Abstract
Ecsit is a cytosolic adaptor protein essential for inflammatory response and embryonic development via the Toll-like and BMP (bone morphogenetic protein) signal transduction pathways, respectively. Here, we demonstrate a mitochondrial function for Ecsit (an evolutionary conserved signaling intermediate in Toll pathways) in the assembly of mitochondrial complex I (NADH:ubiquinone oxidoreductase). An N-terminal targeting signal directs Ecsit to mitochondria, where it interacts with assembly chaperone NDUFAF1 in 500- to 850-kDa complexes as demonstrated by affinity purification and vice versa RNA interference (RNAi) knockdowns. In addition, Ecsit knockdown results in severely impaired complex I assembly and disturbed mitochondrial function. These findings support a function for Ecsit in the assembly or stability of mitochondrial complex I, possibly linking assembly of oxidative phosphorylation complexes to inflammatory response and embryonic development.
Keywords: Mitochondria, oxidative phosphorylation, complex I, NADH:ubiquinone oxidoreductase, Ecsit, NDUFAF1
Regulation of gene expression is one of the cornerstones of biological versatility. It is achieved by feedback mechanisms between different subcellular pathways, often mediated by regulatory adaptor proteins. In 1999, Kopp et al. (1999) described a prominent example of such a protein, termed Ecsit (an evolutionary conserved signaling intermediate in Toll pathways). Ecsit is a cytoplasmic signaling protein that constitutes a molecular link between two pathways: the Toll signaling pathway and the BMP pathway.
The family of Toll-like receptors (TLRs) consists of 10–15 members in most mammalian species, sharing a conserved Toll/IL-1 receptor (TIR) domain (Sims et al. 1988; Greenfeder et al. 1995; Medzhitov et al. 1997; Torigoe et al. 1997; Chaudhary et al. 1998; Rock et al. 1998; Iwasaki and Medzhitov 2004). TLRs bind extracellular ligands (e.g., Gram-negative bacterial lipopolysaccharides) to activate expression of NF-κB and AP-1 transcription factors, which in turn, activate expression of genes involved in the immune response (Medzhitov et al. 1997). Signal transduction can occur in two ways: either via TAK1 and regulators TAB1 and TAB2, or via MEKK1 and subsequent MKK activation (Moustakas and Heldin 2003). Communication between the two cascades is possible, as TAK1 can activate MKKs and their downstream effectors and MEKK1 can activate the TAK1-activated IKK complex. TRAF6 (tumor necrosis factor [TNF] receptor-associated factor 6) is an early component of both cascades, and yeast two hybrid and immunoprecipitation studies have shown the association of Ecsit with TRAF6, thereby directly linking the protein to the Toll pathway (Kopp et al. 1999). In addition, Ecsit interacts with and may facilitate processing of MEKK-1, findings that have placed Ecsit in an immunological context.
In 2003, Xiao et al. (2003) found an additional role for this protein in the BMP (bone morphogenetic protein) pathway in mouse embryogenesis. Null mutation of the Ecsit gene in mice resulted in embryonic lethality with phenotypes that mimic those of a BMP receptor gene (Bmpr1a)-null mutant (reduced epiblast cell proliferation, block of mesoderm formation, and embryonic lethality at the beginning of gastrulation). Bmp4 is known to play an essential role in the gastrulation of the mouse embryo and signals through Bmpr1a, a type I Bmp receptor to induce up-regulation of target genes including Tlx2, a homeobox gene (Tang et al. 1998). Ecsit2 associates constitutively with Smad4, and associates with Smad1 in a Bmp-inducible manner. Furthermore, together with Smad1 and Smad4, it binds to the promoter of specific Bmp target genes (Xiao et al. 2003). Confirmative of its role in the Toll signaling pathway, short hairpin RNA (shRNA) inhibition of Ecsit in a macrophage cell line resulted in drastic inhibition of LPS-induced NF-κB activity (Xiao et al. 2003). This demonstrated that both the BMP and Toll signaling pathways require Ecsit, which herewith represents a link between immunity and embryonic development.
In this study, we show an unexpected additional function for Ecsit in mitochondria, to which it is targeted by an N-terminal targeting sequence and where it interacts with NDUFAF1, a chaperone involved in assembly of mitochondrial complex I (NADH:ubiquinone oxidoreductase) (Janssen et al. 2002; Vogel et al. 2005). Complex I is one of the five enzymatic complexes that comprise the oxidative phosphorylation (OXPHOS) system in the mitochondrial inner membrane, responsible for the generation of ATP from NADH and FADH2 (Brandt 2006; Janssen et al. 2006). Ecsit knockdown using RNA interference (RNAi) results in decreased NDUFAF1 and complex I protein levels, accumulation of complex I subcomplexes, and disturbed mitochondrial function. Herewith, in addition to its cytoplasmic and nuclear functions, these findings point toward a mitochondrial function for Ecsit, and could provide a link between mitochondrial OXPHOS system biogenesis and function, immune response, and mesoderm formation during embryogenesis.
Results
Ecsit copurifies with OXPHOS assembly chaperone NDUFAF1
Mitochondrial ATP production occurs via the OXPHOS system, which consists of five membrane-embedded protein complexes (Janssen et al. 2006). The assembly process of the largest of these, complex I, is a challenging problem (Vogel et al. 2004). It is known to involve assembly chaperone NDUFAF1, but the mechanism of its action remains unclear (Janssen et al. 2002; Vogel et al. 2005). In order to find mitochondrial-binding partners for NDUFAF1 we have performed Tandem Affinity Purification (TAP) of mitochondrial lysates of inducible human embryonic kidney 293 (HEK293) T-REx cells that express TAP-tagged NDUFAF1 protein. FT-MS analysis of the eluate revealed several proteins, which are listed in Supplementary Table 1. Among the peptides found, the analysis specifically identified several peptides corresponding to the Ecsit protein, a previously described cytoplasmic protein involved in immunity and embryonic development (Supplementary Table 1; Kopp et al. 1999; Xiao et al. 2003). Ten Ecsit splice isoforms are predicted, of which only isoforms 1 and 2 have been annotated and encode proteins with predicted molecular masses of 50 and 33 kDa, respectively (Fig. 1A). Analysis of the TAP purified peptides reveals the presence of the peptide DSTGAADPPQPHIVGIQSPDQQAALAR, which crosses the boundary between exons 5 and 6, and thereby identifies the largest, 50 kDa Ecsit isoform 1 (Fig. 1B; Supplementary Table 2).
Figure 1.
Ecsit isoform 1 copurifies with tandem affinity-purified NDUFAF1. (A) Ten Ecsit splice isoforms are predicted by the EBI alternative splicing database, of which two are identified in experimental studies (isoforms 1 and 2). Shown are the exons (vertical bars) and introns (horizontal lines) that comprise the complete transcript. The Swissprot annotation and predicted molecular mass are indicated when available. (B) FT-MS sequence coverage of Ecsit in eluates of tandem affinity-purified NDUFAF1 in HEK293 mitochondria demonstrates the presence of Ecsit isoform 1. Identified peptides are indicated in bold and in boxes, whereas exon 6, unique for Ecsit isoform 1, is indicated in gray.
