Abstract
Nitrogen (N) available to plants mostly originates from N2 fixation carried out by prokaryotes. Certain cyanobacterial species contribute to this energetically expensive process related to carbon (C) metabolism. Several filamentous strains differentiate heterocysts, specialized N2-fixing cells. To understand how C and N metabolism are regulated in photodiazotrophically grown organisms, we investigated the role of sucrose (Suc) biosynthesis in N2 fixation in Anabaena sp. PCC 7120 (also known as Nostoc sp. PCC 7120). The presence of two Suc-phosphate synthases (SPS), SPS-A and SPS-B, directly involved in Suc synthesis with different glucosyl donor specificity, seems to be important in the N2-fixing filament. Measurement of enzyme activity and polypeptide levels plus reverse transcription-polymerase chain reaction experiments showed that total SPS expression is greater in cells grown in N2 versus combined N conditions. Only SPS-B, however, was seen to be active in the heterocyst, as confirmed by analysis of green fluorescent protein reporters. SPS-B gene expression is likely controlled at the transcriptional initiation level, probably in relation to a global N regulator. Metabolic control analysis indicated that the metabolism of glycogen and Suc is likely interconnected in N2-fixing filaments. These findings suggest that N2 fixation may be spatially compatible with Suc synthesis and support the role of the disaccharide as an intermediate in the reduced C flux in heterocyst-forming cyanobacteria.
Nitrogen (N) is the fourth most abundant element in the biosphere. An estimated 80% to 90% of N available to plants in the terrestrial ecosystems originates from the biological conversion of nitrogen gas (N2) to ammonia, an energetically expensive process linked to carbohydrate metabolism (Ludden and Barris, 1986). There are only a few prokaryotic microorganisms, including certain free-living or symbiotically associated bacteria and cyanobacteria that carry out N2 assimilation. They do so through the action of the essentially anaerobic enzyme nitrogenase, an activity that requires reductants and ATP (Peters and Meeks, 1989; Crawford et al., 2000).
Diazotrophic cyanobacteria are the only organisms able to simultaneously and independently fix N2 and produce photosynthetic molecular oxygen. Because nitrogenase is inhibited upon exposure to oxygen, different strains have adaptations that include either temporal or spatial separation of these processes (Berman-Frank et al., 2003). Some filamentous diazotrophic strains can differentiate a photosynthetic vegetative cell into a heterocyst through a variety of structural, biochemical, and genetic changes that allow active nitrogenase (Buikema and Haselkorn, 1991; Wolk, 2000; Yoon and Golden, 2001). To maintain a microaerobic environment, heterocysts have a thick envelope, an oxygen-producing deactivated PSII complex, and active respiration to scavenge any residual oxygen (Wong and Meeks, 2001).
Because heterocysts lack ribulose-1,5-diphosphate carboxylase, a key enzyme of the Calvin cycle, they are limited to heterotrophic metabolism and depend on vegetative cells for the generation of carbon (C) skeletons and reducing power (Wolk, 1968; Wolk et al., 1994; Zhang et al., 2006). The precise C compounds transported from vegetative cells into heterocysts remain to be definitively identified, although several carbohydrates, including Fru, erythrose, and Suc, have been suggested (Privalle and Burris, 1984; Schilling and Ehrnsperger, 1985). To elucidate the structure of the carrier molecule, Schilling and Ehrnsperger (1985) investigated the localization of Suc metabolism enzymes in Anabaena variabilis. They concluded that Suc synthase (SuS; EC 2.4.1.13) was responsible for Suc synthesis in vegetative cells, whereas alkaline invertase, which hydrolyzes Suc into hexoses, was suggested to be present almost exclusively in heterocysts. Based on this, it has been proposed that Suc might be the C transport molecule within the filament cells (Wolk et al., 1994).
