Abstract
Signals can be perceived and amplified at the cell membrane by receptors coupled to the production of a variety of second messengers, including myoinositol 1,4,5-trisphosphate [Ins(1,4,5)P3]. The myoinositol polyphosphate 5-phosphatases (5PTases; EC 3.1.3.56) comprise a large protein family that hydrolyzes 5-phosphates from a variety of myoinositol phosphate (InsP) and phosphoinositide phosphate (PtdInsP) substrates. Arabidopsis thaliana has 15 genes encoding 5PTases. Biochemical analyses of a subgroup of 5PTase enzymes suggest that these enzymes have both overlapping and unique substrate preferences. Ectopic expression of these genes in transgenic plants can reduce Ins(1,4,5)P3 levels and alter abscisic acid (ABA) signaling. To further explore the function of 5PTases in signaling, we have identified and characterized T-DNA insertional mutants for 5PTase1 and 5PTase2 and produced a double mutant. When grown in the dark, the seeds from these mutants germinate faster than wild-type seeds and the mutant seedlings have longer hypocotyls than wild-type seedlings. Seeds from these mutant lines also demonstrate an increase in sensitivity to ABA. These changes in early seedling growth are accompanied by mass increases in Ins(1,4,5)P3, but not by changes in endogenous ABA content. By labeling the endogenous myoinositol pool in 5ptase1 and 5ptase2 mutants, we detected increases in Ins(1,4,5)P3 and a decrease in PtdIns, PtdIns(4)P, and phosphatidylinositol (4,5) bisphosphate. Taken together, these data indicate that the At5PTase1 and At5PTase2 genes have nonredundant roles in hydrolyzing inositol second-messenger substrates and that regulation of Ins(1,4,5)P3 levels is important during germination and early seedling development.
The ability to respond to a variety of biotic and abiotic signals is crucial to plants. Signals outside the cell can be perceived and amplified at the cell membrane by receptors linked to a variety of signaling pathways, including the inositol 1,4,5-trisphosphate [Ins(1,4,5)P3] pathway (Stevenson et al., 2000; Taylor and Thorn, 2001). In this pathway, signals activate phospholipase C (PLC), which catalyzes the hydrolysis of phosphatidylinositol (4,5) bisphosphate [PtdIns(4,5)P2] to form diacylglycerol and Ins(1,4,5)P3. Ins(1,4,5)P3 is thought to stimulate downstream cellular events by triggering intracellular Ca2+ release from various sources within the plant cell, including the vacuole and endoplasmic reticulum (Berridge, 1993).
Because control of second messengers is critical for signaling, there has been recent interest in determining how the plant cell regulates the synthesis and hydrolysis of second messengers, such as Ins(1,4,5)P3. Plants metabolize Ins(1,4,5)P3 by sequential dephosphorylation of Ins(1,4,5)P3 utilizing a set of specific inositol phosphatase enzymes (Majerus et al., 1999). The myoinositol polyphosphate 5-phosphatases (5PTases; EC 3.1.3.56) have the ability to remove a 5-P from several inositol-containing second messengers, including Ins(1,4,5)P3 (Erneux et al., 1998). Eukaryotes have a family of diverse 5PTases, with the Arabidopsis (Arabidopsis thaliana) genome containing 15 genes predicted to encode conserved At5PTases (Berdy et al., 2001). Several of the At5PTase proteins have been examined biochemically and the results reveal that these enzymes vary in their substrate specificity (Berdy et al., 2001; Ercetin and Gillaspy, 2004; Zhong et al., 2004; Zhong and Ye, 2004). For example, At5PTase1 and At5PTase2 enzymes have been reported to hydrolyze water-soluble substrates Ins(1,4,5)P3 and Ins(1,3,4,5)P4 in vitro (Berdy et al., 2001), whereas the At5PTase11 enzyme preferentially hydrolyzes lipid substrates containing a 5-P such as PtdIns(4,5)P2 (Ercetin and Gillaspy, 2004). Other characterized At5PTases have been shown to hydrolyze both types of substrates (Zhong et al., 2004; Zhong and Ye, 2004).
Experiments using plants in which genes encoding 5PTases are mutated or overexpressed help shed light on 5PTase gene function. Both At5PTase1 and At5PTase2 have been shown to alter abscisic acid (ABA) signaling when ectopically expressed in Arabidopsis, a characteristic accompanied by increased hydrolysis of Ins(1,4,5)P3 (Sanchez and Chua, 2001; Burnette et al., 2003). Specifically, plants ectopically expressing At5PTase1 exhibit delays in ABA-induced gene expression and ABA insensitivity in guard cells, but no change in ABA sensitivity in a seed germination assay (Burnette et al., 2003). Seeds from plants ectopically expressing At5PTase2 are deficient in their transcriptional response to ABA, but they differ from those ectopically expressing At5PTase1 in that they are also insensitive to ABA in seed germination assays (Sanchez and Chua, 2001). These gain-of-function experiments suggest that, although At5PTase1 and At5PTase2 enzymes have similar biochemical properties in vitro, these proteins may have unique functions within the plant.
To discern the function of At5PTase1 and At5PTase2 genes, we identified and characterized mutant 5ptase1 and 5ptase2 plants from Arabidopsis. We found alterations in germination and early seedling growth in both 5ptase1 and 5ptase2 mutants that were accompanied by changes in Ins(1,4,5)P3 levels. These results show that both At5PTase1 and At5PTase2 function in hydrolyzing Ins(1,4,5)P3 during early seedling development.
RESULTS
Identification of 5ptase1 and 5ptase2 Mutants
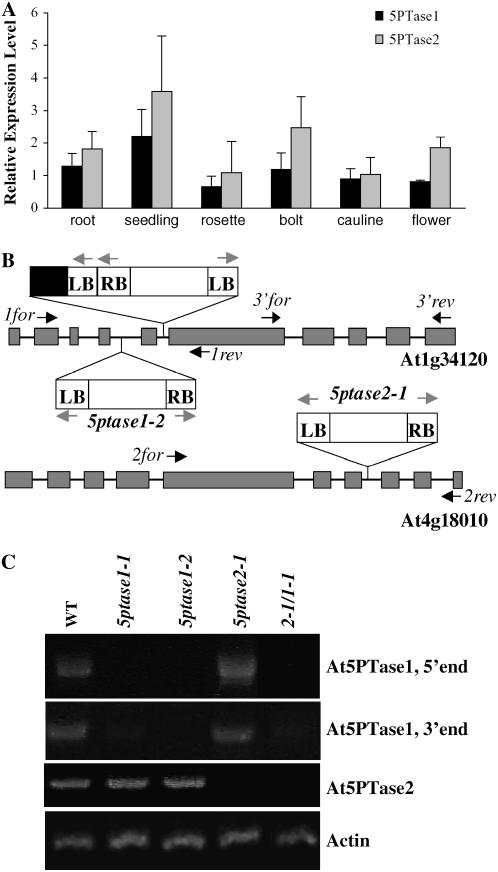
We have previously reported expression of At5PTase1 in seedlings (Burnette et al., 2003), a tissue in which At5PTase2 is also known to be expressed (Sanchez and Chua, 2001). To determine whether At5PTase1 and At5PTase2 expression patterns overlap in other tissues, we examined the relative levels of expression of each gene in Arabidopsis roots, seedlings, rosette leaves, bolts, cauline leaves, and flowers by reverse transcription (RT)-PCR (Fig. 1A). The results indicate that both At5PTase1 and At5PTase2 are expressed in each tissue examined. Further, expression of At5PTase2 was higher than At5PTase1 in most tissues. Microarray data obtained from Genevestigator (Zimmermann et al., 2004) was also examined to determine whether expression patterns of At5PTase1 and At5PTase2 overlapped (Supplemental Fig. S1). Comparison of our RT-PCR data with Genevestigator data indicated good agreement in that both genes were expressed in all tissue examined with At5PTase2 expression being greater in most tissues.
