Abstract
Acinetobacter baumannii has emerged as an important and problematic human pathogen as it is the causative agent of several types of infections including pneumonia, meningitis, septicemia, and urinary tract infections. We explored the pathogenic content of this harmful pathogen using a combination of DNA sequencing and insertional mutagenesis. The genome of this organism was sequenced using a strategy involving high-density pyrosequencing, a novel, rapid method of high-throughput sequencing. Excluding the rDNA repeats, the assembled genome is 3,976,746 base pairs (bp) and has 3830 ORFs. A significant fraction of ORFs (17.2%) are located in 28 putative alien islands, indicating that the genome has acquired a large amount of foreign DNA. Consistent with its role in pathogenesis, a remarkable number of the islands (16) contain genes implicated in virulence, indicating the organism devotes a considerable portion of its genes to pathogenesis. The largest island contains elements homologous to the Legionella/Coxiella Type IV secretion apparatus. Type IV secretion systems have been demonstrated to be important for virulence in other organisms and thus are likely to help mediate pathogenesis of A. baumannii. Insertional mutagenesis generated avirulent isolates of A. baumannii and verified that six of the islands contain virulence genes, including two novel islands containing genes that lacked homology with others in the databases. The DNA sequencing approach described in this study allows the rapid elucidation of the DNA sequence of any microbe and, when combined with genetic screens, can identify many novel genes important for microbial pathogenesis.
Keywords: Ethanol, high-density pyrosequencing, insertional mutagenesis, pathogenesis
Acinetobacter baumannii is a Gram-negative, nonmotile, obligate aerobic coccus that is commonly found in soil, water, and sewage, and in healthcare settings (Baumann et al. 1968a; Juni 1978). Difficulties in containing, controlling, and eliminating the spread of A. baumannii have challenged clinicians and healthcare providers (Bergogne-Berezin and Towner 1996; Bernards et al. 2004; Koulenti and Rello 2006). Recently, drug-resistant A. baumannii was responsible for an outbreak of bacteremia in >240 American troops in Iraq (Centers for Disease Control and Prevention 2004; Abbott 2005), and there is significant concern of a major epidemic involving this organism. This versatile organism can utilize a variety of carbon sources and is able to grow in a range of temperatures and pH conditions (Juni 1978). La Scola and Raoult (2004) isolated A. baumannii from human body lice and speculate that the bacteria may utilize the arthropod host as one means of transmission. This hardiness, combined with its intrinsic resistance to many antimicrobial agents, contributes to the organism’s fitness and has enabled it to thrive in hospital settings worldwide. Mortality in patients suffering A. baumannii infections can be as high as 75% (Chastre and Trouillet 2000).
Alarmingly, little is known about the virulence, antibiotic resistance, or persistence strategies of A. baumannii. The pathogenic determinants that have been reported thus far for A. baumannii include a novel pilus assembly system involved in biofilm formation (Tomaras et al. 2003), an outer membrane protein (Omp38) that causes apoptosis in human epithelial cells (Choi et al. 2005), and a polycistronic siderophore-mediated iron-acquisition system conserved between A. baumannii and Vibrio anguillarum (Dorsey et al. 2003, 2004). This presumably comprises a small fraction of elements involved in A. baumannii pathogenesis, and thus, novel global approaches are essential to comprehensively understand the basic features of this organism in order to ultimately control the spread of A. baumannii infections and to develop effective countermeasures against this harmful pathogen.
In addition to its pathogenesis, the genus Acinetobacter is particularly interesting for other reasons. First, acinetobacters are capable of catabolizing a wide range of carbon sources and metabolites and, as such, were briefly classified as pseudomonads (Stanier et al. 1966). In fact, acinetobacters are among the most widely used microbes for petroleum remediation. Second, representatives of Acinetobacter baylyi, have an extraordinary ability to acquire foreign DNA (Young et al. 2005). It is currently unknown how pervasive natural competence is among acinetobacters. This trait is particularly important for microbial pathogens since it is one important mechanism by which they achieve genetic diversity. Pathogens that can rapidly acquire drug resistance and pathogenicity islands (PAIs) have a selective advantage over those with more static genomes.
Recently a new approach for high-throughput DNA sequencing has been described using pyrophosphate sequencing (Margulies et al. 2005). High-density pyrosequencing involves the clonal amplification of genomic DNA fragments followed by sequencing coupled to two enzymatic reactions (Margulies et al. 2005). The first enzyme, sulfurylase, regenerates ATP from the pyrophosphate released during base incorporation. The second enzyme, luciferase, converts the energy of the regenerated ATP into light. This procedure allows the simultaneous sequencing of hundreds of thousands of short DNA sequences (on average ∼100 base pairs [bp]; see below). However, significant challenges to using high-density pyrosequencing include the short DNA reads generated and a potential loss of accuracy due to long homopolymer stretches or low complexity DNA. High-density pyrosequencing has been used effectively in resequencing efforts but has not been reported in de novo sequencing projects.
In this study we demonstrate that we can determine the DNA sequence of a microbe using pyrophosphate sequencing with reasonable accuracy. Analysis of the A. baumannii ATCC17978 DNA sequence revealed 16 putative alien islands (pAs) predicted to be involved in virulence. Insertional mutagenesis of Acinetobacter coupled with a Caenorhabditis elegans virulence assay that we described previously (Smith et al. 2004) and a new assay using Dictyostelium discoideum described below, confirmed that at least six alien islands are involved in virulence, including four that were not predicted to be involved in pathogenesis by sequence homology. While many of the genes in these alien islands were of previously unknown function, our analysis revealed potential functions for these genes and, by inference, the entire operon. Thus, we were able to assign function to these previously uncharacterized operons. The combined DNA sequence and mutagenesis approach provides rapid and considerable insight into the pathogenicity of this microbe and is an approach that is generally applicable to any pathogenic microbe.
Results
High-density pyrosequencing of the A. baumannii genome
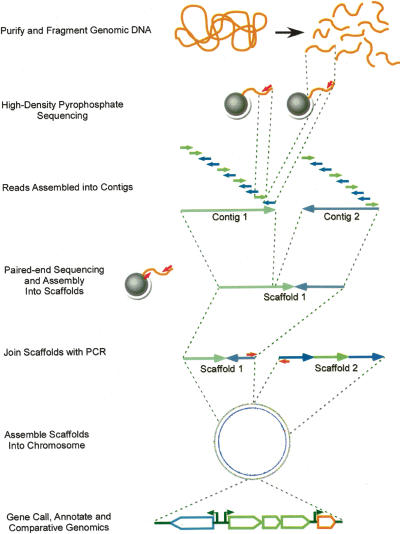
We devised a strategy to determine the DNA sequence of A. baumannii ATCC17978 using high-density pyrophosphate DNA sequencing (Fig. 1). To overcome the limitation of short DNA reads, we sequenced a large excess of genomic DNA. From five DNA sequencing runs, a total of 824,407 sequencing reads and 83,931,609 total base pairs of sequence were obtained; this depth of sequencing proved to be 21.1-fold coverage of the genome. The average read length was 101.8 bp and average G + C content 38.8%. The sequence was assembled into 139 contigs, which ranged in length from 1346 bp to 168,478 bp with an average length of 28,392 bp. We next performed paired-end analysis in which sequence information was recovered from each end of 90,049 DNA fragments. Paired-end sequencing joined the 139 contigs into 22 supercontigs, or scaffolds, which ranged in length from 6199 bp to 1,257,593 bp with an average of 179,384 bp. The predicted size of the A. baumannii chromosome based on these sequences is 3,946,442 bp.
Figure 1.
Flowchart. Steps leading to the completion of the A. baumannii genome.
To complete the assembly, two PCR strategies were employed (Fig. 1). First, several rounds of vectorette PCR were performed from the ends of contigs as described in the Materials and Methods (Riley et al. 1990; Kumar et al. 2002). Over 10,000 PCR and 2200 sequencing reactions were generated using conventional sequencing methods and capillary electrophoresis. This methodology was effective in filling in the gaps between contigs within scaffolds, as well as joining pairs of scaffolds. All remaining gaps were filled by direct PCR using primers that were designed to the ends of each of the remaining contigs. In this manner, the entire genome was sequenced and assembled. Gap filling added 30,304 bp to the genome assembly and indicates that high-density pyrosequencing was effective in returning 99.24% of the total chromosomal sequence. The final A. baumannii ATCC17978 genome sequence we determined is comprised of a single 3,976,746-bp chromosome, not counting tandem repeats (such as might be expected for rDNA), which cannot be detected using the pyrosequencing method.
In addition to the chromosomal DNA sequence, the shotgun approach also revealed the DNA sequence of two plasmids, pAB1 (13,404 bp) and pAB2 (11,520 bp), from this organism. Interestingly, neither of these is identical to the previously sequenced A. baumannii plasmid, pMAC, although all three plasmids share the same replication protein, RepM (Dorsey et al. 2006). pAB1 shares a total of three ORFs with pMAC: RepM, a YPPCP.09C homolog, and a Cro-like protein; however, whereas pMAC contains ORFs involved in peroxide resistance, pAB1 bears genes involved in lysine metabolism. Thus, it is likely that plasmids can readily differ among A. baumannii species.