Ecsit localizes to mitochondria
The interaction between NDUFAF1 and Ecsit is unexpected, since NDUFAF1 is described to be mitochondrial, whereas Ecsit is described as a cytoplasmic protein. However, although a mitochondrial function for Ecsit has not yet been described in the literature, all tested mitochondrial targeting sequence prediction programs (MitoPROT2, TargetP, Predotar, SignalP) clearly indicate a mitochondrial targeting sequence with high probability. MitoPROT2 predicts a 99.3% chance of a cleavable N-terminal target sequence of 49 amino acids, corresponding to ∼5 kDa.
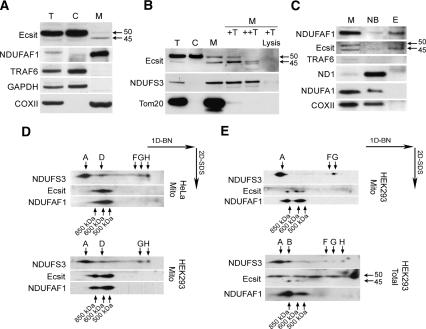
To verify these predictions, we have performed fractionations of HEK293 cells to separate mitochondria from other cellular compartments. Figure 2A shows predominant immunodetection of 50-kDa Ecsit isoform 1 in the cytoplasm/nucleus, in line with its previously described cytoplasmic function (Kopp et al. 1999; Xiao et al. 2003). In contrast to the cytoplasmic fraction, an ∼45-kDa Ecsit signal is specifically observed in mitochondria (Fig. 2A). As controls, Ecsit-binding partners TRAF6 and GAPDH do not show presence in mitochondria and are clearly cytoplasmic, and mitochondrial complex IV subunit COXII localizes in mitochondria.
Figure 2.
Ecsit localizes to mitochondria and interacts with NDUFAF1 in three high-molecular-weight complexes. (A) HEK293 cells were fractionated by pottering and lysates from total cell; mitochondria and cytoplasm were subsequently analyzed by SDS-PAGE and Western blotting. (T) Total cell; (C) cytoplasm; (M) mitochondria. All lanes were immunodecorated with antibodies targeted to Ecsit, NDUFAF1, TRAF6, cytoplasmic control GAPDH, and mitochondrial control COXII. Ecsit is predominantly present in the cytoplasm; however, a smaller band of ∼45 kDa is visible specifically in mitochondria. In contrast, the cytoplasmic Ecsit-binding partner TRAF6 is not detected in mitochondria. (B) Trypsin import assay. (T) Total cell; (C) cytoplasm; (M) mitochondria; (M +T) mitochondria + 30 U trypsin 15 min at 37°C; (M ++T) same as M +T but with 100 U trypsin; (M +T lysis) same as M +T but with 1% Triton to lyse the mitochondria. Increasingly stringent trypsin digestion of mitochondrial outer membrane proteins results in the disappearance of the 50-kDa Ecsit, whereas the 45-kDa Ecsit remains intact. NDUFS3 is used as a mitochondrial matrix control, Tom20 is used as an outer membrane control. (C) Immunoprecipitation using an anti-myc antibody in mitochondria purified from an NDUFAF1-myc-HIS-inducible HEK293 cell line. Shown are mitochondria (M), nonbound (NB), and eluate fractions (E). Myc-immunoprecipitation coelutes the 45-kDa mitochondrial Ecsit together with NDUFAF1, as opposed to mitochondrial controls ND1, NDUFA1, and COXII, and cytoplasmic control TRAF6. (D) Two-dimensional blue-native SDS-PAGE analysis of HeLa and HEK293 mitochondrial lysates demonstrates the colocalization of 45-kDa Ecsit and NDUFAF1 in three complexes of ∼500, 600, and 850 kDa. Complex I subunit NDUFS3 is shown to demonstrate the position of complex I (“A,” 1 MDa). Subcomplexes observed in our previous complex I assembly study (Ugalde et al. 2004) are indicated with A–H when visible. (E) HEK293 mitochondrial Ecsit signal was compared with a total cell preparation. (50) The 50-kDa cytoplasmic Ecsit in the HEK293 total cell lysate, which is absent in HEK293-purified mitochondria. This demonstrates that only the 45-kDa, mitochondrial, Ecsit interacts with NDUFAF1 in complexes of 500–850 kDa.
To confirm the mitochondrial localization of the 45-kDa Ecsit, we have performed trypsin digestion of mitochondrial outer membrane proteins (Fig. 2B). After cell fractionation, performed as for Figure 2A, mitochondria were treated for 15 min at 37°C with either 30 U of trypsin, 100 U of trypsin, or 30 U of trypsin in the presence of 1% Triton, which lyses the mitochondria and should thus result in complete degradation of all mitochondrial proteins. Increasingly, stringent trypsin treatment led to the degradation of the 50-kDa Ecsit, whereas the 45-kDa Ecsit was only degraded upon mitochondrial lysis. This suggests that the 45-kDa Ecsit is shielded by the mitochondrial outer and inner membranes, and most likely is imported into mitochondria. As controls, outer membrane protein Tom20 is rapidly degraded upon trypsin treatment, and matrix protein NDUFS3 is only degraded upon mitochondrial lysis. After digestion using 100 U of trypsin, in addition to the 45-kDa band, smaller bands were detected with the Ecsit antibody. Due to their size, it is unlikely that these bands represent partial tryptic digestion products. However, at this stage, we cannot exclude processing by another (mitochondrial) protease. Both fractionations (Fig. 2A,B) demonstrated that a small fraction of the 50-kDa cytoplasmic Ecsit localizes to mitochondria. This signal may either represent minute cytoplasmic background after pottering or an intermediate in the import process.
To establish whether it is the mitochondrial Ecsit that interacts with NDUFAF1, we performed anti-myc immunoprecipitations in mitochondrial lysates of inducible HEK293 T-REx cells that express myc-tagged NDUFAF1 protein (Fig. 2C). Two bands are visible for Ecsit in the total mitochondrial lysate, of 45 and 50 kDa in size, of which the 45-kDa form specifically copurifies with NDUFAF1. In contrast, mitochondrial controls ND1, NDUFA1, and COXII do not copurify. Cytosolic Ecsit-binding partner TRAF6 is not found in the mitochondrial preparation, demonstrating that the NDUFAF1 interaction is specifically mitochondrial.
Mitochondrial Ecsit and NDUFAF1 colocalize in three high-molecular-weight complexes
To investigate whether Ecsit associates to mitochondrial high-molecular-weight protein complexes, we have performed two-dimensional blue-native PAGE analysis of mitochondria enriched lysates of HEK293 and HeLa cells. Although slightly different between the two cell lines, this shows the specific presence of Ecsit in three complexes of ∼500, 600, and 850 kDa. This pattern strongly resembles that of OXPHOS assembly chaperone NDUFAF1, which accurately comigrates with Ecsit (Fig. 2D). At least for the HEK293 lysates, the two-dimensional resolution shows that instead of the previously reported two NDUFAF1-containing complexes of ∼600 and 700 kDa (Vogel et al. 2005), NDUFAF1 may, in fact, be present in three complexes of ∼500, 600, and 850 kDa. To demonstrate the mitochondrial origin of these protein complexes and to show that this Ecsit signal represents the mitochondrial 45-kDa Ecsit, we have compared a mitochondrial HEK293 lysate to a total cell lysate in the same analysis (Fig. 2E). This analysis revealed the additional presence of the 50-kDa Ecsit in the total cell lysate, confirming that the 45-kDa mitochondrial Ecsit, but not the 50-kDa protein, is present in the three complexes.
