Abstract
Single-stranded small interfering RNA (siRNA) guides the cleavage of homologous mRNA by Argonaute proteins, the catalytic core of the RNA-induced silencing complex (RISC), in the conserved RNA interference (RNAi) pathway. The separation of the siRNA duplex into single strands is essential for the activation of RISC. Previous biochemical studies have suggested that Argonaute proteins cleave and remove the passenger strand of siRNA duplex from RISC, but the in vivo importance of this process and the mechanism for the removal of the nicked passenger strand are not known. Here, we show that in the filamentous fungus Neurospora, the Argonaute homolog QDE-2 and its slicer function are required for the generation of single-stranded siRNA and gene silencing in vivo. Biochemical purification of QDE-2 led to the identification of QIP, a QDE-2-interacting protein, with an exonuclease domain. The disruption of qip in Neurospora impaired gene silencing and siRNA accumulated, mostly in nicked duplex form. Furthermore, our results suggest that QIP acts as an exonuclease that cleaves and removes the nicked passenger strand from siRNA duplex in a QDE-2-dependent manner. Together, these results suggest that both the cleavage and removal of the passenger strand from the siRNA duplex are important steps in RNAi pathways.
Keywords: RNAi, Neurospora, Argonaute, siRNA, exonuclease, RISC
RNA interference (RNAi) is a post-transcriptional gene silencing (PTGS) mechanism conserved from fungi to humans (Hannon 2002; Meister and Tuschl 2004; Mello and Conte 2004; Tomari and Zamore 2005; Catalanotto et al. 2006). RNAi and its related pathways are involved in a wide variety of biological processes such as development, antiviral defense, and maintenance of genomic stability (Brennecke et al. 2003; Sijen and Plasterk 2003; Baulcombe 2004; Lu et al. 2005). The RNAi pathway is initiated by double-stranded RNA (dsRNA) produced from various exogenous or endogenous sources, such as experimental introduction, viral infection, or the presence of multiple copies of transgenes or transposons. dsRNAs are recognized and cleaved by the ribonuclease-III domain-containing enzyme Dicer to generate 20–25 nucleotide (nt) small-interfering RNA (siRNA) duplexes (Zamore et al. 2000; Carmell and Hannon 2004; Meister and Tuschl 2004). The siRNA duplexes are loaded onto the RNAi effector, RNA-induced silencing complex (RISC). An Argonaute (Ago)-family protein forms the catalytic core of RISC (Hammond et al. 2001; Liu et al. 2004; Rand et al. 2004; Song et al. 2004). Before the activation of RISC, the siRNA duplex is dissociated and the passenger strand removed (Nykanen et al. 2001; Okamura et al. 2004; Tomari et al. 2004). The guide strand of the siRNA remains in the RISC and directs cleavage of target mRNA (Hammond et al. 2001; Liu et al. 2004; Rand et al. 2004; Song et al. 2004; Rivas et al. 2005).
The separation of the siRNA duplex and the removal of the passenger strand are critical steps in the activation of RISC. It was previously hypothesized that this process is mediated by an unidentified ATP-dependent RNA helicase (Meister and Tuschl 2004). However, recent elegant in vitro biochemical studies have suggested that in Drosophila, the removal of the passenger strand of the siRNA duplex is mediated by Argonaute 2 (Ago2), which directly binds to the siRNA duplex and cleaves the passenger strand the same way as it cleaves mRNA substrates (Matranga et al. 2005; Miyoshi et al. 2005; Rand et al. 2005). The passenger strand cleavage is important for efficient RISC activation in vitro, since RISC slicer activity is decreased when the cleavage of passenger strand is blocked. Consistent with this model, Drosophila Ago2 mutant embryo lysate failed to generate single-stranded siRNA in vitro (Okamura et al. 2004). On the other hand, it has been shown that a passenger strand cleavage-independent mechanism can ultimately result in the separation of siRNA duplex in vitro (Matranga et al. 2005). There is as yet no evidence that demonstrates the in vivo importance of Ago-2 in the generation of single-stranded siRNA. In addition, the mechanism for the removal of the Ago-cleaved passenger strand from an siRNA duplex is also not known; the nicked siRNA duplex is relatively stable in vitro (Matranga et al. 2005) and can potentially prevent the activation of the RISC. It was proposed that the release of the passenger strand may be facilitated by a cofactor (Haley and Zamore 2004; Matranga et al. 2005). The identity of such a factor(s), if it exists, is not known. A similar conclusion was also drawn in human cells and it was demonstrated that the introduction of a nick in the passenger strand can bypass the requirement of passenger strand cleavage by RISC (Leuschner et al. 2006).
In the filamentous fungus Neurospora crassa, the RNAi pathway is essential for the dsRNA or transgene (quelling)-induced gene silencing (Cogoni and Macino 1997, 1999; Catalanotto et al. 2006). As in higher eukaryotic systems, an Argonaute protein, QDE-2, is the core component of the RISC and is associated with siRNA (Catalanotto et al. 2000, 2002). There are two partially redundant Dicer proteins in Neurospora, DCL-1 and DCL-2, but DCL-2 contributes >90% of the dicer activity (Catalanotto et al. 2004).
To understand the mechanism of RISC activation, we examined the process of single-stranded siRNA generation in Neurospora. We showed that QDE-2, the Neurospora Argonaute homolog, and its slicer function are required for the generation of single-stranded siRNA and gene silencing in vivo. In addition, we identified QIP, a putative exonuclease, as a QDE-2-interacting protein. The disruption of the qip gene in Neurospora resulted in the accumulation of siRNA duplexes and impairment of gene silencing. Furthermore, our results suggest that QIP acts as an exonuclease that removes the nicked passenger strand from the siRNA duplex in a QDE-2-dependent manner.
Results
The slicer function of QDE-2 is required for the generation of single-stranded siRNA in vivo
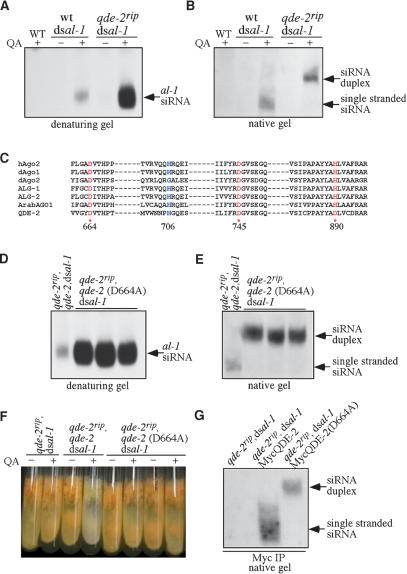
To investigate the role of QDE-2 in the generation of single-stranded siRNA in vivo, we introduced a dsRNA-expressing construct, dsal-1 (∼500-base-pair [bp] inverted repeat sequence specific for albino-1, a gene required for carotenoid biosynthesis), into a wild-type strain and a qde-2 mutant strain (qde-2rip). The qde-2 mutant is a complete loss-of-function mutant with multiple premature stop codons in the QDE-2 ORF (S. Choudhary, H.C. Lee, M. Maiti, Q. He, P. Cheng, Q. Liu, and Y. Liu, in prep.). We and others have shown that the expression of dsRNA from inverted repeat sequences results in gene silencing in Neurospora (Goldoni et al. 2004; Cheng et al. 2005; S. Choudhary, H.C. Lee, M. Maiti, Q. He, P. Cheng, Q. Liu, and Y. Liu, in prep.). The inverted repeats of al-1 are controlled by the quinic acid (QA)-inducible (qa-2) promoter (Giles et al. 1985), so the addition of QA in the medium will lead to the production of dsRNA and inhibition of carotenoid biosynthesis. As shown in Figure 1A, when the siRNA was analyzed by a denaturing gel, addition of QA resulted in al-1-specific siRNA production in both strains, but the level of siRNA was significantly higher in the qde-2rip strain than that in the wild-type strain. To investigate the nature of the siRNA produced in these two strains, RNA from each strain was examined using a native gel (Fig. 1B). As expected, all siRNA produced in the wild-type strain was single stranded. In contrast, there was no singled-stranded siRNA in the qde-2rip mutant, and all of the siRNA from this sample was in the duplex form. The appearance of comparable levels of siRNA in the two strains in a native gel is most likely due to inefficient hybridization of siRNA duplex to the probe during the Northern analysis. These data indicate that QDE-2 is essential for the generation of single-stranded siRNA in vivo. In addition, the high level of siRNA observed in the qde-2rip strain suggests that QDE-2 negatively regulates the levels of siRNA, probably because siRNA duplex is more stable than single-stranded siRNA in cells.
Figure 1.