Sequence verification and annotation
Potential coding sequences were identified using two programs, GLIMMER and RBSfinder. GLIMMER identified 3830 ORFs in the A. baumannii chromosome and an additional 10 and nine ORFs on the two plasmids, respectively (Delcher et al. 1999). Although GLIMMER has a high accuracy rate in calling genes, identifying the precise start site is problematic. Programs, such as, RBSfinder improve this accuracy by identifying potential ribosomal-binding sites and shifting the starting coordinates of predicted ORFs. It was shown that RBSfinder improves the accuracy of start site locations predicted by GLIMMER or Gene Mark from 67%–77% to 90%, and these predictions were validated by N-terminal protein sequencing of representative Escherichia coli proteins (Suzek et al. 2001).
RBSfinder was applied to the A. baumannii sequence, by first computing the consensus ribosomal-binding site, which was then used to identify the start codons in the 3830 ORFs. After the start sites were adjusted by RBSfinder, only 76.6% of the genome encodes protein. While this number is several percentages lower than that typically reported for bacteria, we believe that this is due to the use of RBSfinder, which improves start site prediction accuracy. Indeed, applying the GLIMMER and RBS finding method to the A. baylyi genome revealed a similar protein coding figure and gene homology.
Accuracy of the DNA sequence
One concern with using the high-density pyrosequencing method is its error rate. It was reported that high-density pyrosequencing is 99.96% accurate when compared with DNA sequenced by conventional sequencing methods and capillary electrophoresis (Margulies et al. 2005). To assess the accuracy of the A. baumannii sequence, two methods were used: (1) comparison of the originally assembled contigs to that of 50 PCR fragments sequenced by traditional methods and (2) determination of the frequency of split genes. To compare the sequence determined by pyrosequencing with that of traditional Sanger sequencing, we selected PCR products that did not reside near the ends of the assembled contigs and thus were not likely to contain low complexity DNA, transposons, rDNA, or other repeated sections. These 50 PCR products totaled 33,906 bp and had only 26 bp differences with the sequences assembled by pyrosequencing. Thus, we find that the genomic sequence is 99.92% accurate {Accuracy = 100 − [(26/33906) × 100]}. In all but one case, the base pair differences were in homopolymer run length, which is recognized as the most frequent type of error that high-density pyrosequencing creates (Margulies et al. 2005). In addition, in our efforts to join contigs >750,000 bp of PCR products were generated and sequenced using traditional methods. These sequences corresponded to the lowest complexity, highest repeat-containing, and most problematic sections of the genome. By incorporating the sequences generated by traditional methods into these problematic regions, the accuracy of the DNA sequence is likely to be >99.92% accurate.
Since the most likely sequencing error is a frameshift as a result of inappropriate base calling during homopolymer runs, a second measure of the error rate is the frequency of split genes. We used two criteria to determine the frequency of split genes in our assembly. First, these types of errors are distinguishable as a pair of tandem genes that return the same BLAST target. Second, the sum of the length of the tandem genes should closely approximate the length of the homologous gene. We found 30 instances of split genes in our assembly of the A. baumannii genome. An error rate calculation based on assumptions described in Materials and Methods indicates that the error rate is 0.0014%. Regardless of which accuracy measure is most precise, the sequence obtained from our approach is at least 99.92% accurate, and there are relatively few (<1%) split genes obtained from this sequencing approach.
Annotation of the A. baumannii genome
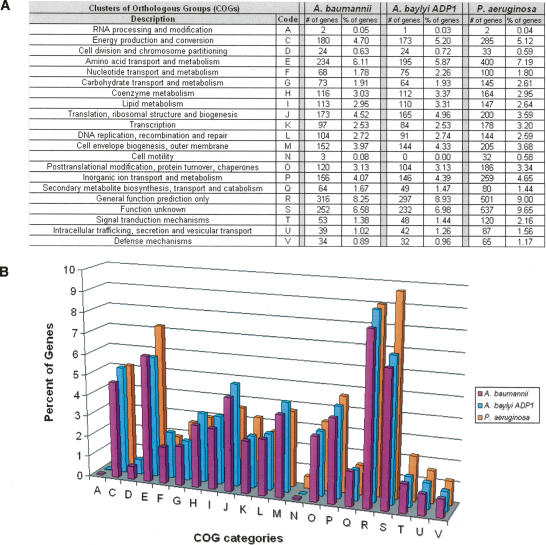
The annotated genes were assigned functions by a combination of BLAST analysis and KEGG annotation (Altschul et al. 1990, 1997; Kanehisa et al. 2006) and then assigned to clusters of orthologous groups (COG) (Tatusov et al. 1997). Approximately 61% of the genes were assigned to a COG functional category (Figs. 2, 3). The most represented classes of genes were those involved in translation, amino acid metabolism, and energy production as would be expected from a member of the catabolically versatile pseudomonad family. We also identified 70 tRNA genes throughout the genome.
Figure 2.
Functional annotation. (A) The functional assignments of all of the called genes in A. baumannii, A. baylyi, and P. aeruginosa. (B) Graphical depiction of the gene annotation. Genes products for A. baumannii (purple), A. baylyi (blue), and P. aeruginosa (orange) are represented by COG assignment (single-letter code): (A) RNA processing and modification; (C) energy production and conversion; (D) cell division and chromosome partitioning; (E) amino acid transport and metabolism; (F) nucleotide transport and metabolism; (G) carbohydrate transport and metabolism; (H) coenzyme metabolism; (I) lipid metabolism; (J) translation, ribosomal structure, and biogenesis; (K) transcription; (L) DNA replication, recombination, and repair; (M) cell envelope biogenesis, outer membrane; (N) cell motility; (O) post-translational modification, protein turnover, chaperones; (P) inorganic ion transport and metabolism; (Q) secondary metabolites biosynthesis, transport, and catabolism; (R) general function prediction only; (S) function unknown; (T) signal transduction mechanisms; (U) intracellular trafficking, secretion, and vesicular transport; (V) defense mechanisms.
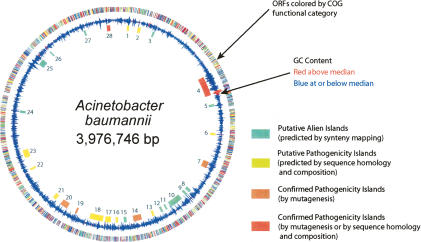
Figure 3.
Circular map of A. baumannii genome. The outermost circle shows genes color-coded by COG assignment: (gold) translation, ribosomal structure and biogenesis; (orange) RNA processing and modification; (dark orange) transcription; (maroon) DNA replication, recombination, and repair; (yellow) cell division and chromosome partitioning; (light pink) defense mechanisms; (purple) signal transduction mechanisms; (peach) cell envelope biogenesis, outer membrane; (medium purple) cell motility and secretion; (dark pink) intracellular trafficking, secretion, and vesicular transport; (light green) post-translational modification, protein turnover, chaperones; (lavender) energy production and conversion; (blue) carbohydrate transport and metabolism; (red) amino acid transport and metabolism; (green) nucleotide transport and metabolism; (light blue) coenzyme metabolism; (cyan) lipid metabolism; (dark purple) inorganic ion transport and metabolism; (sea green) secondary metabolites biosynthesis, transport, and catabolism; (light gray) general function prediction only; (ivory) function unknown; (dark gray) not in COGs. The middle circle represents the G + C percentage, colored red for regions above median GC score (38%) and blue for regions less than or equal to the median. The blue boxes indicate pAs, the yellow boxes indicate islands predicted to be involved in pathogenicity through homology and sequence composition differences, and the red and orange boxes indicate islands confirmed by the screens. The circles were drawn using the program GenomeViz (http://www.uniklinikum-giessen.de/genome/genomeviz/intro.html).
Acinetobacter synteny
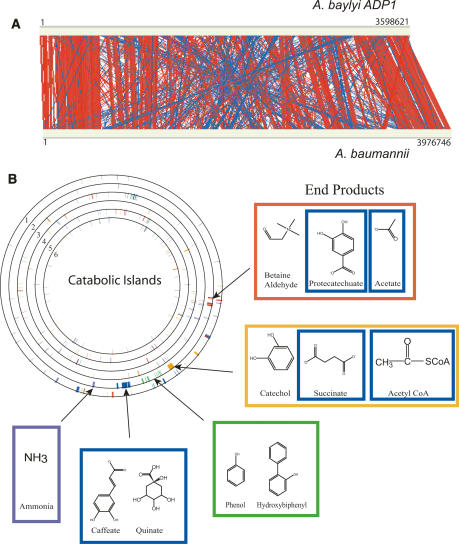
The A. baumannii genome sequence was compared with that of its closest sequenced relative, A. baylyi, using the Artemis Comparison Tool (ACT). This program uses BLAST (either blastn or tblastx) to compare two or more genomes for the arrangement of homologous genes (Carver et al. 2005). Although there are large number of local gene insertions, deletions, and rearrangements, synteny mapping using the Web ACT comparison illustrates that a large amount of synteny exists between the two genomes and that large sections of the two genomes share similar orientations (Fig. 4). Significant genomic similarities are also observed at the protein level in which 2137 of the 3830 (55.79%) predicted A. baumannii proteins returned their top BLAST scores as an A. baylyi gene product.