Ecsit requires its N-terminal targeting sequence for mitochondrial localization
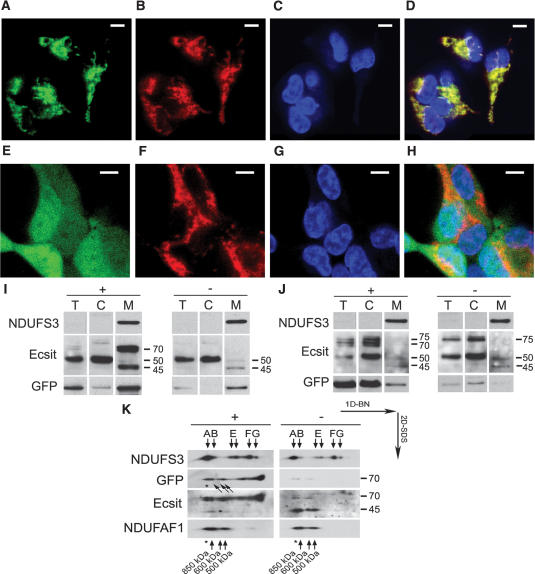
Although only a small percentage of the total cellular Ecsit pool is targeted to mitochondria (Fig. 2A), as discussed, Ecsit is predicted to have an N-terminal mitochondrial targeting sequence. To experimentally verify the requirement of the Ecsit N terminus for mitochondrial targeting, we have analyzed Ecsit subcellular localization by confocal microscopy of C-terminal GFP-tagged Ecsit with and without the first 48 N-terminal amino acids predicted to be required for mitochondrial targeting by MitoProt II (Fig. 3A–H). In living cells, mitochondria (visualized by Mitotracker Red staining) clearly colocalized with the Ecsit-GFP signal, demonstrating mitochondrial targeting of Ecsit-GFP (Fig. 3D). In contrast, Ecsit-GFP lacking its N terminus accumulated in the cytosol (Fig. 3H).
Figure 3.
Ecsit requires its N-terminal targeting sequence for mitochondrial localization. (A–H) Confocal microscopy of HEK293 cells transiently transfected with an inducible Ecsit-GFP construct (A–D) and with an inducible Ecsit-GFP construct lacking the 48-amino-acid Ecsit N terminus (E–H). A and E show GFP signal. B and F show the mitochondrial network using Mitotracker Red. C and G show nuclear staining. D and H show the overlay between the three signals. Bars, 10 μm. Without the N terminus, Ecsit-GFP is no longer targeted to mitochondria. Thus, Ecsit-GFP requires its N-terminal targeting sequence for mitochondrial localization. (I) Cell fractionation of Ecsit-GFP-inducible HEK293 cells. (+) Induction of expression; (−) no induction of expression; (T) total cell; (C) cytoplasm; (M) mitochondria. NDUFS3 is used as mitochondrial control. Other antibodies used are anti-Ecsit (“Ecsit” panel) and anti-GFP (“GFP” panel). The endogenous Ecsit signals are visible at 50 kDa (total cell and cytoplasm) and 45 kDa (mitochondria). The induced Ecsit-GFP targets predominantly to mitochondria and is detected at ∼70 kDa (45 kDa + 24 kDa) in two bands using the anti-Ecsit antibody. Only one of these anti-Ecsit-stained bands can be made visible using the anti-GFP antibody (“GFP” panel). The sensitive anti-GFP antibody also shows minor Ecsit-GFP leakage expression (in the uninduced situation). (J) Cell fractionation of inducible HEK293 cells expressing Ecsit-GFP without N terminus, as performed in I. In the induced situation, Ecsit appears predominantly in total cell and cytoplasm as several bands migrating at ∼70–75 kDa (“Ecsit” panel), of which only one is detected using the anti-GFP antibody (“GFP” panel). Again, minor leakage expression is observed in the uninduced situation. (K) Two-dimensional analysis of Ecsit-GFP incorporation into high-molecular-weight Ecsit/NDUFAF1 protein complexes of 500–850 kDa. The NDUFS3 signal is used as a marker for previously observed complex I subcomplexes A–H when visible (Ugalde et al. 2004). In accordance with A, two types of Ecsit-GFP are visible on the anti-Ecsit incubated blot (“Ecsit” panel), of which only one is detectable using anti-GFP antibody (“GFP” panel). The Ecsit-GFP complexes comigrate with the NDUFAF1 complexes (500–850 kDa). In addition, a larger complex matching the size of complex I (1 MDa) is indicated with an asterisk.
The predominant mitochondrial targeting of Ecsit-GFP (Fig. 3A–D) contrasts with the mainly cytoplasmic localization of endogenous Ecsit (Fig. 2A). To further investigate the mitochondrial targeting of Ecsit-GFP, we have performed biochemical fractionation as was done for endogenous Ecsit in Figure 2A (Fig. 3I). In addition to the cytoplasmic localization of 50-kDa endogenous Ecsit (Fig. 3I, Ecsit panel, 50 kDa), this fractionation confirms the predominant targeting of Ecsit-GFP to mitochondria, in line with the confocal imaging (Fig. 3I, Ecsit panel, 70 kDa). Enriched in mitochondria are two 70-kDa Ecsit-GFP bands, likely representing mitochondrial Ecsit (45 kDa Ecsit + 24 kDa GFP), of which only one is stained using the anti-GFP antibody. Some leakage Ecsit-GFP expression is observed in the uninduced situation using the anti-GFP antibody (Fig. 3I, GFP panel). This minor amount is also predominantly targeted to mitochondria. The same procedure was applied to the inducible HEK293 cells expressing Ecsit-GFP without its N-terminal sequence (Fig. 3J). Using the Ecsit antibody, multiple bands of ∼75 kDa were detected in the induced situation in the total cell and cytoplasmic fractions, but not in the mitochondrial fraction (Fig. 3J, Ecsit panel, 70–75 kDa). Also, the GFP signal was predominantly cytosolic, in line with confocal imaging (Fig. 3J, GFP panel).
To verify whether the Ecsit-GFP (with N terminus) is actually incorporated into mitochondrial protein complexes, we performed two-dimensional blue-native SDS-PAGE analysis of mitochondrial lysates of Ecsit-GFP-inducible cells (Fig. 3K). This analysis shows that Ecsit-GFP is normally incorporated into the endogenous complexes of 500–850 kDa (Fig. 3K, Ecsit and NDUFAF1 panels). As in Figure 3I, two 70-kDa Ecsit-GFP signals can be discerned in Figure 3K, of which only one is detected by the anti-GFP antibody, most likely due to incomplete unfolding of the GFP molecule in one of the two situations.