The slicer function of QDE-2 is required for the generation of single-stranded siRNA in vivo. (A,B) Northern blot analyses of al-1 siRNA by denaturing gel (A) and native gel (B). The cultures were grown with/without QA (1 × 10−3 M). A wild-type (WT) sample without dsal-1 construct was used as a negative control. (C) Amino acid sequence alignment of the residues around the catalytic sites of the PIWI domains in QDE-2 and other Argonaute proteins. The amino acid residues of the DDH motif are highlighted and amino acid numbers of QDE-2 are indicated. Proteins aligned are human AGO-2 (hAgo-2), Drosophila AGO1 and AGO2 (dAgo-1 and dAgo2), C. elegans ALG-1 and ALG-2 (ALG-1 and ALG-2), and Arabidopsis AGO1 (ArabAGO1). (D,E) Northern blot analyses of al-1 siRNA by denaturing gel (D) and native gel (E) for the indicated strains. The cultures were grown in the presence of QA. (F) Pictures of Neurospora slants showing that the gene silencing of al-1 by dsal-1 is abolished in the qde-2rip and qde-2rip, qde-2(D664A) strains but is rescued in the qde-2rip, qde-2,dsal-1 strain. In experiments shown in D–F, several independent qde-2rip,qde-2(D664A),dsal-1 strains were used. (G) Northern blot analysis showing the association of siRNA with MycQDE-2 in the indicated strains.
The identities of these two populations of siRNA were also confirmed by the identical gel mobility behavior of 32P-labeled double- and single-stranded siRNAs in native gel (Supplementary Fig. 1). In addition, both a sense and an antisense al-1 probe could detect al-1 siRNA with similar efficiency (Supplementary Fig. 2), suggesting that siRNA strand selection is not influenced by the presence of complementary target mRNA. The observation of siRNA duplex only in qde-2rip mutant but not in wild-type strain indicates that the observed siRNA duplex is not an artificial product of the RNA extraction process. Furthermore, we showed that the presence of double-stranded siRNA in the qde-2rip mutant was not due to rapid annealing of complementary siRNAs during the RNA extraction process, in which phenol was used (Supplementary Fig. 3). Together, these experiments confirm the single-stranded or duplex nature of siRNA in the wild-type and qde-2rip strains, respectively.
We then examined whether the generation of single-stranded siRNA required the slicer activity of QDE-2 and whether the lack of single-stranded siRNA in the qde-2 mutant was due to siRNA duplex’s inability to be loaded onto the RISC. It has been proposed that the DDH motif within the PIWI domain in the Argonaute proteins are the catalytic residues that hydrolyze RNA (Liu et al. 2004; Parker et al. 2004; Rand et al. 2004; Song et al. 2004; Rivas et al. 2005). The two Asp residues in the DDH motif are conserved in the PIWI domain of QDE-2, whereas the His residue of the DDH motif is not (Fig. 1C). It has been shown that mutation of either the first or second Asp in the DDH motif completely abolishes the slicer activity of human Ago-2, but does not affect its ability to bind siRNA (Liu et al. 2004). We thus mutated Asp-664 (corresponding to the first D in the DDH motif) to Ala in a construct containing the wild-type qde-2 gene (with its own promoter), and both the wild-type qde-2 and the D644A qde-2 constructs were transformed into the qde-2rip dsal-1 strain. As shown in Figure 1, D and E, the level of single-stranded siRNA in the qde-2rip/qde-2,dsal-1 strain was near the wild-type level, indicating the complementation of qde-2 function by the wild-type qde-2 construct. In contrast, the siRNA was at high levels and was in the duplex form in the qde-2ripqde-2(D664A),dsal-1 strain, indicating that the slicer activity of QDE2 was required for separation of siRNA duplex into single strands. The lack of QDE-2 function in the qde-2ripqde-2(D664A),dsal-1 strain was not due to the lack of QDE-2 expression, as the level of expression of the mutant QDE-2 protein was comparable to the QDE-2 level in a qde-2ripqde-2,dsal-1 strain (Supplementary Fig. 4).
We then examined the ability of dsRNA to trigger gene silencing in the qde-2 slicer mutants. As shown in Figure 1F, the addition of QA during growth of the qde-2rip/qde-2,dsal-1 strain led to white aerial hyphae and conidia, the results of al-1 gene silencing. In contrast, the aerial hyphae and conidia remained orange in the qde-2ripqde-2(D664A),dsal-1 strain in the presence of QA, indicating that gene silencing was abolished. Together, these data suggest that the slicer activity of QDE-2 is essential for gene silencing and for the generation of single-stranded siRNA in vivo.
To investigate whether the single-stranded and duplex siRNAs are actually associated with QDE-2 and to examine whether the mutation of QDE-2 catalytic site affected its ability to bind siRNA duplex, we transformed the qde-2rip,dsal-1 strain with a construct expressing Myc-epitoped-tagged QDE-2 (MycQDE-2) or MycQDE-2(D664A). Immunoprecipitation of the cell extracts was performed using Myc monoclonal antibody and the precipitated siRNA was then analyzed by native gel and Northern blot analysis. As shown in Figure 1G, MycQDE-2 was associated with single-stranded siRNA, while MycQDE-2(D664A) was only associated with siRNA duplex. In contrast, no siRNA was pulled down with a control strain lacking the MycQDE-2 construct. This data indicates that the Neurospora RISC is loaded with double-stranded siRNA before the cleavage and removal of the passenger strand.
Identification of QIP, a QDE-2-associated protein with an exonuclease domain
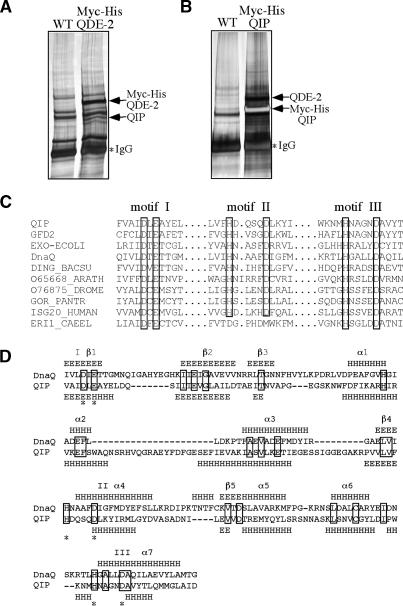
To further understand the mechanism of RISC activation, we tried to identify QDE-2-associated protein by biochemically purifying QDE-2 from Neurospora. A construct that expressed the Myc-His (5 cMyc and 6-His) epitope-tagged QDE-2 (Myc-His-QDE-2) was transformed into a wild-type strain of Neurospora and the Myc-His-QDE-2 was purified through a nickel affinity column and then immunoprecipitated using the monoclonal c-Myc antibody (He et al. 2005). Figure 2A shows a silver-stained SDS-PAGE gel of the final immunoprecipitates. Two specific protein bands (marked by arrows in Fig. 2A) were present in the Myc-His-QDE-2 sample. Mass spectrometry analysis of the excised bands identified one band as Myc-His-QDE-2 and the band at ∼80 kDa as a Neurospora hypothetical protein (NCU00076.2). This protein, named QIP, shows significant sequence homology with budding yeast Gfd2p. Gfd2p was previously identified as a high-copy suppressor of mutation of the DEAD-box RNA helicase gene dbp5 (Estruch and Cole 2003). Dbp5p plays an essential role in mRNA export, suggesting that Gfd2p may have a role in RNA regulation, although neither functional nor biochemical analysis of Gfd2p has been performed. The cloning of the qip cDNA revealed an extra 32 amino acids within the QIP ORF that was not predicted by the database.
Figure 2.
Identification of QIP as a QDE-2-interacting protein with an exonuclease domain. (A,B) Silver-stained SDS-PAGE gels showing the final immunoprecipitation products of the Myc-His-QDE-2 strain (A) and the Myc-His-QIP strain (B) by c-Myc monoclonal antibody. The identity of the indicated proteins was confirmed by mass spectrometry analyses. (C) Amino acid alignment of the three critical motifs in the exonuclease domains in QIP, ERI-1, and other proteins, including GFD2, Saccharomyces cerevisiae, NP_009894; EXO-ECOLI, E. coli, P0AEK0; DnaQ, E. coli, P03007; DING_BACSU, Bacillus subtillis, P54394; O65668_ARATH, Arabidopsis thaliana, O65668; O76875_DROME, Drosophila melanogaster, O76875; GOR_PANTER, Pan troglodytes, P48778; ISG20_HUMAN, Homo sapiens, Q96AZ6; ERI1_CAEEL, C. elegans, O44406. (D) Secondary structure comparison between the exonuclease domains in QIP and DnaQ (E. coli DNA polymerase III ε subunit). The predicted secondary structure of QIP was obtained by JPred (http://www.compbio.dundee.ac.uk/~www-jpred), whereas the secondary structure of DnaQ was based on its crystal structure (Hamdan et al. 2002).
To confirm the QDE-2 and QIP interaction, we purified the Myc-His-tagged QIP from a wild-type strain carrying an expression construct using the same approach as mentioned above (He et al. 2005). Figure 2B shows the silver-stained SDS-PAGE gel of c-Myc immunoprecipitates, which revealed two specific protein bands (indicated by arrows). Mass spectrometry analysis identified the lower band as Myc-His-QIP and the top band as the endogenous QDE-2, confirming the interaction between QDE-2 and QIP. In both experiments, a wild-type strain lacking any Myc-His epitope-tagged construct was used as negative control.