Suc metabolism in Anabaena sp. PCC 7119 and PCC 7120 (also known as Nostoc sp. PCC 7120) has recently been elucidated. It has been demonstrated that Suc is synthesized through two different Suc-P synthases (SPS; EC 2.4.1.14) coupled to Suc-P phosphatase (SPP; EC 3.1.3.24). Suc can either be cleaved by SuS or irreversibly hydrolyzed by two alkaline/neutral invertases (A/N-Inv) when there is high demand for hexoses (Porchia and Salerno, 1996; Cumino et al., 2001, 2002; Curatti et al., 2002; Vargas et al., 2003). Curatti et al. (2002, 2006) showed SuS to be involved in the cleavage of Suc only in vegetative cells in vivo. Besides, in contrast to a previous report, it has recently been shown that A/N-Invs are also expressed in vegetative cells (Schilling and Ehrnsperger, 1985; Curatti et al., 2002; Vargas et al., 2003).
Studies on the relationship between C and N metabolism in heterocyst-forming cyanobacteria have focused on the role of glycogen in N2 fixation (Ernst and Böger, 1985; Jensen et al., 1986; Ernst et al., 1990). During the light phase, most cyanobacteria strains accumulate a high level of glycogen, which is then mobilized to provide reductants and ATP during the night in either the vegetative cells or the heterocysts. N2 fixation can, therefore, also take place at night, even if at a much lower rate (Fay, 1976; Lockau et al., 1978). Glycogen synthesis occurs through ADP-Glc donation of glucosyl for elongation of an α-1,4-glucosidic chain. It is mainly regulated at the level of ADP-Glc synthesis catalyzed by ADP-Glc pyrophosphorylase (AGPase; EC 2.7.7.27), the enzyme encoded by the agp gene. AGPase activity was shown to be allosterically regulated by 3-phosphoglycerate (activator) and inorganic phosphate (Pi; inhibitor) in Anabaena sp. PCC 7120 (Ballicora et al., 2003). Whereas the role of glycogen in N2 fixation has been examined, the role of Suc metabolism still remains to be understood.
The critical role of Suc in C flux modulation in the N2-fixing filaments of Anabaena sp. was recently demonstrated by Curatti et al. (2002), who showed that diazotrophic growth was impaired in a mutant strain overexpressing SuS. Curatti et al. (2006) later showed that expression of SuS and Rubisco, a key enzyme in CO2 fixation during photosynthesis, is similarly down-regulated by a N source-dependent developmental program in the heterocysts.
The presence of two SPSs (SPS-A and SPS-B) with different glucosyl donor specificity (Porchia and Salerno, 1996; Cumino et al., 2002) raised the question of whether each may play a distinct role in Anabaena N2-fixing filaments. We found that, whereas both SPSs contribute to Suc synthesis in vegetative cells, only SPS-B is active in heterocysts. Expression and metabolic analyses were integrated into a model in which a Suc cycle linked to glycogen metabolism and respiration may play a crucial role in the functional support of active heterocysts during the light.
RESULTS
SPS Expression Is Higher in Anabaena Cells Grown in N2 Than in Combined N Conditions
To clarify the physiological role of Suc metabolism in heterocyst-forming cyanobacteria, we built on previous work analyzing the expression of enzymes involved in Suc biosynthesis. We showed that total SPS and SPP are only present in soluble protein fractions (Fig. 1) as reported in plants (Winter and Huber, 2000). Using permeabilized Anabaena filaments, we demonstrated that Suc synthesis occurs under light as well as dark conditions (Fig. 2A).
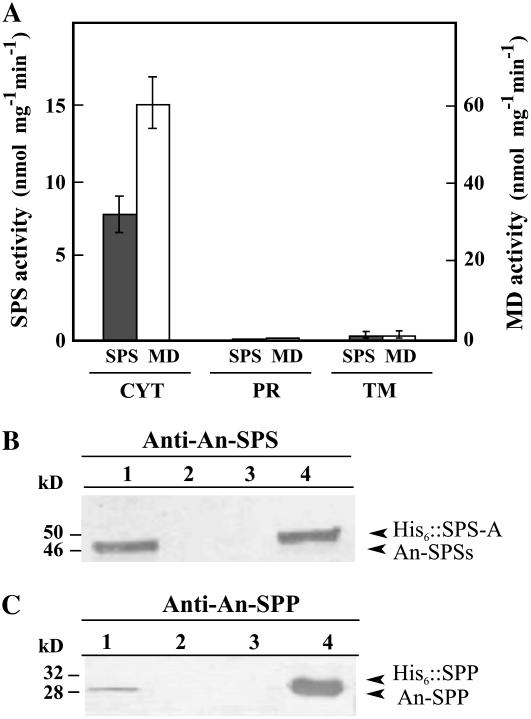
Figure 1.