Ecsit knockdown in HeLa mitochondria results in NDUFAF1 decrease and impaired complex I assembly
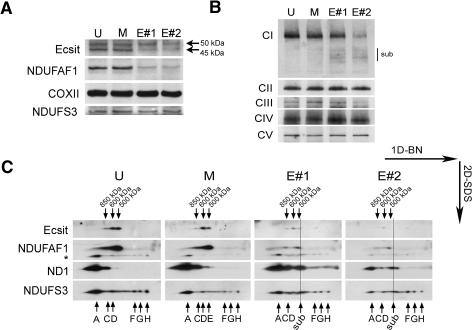
The mitochondrial association with assembly chaperone NDUFAF1 allows the possibility that Ecsit is involved in the assembly of OXPHOS complexes. To address this hypothesis, we investigated the effect of Ecsit knockdown on OXPHOS complex assembly using two small interfering RNAs (siRNAs) against Ecsit mRNA directed against exon 4 (target #1) and against exon 7 (target #2). The effects of knockdown were analyzed by SDS-PAGE analysis (Fig. 4A). Ecsit RNAi effectively knocks down the 45-kDa Ecsit signal, and has substantially reduced the amount of mitochondrial NDUFAF1 (Fig. 4A). Apparently, Ecsit is required for stable presence of NDUFAF1 within the mitochondrion. To analyze the effects of Ecsit knockdown on OXPHOS assembly we performed one-dimensional blue-native PAGE analysis (Fig. 4B). Complex I is severely reduced upon Ecsit knockdown, whereas the amounts of the other OXPHOS complexes appear relatively unchanged. Furthermore, immunodetection for complex I subunit NDUFS3 reveals the accumulation of intermediates of ∼500 kDa in size. To further investigate the nature of these complexes, we performed two-dimensional SDS PAGE analysis (Fig. 4C). As shown using antibodies against complex I subunits NDUFS3 and ND1, RNAi results in accumulation of intermediates smaller than complex I, indicative of disturbed assembly or stability of the holo-complex (Fig. 4C, “sub”). Furthermore, Ecsit knockdown results in a strong decrease in the NDUFAF1 subcomplexes of 500–850 kDa, strongly suggesting that the stability of these complexes relies on the presence of Ecsit and NDUFAF1 in these subcomplexes.
Figure 4.
Ecsit knockdown using RNAi results in disturbed complex I assembly. (A) RNAi was performed using two siRNAs (#1 and #2) against Ecsit mRNA. Following SDS-PAGE and Western blotting, immunodetection was performed for Ecsit, NDUFAF1, COXII, and NDUFS3 in untreated (U), mock-transfected (M), and siRNA-transfected (#1 and #2) HeLa cells. The Ecsit signal is knocked down, which correlates with a severe depression in NDUFAF1 protein. Monomeric COXII and NDUFS3 levels remain unchanged. (B) The effect of Ecsit knockdown on OXPHOS complex assembly was investigated by blue-native PAGE followed by Western blotting and immunodetection of complex I subunit NDUFS3 (CI), complex II subunit SDHA (CII), complex III subunit core2 (CIII), complex IV subunit COXII (CIV), and complex V subunit ATPase α (CV). Arrows indicate accumulated subcomplexes detected with the anti-NDUFS3 antibody. (C) Two-dimensional blue-native SDS-PAGE analysis of samples analyzed in A and B. Shown are immunodetections of Ecsit, NDUFAF1, and complex I (NDUFS3 and ND1 signals), in untreated, mock-transfected, and Ecsit siRNA-transfected (#1 and #2) HeLa cells. Subcomplexes that correspond to previously described complex I subcomplexes (Ugalde et al. 2004) are indicated with A–H. (Sub) Accumulated subcomplexes after both Ecsit siRNA transfections. The Ecsit/NDUFAF1 complexes are indicated with 500–850 kDa. An asterisk indicates signal from a previous NDUFS3 detection.
Ecsit knockdown in HeLa mitochondria results in disturbed mitochondrial function
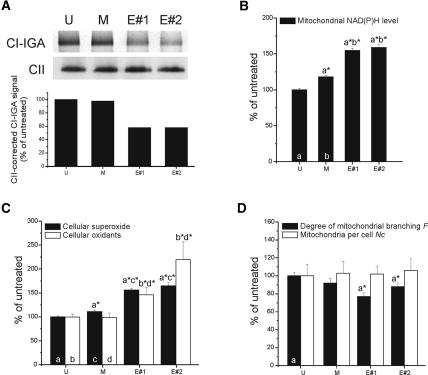
The impaired assembly or stability of complex I upon Ecsit knockdown may have serious consequences for mitochondrial function. For example, complex I deficiency is known to result in a broad spectrum of mitochondrial disorders (Janssen et al. 2006), increases cellular superoxide levels (Koopman et al. 2005a), and affects mitochondrial morphology (Koopman et al. 2005b). To investigate the effects of Ecsit knockdown on mitochondrial physiology and morphology we investigated several parameters. As demonstrated by complex I in-gel activity assay, complex I activity drops to 50%–60% of the control value (Fig. 5A). Mitochondrial NAD(P)H levels were significantly increased in the siRNA-treated cells compared with untreated and mock-treated controls (Fig. 5B), suggesting that NADH oxidation by complex I is decreased. Downstream effects of complex I dysfunction were assayed by measuring superoxide and cytosolic oxidant levels (Koopman et al. 2005a, 2006b) and by determining the degree of mitochondrial branching (F) and number per cell (Nc) (Fig. 5C,D; Koopman et al. 2005a, b, 2006a). This analysis indicated that the level of cellular radical species was increased up to 150%–200% of control values (Fig. 5C). Although the number of mitochondria per cell was not affected by siRNA treatment, mitochondrial branching was reduced (Fig. 5D). Taken together, these data support the notion that Ecsit is required for normal mitochondrial functioning and morphology.
Figure 5.
Ecsit knockdown using RNAi affects mitochondrial and cellular physiology. (A) Complex I in-gel activity (CI-IGA) and complex II (CII) expression following native electrophoresis in untreated (U), mock-treated (M), and two Ecsit RNAi knockdowns (E#1 and E#2). The bottom panel depicts the complex I in-gel activity (CI-IGA) signals corrected for complex II (CII) expression (expressed as percentage of the value in untreated cells) determined by integrated optical density analysis. (B) NAD(P)H levels in untreated (U), mock-treated (M), and siRNA-treated (E#1 and E#2) cells. Bars represent the average of 262 (U), 293 (M), 234 (E#1), and 180 (E#2) cells. (C) Cellular superoxide (filled bars) and oxidant levels (open bars) in untreated (U), mock-treated (M), and siRNA-treated (E#1 and E#2) cells. Bars represent the average of 294 (U), 284 (M), 335 (E#1), and 93 (E#2) cells for superoxide levels, and 47 (U), 33 (M), 41 (E#1), and 25 (E#2) cells for oxidant levels. (D) Degree of mitochondrial branching (F, black bars) and number of mitochondria per cell (Nc, open bars) in untreated (U), mock-treated (M), and siRNA-treated (E#1 and E#2) cells. Bars represent the average of 50 (U), 56 (M), 70 (E#1), and 50 (E#2) cells. In B–D, letters (a, b, c, and d) represent statistically significant differences with the indicated columns. Data was obtained during two independent experiments from multiple cells (N).
Discussion
In previous studies, Ecsit has been described as a cytosolic signaling protein essential for the Toll pathway of innate immunity and the BMP pathway of embryonic development (Kopp et al. 1999; Xiao et al. 2003). In this study, we show N-terminal targeting of Ecsit to mitochondria, where it interacts with mitochondrial complex I-specific assembly chaperone NDUFAF1 in three high-molecular-weight protein complexes of 500–850 kDa. Ecsit is required for correct complex I assembly or stability in particular, and mitochondrial function in general, and may thus represent a link between mitochondrial function, immune response and embryonic development.
