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monticelli S., Ansel K.M., Xiao C., Socci N.D., Krichevsky A.M., Thai T.H., Rajewsky N., Marks D.S., Sander C., Rajewsky K., Ansel K.M., Xiao C., Socci N.D., Krichevsky A.M., Thai T.H., Rajewsky N., Marks D.S., Sander C., Rajewsky K., Xiao C., Socci N.D., Krichevsky A.M., Thai T.H., Rajewsky N., Marks D.S., Sander C., Rajewsky K., Socci N.D., Krichevsky A.M., Thai T.H., Rajewsky N., Marks D.S., Sander C., Rajewsky K., Krichevsky A.M., Thai T.H., Rajewsky N., Marks D.S., Sander C., Rajewsky K., Thai T.H., Rajewsky N., Marks D.S., Sander C., Rajewsky K., Rajewsky N., Marks D.S., Sander C., Rajewsky K., Marks D.S., Sander C., Rajewsky K., Sander C., Rajewsky K., Rajewsky K., et al. MicroRNA profiling of the murine hematopoietic system. Genome Biol. 2005;6:R71. doi: 10.1186/gb-2005-6-8-r71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muljo S.A., Ansel K.M., Kanellopoulou C., Livingston D.M., Rao A., Rajewsky K., Ansel K.M., Kanellopoulou C., Livingston D.M., Rao A., Rajewsky K., Kanellopoulou C., Livingston D.M., Rao A., Rajewsky K., Livingston D.M., Rao A., Rajewsky K., Rao A., Rajewsky K., Rajewsky K. Aberrant T cell differentiation in the absence of Dicer. J. Exp. Med. 2005;202:261–269. doi: 10.1084/jem.20050678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama T., Kasprowicz D.J., Yamashita M., Schubert L.A., Gillard G., Kimura M., Didierlaurent A., Koseki H., Ziegler S.F., Kasprowicz D.J., Yamashita M., Schubert L.A., Gillard G., Kimura M., Didierlaurent A., Koseki H., Ziegler S.F., Yamashita M., Schubert L.A., Gillard G., Kimura M., Didierlaurent A., Koseki H., Ziegler S.F., Schubert L.A., Gillard G., Kimura M., Didierlaurent A., Koseki H., Ziegler S.F., Gillard G., Kimura M., Didierlaurent A., Koseki H., Ziegler S.F., Kimura M., Didierlaurent A., Koseki H., Ziegler S.F., Didierlaurent A., Koseki H., Ziegler S.F., Koseki H., Ziegler S.F., Ziegler S.F. The generation of mature, single-positive thymocytes in vivo is dysregulated by CD69 blockade or overexpression. J. Immunol. 2002;168:87–94. doi: 10.4049/jimmunol.168.1.87. [DOI] [PubMed] [Google Scholar]
- Neely L.A., Patel S., Garver J., Gallo M., Hackett M., McLaughlin S., Nadel M., Harris J., Gullans S., Rooke J., Patel S., Garver J., Gallo M., Hackett M., McLaughlin S., Nadel M., Harris J., Gullans S., Rooke J., Garver J., Gallo M., Hackett M., McLaughlin S., Nadel M., Harris J., Gullans S., Rooke J., Gallo M., Hackett M., McLaughlin S., Nadel M., Harris J., Gullans S., Rooke J., Hackett M., McLaughlin S., Nadel M., Harris J., Gullans S., Rooke J., McLaughlin S., Nadel M., Harris J., Gullans S., Rooke J., Nadel M., Harris J., Gullans S., Rooke J., Harris J., Gullans S., Rooke J., Gullans S., Rooke J., Rooke J. A single-molecule method for the quantitation of microRNA gene expression. Nat. Methods. 2006;3:41–46. doi: 10.1038/nmeth825. [DOI] [PubMed] [Google Scholar]
- Paul M.S., Bass B.L., Bass B.L. Inosine exists in mRNA at tissue-specific levels and is most abundant in brain mRNA. EMBO J. 1998;17:1120–1127. doi: 10.1093/emboj/17.4.1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen C.P., Bordeleau M.E., Pelletier J., Sharp P.A., Bordeleau M.E., Pelletier J., Sharp P.A., Pelletier J., Sharp P.A., Sharp P.A. Short RNAs repress translation after initiation in mammalian cells. Mol. Cell. 2006;21:533–542. doi: 10.1016/j.molcel.2006.01.031. [DOI] [PubMed] [Google Scholar]
- Pillai R.S., Bhattacharyya S.N., Artus C.G., Zoller T., Cougot N., Basyuk E., Bertrand E., Filipowicz W., Bhattacharyya S.N., Artus C.G., Zoller T., Cougot N., Basyuk E., Bertrand E., Filipowicz W., Artus C.G., Zoller T., Cougot N., Basyuk E., Bertrand E., Filipowicz W., Zoller T., Cougot N., Basyuk E., Bertrand E., Filipowicz W., Cougot N., Basyuk E., Bertrand E., Filipowicz W., Basyuk E., Bertrand E., Filipowicz W., Bertrand E., Filipowicz W., Filipowicz W. Inhibition of translational initiation by Let-7 MicroRNA in human cells. Science. 2005;309:1573–1576. doi: 10.1126/science.1115079. [DOI] [PubMed] [Google Scholar]