Amino acid sequence analysis revealed that the predicted QIP contains a 3′–5′ exonuclease domain in the C-terminal part of the protein. Comparison with various 3′–5′ exonuclease domains suggests that the exonuclease domain in QIP belongs to the DEDDh superfamily of 3′–5′ exonucleases. Figure 2C shows the amino acid alignment of three critical motifs of various 3′–5′ exonuclease domains in QIP and other proteins (Hamdan et al. 2002). The three motifs have four invariant acidic residues (D and E in motif I, D in motif II, and H and D in motif III) in the 3′–5′ exonuclease domains; all are conserved in QIP. Furthermore, secondary structure alignment (Fig. 2D) showed that the predicted QIP exonuclease domain was remarkably similar to that in the Escherichia coli DNA polymerase III ε subunit, for which a crystal structure has been solved (Hamdan et al. 2002). Together, these alignment studies suggest that QIP possesses a 3′–5′ DEDDh exonuclease domain. Previous studies showed that the conserved His (H) residue in motif III functions as a general base to direct an activated H2O molecule for nucleophilic attack at the terminal phosphodiester bond and is critical for exonuclease activity (Hamdan et al. 2002). The known RNAi suppressor ERI-1 also contains an exonuclease domain (Kennedy et al. 2004). However, unlike QIP, ERI-1 belongs to the DEMDh subfamily of DEDDh exonucleases, with different active sites in its motif II (Fig. 2C). The similarity between QIP and ERI-1 and QIP’s interaction with QDE-2 suggests that QIP may play a role in the RNAi pathway by processing siRNA.
Disruption of qip results in impairment of RNAi
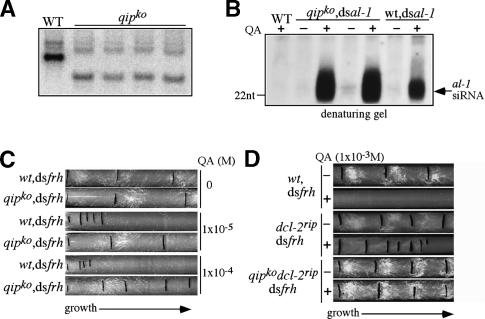
To understand the role of QIP in the Neurospora RNAi pathway in vivo, we generated qip knock-out (qipko) strains by replacing the endogenous qip gene with a hygromycin resistance gene (hph) through homologous recombination. Southern blot analysis (Fig. 3A) showed the disruption of the qip locus and the homokaryotic nature of the mutant strains. To investigate whether QIP, like ERI-1, played a role in regulating the levels of siRNA in vivo, we compared the levels of siRNA in the wt,dsal-1 and qipko,dsal-1 strains and found that siRNA levels were significantly higher in the mutant strains than that in the wild-type strain (Fig. 3B). This result suggests that QIP, like ERI-1, acts as a nuclease and cleaves siRNA.
Figure 3.
Disruption of qip in Neurospora severely impaired RNAi efficiency. (A) Southern blot analysis showing the disruption of the qip locus in several independent qipko strains. (B) Northern blot analyses of al-1 siRNA by denaturing gel showing the increase of al-1 siRNA levels in the qipko/dsal-1 strains compared with the wild-type/dsal-1 strain. Two independent qipko/dsal-1 strains were used. (C,D) Race tube assay showing the growth of the indicated strains in the absence or presence of various concentrations of QA in constant darkness. Each black line marks the growth front marked every 24 h The conidiation bands seen in the race tubes were due to the circadian conidiation rhythms.
We then investigated the role of QIP in the Neurospora RNAi pathway by analysis of dsRNA-induced gene silencing. We compared the gene silencing efficiency of the wild-type and the qipko strains by expressing dsRNA under the control of an inducible promoter. Use of the inducible promoter allowed a controlled level of gene silencing, thus this assay is more quantitative and more reliable than the traditional quelling assay (Cheng et al. 2005). We introduced an inverted repeat construct, under control of a QA-inducible promoter, specific for frh (dsfrh) into each strain. frh is an essential Neurospora gene required for circadian clock function, and silencing by dsfrh results in a QA dose-dependent inhibition of Neurospora growth and the loss of circadian rhythms, which can be easily scored using a race tube assay over the course of days (Cheng et al. 2005).
As shown in Figure 3C, the addition of 1 × 10−5 or 1 × 10−4 M of QA in the race tubes resulted in dramatic inhibition of growth for the wt,dsfrh strain (the distance between two growth front marks corresponds to 24 h of growth). This growth inhibition is QA dosage dependent (compare ±QA race tubes), indicating that this phenotype is not due to any growth defect of the strain itself. In contrast, the same QA treatments only led to modest inhibition of growth for the qipko,dsfrh strain, suggesting that gene silencing is impaired in the absence of QIP.
To further confirm this result, we compared the gene silencing efficiency of the dcl-2rip and dcl-2rip qipko strains by introducing the dsfrh construct. Neurospora has two functionally redundant DCLs (DCL-1 and DCL-2) and DCL-2 contributes >90% of the dicer activity (Catalanotto et al. 2004). Consistent with this, significantly less siRNA was produced in the dcl-2rip qipko strain than in the qipko strain when dsRNA was expressed (Supplementary Fig. 5). Thus, the comparison of RNAi efficiency in the dcl-2rip background should increase the sensitivity of the silencing assay. As shown in Figure 3D, QA, at the 1 × 10−3 M, almost completely inhibited the growth of the wt,dsfrh strain. In the dcl-2rip,dsfrh strain, the frh silencing was impaired compared with the wild type, but QA still led to significant inhibition of growth, indicating a significant reduction of gene silencing due to the deletion of dcl-2. In contrast, QA had no detectable effect on growth of the dcl-2rip/qipko,dsfrh strain as measured by the race tube assay, suggesting an almost complete loss of gene silencing in the double mutant. The lack of silencing in this mutant was also indicated by the persistence of robust circadian conidiation rhythms in the presence of QA (Cheng et al. 2005). In addition, we found that the levels of QDE-2 in the qip mutants were comparable to those in a wild-type strain (data not shown). Thus, the impaired gene silencing we observed in the qip mutants was not due to the lack of QDE-2. Taken together, these in vivo gene silencing analyses indicated that QIP, instead of functioning as an RNAi suppressor, is required for efficient RNAi in Neurospora.
QIP is required for processing of most of the duplex siRNA into single-stranded form
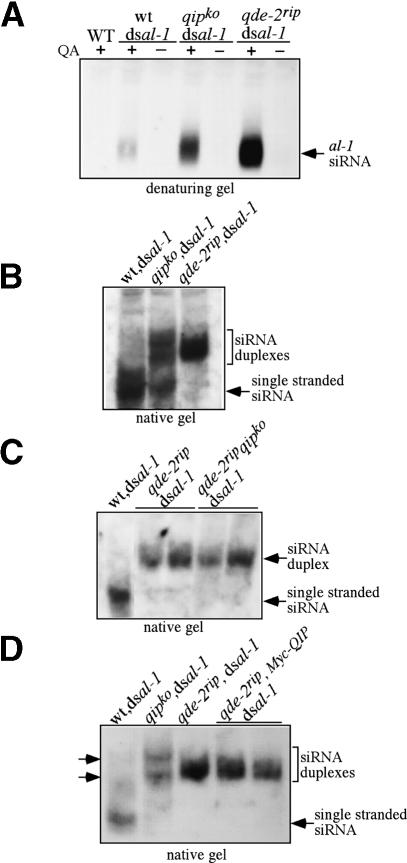
Having established that QIP was important for the Neurospora RNAi pathways, we investigated how QIP functions to mediate gene silencing. First, we found that the dicer activity in the wild-type and qipko strains were comparable (Supplementary Fig. 6). Thus, the impaired RNAi in the qipko strain is unlikely due to its effect on the DCLs. Because of the QIP–QDE-2 interaction and the high levels of siRNA in the qipko,dsal-1 strain, we then compared the levels of siRNA produced in the qipko strain with those in the wild-type and qde-2rip strains. The total siRNA revealed by denaturing gel analysis in the qipko,dsal-1 strain was significantly higher than that in the wt,dsal-1 strain, but lower than that in the qde-2rip,dsal-1 strain (Fig. 4A). When portions of the same siRNA samples were analyzed by a native gel, a small amount of single-stranded siRNA was seen in the qipko, dsal-1 strain, but most of the siRNA species were in duplex forms (Fig. 4B,D). In contrast, two distinct forms of siRNA duplexes with different gel mobility shifts were observed in the qipko,dsal-1 strain: One migrated slightly faster and one slower than the siRNA duplex in the qde-2rip,dsal-1 strain. These results indicate that that QIP is required for efficient processing of siRNA duplex into single-stranded siRNA, and RISC activation is impaired in qipko strain. This defect in the qipko strain explains the impaired RNAi phenotype observed in the dcl-2rip qipko mutant. In addition, the siRNA duplex forms in the qipko strain had different gel mobilities from those observed in the qde-2ripstrain, suggesting that the siRNA duplexes in the qipko strain may have already been processed by QDE-2.