Cytosolic Ecsit associates with TRAF6 and MEKK-1, and is described to facilitate the processing of MEKK-1 (Kopp et al. 1999). MEKK-1, in turn, is able to activate two different pathways leading to transcriptional activation of inflammatory genes (Moustakas and Heldin 2003). It now seems that, while Ecsit may facilitate the processing of another protein such as MEKK-1, Ecsit itself can be processed in the mitochondrion at its N terminus, after which it associates with mitochondrial complexes including complex I assembly chaperone NDUFAF1.
It is yet unclear under which factors determine whether Ecsit is targeted to either the mitochondrion or the cytosol. Whichever the underlying mechanism, we show, at least in HEK293 cells, that only a fraction of the total Ecsit amount is recruited to mitochondria, and that this targeting depends on an N-terminal mitochondrial targeting sequence. In addition, we show that addition of a C terminal GFP tag alters the Ecsit distribution from mainly cytoplasmic to predominantly mitochondrial. This shift in localization is most likely not caused by the overexpression itself, as the (low level) leakage expression of Ecsit-GFP in the uninduced cells is also targeted to the mitochondrion.
Even though, compared with its cytosolic counterpart, only a fraction of the Ecsit pool is present in mitochondria, Ecsit knockdown results in severely depressed NDUFAF1 amounts, demonstrating its requirement for stable mitochondrial presence of a complex I-specific chaperone. Furthermore, Ecsit knockdown results in specifically disturbed complex I assembly or stability and subsequently impaired mitochondrial function. Similar to the complex I-deficient patient and rotenone-treated control cells, NAD(P)H, superoxide (Koopman et al. 2005a), and cytosolic oxidant levels (Koopman et al. 2006b) are increased in Ecsit knockdown cells.
Conversely, NDUFAF1 knockdown is known to result in impaired complex I assembly (Vogel et al. 2005), but only results in a minor decrease in the amount of Ecsit in the 500-kDa subcomplex (data not shown). In contrast to Ecsit knockdown, no accumulation of complex I intermediates can be observed after NDUFAF1 knockdown (R.O. Vogel and R.J.R.J. Janssen, unpubl.), which indicates that although both proteins are present in the 500- to 850-kDa complexes their mechanism of action may be different. Elucidation of the exact composition of these intermediates may clarify the specific significance of the presence of Ecsit in these complexes for complex I assembly or stability.
In conclusion, as Ecsit is required for stabilization of complex I assembly chaperone NDUFAF1 and its absence results in impaired complex I assembly, accumulation of intermediates, and mitochondrial dysfunction, it seems that Ecsit is involved in additional processes apart from its functions in immune response and embryonic development. This putative link between mitochondria and the immune response has recently gained much attention with the discovery of MAVS (mitochondrial antiviral signaling) (Seth et al. 2005; McWhirter et al. 2005). This protein is associated to the mitochondrial outer membrane via a hydrophobic C terminus and acts as a signaling molecule in the immune response via association to TRAF6, similar to what has been described for the Ecsit protein. As Ecsit is now also found inside the mitochondrion, Ecsit may extend the influence of this cascade to the inner-mitochondrial level. Possibly, Ecsit modulates the energetic requirements upon inflammatory response by regulating the rate of complex I synthesis. Alternatively, Ecsit may induce other mitochondrial processes, such as apoptosis, upon microbial or viral infection. Future research will have to verify which of these possibilities is reality.
Materials and methods
Generation of inducible cell lines and cell culture
NDUFAF1-TAP construct—NDUFAF1 was subcloned from the pcDNA4/TO/myc-His A construct (Invitrogen) (Vogel et al. 2005) into pDONR201 (Invitrogen). A Gateway TAP-Destination vector was produced by subcloning the TAP tag of pCTAP-A (Stratagene) in frame behind the Gateway Reading Frame Cassette B (Invitrogen) in pcDNA5/FRT/TO (Invitrogen). To obtain an inducible NDUFAF1-TAP vector, the pDONR201-NDUFAF1 vector was recombined with the TAP-Destination vector using the Gateway LR Clonase II Enzyme Mix (Invitrogen). Ecsit-GFP construct—The Ecsit ORF sequence (BC000193; without stopcodon) flanked by Gateway AttB sites (Invitrogen) was created by PCR following the manufacturer’s instruction and cloned into pDONR201 by using the Gateway BP Clonase II Enzyme Mix (Invitrogen). A Gateway Destination vector was produced by subcloning the BamHI/NotI restriction fragment of pAcGFP1-N1 (Clontech) in frame behind the Gateway Reading Frame Cassette B (Invitrogen) in pcDNA5/FRT/TO (Invitrogen). To obtain an inducible Ecsit-GFP vector, the pDONR201-Ecsit vector was recombined with the AcGFP1-Destination vector using the Gateway LR Clonase II Enzyme Mix (Invitrogen). Ecsit-GFP–N terminus construct—The Ecsit ORF was cloned as for the Ecsit-GFP construct, with the following modification. Ecsit was cloned with an ATG start codon but without the following 141 base pairs (bp), encoding 47 amino acids. Based on the ORF sequence BC000193, the 5′ end of the sequence thus became 5′-ATGAGCTCTGAA. . .-3′. All constructs were transfected into Flp-In T-REx293 cells (Invitrogen) using Superfect Transfection Reagent (Qiagen) following manufacturer’s protocols. The NDUFAF1-mycHIS-inducible HEK293 T-RExtm cell line is previously described in Vogel et al. (2005). All inducible cell lines, HeLa cells, and HEK293 cells were cultured in DMEM (BioWhitaker) supplemented with 10% fetal calf serum (v/v) and 1% penicillin/streptomycin (v/v) (Gibco). The inducible cell lines were treated with 1 μg/mL doxycycline (Sigma-Aldrich) for expression of the transgene.
Preparation of mitochondria, cell fractionation, and trypsin treatment of mitochondria
For blue-native and SDS-PAGE analysis, HEK293 and HeLa mitochondria were purified with the use of digitonin as previously described in Ugalde et al. (2004). Fractionation of HEK293 cells was performed by pottering as described in Vogel et al. (2005). Trypsin treatment of mitochondria was performed by adding 30 U trypsin (Promega), 100 U trypsin, or 30 U trypsin plus 1% v/v Triton X-100 (Roche) to 50 μL of mitochondrial lysate followed by incubation for 15 min at 37°C.
Blue-native and SDS PAGE analysis and complex I in-gel activity assay
Blue-native gradient gels (5%–15%) were cast as earlier described (Nijtmans et al. 2002) and run with 40 or 80 μg of solubilized mitochondrial protein. After electrophoresis, gels were further processed for in-gel activity assays, Western blotting, or second-dimension 10% SDS-PAGE as described in Nijtmans et al. (2002). Proteins were transferred to a PROTAN nitrocellulose membrane (Schleicher & Schuell). One-dimensional 10% SDS-PAGE analysis was performed as described previously (Ugalde et al. 2004).
siRNA transfection
For transfection, HeLa cells were plated in 1.5 mL of DMEM supplemented with 10% FCS (without antibiotics) in six-well plates with a cell density of 2.0 × 105 cells per well. The next day, cells were transfected with siRNA duplex (control: Cyclophilin B [Dharmacon]; Ecsit #1 antisense strand: 5′-UUGAC GUUCAUGAAUCGAG dGdT-3′; #2 antisense strand: 5′-AUUGAUGUCAAACUCGUAG dTdT-3′) in the presence of oligofectamine (7.5 μL) (Invitrogen) and opti-MEM (Invitrogen) to achieve a final concentration of 100 nM siRNA in a total volume of 1.8 mL per well. Cells were incubated at 37°C in a CO2 incubator for 72 h prior to a second, identical round of transfection for 72 h.