Figure 4.
Synteny map. (A) The genomes of A. baylyi (top) and A. baumannii (bottom) were compared by WebACT (http://www.webact.org/WebACT/home) and visualized by ACT (Carver et al. 2005). Red indicates similar genomic organization, whereas blue indicates inversions. (B) The genomes of six bacteria were compared for the distribution of key catabolic enzymes (from the outermost to the innermost ring): A. baumannii, A. baylyi, P. aeruginosa, N. meningitidis, E. coli K12, and B. subtilis. The island clusters were defined by Barbe et al. (2004) based on their location in the A. baylyi genome and are colored gray (−), red (I), orange (II), green (III), blue (IV), and purple (V). The end products of the pathways encoded within each of the catabolic islands are depicted. The boxes surrounding the end products are colored to match the islands from which they were derived. The circles were drawn using the program GenomeViz (http://www.uniklinikum-giessen.de/genome/genomeviz/intro.html).
One of the most interesting features of the A. baylyi genome is the clustering of catabolic operons into an “archipelago of catabolic diversity” (Barbe et al. 2004). Of the genes found within the five islands described by Barbe et al. (2004), we found representatives of only three islands. Furthermore, these genes were not clustered into islands but were scattered throughout the chromosome. We also compared the clustering of the catabolic genes in A. baylyi and A. baumannii with the sequenced genomes of Pseudomonas aeruginosa, Neisseria meningitidis, Bacillus subtilis, and E. coli (Fig. 4). The clustered organization of the catabolic archipelago is found only in A. baylyi and thus is likely to have occurred post-speciation. An interesting feature of the catabolic capacity of A. baumannii is its inability to catabolize glucose, a deficiency shared by many strains of Acinetobacter (Baumann et al. 1968b). We have identified the cause of this deficiency in A. baumannii as the absence of hexokinase, glucokinase, or any other comparable enzyme that can transfer phosphate onto glucose. Thus, the first step of glycolysis cannot be completed.
Nucleic acid translocation
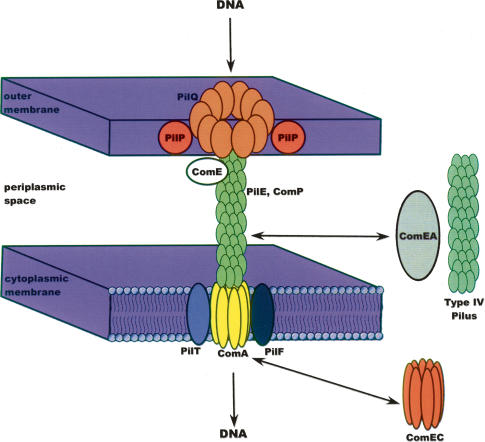
Examination of the A. baumannii genome revealed that it lacks two important genes involved in DNA uptake, comP and comA. However, it does have most others, such as pilQ, comE, and pilF among a total of >20 pilus, fimbrial, and competence genes. In addition, homologs of two genes required for DNA uptake, comEA and comEC, are found in A. baumannii genome but not in that of A. baylyi. Since comEA is a transmembrane protein whose role is to bind external DNA and deliver it to the comEC transporter, this suggests that A. baumannii can potentially compensate for the loss of comP with either comEA and/or its type IV pilus. Also, A. baumannii may be able to compensate for the loss of comA with its homolog, comEC. In doing so it would build a novel nucleic acid transport machine. The presence of these genes may explain, in part, the large amount of foreign DNA found within the A. baumannii chromosome (Fig. 5).
Figure 5.
DNA transport machinery. Adapted from Averhoff and Friedrich (2003) with kind permission from Springer Science and Business Media. In A. baylyi and other Gram-negative bacteria, foreign DNA is delivered to and through the outer membrane transporter PilQ. ComE-bound DNA is transported to the inner membrane transporter ComA (or ComEC) via ComP, PilE and/or the type IV pilus. ComEA may assist in this delivery.
Putative alien islands (pAs)
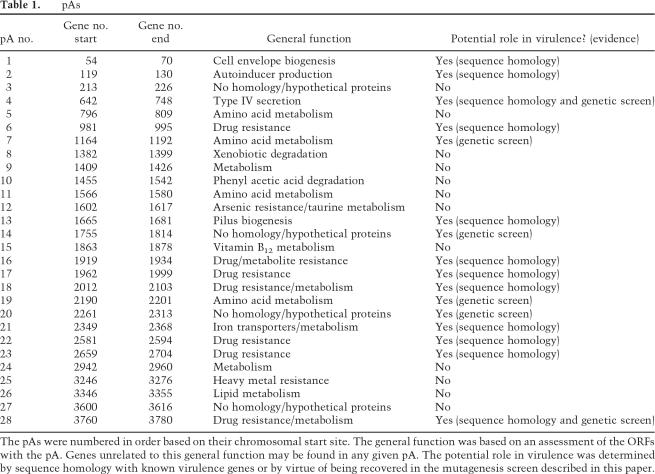
Of particular interest are the potential virulence genes in A. baumannii. Sequence similarities with known virulence genes can reveal the identity of homologs but contribute little new information. We used two approaches to identify A. baumannii virulence genes. Using comparative genomics, our first step was to directly compare the A. baumannii sequence with the nonpathogenic A. baylyi in order to identify A. baumannii specific genes. One of the benefits of the high degree of similarity between these two organisms is that their genomic differences are likely to contribute to their phenotypic differences. We defined our regions of interest as those regions >10 kb that had little homology with A. baylyi. The second approach we utilized was to examine the genome for sequence composition anomalies indicative of pAs (Karlin 2001). Differences in G + C content, amino acid usage, dinucleotide frequency, and codon usage have successfully been used to identify microbial alien islands (Karlin 2001). While pAs may possess genes encoding any number of functions, those determined to be involved in virulence are termed PAIs. There were 28 clusters of genes that fit the criteria as alien islands. Twelve of these possess genes with sequence homology with genes with roles in pathogenesis (Table 1). The largest pA is 133,740 bp, contains several transposons, and integrases and strikingly also contains eight genes homologous to the Legionella/Coxiella Type IV virulence/secretion apparatus and therefore may be a bonafide PAI. Since a separate full complement of genes involved in Type IV mediated conjugation are located elsewhere on the chromosome, we speculate that if the eight Type IV secretion (virulence) genes found in the pA are indeed virulence factors, then in order to build a functional Type IV secretion apparatus for virulence, the structural elements of the conjugation pilus may be utilized during pathogenesis. In addition to the large island described above, seven different pAs contain genes encoding drug-resistance proteins. We also found pAs containing genes encoding heavy metal resistance, iron uptake and metabolism, fimbrial genes, autoinducer processing, and cell envelope biogenesis (Table 1). Of the remaining 16 pAs, 12 islands contained genes involved in various aspects of metabolism, lipid metabolism, amino acid uptake, and processing and xenobiotic degradation. Four of the pAs were difficult to characterize since they contained largely hypothetical genes and mobile elements.
Table 1.
pAs
The pAs were numbered in order based on their chromosomal start site. The general function was based on an assessment of the ORFs with the pA. Genes unrelated to this general function may be found in any given pA. The potential role in virulence was determined by sequence homology with known virulence genes or by virtue of being recovered in the mutagenesis screen described in this paper.
Fournier et al. (2006) recently reported the identification of a hotspot in A. baumannii into which two different genomic islands inserted. The larger of the two islands is an 86,190-bp drug-resistance island containing 45 of the 52 drug-resistance genes in A. baumannii strain AYE (Fournier et al. 2006). The smaller island, found in the drug-sensitive strain SDF, is 19,362 bp and does not contain any drug-resistance genes. We found an insertion similar to the one found in the drug-sensitive SDF strain. It is 13,277 bp, comprising nine genes and found between 5′ and 3′ ends of a putative ATPase. The nine genes include those encoding four hypothetical or unknown proteins, two transposases, one transposition helper, a universal stress protein, and the sulphonamide resistance protein sulI. Although only one drug-resistance gene was found in this insertional hotspot, A. baumannii ATCC17978 possesses an additional 74 potential drug-resistance genes, including 32 efflux pumps (19 RND type, three MFS, and nine others) and 11 permeases of the drug/metabolite transporter (DMT) superfamily. We also identified 26 genes encoding resistance to heavy metals including copper, cadmium, zinc, cobalt, tellurite, and arsenic. Since these genes likely derive from mobile DNA and are at many locations in the genome, clearly the previously identified hotspot is not the only location into which genes have inserted in this organism.