Subcellular localization of Suc biosynthesis enzymes in Anabaena sp. PCC 7120 cells. A, SPS activity from cytoplasmic (CYT), periplasmic (PR), and total membrane (TM) protein extracts. Malate dehydrogenase (MD) activity was used as the cytoplasmic marker. SPS activity was assayed in the presence of UDP-Glc. Data are the mean ± se of five independent experiments. B and C, Immunoblotting of protein extracts. Lanes 1 to 3, CYT, PR, and TM, respectively. Lane 4, Control proteins (His6∷SPS-A [50 kD] or His6∷SPP [32 kD]). Polypeptides were revealed with anti-An-SPS (B) or anti-An-SPP (C). Approximately 200 and 1.5 μg of protein were loaded on lanes 1 to 3 and 4, respectively. Positions of molecular mass markers are indicated on the left (kD); arrowheads indicate the positions of native Anabaena SPSs (An-SPSs, approximately 46 kD), SPP (An-SPP, approximately 28 kD), and the recombinant proteins.
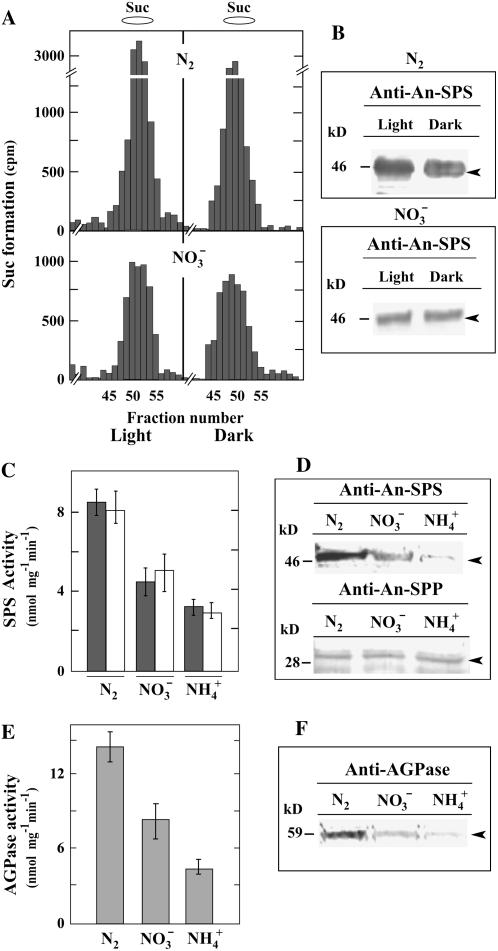
Figure 2.
Effect of N source on SPS, SPP, and AGPase expression in Anabaena sp. PCC 7120 cells. A, Effect of light on the synthesis of Suc. Cells grown in N2-fixing (N2) or nitrate (NO3−) environment (top/bottom) were harvested during midlight or middark, permeabilized with toluene, and incubated with UDP-[U-14C]Glc and Fru-6P. Labeled products were chromatographically separated and radioactivity determined in each fraction. The position of standard Suc is indicated at the top. B, Immunoblot analysis of proteins (100 μg/lane) from cells grown in N2 (top) or NO3− (bottom) revealed with anti-An-SPS. Cells were harvested in the midlight or middark. C to F, Cells grown in N2, NO3−, or ammonium (NH4+) assimilation conditions and harvested during midlight. C, SPS activity assayed in the presence of UDP-Glc (dark gray bars) or ADP-Glc (white bars). Data are the mean ± se of five independent experiments. D, Immunoblot analysis using anti-An-SPS (top, 100 μg/lane) or anti-An-SPP (bottom, 40 μg/lane). Positions of molecular mass markers indicated on the left (kD); arrowheads indicate position of Anabaena SPSs or SPP. E, AGPase activity. Data are the mean ± se of three independent experiments. F, Immunoblot analysis (40 μg/lane) revealed with anti-An-AGPase.