Antibodies and ECL detection
Immunodetection was performed using the following primary antibodies. Complex I: NDUFS3 (Invitrogen), ND1 (a gift from Dr. Anne Lombes, Unite de Recherche INSERM 153, Hôspital de la Salpétrière, Paris, France), NDUFA1 (a gift from Professor Immo Scheffler, Section of Molecular Biology, Division of Biological Sciences, University of California at San Diego, San Diego, CA). NDUFAF1 affinity-purified serum is available from our laboratory (Vogel et al. 2005). Other antibodies used were raised against Ecsit (Abcam), TRAF6 (Abcam), GAPDH (Abcam), COXII (Invitrogen), Core2 (Invitrogen), ATPase α (Invitrogen), SDHA (Invitrogen), Tom20 (BD Biosciences), and GFP (a gift from Dr. Frank van Kuppeveld, Department of Medical Microbiology, Radboud University Nijmegen Medical Center, Nijmegen, The Netherlands). Secondary antibodies that were used are peroxidase-conjugated anti-mouse or anti-rabbit IgGs (Invitrogen). For the detection of immunoprecipitated proteins, ReliaBLOT HRP Conjugate was used (Bethyl Laboratories). The signal was generated using ECL plus (Amersham Biosciences).
Confocal imaging
HEK293 cells expressing inducible Ecsit-GFP (with and without N terminus) were cultured on glass slides, washed with PBS, and incubated with 1 μM Mitotracker Red (Invitrogen) for 15 min and with 10 μM Hoechst 3342 (Invitrogen) for 30 min, both at 37°C. After incubation, cells were washed with PBS and glass slides were mounted in an incubation chamber placed on the stage of an inverted microscope (Nikon Diaphot), attached to an Oz confocal microscope (Noran Instruments). Measurements were performed at 20°C in the dark. The light from an argon–ion laser (488 nm; Omnichrome) was delivered to the cells via a ×40 water immersion fluor objective (NA 1.2; Nikon). GFP and mitotracker fluorescence light were separated by a 565DRLPXR dichroic mirror, directed through 510AF23 and 630DF30 emission filters (all from Omega Optical, Inc.) and quantified using separate photomultiplier tubes (PMTs) at 8-bit resolution (Hamamatsu Photonics). Hoechst 3342 was excited using a 364-nm light generated by a high-power argon–ion laser (Coherent Enterprise) and its fluorescence emission was detected using a 400 nM longpass filter and PMT. Hardware and image acquisition were controlled by Intervision software (version 1.5, Noran Instruments) running under IRIX 6.2 on an Indy workstation (Silicon Graphics, Inc.). Images (512 × 480 pixels) were collected at 30 Hz with a pixel dwell time of 100 nsec and averaged in real time to optimize the signal-to-noise ratio (Koopman et al. 2006a). Image processing and analysis was performed using Image Pro Plus 5.1 (Media Cybernetics).
Quantification of NAD(P)H, superoxide, oxidant levels, and mitochondrial morphology
Mitochondrial NADH levels —NAD(P)H fluorescence intensity was measured using a CoolSNAP HQ CCD camera (Roper Scientific) attached to an inverted microscope (Axiovert 200 M, Carl Zeiss). Prior to recordings, coverslips were washed with PBS and placed in an incubation chamber containing HEPES-Tris medium (132 mM NaCl, 4.2 mM KCl, 1 mM CaCl2, 1 mM MgCl2, 5.5 mM D-glucose, 10 mM HEPES at pH 7.4). NAD(P)H was excited at 360 nm using a monochromator (Polychrome IV, TILL Photonics) and fluorescence emission was directed to the CCD camera using a 415DCLP dichroic mirror and a 510WB40 emission filter (Omega Optical, Inc.). Fields of view were recorded using an image-capturing time of 1 sec. Mean fluorescence intensity was determined in a region of interest (ROI) containing a high density of mitochondrial structures, which was background-corrected using an extracellular ROI of identical size. The imaging setup was controlled using Metafluor 6.0 software (Universal Imaging Corporation). Quantitative image analysis was performed using Metamorph 6.0 (Universal Imaging Corporation). Cellular superoxide and oxidant levels —Cellular superoxide and oxidant levels were quantified using hydroethidine (HEt) and 5-(and-6)-chloromethyl-2′,7′-dichlorodihydrofluorescein diacetate (CM-H2DCFDA) as described previously (Koopman et al. 2005a, 2006b). Mitochondrial morphology—Quantitative analysis of mitochondrial morphology in living cells was performed as described previously (Koopman et al. 2005b, 2006a). Statistics—Numerical results were visualized with Origin Pro 7.5 software (OriginLab) and are presented as means ± SEM (standard error of the mean). Statistical differences were determined with an independent two-sample Student’s t-test (Bonferroni corrected). P values <0.05 (*) were considered significant.
FT-MS analysis
The proteins were in-gel-reduced with 10 mM dithiotreitol and alkylated with 50 mM iodoactamide before in-gel digestion with trypsin. Peptides were extracted from the gel and purified and desalted using Stage tips (Rappsilber et al. 2003). Peptide identification experiments were performed using a nano-HPLC Agilent 1100 nanoflow system connected online to a linear quadrupole ion trap-Fourier transform mass spectrometer (LTQ-FT, Thermo Electron). LC and MS settings are further explained in Supplementary Table 3. Peptides and proteins were identified using the Mascot (Matrix Science) algorithm to search a local version of the NCBInr database (http://www.ncbi.nlm.nih.gov). First-ranked peptides were parsed from Mascot database search HTML files with MSQuant (http://msquant.sourceforge.net) to generate unique first-ranked peptide lists. Protein identifications were evaluated against criteria described in Supplementary Table 3.
Acknowledgments
This work was supported by “Het Prinses Beatrix Fonds” to J.S. and L.v.d.H. (grant no. 02-0104) and the European Community’s sixth Framework Programme for Research, Priority 1 “Life sciences, genomics and biotechnology for health” (contract no. LSHMCT-2004-503116). The Netherlands Organization for Scientific Research supported L.N. with a “Vernieuwingsimpuls” grant. Imaging equipment was financed by grants of ZON (Netherlands Organization for Health Research and Development, no. 903-46-176) and NWO (Netherlands Organization for Scientific Research, no. 911-02-008).