Figure 4.
siRNAs from the qipko strain existed mostly as duplex forms that differed from the siRNA duplex forms from the qde-2rip strain. (A,B) Northern blot analyses of al-1 siRNA by denaturing (A) and native gels (B) showing that siRNA from the qipko/dsal-1 strain exists mostly in duplex forms. (C,D) Northern blot analyses of al-1 siRNA of the indicated strains by native gels showing that the function of QIP in siRNA processing is dependent on QDE-2. Two independent transformants for some of the strains (qde-2rip,dsal-1 and qde-2rip qipko,dsal-1 in C; qde-2rip,MycQIP,dsal-1 in D) were used. Note that the siRNA duplexes from the qipko strain showed different mobility from that of the duplex from the qde-2rip strains (marked by two arrows).
To examine whether the role of QIP in siRNA duplex separation is dependent on QDE-2, as suggested by the QDE-2/QIP interaction, we created a qde-2rip qipko double mutant. As shown in Figure 4C, siRNA produced in the double mutants migrated in one form on a native gel, with identical mobility to that in the qde-2 single mutant, suggesting that QIP acts downstream from QDE-2. When we overexpressed the Myc-tagged QIP in a qde-2rip strain (Fig. 4D), the level and native gel mobilities of siRNA were not altered from those in the qde-2rip strain. As described below, the Myc-QIP is fully functional. Together, these results indicate that QIP requires QDE-2 to process siRNA duplexes, most likely through the QDE-2/QIP interaction.
The predicted exonuclease domain of QIP is important for its function in siRNA duplex separation
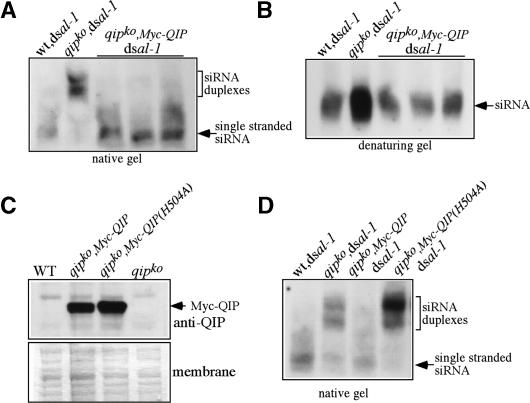
The observation that QIP is involved in siRNA duplex processing raised the obvious question of whether QIP is an exonuclease. To test this hypothesis, we sought in vivo evidence that the predicted exonuclease domain was required for QIP function. As shown in Figure 5, A and B, the expression of the Myc-tagged QIP fully rescued the function of the endogenous QIP in siRNA processing. In the qipko,Myc-QIP,dsal-1 strain, the entire population of siRNA was in the single-stranded form, and its level was similar to that in the wild-type strain. In contrast, the expression of the Myc-tagged QIP (H504A), in which the catalytic histidine in the motif III (Fig. 2C) was mutated to alanine, failed to rescue the siRNA processing defects of the qipko,dsal-1 strain (Fig. 5D). As shown in Figure 5C, the expression levels of the Myc-QIP and Myc-QIP(H504A) were similar (both from the QA-inducible promoter), which were significantly higher than the endogenous QIP level: Our QIP-specific antibody could readily detect the Myc-QIP but could not detect the endogenous QIP protein in Western blot analysis. Despite the overexpression of the Myc-QIP(H504A), most of the siRNA was in duplex forms in these strains (Fig. 5D). In addition, the H504A mutation of QIP did not affect its ability to interact with QDE-2 (data not shown). Therefore, these data suggest that QIP functions as an exonuclease in siRNA processing.
Figure 5.
Mutation of the putative catalytic residue in the QIP exonuclease domain abolished QIP’s function in siRNA processing. (A,B) Northern blot analyses of al-1 siRNA by native (A) and denaturing (B) gels showing that the expression of Myc-QIP complements the siRNA processing defects in the qipko strain. (C) Western blot analysis using an anti-QIP antibody showing the overexpression of Myc-QIP and Myc-QIP(H504A) in the qipko strains. Amido black-stained membrane is shown below. (D) Northern blot analyses of al-1 siRNA in a native gel showing that the expression of Myc-QIP(H504A) cannot complement the siRNA-processing defects in the qipko strain. In A and B, three independent qipko,Myc-QIP,dsal-1 strains and in D, two independent qipko,Myc-QIP(H504A),dsal-1 strains were used.
QIP is involved in removing the nicked passenger strand from the siRNA duplex
Since QIP appeared to function as an exonuclease and required QDE-2 to function, these results suggest that QIP assists QDE-2 in the process of converting siRNA duplex into single-stranded siRNA, thus generating active RISC. However, as an exonuclease, QIP is not expected to have an endonuclease cleavage function as QDE-2 in making the initiate cleavage of the siRNA duplex. The presence of different siRNA duplex species in native gel analysis of siRNA from the qipko and qde-2rip strains (Fig. 4B,D) raises the possibility that the siRNA duplexes in the qipko strain have already been processed by QDE-2. If so, the siRNA duplexes in qipko strain should be nicked: The passenger strand should have been cleaved by QDE-2 and, although the duplex has not been denatured, it has a distinct gel mobility compared with intact siRNA duplex. Consistent with this notion, it has been previously shown that duplexes containing a nicked passenger strand are stable in the early stage of RISC assembly (Matranga et al. 2005). Thus, it is likely that the function of QIP is to digest the nicked passenger strand to generate single-stranded siRNA.
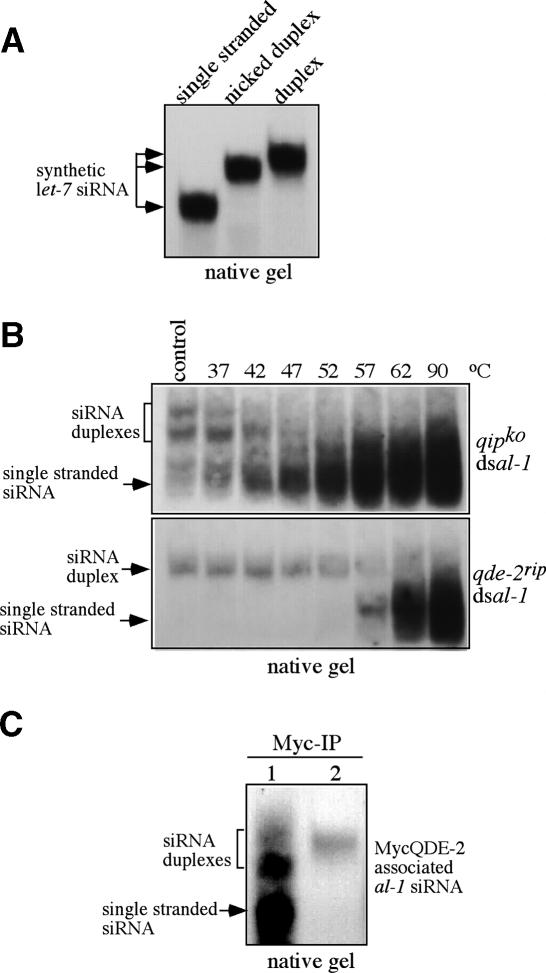
To test this hypothesis, we compared the gel mobility behavior of single-stranded, nicked, and double-stranded synthetic let-7 siRNAs in native gels. As shown in Figure 6A, the nicked let-7 siRNA duplex migrated slightly faster than the intact siRNA duplex. Such a profile is very similar to siRNA found in the qipko dsal-1 and qde-2rip dsal-1 strains, suggesting that the siRNA in the qipko dsal-1 is nicked siRNA duplex. In the qipko dsal-1 strain, there is another form of siRNA with slower mobility, which could be due to nicked siRNA duplex with a different conformation.
Figure 6.
The siRNA in the qipko strain has been cleaved by RISC. (A) Gel mobility profiles of single-stranded, nicked duplex, and intact duplex synthesized let-7 siRNA (25mer). (B) The siRNA duplexes from the qipko strain were significantly less stable than from the qde-2rip strain. The siRNA samples obtained from the qipko and qde-2rip strains (cultured at 25°C) were incubated at the indicated temperatures for 10 min before analysis by Northern blot in native gels by an al-1-specific probe. The control samples were not subjected to the temperature treatment. (C) Northern blot analysis showing the association of siRNA species with QDE-2. Immunoprecipitation was performed using Myc monoclonal antibody. (1) qde-2rip qipko MycQDE- 2. (2) qde-2rip MycQDE-2(D664A).
To further test this hypothesis, we compared the thermostability of siRNA duplexes in the qipko and qde-2rip strains. We reasoned that if the siRNA duplexes in the qipko strain are nicked, they should exhibit less thermostability than the intact siRNA duplex from the qde-2rip strain. The small RNA samples produced in these two strains were incubated at different temperatures (37°C–90°C, control at 4°C) for 10 min before analysis by native gel (Fig. 6B). As predicted, the siRNA duplexes from the qipko strain were significantly less stable than those from the qde-2rip strain: The siRNA duplexes in the qipko strain were beginning to convert into the single-stranded form at 37°C and most of them were single stranded at 57°C, whereas the siRNA duplex in the qde-2rip strain was entirely in the duplex form until 57°C and was fully single stranded only after the 90°C treatment. This result suggests that the siRNA duplexes in the qipko strain have already been processed by QDE-2 and, thus, the passenger strand is cleaved, although the two strands of the duplex have not been separated. We were not able to detect the nicked products in denaturing gel by Northern blot analysis, most likely due to their small sizes, which can result in inefficient transfer and hybridization.