Insertional mutagenesis reveals that many alien islands are important for A. baumannii virulence
Although the genomic sequence analysis revealed many pAs that might be involved in virulence, direct evidence is lacking. We therefore sought to determine whether any of these islands had a role in pathogenesis using a limited insertional mutagenesis approach. A library of 1324 A. baumannii mutants was generated using the EZ∷TN <R6Kγori/KAN-2> Tnp Transposome system from Epicentre (Dorsey et al. 2002). These mutants were then tested for their ability to affect pathogenesis in two assays: reduced brood size of C. elegans and inhibition of D. discoideum. By using two different assays to study the pathogenesis of acinetobacters, we expected to uncover general mediators of virulence, as well as factors that specifically affect a bacterium’s ability to survive in either host.
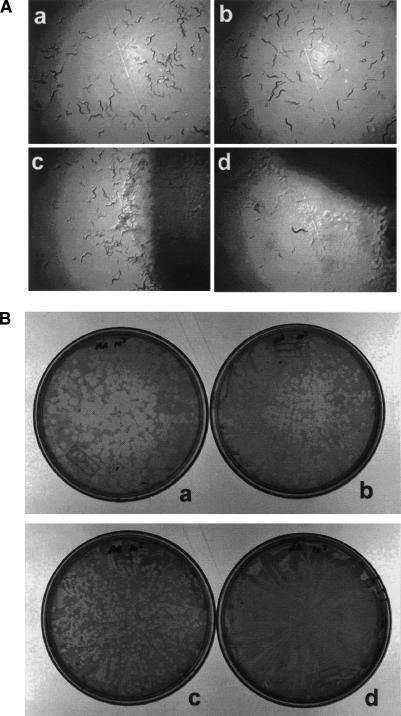
The worm assays were performed by inoculating a lawn of bacteria grown on nematode growth medium (NGM) with a single L4 stage worm. The worm undergoes its final molt to the adult stage and begins laying as many as 300 eggs during the subsequent 24 h. This rapid proliferation of worms results in a brood capable of exhausting the bacterial lawn within 4–5 d. However, when the worms are inoculated onto A. baumannii lawns that have grown on 1% ethanol (which induces virulence) (Smith et al. 2004), worm proliferation is slowed and brood sizes are dramatically diminished (Fig. 6A). Ethanol does not affect the worms directly since worms feeding on E. coli OP50 proliferate equally well on ethanol-containing and ethanol-free media. We screened our A. baumannii library for mutants that would allow worm proliferation at comparable rates on ethanol-containing and ethanol-free media. A total of 114 mutants were able to support the growth of worms on ethanol-containing media and were deemed avirulent to worms (Fig. 7A).
Figure 6.
Ethanol-stimulated virulence of A. baumannii. (A) Bacteria were incubated on NGM plates without (panels a,c) or with (panels b,d) 1% ethanol. A single L4 stage worm was inoculated onto lawns of E. coli OP50 (panels a,b) or A. baumannii (panels c,d) and allowed to proliferate for 4 d. (Panels a,b) The E. coli OP50 lawns are completely consumed by this time regardless of the presence of ethanol. (Panel d) The worm brood is considerably smaller on plates containing NGM + 1% ethanol and A. baumannii. (B) Bacteria and D. discoideum amoebae were incubated on SM/5 plates without (panels a,c) or with (panels b,d) 1% ethanol. (Panels a,b) In lawns containing K. aerogenes amoebae plaques form within 4 d regardless of the presence of ethanol. Amoebae are able to form plaques in A. baumannii (panel c), but plaque formation is completely inhibited when 1% ethanol is added to the media (panel d).
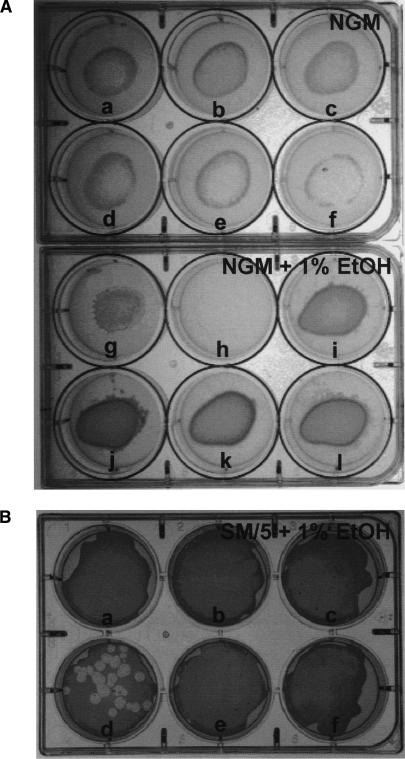
Figure 7.
Ethanol-stimulated virulence mutants of A. baumannii. Mutants were generated as described in Materials and Methods. (A) Six individual A. baumannii mutants were incubated on NGM (wells a–f) or NGM + 1% ethanol (wells g–j). A single L4 stage worm was inoculated onto lawns of each mutant and allowed to proliferate for 4 d. Avirulent bacterial mutants were recovered as those that allowed the worms to consume the bacterial lawns as fast or faster on NGM + 1% ethanol as on NGM alone; e.g., mutant b (b,h). (B) Six individual A. baumannii mutants were mixed with D. discoideum amoebae and incubated on SM/5 + 1% ethanol (wells a–f). These are not the same mutants as shown in A. Avirulent mutants were defined as those that allowed amoebae plaque formation; e.g., mutant d.
We also tested for hostile interactions between A. baumannii and D. discoideum using a simple plating assay that was developed for P. aeruginosa (Pukatzki et al. 2002). Amoebae and bacteria were cultured on media with or without ethanol. Bacteria, due to their greater growth rate, form lawns with the slower-growing amoebae embedded within them. On media without ethanol, the amoebae were able to consume A. baumannii and form plaques in the bacterial lawn. On media supplemented with limited ethanol, no plaques were observed in A. baumannii lawns. As many as 1 × 105 amoebae were plated on a single plate and not a single plaque was formed on lawns of A. baumannii when ethanol was added to the media (Fig. 6B). Ethanol does not appear to affect the amoebae directly since amoebae feeding on Klebsiella.aerogenes form plaques equally well on ethanol-containing and ethanol-free media. Screening the mutant library for bacteria that allow plaque formation by the amoebae in the presence of ethanol revealed 229 avirulent mutants (esv, ethanol-stimulated virulence mutant) out of the 1324 mutants tested (Fig. 7B).
Comparison of the results of the worm and amoebae screens revealed 30 mutants that tested positive in both assays. The identity of the gene disrupted by the insertion mutation in these 30 mutants was determined by rescuing the DNA adjacent to the transposon and DNA sequencing. In addition, we also rescued and sequenced two mutants that were robust in the worm assay but failed in the amoebae assay and three mutants that did not affect amoebae growth but did inhibit worm growth. In all, 35 mutants were sequenced and their genomic locations determined. Fourteen of the 35 mutants had no homology with A. baylyi, seven of these mutants localized to the pAs described above. Two of the six pAs were predicted by sequence homology with encode genes involved in virulence. Of the remaining four pAs, two are predicted to be involved in various aspects of metabolism, so their relation to virulence mechanisms is unclear. The remaining two pAs are particularly interesting as they may represent novel virulence effectors since BLAST analysis revealed little sequence similarity to any known genes and they may represent novel virulence factors.
Further characterization of ethanol-stimulated virulence mutants
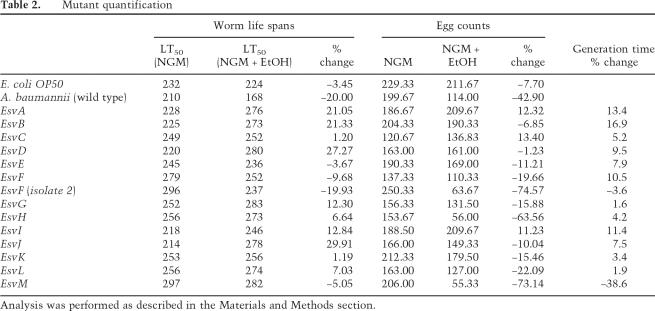
There are two likely mechanisms that would result in the reduction in the worm brood sizes: The worms are dying and/or they are not proliferating. To separate these two possibilities, we determined worm life spans and also counted the number of eggs that worms laid when feeding on E. coli OP50, A. baumannii, or 14 of the avirulent, ethanol-stimulated virulence (Esv) mutants. The results are summarized in Table 2. The life spans of worms feeding on A. baumannii were 20% shorter when ethanol was added to the media. A more dramatic effect is observed when eggs are counted. Worms feeding on ethanol-stimulated A. baumannii lay 42.9% fewer eggs than those feeding on unstimulated bacteria. Thus, it appears that the combined effect of the shortened life span and the reduction in the reproductive capacity of the worms when feeding on ethanol-stimulated A. baumannii causes the diminished brood of worms illustrated in Figure 6A.
Table 2.
Mutant quantification
Analysis was performed as described in the Materials and Methods section.