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.408407
References
- Brandt U. Energy converting NADH:Quinone Oxidoreductase (Complex I) Annu. Rev. Biochem. 2006;75:69–92. doi: 10.1146/annurev.biochem.75.103004.142539. [DOI] [PubMed] [Google Scholar]
- Chaudhary P.M., Ferguson C., Nguyen V., Nguyen O., Massa H.F., Eby M., Jasmin A., Trask B.J., Hood L., Nelson P.S., Ferguson C., Nguyen V., Nguyen O., Massa H.F., Eby M., Jasmin A., Trask B.J., Hood L., Nelson P.S., Nguyen V., Nguyen O., Massa H.F., Eby M., Jasmin A., Trask B.J., Hood L., Nelson P.S., Nguyen O., Massa H.F., Eby M., Jasmin A., Trask B.J., Hood L., Nelson P.S., Massa H.F., Eby M., Jasmin A., Trask B.J., Hood L., Nelson P.S., Eby M., Jasmin A., Trask B.J., Hood L., Nelson P.S., Jasmin A., Trask B.J., Hood L., Nelson P.S., Trask B.J., Hood L., Nelson P.S., Hood L., Nelson P.S., Nelson P.S. Cloning and characterization of two Toll/Interleukin-1 receptor-like genes TIL3 and TIL4: Evidence for a multi-gene receptor family in humans. Blood. 1998;91:4020–4027. [PubMed] [Google Scholar]
- Greenfeder S.A., Nunes P., Kwee L., Labow M., Chizzonite R.A., Ju G., Nunes P., Kwee L., Labow M., Chizzonite R.A., Ju G., Kwee L., Labow M., Chizzonite R.A., Ju G., Labow M., Chizzonite R.A., Ju G., Chizzonite R.A., Ju G., Ju G. Molecular cloning and characterization of a second subunit of the interleukin 1 receptor complex. J. Biol. Chem. 1995;270:13757–13765. doi: 10.1074/jbc.270.23.13757. [DOI] [PubMed] [Google Scholar]
- Iwasaki A., Medzhitov R., Medzhitov R. Toll-like receptor control of the adaptive immune responses. Nat. Immunol. 2004;5:987–995. doi: 10.1038/ni1112. [DOI] [PubMed] [Google Scholar]
- Janssen R., Smeitink J., Smeets R., van Den H.L., Smeitink J., Smeets R., van Den H.L., Smeets R., van Den H.L., van Den H.L. CIA30 complex I assembly factor: A candidate for human complex I deficiency? Hum. Genet. 2002;110:264–270. doi: 10.1007/s00439-001-0673-3. [DOI] [PubMed] [Google Scholar]
- Janssen R.J., Nijtmans L.G., van den Heuvel L.P., Smeitink J.A., Nijtmans L.G., van den Heuvel L.P., Smeitink J.A., van den Heuvel L.P., Smeitink J.A., Smeitink J.A. Mitochondrial complex I: Structure, function and pathology. J. Inherit. Metab. Dis. 2006;29:499–515. doi: 10.1007/s10545-006-0362-4. [DOI] [PubMed] [Google Scholar]
- Koopman W.J., Verkaart S., Visch H.J., Van der Westhuizen F.H., Murphy M.P., van den Heuvel L.W., Smeitink J.A., Willems P.H., Verkaart S., Visch H.J., Van der Westhuizen F.H., Murphy M.P., van den Heuvel L.W., Smeitink J.A., Willems P.H., Visch H.J., Van der Westhuizen F.H., Murphy M.P., van den Heuvel L.W., Smeitink J.A., Willems P.H., Van der Westhuizen F.H., Murphy M.P., van den Heuvel L.W., Smeitink J.A., Willems P.H., Murphy M.P., van den Heuvel L.W., Smeitink J.A., Willems P.H., van den Heuvel L.W., Smeitink J.A., Willems P.H., Smeitink J.A., Willems P.H., Willems P.H. Inhibition of complex I of the electron transport chain causes O2−·-mediated mitochondrial outgrowth. Am. J. Physiol. Cell Physiol. 2005a;288:C1440–C1450. doi: 10.1152/ajpcell.00607.2004. [DOI] [PubMed] [Google Scholar]
- Koopman W.J., Visch H.J., Verkaart S., van den Heuvel L.W., Smeitink J.A., Willems P.H., Visch H.J., Verkaart S., van den Heuvel L.W., Smeitink J.A., Willems P.H., Verkaart S., van den Heuvel L.W., Smeitink J.A., Willems P.H., van den Heuvel L.W., Smeitink J.A., Willems P.H., Smeitink J.A., Willems P.H., Willems P.H. Mitochondrial network complexity and pathological decrease in complex I activity are tightly correlated in isolated human complex I deficiency. Am. J. Physiol. Cell Physiol. 2005b;289:C881–C890. doi: 10.1152/ajpcell.00104.2005. [DOI] [PubMed] [Google Scholar]
- Koopman W.J., Visch H.J., Smeitink J.A., Willems P.H., Visch H.J., Smeitink J.A., Willems P.H., Smeitink J.A., Willems P.H., Willems P.H. Simultaneous quantitative measurement and automated analysis of mitochondrial morphology, mass, potential, and motility in living human skin fibroblasts. Cytometry A. 2006a;69:1–12. doi: 10.1002/cyto.a.20198. [DOI] [PubMed] [Google Scholar]
- Koopman W.J., Verkaart S., van Emst-de Vries S.E., Grefte S., Smeitink J.A., Willems P.H., Verkaart S., van Emst-de Vries S.E., Grefte S., Smeitink J.A., Willems P.H., van Emst-de Vries S.E., Grefte S., Smeitink J.A., Willems P.H., Grefte S., Smeitink J.A., Willems P.H., Smeitink J.A., Willems P.H., Willems P.H. Simultaneous quantification of oxidative stress and cell spreading using 5-(and-6)-chloromethyl-2′,7′-dichlorofluorescein. Cytometry A. 2006b;69:1184–1192. doi: 10.1002/cyto.a.20348. [DOI] [PubMed] [Google Scholar]
- Kopp E., Medzhitov R., Carothers J., Xiao C., Douglas I., Janeway C.A., Ghosh S., Medzhitov R., Carothers J., Xiao C., Douglas I., Janeway C.A., Ghosh S., Carothers J., Xiao C., Douglas I., Janeway C.A., Ghosh S., Xiao C., Douglas I., Janeway C.A., Ghosh S., Douglas I., Janeway C.A., Ghosh S., Janeway C.A., Ghosh S., Ghosh S. ECSIT is an evolutionarily conserved intermediate in the Toll/IL-1 signal transduction pathway. Genes & Dev. 1999;13:2059–2071. doi: 10.1101/gad.13.16.2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McWhirter S.M., Tenoever B.R., Maniatis T., Tenoever B.R., Maniatis T., Maniatis T. Connecting mitochondria and innate immunity. Cell. 2005;122:645–647. doi: 10.1016/j.cell.2005.08.026. [DOI] [PubMed] [Google Scholar]
- Medzhitov R., Preston-Hurlburt P., Janeway C.A., Jr., Preston-Hurlburt P., Janeway C.A., Jr., Janeway C.A., Jr. A human homologue of the Drosophila Toll protein signals activation of adaptive immunity. Nature. 1997;388:394–397. doi: 10.1038/41131. [DOI] [PubMed] [Google Scholar]
- Moustakas A., Heldin C.H., Heldin C.H. Ecsit-ement on the crossroads of Toll and BMP signal transduction. Genes & Dev. 2003;17:2855–2859. doi: 10.1101/gad.1161403. [DOI] [PubMed] [Google Scholar]