We then examined whether the nicked siRNA forms observed in qipko strain are associated with QDE-2. The MycQDE-2 construct was transformed into the qde-2rip qipko dsal-1 strain. Immunoprecipitation of the cell extracts using Myc monoclonal antibody and the Northern blot of the precipitated siRNA were performed. As shown in Figure 6C, the nicked siRNA duplexes and single-stranded siRNA were associated with MycQDE-2 in the qipko strain. In contrast, only the duplex siRNAs were associated with MycQDE-2(D664A) in the qde-2rip strain. These data suggest that after the cleavage of the passenger siRNA strand by RISC, the nicked siRNA duplex is associated with QDE-2. Taken together, our results suggest that QIP functions in the RISC activation process by removing the nicked passenger strand from the siRNA duplexes.
Discussion
Separation of the two strands of siRNA duplex is a critical step for the activation of the RISC in the RNAi pathways. In this study, we demonstrated that the Neurospora Argonaute protein, QDE-2, and its slicer function are essential for the generation of single-stranded siRNA in vivo. In addition, we showed that QIP, a putative exonuclease that interacts with QDE-2, was important for the separation of cleaved passenger strand from the siRNA guide strand and for efficient RNAi. Therefore, both the initial cleavage of passenger strand and its removal are important steps in the activation of a RISC.
QDE-2 is required for the cleavage of the siRNA passenger strand and for the generation of single-stranded siRNA in vivo
Argonaute proteins are the central players in RNAi and related pathways, and they all contain two signature domains: PAZ and PIWI. The PAZ domain is involved in the binding of siRNA (Song et al. 2003), whereas the PIWI domain acts as the nuclease to cleave mRNA substrates when guided by single-stranded siRNA (Liu et al. 2004; Rand et al. 2004; Song et al. 2004; Rivas et al. 2005). Previous biochemical studies have suggested a model in which the Argonautes are also responsible for the generation of single-stranded siRNA through cleavage of the passenger strand of siRNA duplex (Matranga et al. 2005; Miyoshi et al. 2005; Rand et al. 2005; Leuschner et al. 2006). Here, we presented strong in vivo evidence to support this model. We showed that siRNA exists only as siRNA duplex in the qde-2 mutant (Fig. 1). In addition, the siRNA levels in the qde-2 mutant were significantly higher than those in the wild-type stain, suggesting that the siRNA duplexes in the mutant strain were more stable in the cell. Furthermore, when the critical catalytic residue of the QDE-2 PIWI domain was mutated, siRNA remained as duplex and gene silencing was completely abolished. These results suggest that the slicer function of QDE-2 is required for the generation of single-stranded siRNA in vivo.
Interestingly, it was previously shown in Drosophila extracts that when RISC is loaded with siRNA containing a noncleavable bond at the cleavage site of the passenger strand, a bypass mechanism separates the siRNA duplex (Matranga et al. 2005). In contrast, our data showed that in Neurospora, passenger strand cleavage by QDE-2 was absolutely required for the production of single-stranded siRNA in vivo, indicating an essential role of passenger strand cleavage by QDE-2 in RISC activation in this organism. Since the submission of our paper, a recent study in Drosophila also showed that the cleavage activity of Ago2 is required for the removal of the passenger strand from RISC (Kim et al. 2007).
QIP, a putative exonuclease, removes cleaved passenger strand from siRNA duplex by interacting with QDE-2
It was previously proposed that the cleaved passenger strand is actively removed from siRNA duplex by an unknown factor (Haley and Zamore 2004; Matranga et al. 2005). Our study confirmed this hypothesis and identified QIP as an important player in this mechanism. A current model of the RNAi pathway in Neurospora is shown in Figure 7. In this model, the siRNA duplex is first loaded onto the RISC, then QDE-2 cleaves passenger strand of the siRNA duplex. Subsequently, QIP, in association with QDE-2, acts as an exonuclease to digest the passenger strand, leaving the guide strand in the complex and resulting in the activation of the RISC.
Figure 7.
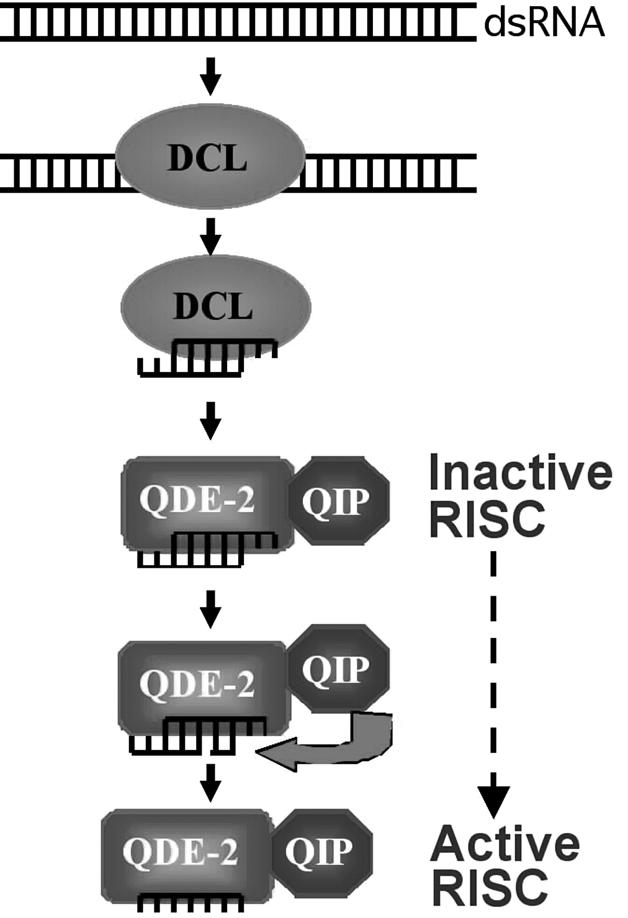
An updated model for the RNAi pathway in Neurospora explaining the roles of QDE-2 and QIP in generation of active RISC-containing single-stranded siRNA.
This model is supported by several lines of evidence. First, QIP was identified as a QDE-2-interacting protein in vivo. Second, the disruption of qip in Neurospora resulted in high levels of siRNA, which were mostly in duplex forms. Third, gene silencing efficiency was impaired in the qipko strain and was almost completely abolished in the qip dcl-2 double mutant, providing further evidence for the importance of generation of single-stranded siRNA in the RNAi pathway. Fourth, the distinct mobility shifts of the siRNA duplexes in qipko and qde-2rip strains suggested that the siRNA duplexes in the qipko strain are nicked siRNA duplex. Further supporting this notion, we showed that the siRNA duplexes in the qipko strain, which are associated with QDE-2, were significantly less thermodynamically stable than the intact duplex, suggesting that the passenger strand had been cleaved in these siRNA duplexes. Finally, by mutating a critical residue in the putative exonuclease domain of QIP, we showed that the exonuclease domain of QIP was required for its function in siRNA processing. Thus, it is very likely that QIP functions as an exonuclease and can digest the previously cleaved passenger strand in siRNA duplexes. In the qipko strain, most of the siRNA was in duplex form, although some single-stranded siRNA was observed, which was presumably responsible for the partial gene silencing we observed in the qipko strain. These results suggest that additional exonucleases or natural dissociation of the nicked duplexes are also responsible for the production of some single-stranded siRNA.
Based on the evidence we presented, it appears that QIP cannot function independently of QDE-2. Consistent with this notion, we found that the recombinant QIP expressed in sf9 insect cells failed to exhibit significant exonuclease activity toward siRNA in vitro (data not shown). In addition, the overexpression of QIP in the qde-2 mutant background did not change the level or nature of siRNA complex. Thus, QIP appears rely on its interaction with QDE-2 to find its substrates.
QDE-2 functions as more than just a substrate-recruiting subunit for QIP, it creates the substrates for QIP. In the qde-2rip qde-2(D664A) strain, which expresses a QDE-2 with a mutation in the catalytic site of the PIWI domain and a wild-type copy of qip, the level and the nature of the siRNA remained the same as in the qde-2rip strain (Fig. 1E). In addition, the overexpression of QIP in this mutant strain did not have any detectable impact on siRNA processing (data not shown). Since mutation of the catalytic site of QDE-2 did not affect the QDE-2/QIP interaction (data not shown), these data suggest that the substrates of QIP are previously cleaved siRNA strands rather than an intact siRNA duplex. Although we do not yet know how QIP distinguishes the intact siRNA duplex from the nicked duplex associated with QDE-2, it is possible that QIP recognizes a nicked duplex due to its unique conformation. Alternatively, the specificity may be determined by the structural position of QIP in the QDE-2-siRNA complex. It is possible that within the RISC, QIP is positioned to degrade only the precleaved passenger strand.
Because QIP interacts with QDE-2, QIP may also have a role in siRNA loading onto the RISC. Although we cannot completely eliminate this possibility, our results showed that the siRNA in the qip mutant had been processed, suggesting that QIP functions downstream from the siRNA loading step and does not have a major role in this process. Furthermore, the association of siRNA with MycQDE-2 in the qipko:qde-2rip double-mutant strain suggests that the siRNA loading process was not impaired in the qipko strain.