Figure 1.
At5PTase expression and T-DNA insertions and gene expression in 5ptase single- and double-mutant lines. A, RT-PCR expression analysis of At5PTase1 and At5PTase2 genes. cDNA was synthesized from RNA from the indicated tissues and PCR reactions with At5PTase1-, At5PTase2-, and actin-specific primers were performed as described in “Materials and Methods.” At5PTase1 and At5PTase2 expression was normalized to actin expression levels. Values are means ± se of three independent experiments. B, Schematic of the T-DNA insertion sites in the At5PTase1 and At5PTase2 genes. Exons are shown as dark gray boxes; gray arrows indicate primers used to amplify the RB (right border) and LB of the T-DNA; black arrows indicate the positions of gene-specific primers. The black box indicates truncated T-DNA found in the 5ptase1-1 mutant. C, Verification of At5PTase 1 or At5PTase2 expression in mutant lines. Total RNA was isolated from leaves of soil-grown plants. RT-PCR was carried out with gene-specific primers (black arrows) for the At5PTase and actin genes. Primer sequences can be found in “Materials and Methods.”
To further examine the physiological function of these genes, we isolated T-DNA insertional mutants for each gene. Two mutants in the At5PTase1 gene (At1g34120; SAIL_171A10, named 5ptase1-1 and SALK_123083, named 5ptase1-2) were identified and characterized and contain a T-DNA insertion within the fifth and fourth introns, respectively (Fig. 1B). We also obtained a T-DNA insertion mutant in the At5PTase2 gene (At4g18010; SAIL_138G01, named 5ptase2-1) containing a T-DNA insertion in the seventh intron (Fig. 1B). In each case, the presence of the T-DNA insertion was verified by diagnostic PCR reactions using genomic DNA and primers specific for the T-DNA left border (LB) and gene-specific primers that flank the T-DNA insertion. The resulting LB gene-specific fragments were sequenced to determine the precise location within the genome. In 5ptase1-1 mutants, a second partial T-DNA containing a LB is found in tandem in the fifth intron (Fig. 1B).
To obtain a double At5PTase1/At5PTase2 mutant, we emasculated a flower from a 5ptase1-1 homozygous parent plant and pollinated it with pollen from a 5ptase2-1 plant. Homozygous double mutants (2-1/1-1) were identified in the F3 generation. A decrease or lack of expression of At5PTase1 or At5PTase2 expression was verified in the single and double mutants by RT-PCR (Fig. 1C). Primers specific for an actin gene (ACT2) were used as a positive control (see Fig. 1C). Using primers specific for the 5′ end of At5PTase1 (1for and 1rev), we detected a PCR product corresponding to At5PTase1 in wild-type and 5ptase2-1 plants, but not in 5ptase1-1, 5ptase1-2, and 2-1/1-1 mutants (Fig. 1C). Using primers that amplify the 3′ end of At5PTase1 (3′for and 3′rev), we detected a 1,315-bp product in wild-type and 5ptase2-1 mutants, as expected. Whereas this PCR fragment was not detectable in the 5ptase1-2 mutants, 5ptase1-1 and 2-1/1-1 mutants did contain very small amounts of this PCR product.
We conclude that the 5ptase1-2 mutants are totally lacking At5PTase1 expression, whereas the 5ptase1-1 and 2-1/1-1 mutants have greatly reduced At5PTase1 expression. We also analyzed these plants for At5PTase2 expression, detecting a band corresponding to At5PTase2 in cDNA made from wild-type, 5ptase1-1, and 5ptase1-2 plants, but not in the 5ptase2-1 or 2-1/1-1 mutant plants. From this analysis, we conclude that these mutants are suitable for examining the consequences of eliminating or reducing At5PTase1 and/or At5PTase2 expression. It should be noted that we also identified other potential 5ptase2 mutants from the SALK collection, but found that these mutant lines did not contain reduced At5PTase2 expression (data not shown).
Growth of 5ptase1 and 5ptase2 Mutants
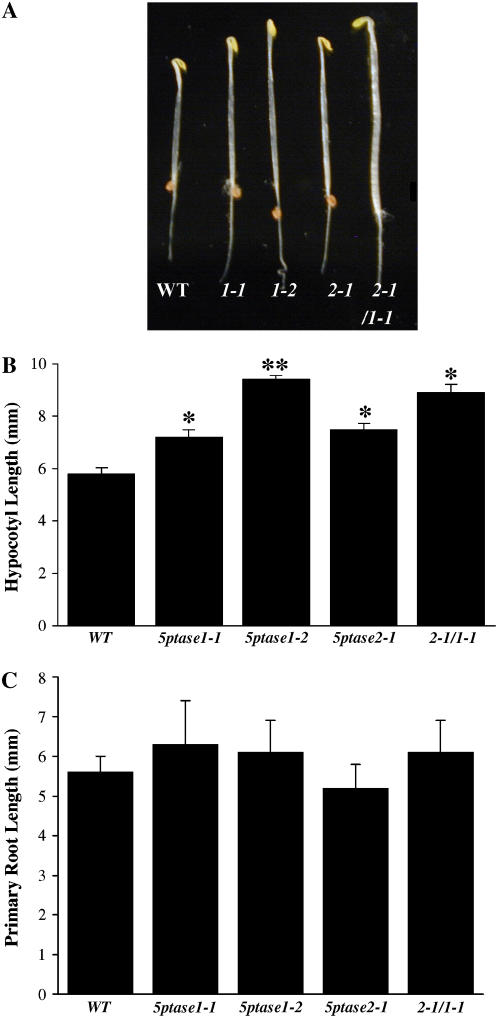
We examined the growth and development of all single- and double-mutant plants grown in soil. Under standard laboratory conditions, 5ptase mutants did not exhibit any abnormalities in plant growth or development. To detect more subtle changes in growth that might result from reduction or loss of 5PTase gene function, we produced age-matched seed populations that had been harvested from plants grown at the same time. 5ptase age-matched single- and double-mutant seeds were plated on Murashige and Skoog medium, stratified for 3 d at 4°C in the dark, and then grown in the light. After 3 to 7 d, no apparent differences were noted in the 5ptase mutant lines as compared to wild-type seedlings. We next examined the growth of 5ptase single and double mutants grown in the dark. Seeds from wild-type and 5ptase mutants were plated on Murashige and Skoog medium, stratified for 3 d at 4°C in the dark, and then grown for 3 more days in the dark at ambient temperature. We found that hypocotyls from 5ptase1-1, 5ptase1-2, 5ptase2-1, and the double mutant 2-1/1-1, were longer as compared to wild-type hypocotyls (Fig. 2A). There was variation among the different 5ptase mutant lines, but, in several independent experiments, dark-grown seedlings from all three single-mutant lines and the double-mutant line contained statistically significant longer hypocotyls than wild-type seedlings (P value < 0.05; Fig. 2B). We conclude that At5PTase1 and At5PTase2 genes play a role in regulating the hypocotyl length of seedlings grown in the dark. In addition, 5ptase1-2 mutants contained significantly longer hypocotyls than either 5ptase1-1 or 5ptase2-1 mutants (P value < 0.05; Fig. 2B). The difference in hypocotyl length between the 5ptase1-1 and 5ptase1-2 mutants correlates well with At5PTase1 expression levels (Fig. 1C), suggesting that residual At5PTase1 expression in 5ptase1-1 mutants impacts hypocotyl growth. The difference in hypocotyl length between the 5ptase1-2 and 5ptase2-1 mutants suggests that the At5PTase1 gene has a greater overall impact on hypocotyl growth than the At5PTase2 gene. Interestingly, there was not a statistically significant difference in the length of hypocotyls between the double mutant 2-1/1-1 and either parent (5ptase2-1 and 5ptase1-1), suggesting that the effects of At5PTase1 and At5PTase2 mutations are not additive.