Several of the bacterial mutants are able to reverse the deleterious effects of ethanol-stimulated A. baumannii on worm proliferation by increasing both the worm life spans and egg-laying capacity. EsvA, encodes a transcription factor in the AraC/XylS family. EsvC encodes the γ subunit of urease. EsvI is a likely multidrug efflux transport protein. Some of the mutants were able to sustain prolonged worm life spans but had lesser effects on egg production. Two such examples are EsvB, which encodes a protein in the MarR class of transcription factors, and EsvD, which encodes an ABC-type membrane transporter. Two of the mutants, EsvF and EsvM, appeared to remain virulent and will not be studied further.
To investigate whether the mutations caused a slow growth phenotype that might indirectly lead to avirulence, we measured the generation times of each mutant. Each mutant strain doubled at a rate equal to or faster than wild-type A. baumannii except for EsvF andEsvM (Table 2). Thus, slow growth is not responsible for the avirulent phenotypes observed in most of these mutants.
Biofilm formation is a well-characterized developmental pathway utilized by many bacteria in their pathogenesis. Wild-type A. baumannii form biofilms on abiotic surfaces (Tomaras et al. 2003). We examined the effect of ethanol on biofilm formation and the ability of our mutants to form biofilms on plastic surfaces. Ethanol has no apparent effect on biofilm formation; the cells adhere to plastic surfaces equally well in the presence or absence of ethanol (data not shown). Furthermore, all of the mutants tested retained the ability to form biofilms, suggesting that, in this strain, biofilms are not essential for virulence (data not shown). Thus, these genes are likely involved in other virulence processes.
Discussion
This paper reports the first de novo sequencing effort using a novel high-throughput method for genomic DNA sequence. Examination of this microbial genome revealed little nucleotide sequence identity between A. baumannii and A. baylyi, the only previously sequenced member of the Acinetobacter genus. Genomic similarities are seen at the protein level in which 2137 of the 3830 (55.79%) predicted A. baumannii share homology with A. baylyi gene products. The most interesting differences between these two organisms lies in the 28 pAs identified in A. baumannii. Many of the drug-resistance and potential virulence factors found in the A. baumannii genome reside on these islands, indicating that a large number of them are important for the pathogenesis of this organism.
High-density pyrophosphate sequencing is a rapid, cost-efficient method for large sequencing projects without the labor or potential bias of cloning steps. By virtue of large numbers of parallel sequencing runs, genomic sequence coverage is high, in our case 21.1 times. This powerful method has been used effectively in genome resequencing efforts and for sequencing microbial strain variants (Margulies et al. 2005; Hofreuter et al. 2006). The primary drawbacks of this methodology are the short sequencing runs, which result in difficulties in assembly of sequences in low complexity or repeated regions, and errors in base calling in stretches of homopolymers, which can result in frameshifts. These limitations were overcome by not only the high sequence coverage, but also the use of paired-end analysis and gap closures by traditional and random-ended PCR.
We found that, when combined with conventional gap filling, the accuracy of the sequence and assembly are comparable to the whole-genome shotgun sequencing methods that have become the gold standard of bacterial genomic sequencing. In contrast to a recent report, which suggests that high-density pyrosequencing is unable to replace Sanger sequencing for de novo microbial genome projects (Goldberg et al. 2006), we find that high-density pyrosequencing can be a suitable replacement for the sequencing of microbial genomes. We found that an initial assembly of 21.1 times high-density pyrosequencing data is sufficient to determine the genome size and build a working draft that could be used for almost any genomic analysis. Furthermore, our initial draft assembly predicted a genome of 3,946,442 bp, whereas the final assembly predicts a genome of 3,976,746 bp. Therefore the initial draft provided coverage for >99.24% of the completed genome, and only 30,302 bp were missing. One factor that may have negatively impacted the evaluation by Goldberg et al. (2006) was the omission of paired-end analysis. Our assembly was greatly facilitated by paired-end sequencing and resulted in the joining of 139 contigs into a considerably more manageable 22 scaffolds. The ability to obtain longer reads will likely facilitate contig assembly; however, paired-end reads will still be useful for identification of large tandem repeats.
The sequence of A. baumannii reveals that this organism has acquired a number of genes from its environment and that these genes likely play a direct role in its virulence. We identified 28 pAs based on sequence characteristics and also sequence comparisons with a nonpathogenic relative. One-fourth of these islands appear to be involved in drug resistance. The strain used for these studies is resistant to β-lactams but shows only weak resistance to tetracycline and is aminoglycoside (kanamycin) sensitive (M.G. Smith and M. Snyder, unpubl.). Thus, the presence of so many drug-resistance islands is surprising. This strain was isolated in or around 1951 (Piechaud and Second 1951; Baumann et al. 1968b), prior to the development of the macrolides (erythromycin), glycopeptides (vancomycin), and cephalosporins and other latter generations of β-lactams. It is possible that this strain was never exposed to chloramphenicol, which was first used as a therapeutic in 1949. For these reasons, it would be interesting to subject ATCC17978 to a full panel of antibiotics and assess its resistance capacity. Potentially more interesting will be comparative analysis with more recent isolates that can assess the evolution of antibiotic resistance over the last 50–60 yr. In this respect a recent clinical isolate, resistant to several β-lactams, aminoglycosides, fluoroquinolones, chloramphenicol, tetracycline, and rifampin was found to contain a single large drug-resistance island containing 45 of the 52 drug-resistance genes in its genome (Fournier et al. 2006). This impressive clustering of drug-resistance genes was not observed in the ATCC17978 strain that we sequenced. Given the differences in the resistance capacities of these two strains, it appears that the more recent isolate has dispensed with the many less effective resistance genes in favor of a single, highly potent cassette of drug-resistance genes.
Other pAs that appear to play a role in virulence were identified using a random insertion mutagenesis protocol. Six different islands were identified as virulent by two different pathogenesis screens. Of these six, two have genes thought to be involved in virulence, namely, the type IV secretion apparatus and drug-resistance genes. Two are predicted to be involved in metabolism. The fact that they were identified in this screen may be related to the use of ethanol as a virulence stimulus and will be investigated further. The final pair of pAs identified by the mutagenesis contained many hypothetical genes and nucleic acid mobility related genes such as transposons, integrases, and phage proteins. Thus their potential function assigned by sequence homology is speculative. However, since insertion in these sites results in avirulent mutants, the function of these islands is important for pathogenesis. Thus, the combination of genome sequencing and mutagenesis is a powerful approach for identifying and validating pathogenic genes in A. baumannii and will be a useful general method for finding such genes.
Many host–pathogen interactions have evolved in the environment where bacteria interact with predators and competitors. C. elegans and D. discoideum have been successfully used as host models for bacterial infection (Pukatzki et al. 2002; Alegado et al. 2003). These two organisms consume their bacterial prey in different ways. C. elegans has a digestive tract in which the bacteria are crushed, lysed, and enzymatically digested, and the subsequent nutrients are absorbed by the cells comprising the intestine (Avery and Thomas 1997). This process is almost entirely extracellular. Conversely, the unicellular D. discoideum uses phagocytosis, followed by vesicle fission and fusion resulting in a phagolysosome to digest bacteria; this process is largely intracellular (Cardelli 2001). Escape from either the worm gut or amoebae vesicles presents two distinct challenges to the bacteria, and while some of the tools required may overlap, many must be different. This is demonstrated by the fact that only 30 of the mutants overlapped between the two screens. Therefore, using both assays to study pathogenesis will uncover global regulators of virulence required in both instances, as well as factors specific to either host. This approach was validated by the recovery of two transcription factors that are potentially global regulators of virulence. Future studies will identify their downstream effectors. One interesting gene that was uncovered by our screen was urease. The enzymatic function of urease is to produce ammonia and carbon dioxide from urea. However, recent studies on plant ureases suggest that they play a role in defense mechanisms and have biological function independent of their enzymatic activity (Olivera-Severo et al. 2006). The A. baumannii urease gene may play a similar role.
In summary, the DNA sequence of A. baumannii has revealed several new insights in the biology and pathogenesis of this organism. We have demonstrated that the coupling of a mutagenesis screen with genomic sequencing can quickly uncover and validate several pathogenicity factors. We anticipate this flexible framework will be broadly applicable to the study of other microbes. These studies lay the foundation for a further understanding of both the biology and pathogenicity of this organism, providing many interesting avenues for further investigation.
Materials and methods
Whole-genome sequencing
Genomic DNA from A. baumannii (American Type Culture Collection [ATCC] 17,978) was extracted from cells grown overnight at 30°C in LB liquid cultures using the MasterPure DNA kit (Epicentre). Fifty micrograms of DNA were then sequenced by 454 Life Sciences using high-density pyrosequencing methodology (Margulies et al. 2005).