In the Caenorhabditis elegans eri-1 mutants, the RNAi efficiency for foreign dsRNA is up-regulated due to the accumulation of siRNA (Kennedy et al. 2004). Interestingly, a recent study indicates that, instead of functioning as a suppressor for all RNAi responses in C. elegans, ERI-1 is required for the endogenously produced siRNA-mediated gene silencing (Duchaine et al. 2006). Thus, ERI-1, which contains an exonuclease domain, is a required component of the endogenous gene silencing machinery. MUT-7, another 3′–5′ exonuclease from the RNase D subfamily, has also been shown to be important for the RNAi pathway in C. elegans (Ketting et al. 1999). How ERI-1 and MUT-7 function to promote gene silencing is not clear. The results presented here suggest that like QIP, these exonucleases may be involved in the processing siRNA at a step that is important for RNAi. Our study also suggests that exonucleases in other organisms may be also important for the removal of the passenger strand from siRNA duplexes, a critical step in RISC activation.
Materials and methods
Strains and growth conditions
The wild-type strain used in this study was FGSC 4200(a). FGSC7088 (his-3, a) was the host strain used for his-3 targeting constructs. The following mutant strains, in the wild-type as well as his-3 backgrounds, were created for this study: qipko, qipko dcl-2rip, and qipko qde-2rip. The qde-2rip and dcl-2rip strains were generated in a separate study (S. Choudhary, H.C. Lee, M. Maiti, Q. He, P. Cheng, Q. Liu, and Y. Liu, in prep.). Culture conditions were the same as described previously (Cheng et al. 2001b). For liquid cultures containing QA, 0.01 M QA (pH 5.8) was added to the liquid culture medium containing 1× Vogel’s, 0.1% glucose, and 0.17% arginine.
Creation of mutant strains
Gene replacement was used to replace the entire Neurospora qip gene with the hygromycin resistance gene (hph) using a protocol described previously (Colot et al. 2006). A plasmid carrying the gene replacement cassette was introduced into a wild-type strain by electroporation (Margolin et al. 1999). Southern blot analysis was performed to identify the positive transformants. Positive transformants were crossed with a wild-type strain and sexual spores were picked individually and germinated on slants containing hygromycin. Southern blot analysis was performed to identify the homokaryon mutant strains.
Antibody generation
The GST-QIP (containing QIP amino acids 181–567) fusion protein was expressed in BL21 cells and the inclusion bodies containing the recombinant proteins were purified and used as the antigen to generate rabbit polyclonal antiserum, as described previously (Cheng et al. 2001a). The QDE-2-specific antibody was generated in a separate study (S. Choudhary, H.C. Lee, M. Maiti, Q. He, P. Cheng, Q. Liu, and Y. Liu, in prep.).
Protein analyses
Protein extraction, quantification, and Western blot analysis were performed as previously described (Cheng et al. 2001a). Equal amounts of total protein (50 μg) were loaded in each lane of the SDS-PAGE gel and, after electrophoresis, proteins were transferred onto PVDF membrane and Western blot analysis was performed.
Purification of small RNA and Northern blot analysis
Total RNA was extracted as described previously (Aronson et al. 1994). From this total RNA, low-molecular-weight RNA was enriched using 5% polyethylene glycol (MW8000) and 500 mM NaCl as previously described (Catalanotto et al. 2002). Low-molecular-weight RNA was quantified by spectrophotometric analysis and was separated by electrophoresis through 16% polyacrylamide, 7 M urea, 0.5× tris-borate EDTA (TBE) gels. Equal amounts of RNA (25 μg) were loaded in each lane. The RNA was then transferred onto Hybond-N+ filters (Amersham Biosciences) and fixed by UV cross-linking. For native gel analysis, RNA was separated through 16% TBE (without urea) gels. The size of RNAs was verified by oligonucleotide markers. Prehybridization and hybridization were performed at 42°C using ULTRAhybTM buffer (Ambion). For hybridization, a single-stranded RNA probe, transcribed from a DNA template in antisense orientation in the presence of 32P-labeled uridine triphosphate (New England Nuclear) using MAXIscript T7 kit (Ambion), was generated. The RNA probe was then treated with TURBO DNase (Ambion) and hydrolyzed to an average size of 50 nt using 80 mM sodium bicarbonate and 120 mM sodium carbonate as described (Catalanotto et al. 2002). The hydrolyzed probe was then added to hybridization solution for overnight hybridization. After hybridization, the membrane was washed three times with 2× SSC and 2% SDS buffer for 30 min at 42°C before it was exposed to X-ray film.
Dicer assay
Uniformly radiolabeled dsRNA substrates were prepared and dicer assay were performed as described previously (Liu et al. 2003). The products of dicer reaction from wild-type, qipko, and dcl-1ko:dcl-2rip strains were compared in 16% denaturing (7 M urea) polyacrylamide gel.
Synthetic siRNAs
The sequences of the two strands of let-7 siRNA are siRNA01(for), UGAGGUAGUAGGUUGUAUAGUGAUU; siRNA02 (rev), UCACUAUACAACCUACUACCUCAUU. Nicked sequences are UCACUAUACA and ACCUACUAC CUCAUU. siRNA01 was radiolabeled at the 5′ end with [γ-32p]ATP using T4 polynucleotide kinase. siRNA duplex was prepared by annealing the siRNA02 with the radiolabeled siRNA01 in 1× annealing buffer (10 mM HEPES at pH 7.4, 5 Mm MgCl2). For the nicked duplex, radiolabeled siRNA01 was annealed with the two complementary nicked sequences. Afterward, the radiolabeled single-stranded, intact duplex and nicked duplex siRNAs were passed through a Micro Bio-Spin 6 column (Bio-Rad) and analyzed in 16% native polyacrylamide gel.
Coimmunoprecipitation of siRNAs
Cell extracts from strains expressing either MycQDE-2 or MycQDE-2 (D664A) were prepared in a buffer containing 25 mM Tris (pH 7.5), 150 mM NaCl, 1.5 mM MgCl2, 1% NP40, 1 mM DTT, protease inhibitors, and 100 U/mL RNase inhibitor. Cell extracts were precleared by incubation with 20 μL of Protein G Sepharose (GE Healthcare) for 1 h at 4°C. For immunoprecipitation of MycQDE-2, precleared extracts were incubated with Myc monoclonal antibody overnight at 4°C and then with 25 μL of Protein G Sepharose for 2 h at 4°C. Immunoprecipitates were washed five times using the extraction buffer. To recover coprecipitated RNAs, 150 μL of TE (10 mM Tris, 1 mM EDTA at pH 7) was added to the precipitates, followed by phenol/chloroform extraction and ethanol precipitation. The precipitated RNAs were analyzed by 16% native polyacrylamide gel and Northern blot analysis using single-stranded RNA probe.