Figure 2.
Alterations in seedling growth in 5ptase mutants. A, Phenotypes of 3-d-old dark-grown seedlings from wild-type and the indicated 5ptase mutant lines. Seeds were germinated and grown in the dark for 3 d on 0.5× Murashige and Skoog salts, 0.8% agar, and 1% Suc. B, Hypocotyls from wild type and 5ptase mutants in A were measured; values are means ± se (n = 50) from one of many independent experiments that gave similar results. Student's t-test comparisons between each mutant and the wild type were performed. *, P value was <0.05; **, P value of <0.05 when compared to wild-type and single mutants. C, Primary roots from A were measured. Values are means ± se (n = 50) from one of three independent experiments. [See online article for color version of this figure.]
We also examined roots from 5ptase mutant seeds germinated and grown in the dark for 3 d. The results shown in Figure 2C indicate no differences in root lengths between any mutant line and wild-type seedlings. We conclude that seedling root growth is not impacted by a reduction or loss of function in the At5PTase1 or At5PTase2 genes.
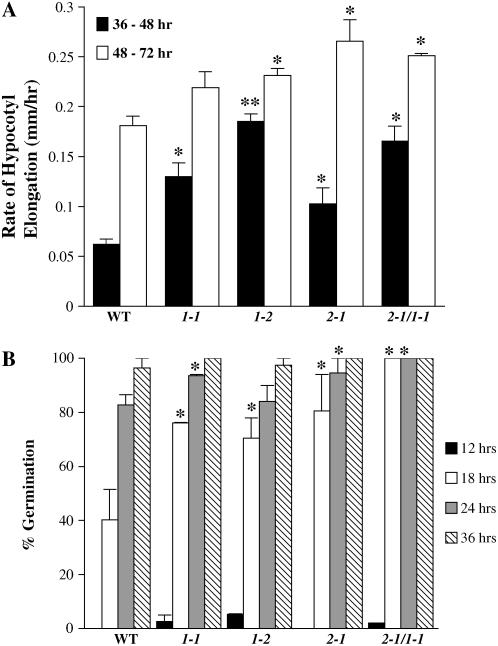
To determine whether the observed increases in hypocotyl length are due to an increase in the rate of hypocotyl elongation, we measured the hypocotyl length of 50 seedlings of each mutant line at 37, 48, and 72 h of growth in the dark. The rate of elongation is presented in Figure 3A for each mutant line and for wild-type seedlings. The results indicate that 5ptase mutants have a statistically significant increase in hypocotyl elongation rate. In each mutant, except for 5ptase1-1, the increase in elongation rate occurs at both time intervals tested. We conclude that the observed differences in hypocotyl length of 3-d-old dark-grown seedlings of 5ptase1-1, 5ptase1-2, 5ptase2-1, and the 2-1/1 double mutant are due, in part, to an increase in the hypocotyl elongation rate. The elongation rate from 37 to 48 h is significantly increased in the 5ptase1-2 mutants as compared to the 5ptase1-1 and 5ptase2-1 mutants, correlating well with the increased hypocotyl length noted in 5ptase1-2 mutants (Fig. 2B). This suggests that the impact of At5PTase1 as compared to At5PTase2 is slightly greater on hypocotyl growth.
Figure 3.
Rate of hypocotyl elongation and seed germination in 5ptase mutants. A, Seeds from wild type and 5ptase mutants were germinated in the dark and grown for 36, 48, or 72 h. Hypocotyl length was measured and the rate of increase in length is presented for two time intervals; values are means ± se (n = 50). *, P value of <0.05 as described in Figure 2; **, P value of <0.05 when compared to wild-type and single mutants. B, Seeds were germinated in the dark as described and scored for germination (as judged by radical extrusion) at 12, 18, 24, or 36 h. Values are mean ± se (n = 50); * as in A. The experiment was independently repeated two times.
We also examined whether 5ptase single and double mutants were altered in the timing of germination because this could also affect the final hypocotyl length. We plated age-matched 5ptase mutant and wild-type seed on Murashige and Skoog plates and compared the germination rate in the absence of a stratification treatment. There were no significant differences in germination rates between the 5ptase mutant and wild-type seed (data not shown). We next tested whether there were differences in germination rate when seeds are stratified for 3 d at 4°C. We found that a small fraction (2%–5%) of 5ptase1-1, 5ptase1-2, and double-mutant seed germinated after 12 h as compared to none for 5ptase2-1 and wild-type seed (Fig. 3B). In addition, by 18 h there was a significant increase in the germination rate for each mutant as compared to the wild-type seed population. From these data, we conclude that seeds from 5ptase1-1, 5ptase1-2, 5ptase2-1, and the 2-1/1 double mutants have the ability to germinate faster than wild-type seed when stratified. Faster germination could also contribute to the final increase in hypocotyl length seen in mutants (Fig. 2A). Overall, the results of these experiments indicate that 5ptase mutant seeds do not differ in seed dormancy, but germinate faster and undergo faster hypocotyl elongation as compared to wild-type seed when stratified and grown in the dark.