Sequence assembly and validation
Initial assembly efforts were performed by 454 Life Sciences and resulted in the formation of 139 contigs assembled into 22 scaffolds. Contigs were joined using Vectorette PCR (Riley et al. 1990; Kumar et al. 2002). Briefly, the primers ABP1 and ABP2 (ABP1, 5′-GAAGGAGAGGACGCTGTCTGTCGAAGGTAAG GAACGGACGAGAGAAGGGAGAG-3′; ABP2, 5′-GACTCTC CCTTCTCGAATCGTAACCGTTCGTACGAGAATCGCTGT CCTCTCCTTC-3′) were denatured for 5 min at 95°C and annealed together at 5 nM each in annealing buffer (10 mM Tris at pH 8.0, 10 mM MgCl2, 50 mM NaCl). The primers were then ligated to genomic DNA previously digested with a blunt-ended restriction endonuclease. The ligation mixture was then used as a template for HotStart PCR (Qiagen). The PCR primer UV was used in all reactions (UV, 5′-CGAATCGTAACCGTTCGTAC GAGAATCGCT-3′), while the reverse primer was designed to each of the two ends of the 139 contigs. In all, nine different restriction enzymes/enzyme combinations were used (DraI, HaeIII, HincII, PsiI, ScaI, SspI, XmnI, HincII/EcoRV, and MscI/HpaI/PvuII). PCR products were gel-purified and sequenced using the same primers from which they were amplified and standard methods and capillary electrophoresis using an ABI 3730 DNA sequencing instrument. Contigs and PCR products were assembled using the Sequencher assembly program (Gene Codes).
Genome analysis and annotation
A gene prediction strategy was applied on the assembled sequences using GLIMMER. Putative ribosomal-binding sites and tRNA genes were identified with RBSfinder (Suzek et al. 2001) and tRNAscan-SE (Schattner et al. 2005), respectively. Prior to the manual annotation of each predicted gene, an automatic functional annotation was computed based on different analyses. Similarity searches were performed against different databases, including BLASTnr and KEGG. Finally, each gene was functionally classified by assigning a COG number and corresponding COG category.
Calculating error frequency
To calculate an error frequency, we divided the number of errors resulting in a split gene by the fraction of protein coding regions whose frameshift is likely to produce two ORFs (≥100 bp) that can be detected by our BLAST searches. This amounts to 2,853,379 bp of total protein coding sequence minus 200 bp for each gene (723,181 bp), since insertions in this regions will not generate new ORFs. This calculation assumes that an error will result in a nearby translation termination. Thus, the final error rate from this method is 30/2,130,198 bp or 0.0014%.
Genomic comparison
For comparative analyses, the annotated genome sequence of A. baylyi (GenBank accession no. CR543861) was accessed. Homology searches were conducted on the nucleotide and amino acid sequence level using BLAST. Comparisons of chromosomal sequences were carried out with ACT (http://www.sanger.ac.uk/Software/ACT).
Detection of regions with atypical G + C content
Genomic regions with atypical G + C content were identified using a “sliding window” technique with a window size of 1000 bp. For this, the G + C content was treated as a Gaussian distribution, and regions with differences of at least 1.5 standard deviations from the mean were calculated.
Detection of putative PAIs
The four methods outlined by Karlin (2001) were implemented for detection of anomalous regions. These included atypical G + C content, dinucleotide signature, codon bias, and amino acid bias. In each case, a “sliding window” technique with a window size of 1000 bp was used. Assuming a Gaussian distribution, those regions with a difference of at least 1.5 standard deviations from the mean were identified as anomalous regions. Homology searches on these regions were carried out to identify putative PAIs.
A. baumannii mutagenesis
Mutagenesis of A. baumannii was performed as described previously (Dorsey et al. 2002). Briefly, electrocompetent cells were generated as follows: Overnight cultures of A. baumannii were diluted 1:100 and grown to a cell density of 0.5 × 108 to 0.8 × 108 cells/mL. The cells were collected by centrifugation and washed with ice-cold ddH2O three times. The cells were washed once in ice-cold 10% glycerol and resuspended in 10% glycerol to a final concentration of 2.0 × 1010 to 2.5 × 1010 cells/mL. These electrocompetent cells were electroporated with the EZ∷Tn5 <R6Kγori/KAN-2> Tnp Transposome (Epicentre), and kanamycin-resistant transformants were selected. The electroporation was performed using a Gene Pulser II (Bio-Rad Laboratories) at settings of 25 μFD, 200 Ω, and 1.8 kV. Mutants were rescue-cloned according to Epicentre’s protocol. Briefly, mutant bacteria were grown, and their genomic DNA was purified. The DNA was cut using EcoRI, ligated to itself, and transformed into pir-116 electrocompetent E. coli. Kanamycin-resistant colonies were harvested, and plasmids containing the insertion cassette and flanking A. baumannii sequence were purified and sequenced using primers complementary to the insertion cassette.
A. baumannii generation times
Three colonies of each mutant as well as the wild-type A. baumannii were inoculated into LB and incubated overnight at 37°C with shaking. Cultures were diluted to a cell density of 1.0 × 107 cells/mL in fresh LB and incubated at 37°C with shaking. Cell densities were measured by optical density at 600 nm at 1- to 2-h intervals for 36 h. Generation times were determined during the exponential phase of growth for each isolate, and the average for each strain was determined. Generation times of the mutants were then compared with the wild-type strain.
C. elegans killing assay
E. coli (strain OP50) grown on NGM media (Sulston and Hodgkin 1988) were fed to C. elegans (strain N2). L3/L4 stage worms were placed onto lawns of A. baumannii grown on NGM or NGM + 1% ethanol. Plates were incubated at 25°C. Viability was tested every 24 h by visual examination. Worms were considered dead if they no longer moved or responded to touch. For each strain of bacteria tested, 60 worms were assayed. The LT50 is defined as the time it takes for half of the worms to die.
C. elegans egg count assay
One L4 stage worm was placed onto lawns of E. coli OP50 or A. baumannii grown on NGM or NGM + 1% ethanol. Each day for five consecutive days, the worm was moved to a fresh bacterial lawn. The plates were examined for the presence of eggs/L1 stage worm for each of the five plates. Egg production typically peaked at day 2 or 3 and was exhausted by day 5. Plates were incubated at 25°C. For all strains of bacteria tested, the egg production of nine worms were assayed.
D. discoideum plaque assay
This assay was performed essentially as described by Pukatzki et al. (2002). Briefly, D. discoideum AX3 were grown axenically in HL/5 media (Sussman 1987) at 20°C. D. discoideum cells from mid-logarithmic cultures were collected by centrifugation (1000g; 4 min), washed once with SM/5 medium (Sussman 1987), and added to the bacterial suspensions at a final concentration of 5 × 102cells/mL suspension; 0.2 mL of this mixture was plated out on SM/5 plates. Plates were incubated for 3–5 d and examined for plaques formed by amoebae.
Database submission
The nucleotide sequences of the chromosome of A. baumannii ATCC17978 and its two plasmids, pAB1 and pAB2, were submitted to GenBank. Accession numbers: CP000521–CP000523.