- Nijtmans L.G., Henderson N.S., Holt I.J., Henderson N.S., Holt I.J., Holt I.J. Blue Native electrophoresis to study mitochondrial and other protein complexes. Methods. 2002;26:327–334. doi: 10.1016/S1046-2023(02)00038-5. [DOI] [PubMed] [Google Scholar]
- Rappsilber J., Ishihama Y., Mann M., Ishihama Y., Mann M., Mann M. Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal. Chem. 2003;75:663–670. doi: 10.1021/ac026117i. [DOI] [PubMed] [Google Scholar]
- Rock F.L., Hardiman G., Timans J.C., Kastelein R.A., Bazan J.F., Hardiman G., Timans J.C., Kastelein R.A., Bazan J.F., Timans J.C., Kastelein R.A., Bazan J.F., Kastelein R.A., Bazan J.F., Bazan J.F. A family of human receptors structurally related to Drosophila Toll. Proc. Natl. Acad. Sci. 1998;95:588–593. doi: 10.1073/pnas.95.2.588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seth R.B., Sun L., Ea C.K., Chen Z.J., Sun L., Ea C.K., Chen Z.J., Ea C.K., Chen Z.J., Chen Z.J. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-κB and IRF 3. Cell. 2005;122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- Sims J.E., March C.J., Cosman D., Widmer M.B., MacDonald H.R., McMahan C.J., Grubin C.E., Wignall J.M., Jackson J.L., Call S.M., March C.J., Cosman D., Widmer M.B., MacDonald H.R., McMahan C.J., Grubin C.E., Wignall J.M., Jackson J.L., Call S.M., Cosman D., Widmer M.B., MacDonald H.R., McMahan C.J., Grubin C.E., Wignall J.M., Jackson J.L., Call S.M., Widmer M.B., MacDonald H.R., McMahan C.J., Grubin C.E., Wignall J.M., Jackson J.L., Call S.M., MacDonald H.R., McMahan C.J., Grubin C.E., Wignall J.M., Jackson J.L., Call S.M., McMahan C.J., Grubin C.E., Wignall J.M., Jackson J.L., Call S.M., Grubin C.E., Wignall J.M., Jackson J.L., Call S.M., Wignall J.M., Jackson J.L., Call S.M., Jackson J.L., Call S.M., Call S.M. cDNA expression cloning of the IL-1 receptor, a member of the immunoglobulin superfamily. Science. 1988;241:585–589. doi: 10.1126/science.2969618. [DOI] [PubMed] [Google Scholar]
- Tang S.J., Hoodless P.A., Lu Z., Breitman M.L., McInnes R.R., Wrana J.L., Buchwald M., Hoodless P.A., Lu Z., Breitman M.L., McInnes R.R., Wrana J.L., Buchwald M., Lu Z., Breitman M.L., McInnes R.R., Wrana J.L., Buchwald M., Breitman M.L., McInnes R.R., Wrana J.L., Buchwald M., McInnes R.R., Wrana J.L., Buchwald M., Wrana J.L., Buchwald M., Buchwald M. The Tlx-2 homeobox gene is a downstream target of BMP signalling and is required for mouse mesoderm development. Development. 1998;125:1877–1887. doi: 10.1242/dev.125.10.1877. [DOI] [PubMed] [Google Scholar]
- Torigoe K., Ushio S., Okura T., Kobayashi S., Taniai M., Kunikata T., Murakami T., Sanou O., Kojima H., Fujii M., Ushio S., Okura T., Kobayashi S., Taniai M., Kunikata T., Murakami T., Sanou O., Kojima H., Fujii M., Okura T., Kobayashi S., Taniai M., Kunikata T., Murakami T., Sanou O., Kojima H., Fujii M., Kobayashi S., Taniai M., Kunikata T., Murakami T., Sanou O., Kojima H., Fujii M., Taniai M., Kunikata T., Murakami T., Sanou O., Kojima H., Fujii M., Kunikata T., Murakami T., Sanou O., Kojima H., Fujii M., Murakami T., Sanou O., Kojima H., Fujii M., Sanou O., Kojima H., Fujii M., Kojima H., Fujii M., Fujii M., et al. Purification and characterization of the human interleukin-18 receptor. J. Biol. Chem. 1997;272:25737–25742. doi: 10.1074/jbc.272.41.25737. [DOI] [PubMed] [Google Scholar]
- Ugalde C., Vogel R., Huijbens R., van den Heuvel L.P., Smeitink J., Nijtmans L., Vogel R., Huijbens R., van den Heuvel L.P., Smeitink J., Nijtmans L., Huijbens R., van den Heuvel L.P., Smeitink J., Nijtmans L., van den Heuvel L.P., Smeitink J., Nijtmans L., Smeitink J., Nijtmans L., Nijtmans L. Human mitochondrial complex I assembles through the combination of evolutionary conserved modules: A framework to interpret complex I deficiencies. Hum. Mol. Genet. 2004;13:2461–2472. doi: 10.1093/hmg/ddh262. [DOI] [PubMed] [Google Scholar]
- Vogel R., Nijtmans L., Ugalde C., van den Heuvel L.P., Smeitink J., Nijtmans L., Ugalde C., van den Heuvel L.P., Smeitink J., Ugalde C., van den Heuvel L.P., Smeitink J., van den Heuvel L.P., Smeitink J., Smeitink J. Complex I assembly: A puzzling problem. Curr. Opin. Neurol. 2004;17:179–186. doi: 10.1097/00019052-200404000-00016. [DOI] [PubMed] [Google Scholar]
- Vogel R.O., Janssen R.J., Ugalde C., Grovenstein M., Huijbens R.J., Visch H.J., van den Heuvel L.P., Willems P.H., Zeviani M., Smeitink J.A., Janssen R.J., Ugalde C., Grovenstein M., Huijbens R.J., Visch H.J., van den Heuvel L.P., Willems P.H., Zeviani M., Smeitink J.A., Ugalde C., Grovenstein M., Huijbens R.J., Visch H.J., van den Heuvel L.P., Willems P.H., Zeviani M., Smeitink J.A., Grovenstein M., Huijbens R.J., Visch H.J., van den Heuvel L.P., Willems P.H., Zeviani M., Smeitink J.A., Huijbens R.J., Visch H.J., van den Heuvel L.P., Willems P.H., Zeviani M., Smeitink J.A., Visch H.J., van den Heuvel L.P., Willems P.H., Zeviani M., Smeitink J.A., van den Heuvel L.P., Willems P.H., Zeviani M., Smeitink J.A., Willems P.H., Zeviani M., Smeitink J.A., Zeviani M., Smeitink J.A., Smeitink J.A., et al. Human mitochondrial complex I assembly is mediated by NDUFAF1. FEBS J. 2005;272:5317–5326. doi: 10.1111/j.1742-4658.2005.04928.x. [DOI] [PubMed] [Google Scholar]
- Xiao C., Shim J.H., Kluppel M., Zhang S.S., Dong C., Flavell R.A., Fu X.Y., Wrana J.L., Hogan B.L., Ghosh S., Shim J.H., Kluppel M., Zhang S.S., Dong C., Flavell R.A., Fu X.Y., Wrana J.L., Hogan B.L., Ghosh S., Kluppel M., Zhang S.S., Dong C., Flavell R.A., Fu X.Y., Wrana J.L., Hogan B.L., Ghosh S., Zhang S.S., Dong C., Flavell R.A., Fu X.Y., Wrana J.L., Hogan B.L., Ghosh S., Dong C., Flavell R.A., Fu X.Y., Wrana J.L., Hogan B.L., Ghosh S., Flavell R.A., Fu X.Y., Wrana J.L., Hogan B.L., Ghosh S., Fu X.Y., Wrana J.L., Hogan B.L., Ghosh S., Wrana J.L., Hogan B.L., Ghosh S., Hogan B.L., Ghosh S., Ghosh S. Ecsit is required for Bmp signaling and mesoderm formation during mouse embryogenesis. Genes & Dev. 2003;17:2933–2949. doi: 10.1101/gad.1145603. [DOI] [PMC free article] [PubMed] [Google Scholar]