ABA Content of 5ptase Mutant Seed
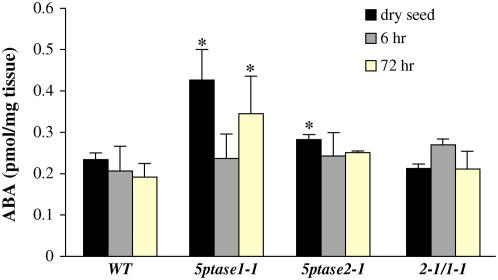
To determine whether alteration in timing of seed germination after stratification is due to decreases in seed ABA levels, we used ELISA to quantify ABA levels in seed from 5ptase single and double mutants. We assayed dry seeds as well as seeds that had been imbibed for 6 or 72 h. Most mutant seed samples analyzed were similar to wild-type seed in ABA content (Fig. 4). 5ptase1-1 mutant seed contained a 2- and 1.4-fold increase in ABA levels in dry and 72-h imbibed seeds as compared to similarly treated wild-type seed. However, 5ptase1-1 mutant seed imbibed for 6 h had similar ABA content to wild-type seed (Fig. 4). Given this, plus the fact that the se in ABA measurements from 5ptase1-1 mutant dry and 72-h imbibed seed is large, we reject the hypothesis that 5ptase mutant seeds germinate faster because of lower seed ABA content.
Figure 4.
ABA levels in wild-type and 5ptase mutant seed. Wild-type and mutant dry seeds were stored at room temperature or imbibed for 6 or 72 h at 4°C on filter paper. ABA was measured in approximately 10 mg of seeds that were ground in liquid nitrogen. Bars represent the mean ± se of two independent experiments. *, P value <0.05. [See online article for color version of this figure.]
Response of 5ptase Mutants to ABA
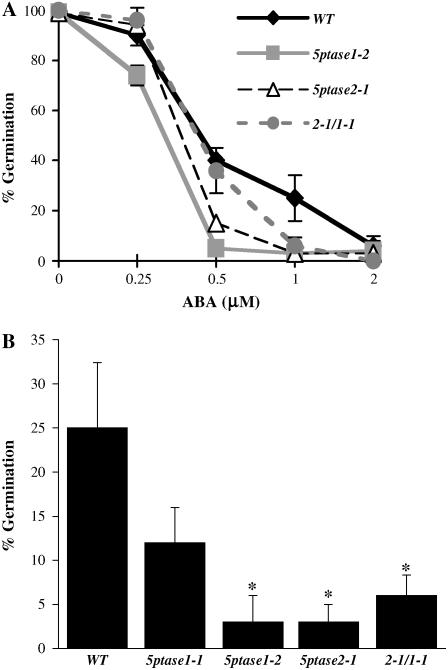
One possibility to explain the faster germination of 5ptase mutant seed is that these mutants contain a decrease in sensitivity to ABA. Previously, it has been shown that ectopic expression of At5PTase2 results in ABA insensitivity in seeds (Sanchez and Chua, 2001). To determine whether a reduction or loss of function in either At5PTase1 or At5PTase2 alters ABA sensitivity, we germinated both 5ptase single and double mutants in the presence of 0, 0.25, 0.5, 1, and 2 μm ABA and measured the impact on germination after 5 or 8 d. In contrast to our expectation that the faster germination phenotype might be related to a lack of ABA sensitivity, seeds from 5ptase single and double mutants were hypersensitive to small amounts of ABA (Fig. 5A). At day 5, the 5ptase1-2, 5ptase2-1, and 5ptase1-1/2-1 mutants have a significant decrease in seed germination in the presence of 1 μm ABA (Fig. 5A). The 5ptase1-1 mutants also have a decrease in seed germination in the presence of ABA; however, the decrease is not as great as that of the other 5ptase mutants, suggesting that residual expression of At5PTase1 in this mutant lessens the impact on ABA hypersensitivity. We conclude that the 5ptase1, 5ptase2, and 5ptase1-1/2-1 double mutants are hypersensitive to ABA. As noted with the hypocotyl growth and seed germination experiments, no statistically significant difference in ABA sensitivity exists between the double mutant 2-1/1-1 and either parent (5ptase2-1 and 5ptase1-1), suggesting that the effects of At5PTase1 and At5PTase2 mutations are not additive with regard to ABA sensitivity during seed germination.
Figure 5.
5ptase mutant seeds are hypersensitive to ABA. A, Seeds were surface sterilized and stratified at 4°C for 3 d before being placed at 23°C for germination. A, Germination was measured on increasing doses of ABA for 5 d. Values represent the mean ± se of three independent experiments. B, Seed germination was measured after 5 d of growth on 1 μm ABA. *, P value <0.05.
InsP and PtdInsP Levels in 5ptase Mutant Seedlings
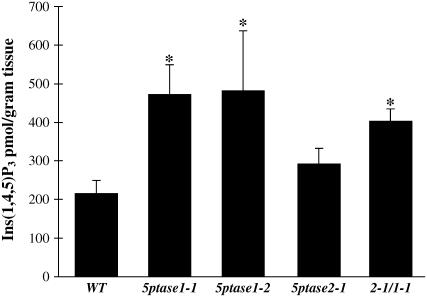
To determine whether alterations in seed germination and seedling growth are accompanied by alterations in inositol-containing second messengers, we first measured mass Ins(1,4,5)P3 levels in 3-d-old dark-grown 5ptase mutants and wild-type seedlings using a commercially available Ins(1,4,5)P3 mass assay kit (Fig. 6). In this assay, radioactive Ins(1,4,5)P3 is incubated with plant extracts and a partially purified Ins(1,4,5)P3 receptor is used in a competitive binding assay. Values from the competition assay are then compared to a standard curve and mass Ins(1,4,5)P3 can be calculated. The results in Figure 6 indicate that both 5ptase1-1 and 5ptase1-2 mutant lines contain an increase in Ins(1,4,5)P3 as compared to wild-type seedlings. This indicates that reduction or loss of function in the 5PTase1 gene results in less Ins(1,4,5)P3 hydrolysis under these conditions. Mass Ins(1,4,5)P3 levels are also increased in 5ptase2-1 mutant seedlings, although to a lesser extent than in 5ptase1 mutants. Also, 5ptase2-1/1-1 double mutants contain an increase in Ins(1,4,5)P3, and this increase is intermediate between that observed for 5ptase1 and 5ptase2 mutants. We conclude that a reduction or loss of function in either At5PTase1 or At5PTase2 impacts Ins(1,4,5)P3 hydrolysis in dark-grown seedlings.
Figure 6.
Mass Ins(1,4,5)P3 levels in wild-type and 5ptase mutant seed. Three-day-old dark-grown seedlings were frozen in liquid nitrogen, ground, and analyzed for mass Ins (1,4,5)P3 levels as described in “Materials and Methods.” Bars represent the mean ± se of five independent experiments. *, P value <0.05.