Acknowledgments
We thank 454 Life Sciences, in particular Michael Egholm, Lei Du, and Jennifer Tsai for their assistance throughout this project. This research was supported by funds from the National Insitutes of Health as well as the Burroughs Wellcome Foundation. M.G.S. was supported by a post-doctoral fellowship from the American Cancer Society (PF-03-018-01-CSM). We also thank Nicholas Carrerio and the Yale Center for High Performance Computation in Biology and Biomedicine for the use of computing power during the annotation process, as well as NIH grant RR19895, which funded the instrumentation.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1510307
References
- Abbott A. Medics braced for fresh superbug. Nature. 2005;436:758. doi: 10.1038/436758a. [DOI] [PubMed] [Google Scholar]
- Alegado R.A., Campbell M.C., Chen W.C., Slutz S.S., Tan M.W., Campbell M.C., Chen W.C., Slutz S.S., Tan M.W., Chen W.C., Slutz S.S., Tan M.W., Slutz S.S., Tan M.W., Tan M.W. Characterization of mediators of microbial virulence and innate immunity using the Caenorhabditis elegans host-pathogen model. Cell. Microbiol. 2003;5:435–444. doi: 10.1046/j.1462-5822.2003.00287.x. [DOI] [PubMed] [Google Scholar]
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J., Gish W., Miller W., Myers E.W., Lipman D.J., Miller W., Myers E.W., Lipman D.J., Myers E.W., Lipman D.J., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J., Zhang J., Zhang Z., Miller W., Lipman D.J., Zhang Z., Miller W., Lipman D.J., Miller W., Lipman D.J., Lipman D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Averhoff B., Friedrich A., Friedrich A. Type IV pili-related natural transformation systems: DNA transport in mesophilic and thermophilic bacteria. Arch. Microbiol. 2003;180:385–393. doi: 10.1007/s00203-003-0616-6. [DOI] [PubMed] [Google Scholar]
- Avery L., Thomas J.H., Thomas J.H. Feeding and defecation. In: Riddle D.L., et al., editors. C. elegans II. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1997. pp. 679–716. [PubMed] [Google Scholar]
- Barbe V., Vallenet D., Fonknechten N., Kreimeyer A., Oztas S., Labarre L., Cruveiller S., Robert C., Duprat S., Wincker P., Vallenet D., Fonknechten N., Kreimeyer A., Oztas S., Labarre L., Cruveiller S., Robert C., Duprat S., Wincker P., Fonknechten N., Kreimeyer A., Oztas S., Labarre L., Cruveiller S., Robert C., Duprat S., Wincker P., Kreimeyer A., Oztas S., Labarre L., Cruveiller S., Robert C., Duprat S., Wincker P., Oztas S., Labarre L., Cruveiller S., Robert C., Duprat S., Wincker P., Labarre L., Cruveiller S., Robert C., Duprat S., Wincker P., Cruveiller S., Robert C., Duprat S., Wincker P., Robert C., Duprat S., Wincker P., Duprat S., Wincker P., Wincker P., et al. Unique features revealed by the genome sequence of Acinetobacter sp. ADP1, a versatile and naturally transformation competent bacterium. Nucleic Acids Res. 2004;32:5766–5779. doi: 10.1093/nar/gkh910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann P., Doudoroff M., Stanier R.Y., Doudoroff M., Stanier R.Y., Stanier R.Y. Study of the Moraxella group. I. Genus Moraxella and the Neisseria catarrhalis group. J. Bacteriol. 1968a;95:58–73. doi: 10.1128/jb.95.1.58-73.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann P., Doudoroff M., Stanier R.Y., Doudoroff M., Stanier R.Y., Stanier R.Y. A study of the Moraxella group. II. Oxidative-negative species (genus Acinetobacter) J. Bacteriol. 1968b;95:1520–1541. doi: 10.1128/jb.95.5.1520-1541.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergogne-Berezin E., Towner K.J., Towner K.J. Acinetobacter spp. as nosocomial pathogens: Microbiological, clinical, and epidemiological features. Clin. Microbiol. Rev. 1996;9:148–165. doi: 10.1128/cmr.9.2.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernards A.T., Harinck H.I., Dijkshoorn L., van der Reijden T.J., van den Broek P.J., Harinck H.I., Dijkshoorn L., van der Reijden T.J., van den Broek P.J., Dijkshoorn L., van der Reijden T.J., van den Broek P.J., van der Reijden T.J., van den Broek P.J., van den Broek P.J. Persistent Acinetobacter baumannii? Look inside your medical equipment. Infect. Control Hosp. Epidemiol. 2004;25:1002–1004. doi: 10.1086/502335. [DOI] [PubMed] [Google Scholar]
- Cardelli J. Phagocytosis and macropinocytosis in Dictyostelium: Phosphoinositide-based processes, biochemically distinct. Traffic. 2001;2:311–320. doi: 10.1034/j.1600-0854.2001.002005311.x. [DOI] [PubMed] [Google Scholar]
- Carver T.J., Rutherford K.M., Berriman M., Rajandream M.A., Barrell B.G., Parkhill J., Rutherford K.M., Berriman M., Rajandream M.A., Barrell B.G., Parkhill J., Berriman M., Rajandream M.A., Barrell B.G., Parkhill J., Rajandream M.A., Barrell B.G., Parkhill J., Barrell B.G., Parkhill J., Parkhill J. ACT: The Artemis Comparison Tool. Bioinformatics. 2005;21:3422–3423. doi: 10.1093/bioinformatics/bti553. [DOI] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention Acinetobacter baumannii infections among patients at military medical facilities treating injured U.S. service members, 2002–2004. MMWR Morb. Mortal Wkly. Rep. 2004;53:1063–1066. [PubMed] [Google Scholar]
- Chastre J., Trouillet J.L., Trouillet J.L. Problem pathogens (Pseudomonas aeruginosa and Acinetobacter) Semin. Respir. Infect. 2000;15:287–298. doi: 10.1053/srin.2000.20944. [DOI] [PubMed] [Google Scholar]
- Choi C.H., Lee E.Y., Lee Y.C., Park T.I., Kim H.J., Hyun S.H., Kim S.A., Lee S.K., Lee J.C., Lee E.Y., Lee Y.C., Park T.I., Kim H.J., Hyun S.H., Kim S.A., Lee S.K., Lee J.C., Lee Y.C., Park T.I., Kim H.J., Hyun S.H., Kim S.A., Lee S.K., Lee J.C., Park T.I., Kim H.J., Hyun S.H., Kim S.A., Lee S.K., Lee J.C., Kim H.J., Hyun S.H., Kim S.A., Lee S.K., Lee J.C., Hyun S.H., Kim S.A., Lee S.K., Lee J.C., Kim S.A., Lee S.K., Lee J.C., Lee S.K., Lee J.C., Lee J.C. Outer membrane protein 38 of Acinetobacter baumannii localizes to the mitochondria and induces apoptosis of epithelial cells. Cell. Microbiol. 2005;7:1127–1138. doi: 10.1111/j.1462-5822.2005.00538.x. [DOI] [PubMed] [Google Scholar]
- Delcher A.L., Harmon D., Kasif S., White O., Salzberg S.L., Harmon D., Kasif S., White O., Salzberg S.L., Kasif S., White O., Salzberg S.L., White O., Salzberg S.L., Salzberg S.L. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 1999;27:4636–4641. doi: 10.1093/nar/27.23.4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsey C.W., Tomaras A.P., Actis L.A., Tomaras A.P., Actis L.A., Actis L.A. Genetic and phenotypic analysis of Acinetobacter baumannii insertion derivatives generated with a transposome system. Appl. Environ. Microbiol. 2002;68:6353–6360. doi: 10.1128/AEM.68.12.6353-6360.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsey C.W., Beglin M.S., Actis L.A., Beglin M.S., Actis L.A., Actis L.A. Detection and analysis of iron uptake components expressed by Acinetobacter baumannii clinical isolates. J. Clin. Microbiol. 2003;41:4188–4193. doi: 10.1128/JCM.41.9.4188-4193.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsey C.W., Tomaras A.P., Connerly P.L., Tolmasky M.E., Crosa J.H., Actis L.A., Tomaras A.P., Connerly P.L., Tolmasky M.E., Crosa J.H., Actis L.A., Connerly P.L., Tolmasky M.E., Crosa J.H., Actis L.A., Tolmasky M.E., Crosa J.H., Actis L.A., Crosa J.H., Actis L.A., Actis L.A. The siderophore-mediated iron acquisition systems of Acinetobacter baumannii ATCC 19606 and Vibrio anguillarum 775 are structurally and functionally related. Microbiol. 2004;150:3657–3667. doi: 10.1099/mic.0.27371-0. [DOI] [PubMed] [Google Scholar]
- Dorsey C.W., Tomaras A.P., Actis L.A., Tomaras A.P., Actis L.A., Actis L.A. Sequence and organization of pMAC, an Acinetobacter baumannii plasmid harboring genes involved in organic peroxide resistance. Plasmid. 2006;56:112–123. doi: 10.1016/j.plasmid.2006.01.004. [DOI] [PubMed] [Google Scholar]