- Porritt H.E., Rumfelt L.L., Tabrizifard S., Schmitt T.M., Zuniga-Pflucker J.C., Petrie H.T., Rumfelt L.L., Tabrizifard S., Schmitt T.M., Zuniga-Pflucker J.C., Petrie H.T., Tabrizifard S., Schmitt T.M., Zuniga-Pflucker J.C., Petrie H.T., Schmitt T.M., Zuniga-Pflucker J.C., Petrie H.T., Zuniga-Pflucker J.C., Petrie H.T., Petrie H.T. Heterogeneity among DN1 prothymocytes reveals multiple progenitors with different capacities to generate T cell and non-T cell lineages. Immunity. 2004;20:735–745. doi: 10.1016/j.immuni.2004.05.004. [DOI] [PubMed] [Google Scholar]
- Reinhart B.J., Slack F.J., Basson M., Pasquinelli A.E., Bettinger J.C., Rougvie A.E., Horvitz H.R., Ruvkun G., Slack F.J., Basson M., Pasquinelli A.E., Bettinger J.C., Rougvie A.E., Horvitz H.R., Ruvkun G., Basson M., Pasquinelli A.E., Bettinger J.C., Rougvie A.E., Horvitz H.R., Ruvkun G., Pasquinelli A.E., Bettinger J.C., Rougvie A.E., Horvitz H.R., Ruvkun G., Bettinger J.C., Rougvie A.E., Horvitz H.R., Ruvkun G., Rougvie A.E., Horvitz H.R., Ruvkun G., Horvitz H.R., Ruvkun G., Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- Shiow L.R., Rosen D.B., Brdickova N., Xu Y., An J., Lanier L.L., Cyster J.G., Matloubian M., Rosen D.B., Brdickova N., Xu Y., An J., Lanier L.L., Cyster J.G., Matloubian M., Brdickova N., Xu Y., An J., Lanier L.L., Cyster J.G., Matloubian M., Xu Y., An J., Lanier L.L., Cyster J.G., Matloubian M., An J., Lanier L.L., Cyster J.G., Matloubian M., Lanier L.L., Cyster J.G., Matloubian M., Cyster J.G., Matloubian M., Matloubian M. CD69 acts downstream of interferon-α/β to inhibit S1P1 and lymphocyte egress from lymphoid organs. Nature. 2006;440:540–544. doi: 10.1038/nature04606. [DOI] [PubMed] [Google Scholar]
- Stark A., Brennecke J., Bushati N., Russell R.B., Cohen S.M., Brennecke J., Bushati N., Russell R.B., Cohen S.M., Bushati N., Russell R.B., Cohen S.M., Russell R.B., Cohen S.M., Cohen S.M. Animal MicroRNAs confer robustness to gene expression and have a significant impact on 3′UTR evolution. Cell. 2005;123:1133–1146. doi: 10.1016/j.cell.2005.11.023. [DOI] [PubMed] [Google Scholar]
- Starr T.K., Jameson S.C., Hogquist K.A., Jameson S.C., Hogquist K.A., Hogquist K.A. Positive and negative selection of T cells. Annu. Rev. Immunol. 2003;21:139–176. doi: 10.1146/annurev.immunol.21.120601.141107. [DOI] [PubMed] [Google Scholar]
- Suzuki R., Shimodaira H., Shimodaira H. Pvclust: An R package for assessing the uncertainty in hierarchical clustering. Bioinformatics. 2006;22:1540–1542. doi: 10.1093/bioinformatics/btl117. [DOI] [PubMed] [Google Scholar]
- Yang J.H., Luo X., Nie Y., Su Y., Zhao Q., Kabir K., Zhang D., Rabinovici R., Luo X., Nie Y., Su Y., Zhao Q., Kabir K., Zhang D., Rabinovici R., Nie Y., Su Y., Zhao Q., Kabir K., Zhang D., Rabinovici R., Su Y., Zhao Q., Kabir K., Zhang D., Rabinovici R., Zhao Q., Kabir K., Zhang D., Rabinovici R., Kabir K., Zhang D., Rabinovici R., Zhang D., Rabinovici R., Rabinovici R. Widespread inosine-containing mRNA in lymphocytes regulated by ADAR1 in response to inflammation. Immunology. 2003;109:15–23. doi: 10.1046/j.1365-2567.2003.01598.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W., Chendrimada T.P., Wang Q., Higuchi M., Seeburg P.H., Shiekhattar R., Nishikura K., Chendrimada T.P., Wang Q., Higuchi M., Seeburg P.H., Shiekhattar R., Nishikura K., Wang Q., Higuchi M., Seeburg P.H., Shiekhattar R., Nishikura K., Higuchi M., Seeburg P.H., Shiekhattar R., Nishikura K., Seeburg P.H., Shiekhattar R., Nishikura K., Shiekhattar R., Nishikura K., Nishikura K. Modulation of microRNA processing and expression through RNA editing by ADAR deaminases. Nat. Struct. Mol. Biol. 2006;13:13–21. doi: 10.1038/nsmb1041. [DOI] [PMC free article] [PubMed] [Google Scholar]