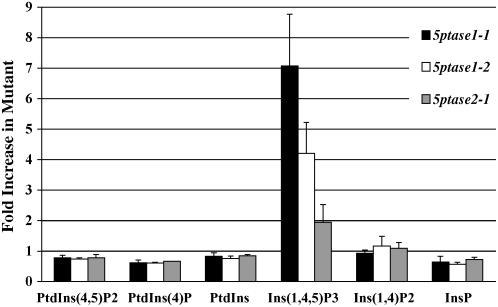
To examine the impact of 5ptase loss of function on other known Ins(1,4,5)P3 pathway metabolites, we incubated dark-grown 5ptase mutant and wild-type seedlings in 3H-myoinositol and then extracted and analyzed relevant InsPs and PtdInsPs by HPLC. Because these experiments involve challenging and rigorous analysis, we focused our investigation on the single 5ptase mutant lines and metabolites within the pathway that we could measure accurately. PtdIns(4,5)P2 is the substrate for activated PLC action, which generates Ins(1,4,5)P3. Levels of PtdIns(4,5,)P2 are slightly decreased in both 5ptase1 mutants and in the 5ptase2-1 mutant (Fig. 7). PtdIns(4)P and PtdIns are also slightly decreased in both 5ptase1 mutants and in the 5ptase2-1 mutant. In contrast and, as expected, levels of Ins(1,4,5)P3 are increased in 5ptase1-1, 5ptase1-2, and 5ptase2-1 mutants (Fig. 7), confirming our results seen with the Ins(1,4,5)P3 mass assays (Fig. 6). 5PTase catalysis of Ins(1,4,5)P3 yields Ins(1,4)P2. This metabolite is not altered in any of the mutant lines as seen in Figure 7. The next metabolite is Ins(1 or 4)P; levels of this metabolite were slightly decreased in 5ptase mutants as compared to wild-type seedlings. From these data, we conclude that the most dramatic change in 5ptase1 and 5ptase2 mutants is the increase in Ins(1,4,5)P3, with smaller accompanying decreases in PtdIns(4,5)P2, PtdIns(4)P, PtdIns, and InsP. It should be noted that, using our methodology, we could not detect a peak corresponding to Ins(1,3,4,5)P4 or any other InsP4 species within seedlings. Therefore, we cannot determine whether this potential substrate is also altered in the 5ptase mutants we examined. The changes we documented in Ins(1,4,5)P3 are, however, temporally associated with the faster germination and increased hypocotyl elongation noted previously for dark-grown 5ptase1 and 5ptase2 mutants (Figs. 2 and 3).
Figure 7.
Anion-exchange HPLC analysis of Ins(1,4,5)P3 pathway molecules in wild-type and 5ptase mutant seedlings; 2.5-d-old wild-type and 5ptase mutant seedlings were labeled with myo-[2-3H]inositol for 12 h in the dark. Fractions containing InsPs and deacylated lipid fractions containing PtdInsPs were isolated and an equal number of cpm from each was analyzed by HPLC using an ammonium phosphate gradient as described in “Materials and Methods.” Data for InsP and PtdInsP species are presented as fold increase as compared to wild type. Bars represent the mean ± se of three independent experiments. Raw values for each molecular species in wild-type seedlings are presented in “Materials and Methods.”
DISCUSSION
General interest in controlling plant growth and signaling have prompted efforts to identify genetic targets useful for altering second messengers. PLC signaling, which produces an Ins(1,4,5)P3 second messenger, has been implicated in several plant stress responses (Shacklock et al., 1992; Lee et al., 1996; Perera et al., 1999, 2001, 2005; Kashem et al., 2000; Reggiani and Laoreti, 2000; Shigaki and Bhattacharyya, 2000; DeWald et al., 2001; Ortega and Perez, 2001; Liu et al., 2006). Because some 5PTase enzymes can act to hydrolyze this second messenger, the 5PTase genes are logical targets for altering Ins(1,4,5)P3 levels and stress responses in plants. However, the diversity of the 5PTase protein family, which is encoded by 15 genes in Arabidopsis (Berdy et al., 2001) and more in crop species (G. Gillaspy, unpublished data), suggests either major redundancy within this protein family or very specific developmental roles for individual 5PTases. Data presented here, plus that from other recent studies, supports the latter scenario.
Three 5PTases that function in plant development have been identified through genetic or microarray screens. The cotyledon vascular pattern2 (cvp2; At5PTase6; At1g05470), fragile fiber3 (fra3; At1g65580), and root hair morphogenesis3 (mrh3; At5PTase5; At5g65090) mutants are altered in cotyledon vascular patterning (Carland and Nelson, 2004), fiber cell development (Zhong et al., 2004), and root hair initiation (Jones et al., 2006), respectively. In wild-type plants, CVP2 expression is restricted to developing vascular elements within seedlings (Carland and Nelson, 2004), whereas FRA3 expression is restricted to fiber cells and developing vasculature within the stem (Zhong et al., 2004). In addition, the mrh3 phenotype also suggests a specific role in root hair development for this gene (Jones et al., 2006). Both cvp2 and fra3 mutants contain higher levels of inositol second-messenger signaling molecules with cvp2 mutants containing an approximately 3-fold increase in Ins(1,4,5)P3 levels (Carland and Nelson, 2004), and fra3 mutant stems containing a 1.8- and 1.7-fold increase in Ins(1,4,5,)P3 and PtdIns(4,5)P2, respectively (Zhong et al., 2004). Substrate specificity of the CVP2 enzyme has not been determined; however, results from the cvp 2 mutants suggest that CVP hydrolyzes Ins(1,4,5)P3 primarily in the developing vascular system of seedlings. FRA3, in contrast, encodes a WD40-containing 5PTase, which hydrolyzes Ins(1,4,5)P3, PtdIns(4,5)P2, and PtdIns(3,4,5)P3 (Zhong et al., 2004). Thus, CVP2 and FRA3 are expressed in specific tissues in which hydrolysis of Ins(1,4,5)P3 provides critical information for proper development.
A major question underlying our work is how At5PTase1 and At5PTase2 function and whether these genes are redundant. One indicator of potential redundancy is whether two genes are expressed in the same tissues. Our experimental (Fig. 1) and Genevestigator analyses (Supplemental Fig. S1) indicate potential for redundant function in that both At5PTase1 and At5PTase2 are coexpressed in almost all Arabidopsis tissues. Another indicator of redundancy is whether or not loss-of-function mutants exhibit a phenotype. Both 5ptase1 and 5ptase2 mutants exhibit mild phenotypes that differ from the previously characterized 5PTase mutant phenotypes. Therefore, we conclude that At5PTase1 and At5PTase2 are not functionally redundant, although the loss of either impacts the seedling in a similar way. Both 5ptase1 and 5ptase2 mutants undergo faster seed germination and increased hypocotyl elongation when grown in the dark (Figs. 2 and 3) and both have increased sensitivity to exogenously supplied ABA (Fig. 5). From our analyses of the double mutant (2-1/1-1), we can conclude that the effects of mutating At5PTase1 and At5PTase2 together do not result in an additive effect. There was no significant difference in mass Ins(1,4,5)P3 levels, hypocotyl elongation, seed germination, and ABA sensitivity of the 2-1/1-1 double mutant as compared to either parent 5ptase1-1 and 5ptase2-1 mutants. This may indicate that a compensatory mechanism is in place to limit alterations in Ins(1,4,5)P3 within the seedling.