Acknowledgments
We thank Lixin Wang, Ping Cheng, Swati Choudhary, and Qiyang He for technical assistance. We thank Qinghua Liu and his laboratory for technical assistance and Yue Chen from Southwestern Protein Chemistry Core facility for protein identification. Y.L. is an endowed scholar in Biomedical Research at University of Texas Southwestern Medical Center.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.1497607
References
- Aronson B.D., Johnson K.A., Loros J.J., Dunlap J.C., Johnson K.A., Loros J.J., Dunlap J.C., Loros J.J., Dunlap J.C., Dunlap J.C. Negative feedback defining a circadian clock: Autoregulation of the clock gene frequency. Science. 1994;263:1578–1584. doi: 10.1126/science.8128244. [DOI] [PubMed] [Google Scholar]
- Baulcombe D. RNA silencing in plants. Nature. 2004;431:356–363. doi: 10.1038/nature02874. [DOI] [PubMed] [Google Scholar]
- Brennecke J., Hipfner D.R., Stark A., Russell R.B., Cohen S.M., Hipfner D.R., Stark A., Russell R.B., Cohen S.M., Stark A., Russell R.B., Cohen S.M., Russell R.B., Cohen S.M., Cohen S.M. bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell. 2003;113:25–36. doi: 10.1016/s0092-8674(03)00231-9. [DOI] [PubMed] [Google Scholar]
- Carmell M.A., Hannon G.J., Hannon G.J. RNase III enzymes and the initiation of gene silencing. Nat. Struct. Mol. Biol. 2004;11:214–218. doi: 10.1038/nsmb729. [DOI] [PubMed] [Google Scholar]
- Catalanotto C., Azzalin G., Macino G., Cogoni C., Azzalin G., Macino G., Cogoni C., Macino G., Cogoni C., Cogoni C. Gene silencing in worms and fungi. Nature. 2000;404:245. doi: 10.1038/35005169. [DOI] [PubMed] [Google Scholar]
- Catalanotto C., Azzalin G., Macino G., Cogoni C., Azzalin G., Macino G., Cogoni C., Macino G., Cogoni C., Cogoni C. Involvement of small RNAs and role of the qde genes in the gene silencing pathway in Neurospora. Genes & Dev. 2002;16:790–795. doi: 10.1101/gad.222402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catalanotto C., Pallotta M., ReFalo P., Sachs M.S., Vayssie L., Macino G., Cogoni C., Pallotta M., ReFalo P., Sachs M.S., Vayssie L., Macino G., Cogoni C., ReFalo P., Sachs M.S., Vayssie L., Macino G., Cogoni C., Sachs M.S., Vayssie L., Macino G., Cogoni C., Vayssie L., Macino G., Cogoni C., Macino G., Cogoni C., Cogoni C. Redundancy of the two dicer genes in transgene-induced posttranscriptional gene silencing in Neurospora crassa. Mol. Cell. Biol. 2004;24:2536–2545. doi: 10.1128/MCB.24.6.2536-2545.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catalanotto C., Nolan T., Cogoni C., Nolan T., Cogoni C., Cogoni C. Homology effects in Neurospora crassa. FEMS Microbiol. Lett. 2006;254:182–189. doi: 10.1111/j.1574-6968.2005.00037.x. [DOI] [PubMed] [Google Scholar]
- Cheng P., Yang Y., Heintzen C., Liu Y., Yang Y., Heintzen C., Liu Y., Heintzen C., Liu Y., Liu Y. Coiled-coil domain-mediated FRQ-FRQ interaction is essential for its circadian clock function in Neurospora. EMBO J. 2001a;20:101–108. doi: 10.1093/emboj/20.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng P., Yang Y., Liu Y., Yang Y., Liu Y., Liu Y. Interlocked feedback loops contribute to the robustness of the Neurospora circadian clock. Proc. Natl. Acad. Sci. 2001b;98:7408–7413. doi: 10.1073/pnas.121170298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng P., He Q., He Q., Wang L., Liu Y., He Q., He Q., Wang L., Liu Y., He Q., Wang L., Liu Y., Wang L., Liu Y., Liu Y. Regulation of the Neurospora circadian clock by an RNA helicase. Genes & Dev. 2005;19:234–241. doi: 10.1101/gad.1266805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cogoni C., Macino G., Macino G. Isolation of quelling-defective (qde) mutants impaired in posttranscriptional transgene-induced gene silencing in Neurospora crassa. Proc. Natl. Acad. Sci. 1997;94:10233–10238. doi: 10.1073/pnas.94.19.10233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cogoni C., Macino G., Macino G. Gene silencing in Neurospora crassa requires a protein homologous to RNA-dependent RNA polymerase. Nature. 1999;399:166–169. doi: 10.1038/20215. [DOI] [PubMed] [Google Scholar]
- Colot H.V., Park G., Turner G.E., Ringelberg C., Crew C.M., Litvinkova L., Weiss R.L., Borkovich K.A., Dunlap J.C., Park G., Turner G.E., Ringelberg C., Crew C.M., Litvinkova L., Weiss R.L., Borkovich K.A., Dunlap J.C., Turner G.E., Ringelberg C., Crew C.M., Litvinkova L., Weiss R.L., Borkovich K.A., Dunlap J.C., Ringelberg C., Crew C.M., Litvinkova L., Weiss R.L., Borkovich K.A., Dunlap J.C., Crew C.M., Litvinkova L., Weiss R.L., Borkovich K.A., Dunlap J.C., Litvinkova L., Weiss R.L., Borkovich K.A., Dunlap J.C., Weiss R.L., Borkovich K.A., Dunlap J.C., Borkovich K.A., Dunlap J.C., Dunlap J.C. A high-throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proc. Natl. Acad. Sci. 2006;103:10352–10357. doi: 10.1073/pnas.0601456103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duchaine T.F., Wohlschlegel J.A., Kennedy S., Bei Y., Conte D., Jr., Pang K., Brownell D.R., Harding S., Mitani S., Ruvkun G., Wohlschlegel J.A., Kennedy S., Bei Y., Conte D., Jr., Pang K., Brownell D.R., Harding S., Mitani S., Ruvkun G., Kennedy S., Bei Y., Conte D., Jr., Pang K., Brownell D.R., Harding S., Mitani S., Ruvkun G., Bei Y., Conte D., Jr., Pang K., Brownell D.R., Harding S., Mitani S., Ruvkun G., Conte D., Jr., Pang K., Brownell D.R., Harding S., Mitani S., Ruvkun G., Pang K., Brownell D.R., Harding S., Mitani S., Ruvkun G., Brownell D.R., Harding S., Mitani S., Ruvkun G., Harding S., Mitani S., Ruvkun G., Mitani S., Ruvkun G., Ruvkun G., et al. Functional proteomics reveals the biochemical niche of C. elegans DCR-1 in multiple small-RNA-mediated pathways. Cell. 2006;124:343–354. doi: 10.1016/j.cell.2005.11.036. [DOI] [PubMed] [Google Scholar]
- Estruch F., Cole C.N., Cole C.N. An early function during transcription for the yeast mRNA export factor Dbp5p/Rat8p suggested by its genetic and physical interactions with transcription factor IIH components. Mol. Biol. Cell. 2003;14:1664–1676. doi: 10.1091/mbc.E02-09-0602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giles N.H., Case M.E., Baum J., Geever R., Huiet L., Patel V., Tyler B., Case M.E., Baum J., Geever R., Huiet L., Patel V., Tyler B., Baum J., Geever R., Huiet L., Patel V., Tyler B., Geever R., Huiet L., Patel V., Tyler B., Huiet L., Patel V., Tyler B., Patel V., Tyler B., Tyler B. Gene organization and regulation in the qa (quinic acid) gene cluster of Neurospora crassa. Microbiol. Rev. 1985;49:338–358. doi: 10.1128/mr.49.3.338-358.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldoni M., Azzalin G., Macino G., Cogoni C., Azzalin G., Macino G., Cogoni C., Macino G., Cogoni C., Cogoni C. Efficient gene silencing by expression of double stranded RNA in Neurospora crassa. Fungal Genet. Biol. 2004;41:1016–1024. doi: 10.1016/j.fgb.2004.08.002. [DOI] [PubMed] [Google Scholar]
- Haley B., Zamore P.D., Zamore P.D. Kinetic analysis of the RNAi enzyme complex. Nat. Struct. Mol. Biol. 2004;11:599–606. doi: 10.1038/nsmb780. [DOI] [PubMed] [Google Scholar]
- Hamdan S., Carr P.D., Brown S.E., Ollis D.L., Dixon N.E., Carr P.D., Brown S.E., Ollis D.L., Dixon N.E., Brown S.E., Ollis D.L., Dixon N.E., Ollis D.L., Dixon N.E., Dixon N.E. Structural basis for proofreading during replication of the Escherichia coli chromosome. Structure. 2002;10:535–546. doi: 10.1016/s0969-2126(02)00738-4. [DOI] [PubMed] [Google Scholar]
- Hammond S.M., Boettcher S., Caudy A.A., Kobayashi R., Hannon G.J., Boettcher S., Caudy A.A., Kobayashi R., Hannon G.J., Caudy A.A., Kobayashi R., Hannon G.J., Kobayashi R., Hannon G.J., Hannon G.J. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science. 2001;293:1146–1150. doi: 10.1126/science.1064023. [DOI] [PubMed] [Google Scholar]
- Hannon G.J. RNA interference. Nature. 2002;418:244–251. doi: 10.1038/418244a. [DOI] [PubMed] [Google Scholar]
- He Q., Cheng P., He Q., Liu Y., Cheng P., He Q., Liu Y., He Q., Liu Y., Liu Y. The COP9 signalosome regulates the Neurospora circadian clock by controlling the stability of the SCFFWD-1 complex. Genes & Dev. 2005;19:1518–1531. doi: 10.1101/gad.1322205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy S., Wang D., Ruvkun G., Wang D., Ruvkun G., Ruvkun G. A conserved siRNA-degrading RNase negatively regulates RNA interference in C. elegans. Nature. 2004;427:645–649. doi: 10.1038/nature02302. [DOI] [PubMed] [Google Scholar]