- Fournier P.E., Vallenet D., Barbe V., Audic S., Ogata H., Poirel L., Richet H., Robert C., Mangenot S., Abergel C., Vallenet D., Barbe V., Audic S., Ogata H., Poirel L., Richet H., Robert C., Mangenot S., Abergel C., Barbe V., Audic S., Ogata H., Poirel L., Richet H., Robert C., Mangenot S., Abergel C., Audic S., Ogata H., Poirel L., Richet H., Robert C., Mangenot S., Abergel C., Ogata H., Poirel L., Richet H., Robert C., Mangenot S., Abergel C., Poirel L., Richet H., Robert C., Mangenot S., Abergel C., Richet H., Robert C., Mangenot S., Abergel C., Robert C., Mangenot S., Abergel C., Mangenot S., Abergel C., Abergel C., et al. Comparative genomics of multidrug resistance in Acinetobacter baumannii. PLoS Genet. 2006;2:e7. doi: 10.1371/journal.pgen.0020007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg S.M., Johnson J., Busam D., Feldblyum T., Ferriera S., Friedman R., Halpern A., Khouri H., Kravitz S.A., Lauro F.M., Johnson J., Busam D., Feldblyum T., Ferriera S., Friedman R., Halpern A., Khouri H., Kravitz S.A., Lauro F.M., Busam D., Feldblyum T., Ferriera S., Friedman R., Halpern A., Khouri H., Kravitz S.A., Lauro F.M., Feldblyum T., Ferriera S., Friedman R., Halpern A., Khouri H., Kravitz S.A., Lauro F.M., Ferriera S., Friedman R., Halpern A., Khouri H., Kravitz S.A., Lauro F.M., Friedman R., Halpern A., Khouri H., Kravitz S.A., Lauro F.M., Halpern A., Khouri H., Kravitz S.A., Lauro F.M., Khouri H., Kravitz S.A., Lauro F.M., Kravitz S.A., Lauro F.M., Lauro F.M., et al. A Sanger/pyrosequencing hybrid approach for the generation of high-quality draft assemblies of marine microbial genomes. Proc. Natl. Acad. Sci. 2006;103:11240–11245. doi: 10.1073/pnas.0604351103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofreuter D., Tsai J., Watson R.O., Novik V., Altman B., Benitez M., Clark C., Perbost C., Jarvie T., Du L., Tsai J., Watson R.O., Novik V., Altman B., Benitez M., Clark C., Perbost C., Jarvie T., Du L., Watson R.O., Novik V., Altman B., Benitez M., Clark C., Perbost C., Jarvie T., Du L., Novik V., Altman B., Benitez M., Clark C., Perbost C., Jarvie T., Du L., Altman B., Benitez M., Clark C., Perbost C., Jarvie T., Du L., Benitez M., Clark C., Perbost C., Jarvie T., Du L., Clark C., Perbost C., Jarvie T., Du L., Perbost C., Jarvie T., Du L., Jarvie T., Du L., Du L., et al. Unique features of a highly pathogenic Campylobacter jejuni strain. Infect. Immun. 2006;74:4694–4707. doi: 10.1128/IAI.00210-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juni E. Genetics and physiology of Acinetobacter. Annu. Rev. Microbiol. 1978;32:349–371. doi: 10.1146/annurev.mi.32.100178.002025. [DOI] [PubMed] [Google Scholar]
- Kanehisa M., Goto S., Hattori M., Aoki-Kinoshita K.F., Itoh M., Kawashima S., Katayama T., Araki M., Hirakawa M., Goto S., Hattori M., Aoki-Kinoshita K.F., Itoh M., Kawashima S., Katayama T., Araki M., Hirakawa M., Hattori M., Aoki-Kinoshita K.F., Itoh M., Kawashima S., Katayama T., Araki M., Hirakawa M., Aoki-Kinoshita K.F., Itoh M., Kawashima S., Katayama T., Araki M., Hirakawa M., Itoh M., Kawashima S., Katayama T., Araki M., Hirakawa M., Kawashima S., Katayama T., Araki M., Hirakawa M., Katayama T., Araki M., Hirakawa M., Araki M., Hirakawa M., Hirakawa M. From genomics to chemical genomics: New developments in KEGG. Nucleic Acids Res. 2006;34:D354–D357. doi: 10.1093/nar/gkj102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlin S. Detecting anomalous gene clusters and pathogenicity islands in diverse bacterial genomes. Trends Microbiol. 2001;9:335–343. doi: 10.1016/s0966-842x(01)02079-0. [DOI] [PubMed] [Google Scholar]
- Koulenti D., Rello J., Rello J. Gram-negative bacterial pneumonia: Aetiology and management. Curr. Opin. Pulm. Med. 2006;12:198–204. doi: 10.1097/01.mcp.0000219269.73180.5d. [DOI] [PubMed] [Google Scholar]
- Kumar A., Vidan S., Snyder M., Vidan S., Snyder M., Snyder M. Insertional mutagenesis: Transposon-insertion libraries as mutagens in yeast. Methods Enzymol. 2002;350:219–229. doi: 10.1016/s0076-6879(02)50965-4. [DOI] [PubMed] [Google Scholar]
- La Scola B., Raoult D., Raoult D. Acinetobacter baumannii in human body louse. Emerg. Infect. Dis. 2004;10:1671–1673. doi: 10.3201/eid1009.040242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulies M., Egholm M., Altman W.E., Attiya S., Bader J.S., Bemben L.A., Berka J., Braverman M.S., Chen Y.J., Chen Z., Egholm M., Altman W.E., Attiya S., Bader J.S., Bemben L.A., Berka J., Braverman M.S., Chen Y.J., Chen Z., Altman W.E., Attiya S., Bader J.S., Bemben L.A., Berka J., Braverman M.S., Chen Y.J., Chen Z., Attiya S., Bader J.S., Bemben L.A., Berka J., Braverman M.S., Chen Y.J., Chen Z., Bader J.S., Bemben L.A., Berka J., Braverman M.S., Chen Y.J., Chen Z., Bemben L.A., Berka J., Braverman M.S., Chen Y.J., Chen Z., Berka J., Braverman M.S., Chen Y.J., Chen Z., Braverman M.S., Chen Y.J., Chen Z., Chen Y.J., Chen Z., Chen Z., et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivera-Severo D., Wassermann G.E., Carlini C.R., Wassermann G.E., Carlini C.R., Carlini C.R. Ureases display biological effects independent of enzymatic activity: Is there a connection to diseases caused by urease-producing bacteria? Braz. J. Med. Biol. Res. 2006;39:851–861. doi: 10.1590/s0100-879x2006000700002. [DOI] [PubMed] [Google Scholar]
- Piechaud M., Second L., Second L. [Studies of 26 strains of Moraxella Iwoffi.] Ann. Inst. Pasteur (Paris) 1951;80:97–99. [PubMed] [Google Scholar]
- Pukatzki S., Kessin R.H., Mekalanos J.J., Kessin R.H., Mekalanos J.J., Mekalanos J.J. The human pathogen Pseudomonas aeruginosa utilizes conserved virulence pathways to infect the social amoeba Dictyostelium discoideum. Proc. Natl. Acad. Sci. 2002;99:3159–3164. doi: 10.1073/pnas.052704399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley J., Butler R., Ogilvie D., Finniear R., Jenner D., Powell S., Anand R., Smith J.C., Markham A.F., Butler R., Ogilvie D., Finniear R., Jenner D., Powell S., Anand R., Smith J.C., Markham A.F., Ogilvie D., Finniear R., Jenner D., Powell S., Anand R., Smith J.C., Markham A.F., Finniear R., Jenner D., Powell S., Anand R., Smith J.C., Markham A.F., Jenner D., Powell S., Anand R., Smith J.C., Markham A.F., Powell S., Anand R., Smith J.C., Markham A.F., Anand R., Smith J.C., Markham A.F., Smith J.C., Markham A.F., Markham A.F. A novel, rapid method for the isolation of terminal sequences from yeast artificial chromosome (YAC) clones. Nucleic Acids Res. 1990;18:2887–2890. doi: 10.1093/nar/18.10.2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schattner P., Brooks A.N., Lowe T.M., Brooks A.N., Lowe T.M., Lowe T.M. The tRNAscan-SE, snoscan and snoGPS Web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 2005;33:W686–W689. doi: 10.1093/nar/gki366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M.G., Des Etages S.G., Snyder M., Des Etages S.G., Snyder M., Snyder M. Microbial synergy via an ethanol-triggered pathway. Mol. Cell. Biol. 2004;24:3874–3884. doi: 10.1128/MCB.24.9.3874-3884.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanier R.Y., Palleroni N.J., Doudoroff M., Palleroni N.J., Doudoroff M., Doudoroff M. The aerobic pseudomonads: A taxonomic study. J. Gen. Microbiol. 1966;43:159–271. doi: 10.1099/00221287-43-2-159. [DOI] [PubMed] [Google Scholar]
- Sulston J., Hodgkin J., Hodgkin J. Methods. In: Wood W.B., editor. The nematode Caenorhabditis elegans. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York: 1988. pp. 587–606. [Google Scholar]
- Sussman M. Cultivation and synchronous morphogenesis of Dictyostelium under controlled experimental conditions. Methods Cell Biol. 1987;28:9–29. doi: 10.1016/s0091-679x(08)61635-0. [DOI] [PubMed] [Google Scholar]
- Suzek B.E., Ermolaeva M.D., Schreiber M., Salzberg S.L., Ermolaeva M.D., Schreiber M., Salzberg S.L., Schreiber M., Salzberg S.L., Salzberg S.L. A probabilistic method for identifying start codons in bacterial genomes. Bioinformatics. 2001;17:1123–1130. doi: 10.1093/bioinformatics/17.12.1123. [DOI] [PubMed] [Google Scholar]
- Tatusov R.L., Koonin E.V., Lipman D.J., Koonin E.V., Lipman D.J., Lipman D.J. A genomic perspective on protein families. Science. 1997;278:631–637. doi: 10.1126/science.278.5338.631. [DOI] [PubMed] [Google Scholar]
- Tomaras A.P., Dorsey C.W., Edelmann R.E., Actis L.A., Dorsey C.W., Edelmann R.E., Actis L.A., Edelmann R.E., Actis L.A., Actis L.A. Attachment to and biofilm formation on abiotic surfaces by Acinetobacter baumannii: Involvement of a novel chaperone-usher pili assembly system. Microbiol. 2003;149:3473–3484. doi: 10.1099/mic.0.26541-0. [DOI] [PubMed] [Google Scholar]
- Young D.M., Parke D., Ornston L.N., Parke D., Ornston L.N., Ornston L.N. Opportunities for genetic investigation afforded by Acinetobacter baylyi, a nutritionally versatile bacterial species that is highly competent for natural transformation. Annu. Rev. Microbiol. 2005;59:519–551. doi: 10.1146/annurev.micro.59.051905.105823. [DOI] [PubMed] [Google Scholar]