Growth alterations in 5ptase1 and 5ptase2 mutant seedlings correlate with changes in mass Ins(1,4,5)P3. Because biochemical studies have shown that the At5PTase1 and At5PTase2 enzymes have a substrate preference for the water-soluble InsPs, including Ins(1,4,5)P3 and Ins(1,3,4,5)P4 (Berdy et al., 2001; Sanchez and Chua, 2001), we expected to find increases in these second messengers. Our mass measurements of Ins(1,4,5)P3 levels indicate a 2.5-fold increase in mass Ins(1,4,5)P3 in 5ptase1 mutants and smaller increases in the 5ptase2 mutant (1.4-fold) and double 1-1/2-1 mutant (1.8-fold). The change in mass Ins(1,4,5)P3 levels in 5ptase1 mutants is similar in magnitude to the fold change we saw when the At5PTase1 gene was ectopically expressed in transgenic plants (Burnette et al., 2003) and to results from cvp2 and fra3 mutants (Carland and Nelson, 2004; Zhong et al., 2004). Our results from radiolabeling and HPLC analyses correlate well with mass Ins(1,4,5)P3 increases, indicating a slightly higher Ins(1,4,5)P3 increase of 3.8-fold for 5ptase1 and 3.3-fold for 5ptase2 mutants (Figs. 6 and 7).
We did not observe a peak corresponding to Ins(1,3,4,5)P4 or other InsP4 molecules in any of our plant extracts; therefore, we conclude that this molecule is below our detection limit. Stevenson-Paulik et al. (2005) reported the presence of InsP4 in Arabidopsis inositol polyphosphate mutant seedlings using a similar radiolabeling approach; however, InsP4 was barely detectable in wild-type seedlings.
To understand how an increase in Ins(1,4,5)P3 could impact the speed of seed germination and the hypocotyl elongation rate, we favor the idea that the elevated levels of Ins(1,4,5)P3 found in these plants increases the rate of cellular growth within seeds and seedlings grown in the dark. This impact on growth may be similar in nature to the one observed in mrh3 mutants (Jones et al., 2006). The mrh3 mutants have increased root hair initiation sites and root hairs early in development. It is interesting to note that increases in Ins(1,4,5)P3 have been correlated with the gravitropic bending response (Perera et al., 1999, 2001, 2005). Here, Ins(1,4,5)P3 is hypothesized to function upstream of auxin redistribution (Perera et al., 2005). Recently, overexpression of the human type I 5PTase in Arabidopsis was shown to dramatically reduce Ins(1,4,5)P3 and impair gravitropic responses, although no other general growth effects were noted (Perera et al., 2005). Together, these data suggest that Ins(1,4,5)P3 is an important regulator for specific types of growth within plants.
We found that 5ptase1 and 5ptase2 mutants are increased in their sensitivity to ABA in a seed germination assay (Fig. 5), which is consistent with the proposed role of Ins(1,4,5)P3 in the ABA response-signaling pathway. ABA hypersensitivity has also been noted for mutants containing altered Ins(1,4,5)P3 levels, specifically the cvp2 mutants mentioned previously (Carland and Nelson, 2004) and fiery1 mutants (Xiong et al., 2001). FIERY1 protein is most similar to the inositol bisphosphatases and fiery1 mutants contain a 10-fold increase in mass Ins(1,4,5)P3 levels, perhaps due to substrate feedback inhibition (Xiong et al., 2001). In contrast, fra3 mutants are not ABA hypersensitive, most likely because the Ins(1,4,5)P3 increase noted in these mutants occurs only within the inflorescence stems (Zhong et al., 2004). Similarly, alterations in the sac9 mutant, which result in elevated Ins(1,4,5)P3 and PtdInsP2 levels in the root only, do not result in ABA hypersensitivity (Williams et al., 2005). Thus, it appears that Ins(1,4,5)P3 increases may only be associated with ABA hypersensitivity when the second-messenger increase occurs within the majority of cells within the seedling.
Last, we note that signaling pathways besides the ABA-signaling pathway may utilize PtdInsPs and Ins(1,4,5)P3 as second messengers. The 5ptase mutants described here will provide useful tools to address this issue.
MATERIALS AND METHODS
Mutant Isolation
Arabidopsis (Arabidopsis thaliana) ecotype Columbia was used for all experiments. Growth conditions of soil-grown plants have been previously described (Berdy et al., 2001). Potential 5ptase1 (At1g34120) and 5ptase2 (At4g18010) mutants were identified from the Salk T-DNA lines (Alonso et al., 2003) through the analysis of the SiGnAL database (http://www.signal.salk.edu/cgi-bin/tdnaexpress). Seeds for 5ptase1-1 (SAIL_171A10), 5ptase1-2 (SALK_123083), and 5ptase2-1 (SAIL_138G01) were obtained from the Ohio State University Arabidopsis Biological Resource Center. Genomic DNA was isolated from soil-grown plant leaves. DNA from segregating plants was screened with PCR utilizing the SALK and SAIL LB primers 5′-TTCATAACCAATCTCGATACAC-3′ and 5′-GCGTGGACCGCTTGCTGCAACT-3′, respectively, and At5PTase gene-specific primers. To amplify At5PTase1, annealing at 55°C was performed with the LB primer and the following forward and reverse primers: 1for, 5′-ACTGGGCGCGTATTGTTCT-3′ and 1rev, 5′-ACTCGGTTTAAGGCATCACG-3′. For 5PTase2 amplification, a 60°C annealing temperature was used with the following forward and reverse primers: 2for, 5′-CAGACGGTGAAGTAAGAAAGC-3′ and 2rev, 5′-GCTCTTTGAAGTTTCCGGTGATCG-3′. The resulting PCR fragments were sequenced and compared to the genomic sequence for each gene to map the T-DNA insertion. In the case of 5ptase1-1, a second LB-forward primer PCR product was also apparent. The DNA from this fragment was sequenced to confirm that a second, truncated T-DNA was present in tandem in the 5PTase1 gene. To generate the 5ptase2-1/1-1 double mutant, homozygous 5ptase2-1 and 5ptase1-1 single mutants were crossed. F1 plants were self pollinated and individual F2 plants were screened for homozygous double mutants. These plants were self pollinated and double homozygous 5ptase2-1/1-1 plants were obtained in the F3 generation. We examined 87 seeds from our segregating F2 progeny from the 2-1 × 1-1 cross to determine the frequency of increased hypocotyl growth in the dark. Of these F2 progeny, 43/87 (0.49%) developed hypocotyls longer than 7 mm after 3 d. The expected frequency of increased hypocotyl growth is 7/16 (0.44%) if the At5PTase1 and At5PTase2 genes are independent and nonadditive genes, which, when present in the recessive state, give rise to increased hypocotyl growth.
RT-PCR
Total RNA was extracted from leaf tissue of 5-week-old soil-grown wild-type and 5ptase mutant plants using the RNeasy plant mini kit (Qiagen). One microgram of total RNA was reverse transcribed using the Qiagen Omniscript RT kit according to the manufacturer's instructions. Approximately one-tenth of the resulting mRNA eluate was used as template in each PCR reaction, which was prepared in a 25-μL mixture. Conditions for At5PTase1 and actin amplification have been described (Berdy et al., 2001; Burnette et al., 2003). Amplification of At5PTase2 was achieved using the same PCR conditions as for At5PTase1, only the annealing temperature was 60°C and the 2for and 2rev primers described previously were used. PCR products were separated by agarose gel electrophoresis and stained with ethidium bromide. To compare At5PTase1 and At5PTase relative expression levels, the intensity of PCR products was quantified with Quantity One software (Bio-Rad). Data are presented as the relative expression of individual At5PTase genes normalized to actin expression. Standard error was determined from three independent experiments. The 3′ end of At5PTase1 was amplified with a 3′for primer (5′-AGRGATCCCCCTTCTCCATC-3′) and a 3′rev primer (5′-CCATGGCGTCAAGGCCTTGAAT-3′) for 30 cycles and annealing temperature of 55°C resulting in a 1,315-bp fragment.