- Ketting R.F., Haverkamp T.H., van Luenen H.G., Plasterk R.H., Haverkamp T.H., van Luenen H.G., Plasterk R.H., van Luenen H.G., Plasterk R.H., Plasterk R.H. Mut-7 of C. elegans, required for transposon silencing and RNA interference, is a homolog of Werner syndrome helicase and RNaseD. Cell. 1999;99:133–141. doi: 10.1016/s0092-8674(00)81645-1. [DOI] [PubMed] [Google Scholar]
- Kim K., Lee Y.S., Carthew R.W., Lee Y.S., Carthew R.W., Carthew R.W. Conversion of pre-RISC to holo-RISC by Ago2 during assembly of RNAi complexes. RNA. 2007;13:22–29. doi: 10.1261/rna.283207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leuschner P.J., Ameres S.L., Kueng S., Martinez J., Ameres S.L., Kueng S., Martinez J., Kueng S., Martinez J., Martinez J. Cleavage of the siRNA passenger strand during RISC assembly in human cells. EMBO Rep. 2006;7:314–320. doi: 10.1038/sj.embor.7400637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q., Rand T.A., Kalidas S., Du F., Kim H.E., Smith D.P., Wang X., Rand T.A., Kalidas S., Du F., Kim H.E., Smith D.P., Wang X., Kalidas S., Du F., Kim H.E., Smith D.P., Wang X., Du F., Kim H.E., Smith D.P., Wang X., Kim H.E., Smith D.P., Wang X., Smith D.P., Wang X., Wang X. R2D2, a bridge between the initiation and effector steps of the Drosophila RNAi pathway. Science. 2003;301:1921–1925. doi: 10.1126/science.1088710. [DOI] [PubMed] [Google Scholar]
- Liu J., Carmell M.A., Rivas F.V., Marsden C.G., Thomson J.M., Song J.J., Hammond S.M., Joshua-Tor L., Hannon G.J., Carmell M.A., Rivas F.V., Marsden C.G., Thomson J.M., Song J.J., Hammond S.M., Joshua-Tor L., Hannon G.J., Rivas F.V., Marsden C.G., Thomson J.M., Song J.J., Hammond S.M., Joshua-Tor L., Hannon G.J., Marsden C.G., Thomson J.M., Song J.J., Hammond S.M., Joshua-Tor L., Hannon G.J., Thomson J.M., Song J.J., Hammond S.M., Joshua-Tor L., Hannon G.J., Song J.J., Hammond S.M., Joshua-Tor L., Hannon G.J., Hammond S.M., Joshua-Tor L., Hannon G.J., Joshua-Tor L., Hannon G.J., Hannon G.J. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- Lu R., Maduro M., Li F., Li H.W., Broitman-Maduro G., Li W.X., Ding S.W., Maduro M., Li F., Li H.W., Broitman-Maduro G., Li W.X., Ding S.W., Li F., Li H.W., Broitman-Maduro G., Li W.X., Ding S.W., Li H.W., Broitman-Maduro G., Li W.X., Ding S.W., Broitman-Maduro G., Li W.X., Ding S.W., Li W.X., Ding S.W., Ding S.W. Animal virus replication and RNAi-mediated antiviral silencing in Caenorhabditis elegans. Nature. 2005;436:1040–1043. doi: 10.1038/nature03870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolin B.S., Freitag M., Selker E.U., Freitag M., Selker E.U., Selker E.U. Improved plasmids for gene targeting at the his-3 locus of Neurospora crassa by electroporation. Fungal Genet. Newsl. 1999;44:24–36. [Google Scholar]
- Matranga C., Tomari Y., Shin C., Bartel D.P., Zamore P.D., Tomari Y., Shin C., Bartel D.P., Zamore P.D., Shin C., Bartel D.P., Zamore P.D., Bartel D.P., Zamore P.D., Zamore P.D. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell. 2005;123:607–620. doi: 10.1016/j.cell.2005.08.044. [DOI] [PubMed] [Google Scholar]
- Meister G., Tuschl T., Tuschl T. Mechanisms of gene silencing by double-stranded RNA. Nature. 2004;431:343–349. doi: 10.1038/nature02873. [DOI] [PubMed] [Google Scholar]
- Mello C.C., Conte D., Jr., Conte D., Jr. Revealing the world of RNA interference. Nature. 2004;431:338–342. doi: 10.1038/nature02872. [DOI] [PubMed] [Google Scholar]
- Miyoshi K., Tsukumo H., Nagami T., Siomi H., Siomi M.C., Tsukumo H., Nagami T., Siomi H., Siomi M.C., Nagami T., Siomi H., Siomi M.C., Siomi H., Siomi M.C., Siomi M.C. Slicer function of Drosophila Argonautes and its involvement in RISC formation. Genes & Dev. 2005;19:2837–2848. doi: 10.1101/gad.1370605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nykanen A., Haley B., Zamore P.D., Haley B., Zamore P.D., Zamore P.D. ATP requirements and small interfering RNA structure in the RNA interference pathway. Cell. 2001;107:309–321. doi: 10.1016/s0092-8674(01)00547-5. [DOI] [PubMed] [Google Scholar]
- Okamura K., Ishizuka A., Siomi H., Siomi M.C., Ishizuka A., Siomi H., Siomi M.C., Siomi H., Siomi M.C., Siomi M.C. Distinct roles for Argonaute proteins in small RNA-directed RNA cleavage pathways. Genes & Dev. 2004;18:1655–1666. doi: 10.1101/gad.1210204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker J.S., Roe S.M., Barford D., Roe S.M., Barford D., Barford D. Crystal structure of a PIWI protein suggests mechanisms for siRNA recognition and slicer activity. EMBO J. 2004;23:4727–4737. doi: 10.1038/sj.emboj.7600488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rand T.A., Ginalski K., Grishin N.V., Wang X., Ginalski K., Grishin N.V., Wang X., Grishin N.V., Wang X., Wang X. Biochemical identification of Argonaute 2 as the sole protein required for RNA-induced silencing complex activity. Proc. Natl. Acad. Sci. 2004;101:14385–14389. doi: 10.1073/pnas.0405913101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rand T.A., Petersen S., Du F., Wang X., Petersen S., Du F., Wang X., Du F., Wang X., Wang X. Argonaute2 cleaves the anti-guide strand of siRNA during RISC activation. Cell. 2005;123:621–629. doi: 10.1016/j.cell.2005.10.020. [DOI] [PubMed] [Google Scholar]
- Rivas F.V., Tolia N.H., Song J.J., Aragon J.P., Liu J., Hannon G.J., Joshua-Tor L., Tolia N.H., Song J.J., Aragon J.P., Liu J., Hannon G.J., Joshua-Tor L., Song J.J., Aragon J.P., Liu J., Hannon G.J., Joshua-Tor L., Aragon J.P., Liu J., Hannon G.J., Joshua-Tor L., Liu J., Hannon G.J., Joshua-Tor L., Hannon G.J., Joshua-Tor L., Joshua-Tor L. Purified Argonaute2 and an siRNA form recombinant human RISC. Nat. Struct. Mol. Biol. 2005;12:340–349. doi: 10.1038/nsmb918. [DOI] [PubMed] [Google Scholar]
- Sijen T., Plasterk R.H., Plasterk R.H. Transposon silencing in the Caenorhabditis elegans germ line by natural RNAi. Nature. 2003;426:310–314. doi: 10.1038/nature02107. [DOI] [PubMed] [Google Scholar]
- Song J.J., Liu J., Tolia N.H., Schneiderman J., Smith S.K., Martienssen R.A., Hannon G.J., Joshua-Tor L., Liu J., Tolia N.H., Schneiderman J., Smith S.K., Martienssen R.A., Hannon G.J., Joshua-Tor L., Tolia N.H., Schneiderman J., Smith S.K., Martienssen R.A., Hannon G.J., Joshua-Tor L., Schneiderman J., Smith S.K., Martienssen R.A., Hannon G.J., Joshua-Tor L., Smith S.K., Martienssen R.A., Hannon G.J., Joshua-Tor L., Martienssen R.A., Hannon G.J., Joshua-Tor L., Hannon G.J., Joshua-Tor L., Joshua-Tor L. The crystal structure of the Argonaute2 PAZ domain reveals an RNA binding motif in RNAi effector complexes. Nat. Struct. Biol. 2003;10:1026–1032. doi: 10.1038/nsb1016. [DOI] [PubMed] [Google Scholar]
- Song J.J., Smith S.K., Hannon G.J., Joshua-Tor L., Smith S.K., Hannon G.J., Joshua-Tor L., Hannon G.J., Joshua-Tor L., Joshua-Tor L. Crystal structure of Argonaute and its implications for RISC slicer activity. Science. 2004;305:1434–1437. doi: 10.1126/science.1102514. [DOI] [PubMed] [Google Scholar]
- Tomari Y., Zamore P.D., Zamore P.D. Perspective: Machines for RNAi. Genes & Dev. 2005;19:517–529. doi: 10.1101/gad.1284105. [DOI] [PubMed] [Google Scholar]
- Tomari Y., Du T., Haley B., Schwarz D.S., Bennett R., Cook H.A., Koppetsch B.S., Theurkauf W.E., Zamore P.D., Du T., Haley B., Schwarz D.S., Bennett R., Cook H.A., Koppetsch B.S., Theurkauf W.E., Zamore P.D., Haley B., Schwarz D.S., Bennett R., Cook H.A., Koppetsch B.S., Theurkauf W.E., Zamore P.D., Schwarz D.S., Bennett R., Cook H.A., Koppetsch B.S., Theurkauf W.E., Zamore P.D., Bennett R., Cook H.A., Koppetsch B.S., Theurkauf W.E., Zamore P.D., Cook H.A., Koppetsch B.S., Theurkauf W.E., Zamore P.D., Koppetsch B.S., Theurkauf W.E., Zamore P.D., Theurkauf W.E., Zamore P.D., Zamore P.D. RISC assembly defects in the Drosophila RNAi mutant armitage. Cell. 2004;116:831–841. doi: 10.1016/s0092-8674(04)00218-1. [DOI] [PubMed] [Google Scholar]
- Zamore P.D., Tuschl T., Sharp P.A., Bartel D.P., Tuschl T., Sharp P.A., Bartel D.P., Sharp P.A., Bartel D.P., Bartel D.P. RNAi: Double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell. 2000;101:25–33. doi: 10.1016/S0092-8674(00)80620-0. [DOI] [PubMed] [Google Scholar]