Seedling Growth and Seed Germination Assays
Age-matched seeds used for assays were harvested from plants grown in parallel on the same shelf in a growth room and seeds were harvested on the same day and ripened for 6 weeks at room temperature. Seeds were surface sterilized and plated on 0.5× Murashige and Skoog salt solution (pH 5.8) containing 0.8% agarose and 1% Suc. Seeds were stratified on plates at 4°C for 3 d and germinated at 23°C in the light or dark. Seedlings were removed from plates at the indicated times and hypocotyl measurements were made with photographs taken with a Zeiss dissecting microscope and a synchroscopy imaging system. To calculate the rate of hypocotyl elongation, the difference in length at the indicated time intervals was determined and divided by the number of hours in the interval. For root length measurements, seeds were sown in a straight line and, after stratification, plates were grown vertically. Measurements were made at 3 d of growth in the dark or light. For seed germination assays, age-matched seeds were sterilized and stratified as before and incubated in the light at 23°C for 12, 18, 24, or 36 h. Dormancy tests were performed with unstratified seeds. Germination was scored as positive when the radicle protruded through the seed coat. For ABA sensitivity experiments, ABA (Sigma) was dissolved in 100% ethanol and added to cooled, sterile medium at a final concentration of 0.25, 0.5, 1, or 2 μm ABA. Hormone treatment experiments were repeated three times.
ABA Measurements
Age-matched wild-type and 5ptase single- and double-mutant dry seeds were stored at room temperature or imbibed for 6 or 72 h at 4°C on filter paper. Seeds (approximately 10 mg for each sample) were harvested and ground in liquid nitrogen, transferred to a preweighed tube, vacuum dried, and reweighed to obtain the dry weight of the ground seeds. After the dry weight was determined, ABA was extracted from the powder by stirring overnight at 4°C in extraction solution containing 80% methanol, 100 mg/L butylated hydroxytoluene, and 0.5g/L citric acid monohydrate. The sample was centrifuged at 1,000g for 20 min. The supernatant was vacuum dried. The dry residue was dissolved in 100 μL of methanol plus 900 μL of Tris-buffered saline (50 mm Tris, 0.1 mm MgCl2, 6H2O, and 0.15 mm NaCl, pH 7.8). ABA levels in the samples were determined by competitive ELISA using the AGDIA immunodetection kit according to the manufacturer's instructions. The experiment was repeated twice.
Extraction and Measurement of Mass Ins(1,4,5)P3
Three-day-old seedlings grown on filter paper in the dark were used for the measurement of Ins(1,4,5)P3. Tissues were ground into a fine powder in liquid nitrogen. The fresh weight of the ground tissue was determined in preweighed tubes. For extraction of Ins(1,4,5)P3, 500 mg of freshly ground tissue were used. The procedure of extraction of tissue was as described by Burnette et al. (2003). Ins(1,4,5)P3 was measured using the InsP3 mass measurement kit (Amersham-Pharmacia Biotech) according to the manufacturer's protocol. The amount of Ins(1,4,5)P3 in the samples was interpolated from a standard curve generated with known amounts of Ins(1,4,5)P3. Assays were performed in duplicate and the experiment was repeated five times.
Extraction of InsPs and PtdInsPs
Seeds (20 mg) were surface sterilized, plated on filter paper, and placed in a 2-cm-deep petri dish. Plates were wrapped and kept in the dark at 4°C for 3 d and then grown for 2.5 d in the dark at 23°C; 150 μCi of myo-[2-3H]inositol (Perkin-Elmer) in 500 μL of sterile water was added and, after 12 h, 10 mL of a 5% ice-cold TCA was added to the filter paper to cover the growing seedlings. Seedlings were transferred to a 50-mL conical tube containing 3 mL of an ice-cold TCA solution and incubated for 1 h. The TCA solution (containing InsPs) was removed, dried in vacuo, and extracted once, using 1 n HCl:chloroform:methanol (1.33:1:1) and stored at −80°C for HPLC analysis. The seedlings were washed three times with distilled deionized water and then homogenized in 1 n HCl:chloroform:methanol (1.33:1:1). Homogenates were centrifuged and the bottom (organic) phase was recovered and aqueous phase reextracted once more with a 1:1 mixture of chloroform:methanol. The aqueous phase containing InsPs was dried in vacuo and resuspended in 100 μL of water for HPLC analysis. A detailed description of the lipid extraction has been described elsewhere (Hama et al., 2000, 2004). Briefly, the organic phase containing PtdInsPs was dried in vacuo and deacylated using 33% methylamine:methanol:n-butanol:water (32.3:45.7:11.4:10.4) by bath sonication. The deacylated glycerophosphoinositol head groups (gPtdIns) were extracted with 0.75 mL of water and 0.5 mL of an n-butanol:petroleum ether:ethyl formate (20:4:1) solution three times and dried in vacuo. After resuspension in water, a small aliquot of each TCA, aqueous, and organic fraction was assayed by liquid scintillation counting before HPLC analysis.
HPLC Analyses
Equal amounts of cpm from TCA, organic, and aqueous phase fractions were resolved by anion-exchange HPLC using a Beckman System Gold instrument (Beckman Instruments) equipped with a Partisil 10 SAX column (Whatman). An in-line Beckman 171 radioisotope detector and 32K software (Beckman) were used to determine the radioactivity present in each fraction. For both gPtdInsP and InsP analyses, the gradient condition was set for 5 mL of isocratic 10 mm ammonium phosphate (pH 3.8), followed by a linear gradient from 10 mm to 0.8 m in 60 mL (DeWald et al., 2001). Comigration with standards was used to verify peaks: [3H]Ins(1,3,4,5)P4, [3H]Ins(1,4,5)P3, 3H-Ins(1,4)P2, 3H-Ins, 3H-PtdIns(4,5)P2 (Perkin-Elmer), 14C-Ins(1)P, and 3H-PtdIns(4)P (American Radiolabeled Chemicals). Values from integrated areas of identified peaks were analyzed with Excel software (Microsoft), and expressed as arbitrary units. HPLC traces from the authors have been previously published (DeWald et al., 2001; Ercetin and Gillaspy, 2004). The average number of units found in wild-type samples was PtdIns(4,5)P2: 1,316,598; PtdIns(4)P: 24,651,285; PtdIns: 436,616,367; Ins(1,4,5)P3: 81,551; Ins(1,4)P2: 15,440,759; and InsP: 23,320,173. The fold increase of each molecular species in the mutant lines was calculated by comparison to the corresponding peak from the wild-type sample. The figures are representative of three independent replications.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Genevestigator expression analysis.
Supplementary Material
Acknowledgments
We are grateful to SIGnAL and the Arabidopsis Biological Resource Center for supplying mutant seeds and to Ryan Burnette and Rachel Kerwin for assistance in isolating mutants.
This work was supported by the U.S. Department of Agriculture (grant no. 2003–35318–13690 to G.E.G.) and by the Hatch project (grant no. VA–135583).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Glenda Gillaspy (gillaspy@vt.edu).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657 [DOI] [PubMed] [Google Scholar]
- Berdy S, Kudla J, Gruissem W, Gillaspy G (2001) Molecular characterization of At5PTase1, an inositol phosphatase capable of terminating IP3 signaling. Plant Physiol 126 801–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berridge MJ (1993) Inositol trisphosphate and calcium signaling. Nature 361 315–325 [DOI] [PubMed] [Google Scholar]
- Burnette RN, Gunesekera BM, Gillaspy GE (2003) An Arabidopsis inositol 5-phosphatase gain-of-function alters abscisic acid signaling. Plant Physiol 132 1011–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carland FM, Nelson T (2004) Cotyledon vascular pattern 2-mediated inositol (1,4,5) triphosphate signal transduction is essential for closed venation patterns of Arabidopsis foliar organs. Plant Cell 16 1263–1275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeWald DB, Torabinejad J, Jones CA, Shope JC, Cangelosi AR, Thompson JE, Prestwich GD, Hama H (2001) Rapid accumulation of phosphatidylinositol 4,5-bisphosphate and inositol 1,4,5-trisphosphate correlates with calcium mobilization in salt-stressed Arabidopsis. Plant Physiol 126 759–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ercetin ME, Gillaspy GE (2004) Molecular characterization of an Arabidopsis gene encoding a phospholipid-specific inositol polyphosphate 5-phosphatase. Plant Physiol 135 938–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erneux C, Govaerts C, Communi D, Pesesse X (1998) The diversity and possible functions of the inositol 5-polyphosphatases. Biochim Biophys Acta 1436 185–199 [DOI] [PubMed] [Google Scholar]
- Hama H, Takemoto JY, DeWald DB (2000) Analysis of phosphoinositides in protein trafficking. Methods 20 465–473 [DOI] [PubMed] [Google Scholar]
- Hama H, Torabinejad J, Prestwich GD, DeWald DB (2004) Measurement and immunofluorescence of cellular phosphoinositides. Methods Mol Biol 284 243–258 [DOI] [PubMed] [Google Scholar]
- Jones MA, Raymond MJ, Smirnoff N (2006) Analysis of the root-hair morphogenesis transcriptome reveals the molecular identity of six genes with roles in root-hair development in Arabidopsis. Plant J 45 83–100 [DOI] [PubMed] [Google Scholar]
- Kashem MA, Itoh K, Iwabuchi S, Hori H, Mitsui T (2000) Possible involvement of phosphoinositide-Ca2+ signaling in the regulation of alpha-amylase expression and germination of rice seed (Oryza sativa L.). Plant Cell Physiol 41 399–407 [DOI] [PubMed] [Google Scholar]
- Lee YL, Coi YB, Suh S, Lee JD, Assmann SM, Joe CO, Kelleher JF, Crain RC (1996) Abscisic acid-induced phosphoinositide turnover in guard cell protoplasts of Vicia faba. Plant Physiol 110 987–996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu HT, Gao F, Cui SJ, Han JL, Sun DY, Zhou RG (2006) Primary evidence for involvement of IP3 in heat-shock signal transduction in Arabidopsis. Cell Res 16 394–400 [DOI] [PubMed] [Google Scholar]
- Majerus PW, Kisseleva MV, Norris FA (1999) The role of phosphatases in inositol signaling reactions. J Biol Chem 274 10669–10672 [DOI] [PubMed] [Google Scholar]
- Ortega X, Perez LM (2001) Participation of the phosphoinositide metabolism in the hypersensitive response of Citrus limon against Alternaria alternata. Biol Res 34 43–50 [DOI] [PubMed] [Google Scholar]
- Perera IY, Heilmann I, Boss WF (1999) Transient and sustained increases in inositol 1,4,5-trisphosphate precede the differential growth response in gravistimulated maize pulvini. Proc Natl Acad Sci USA 96 5838–5843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera IY, Heilmann I, Chang SC, Boss WF, Kaufman PB (2001) A role for inositol 1,4,5-trisphosphate in gravitropic signaling and the retention of cold-perceived gravistimulation of oat shoot pulvini. Plant Physiol 125 1499–1507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera IY, Hung CY, Brady S, Muday GK, Boss WF (2005) A universal role for inositol 1,4,5-trisphosphate-mediated signaling in plant gravitropism. Plant Physiol 140 746–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiani R, Laoreti P (2000) Evidence for the involvement of phospholipase C in the anaerobic signal transduction. Plant Cell Physiol 41 1392–1396 [DOI] [PubMed] [Google Scholar]
- Sanchez JP, Chua NH (2001) Arabidopsis plc1 is required for secondary responses to abscisic acid signals. Plant Cell 13 1143–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shacklock PS, Read ND, Trewavas AJ (1992) Cytosolic free calcium mediated red light-induced photomorphogenesis. Nature 358 753–755 [Google Scholar]
- Shigaki T, Bhattacharyya MK (2000) Decreased inositol 1,4,5-trisphosphate content in pathogen-challenged soybean cells. Mol Plant Microbe Interact 13 563–567 [DOI] [PubMed] [Google Scholar]
- Stevenson JM, Perera IY, Heilmann II, Persson S, Boss WF (2000) Inositol signaling and plant growth. Trends Plant Sci 5 357. [DOI] [PubMed] [Google Scholar]
- Stevenson-Paulik J, Bastidas RJ, Chiou ST, Frye RA, York JD (2005) Generation of phytate-free seeds in Arabidopsis through disruption of inositol polyphosphate kinases. Proc Natl Acad Sci USA 102 12612–12617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor CW, Thorn P (2001) Calcium signalling: IP3 rises again…and again. Curr Biol 11 R352–355 [DOI] [PubMed] [Google Scholar]
- Williams ME, Torabinejad J, Cohick E, Parker K, Drake EJ, Thompson JE, Hortter M, Dewald DB (2005) Mutations in the Arabidopsis phosphoinositide phosphatase gene SAC9 lead to overaccumulation of PtdIns(4,5)P2 and constitutive expression of the stress-response pathway. Plant Physiol 138 686–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong L, Lee B, Ishitani M, Lee H, Zhang C, Zhu JK (2001) FIERY1 encoding an inositol polyphosphate 1-phosphatase is a negative regulator of abscisic acid and stress signaling in Arabidopsis. Genes Dev 15 1971–1984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, Burk DH, Morrison WH, Ye ZH (2004) FRAGILE FIBER3, an Arabidopsis gene encoding a type II inositol polyphosphate 5-phosphatase, is required for secondary wall synthesis and actin organization in fiber cells. Plant Cell 16 3242–3259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, Ye ZH (2004) Molecular and biochemical characterization of three WD-repeat-domain-containing inositol polyphosphate 5-phosphatases in Arabidopsis thaliana. Plant Cell Physiol 45 1720–1728 [DOI] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W (2004) GENEVESTIGATOR: Arabidopsis microarray database and analysis toolbox. Plant Physiol 136 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.