This study aimed to relate Suc synthesis to N2 fixation. The experiments were performed in illuminated cells because most N2 fixation takes place under light conditions (Stal, 2003). Analysis of the effect of N availability on SPS expression showed that total enzyme activity was maximal in N2-fixing cells (Fig. 2, A and C). Immunoanalysis of the SPS polypeptide level showed a similar pattern of SPS expression (Fig. 2, B and D). Enzyme activity was also higher in the presence of ADP-Glc (SPS-B activity) in cells grown in a diazotrophic versus a N combined environment (Fig. 2C).
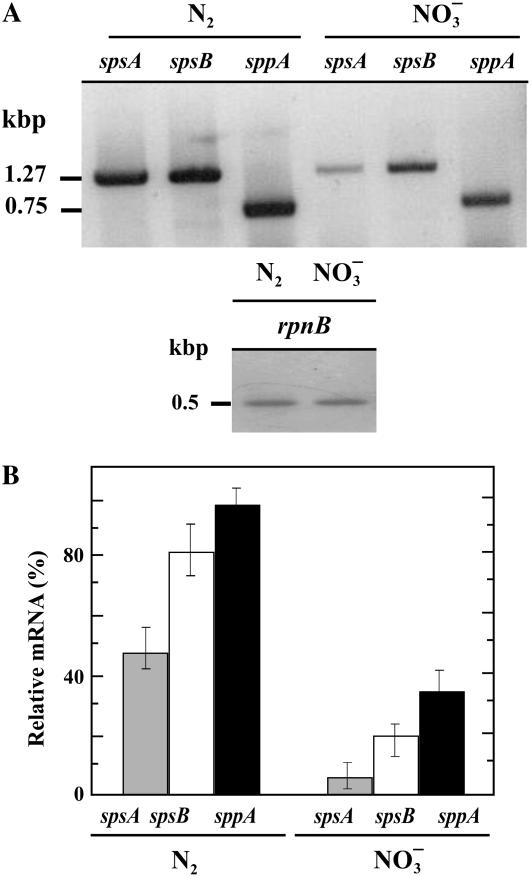
Transcriptional analysis of spsA and spsB, determined by reverse transcription (RT)-PCR, likewise showed maximal transcript level in N-deficient cells for both genes; the total amount in N-deficient cells was approximately 4 times higher than in the combined N-grown cells (Fig. 3). It was also found that spsB mRNA is likely to be predominant during N2 fixation. Independent of the N source, the level of sppA transcripts was in accordance with the increase in sps gene expression (Fig. 3). SPP polypeptide levels, however, were similar in the different N sources provided in the medium (Fig. 2D).
Figure 3.
Expression of spsA, spsB, and sppA in Anabaena sp. PCC 7120 cells grown in N2 or NO3−. A, RT-PCR analysis from total RNA. Amplification of Anabaena rpnB used as loading control. Position of molecular size markers indicated on the left. B, Densitometry of mRNA levels corresponding to detection in A. Values are the mean ± se of five independent experiments.
In contrast to the high Suc-synthesizing capacity seen in N2-fixing filaments, the Suc content of the N-2deficient-grown cells was approximately one-half that of the combined N-grown cells (6–10 nmol mg fresh weight−1). Because both UDP-Glc and ADP-Glc can be substrates for Anabaena SPS reactions, expression of the enzymes responsible for their synthesis was also investigated. It was seen that UDP-Glc pyrophosphorylase (UGPase; EC 2.7.7.9) activity was similar in cells grown in N2 and in combined N (approximately 55 nmol UDP-Glc min−1 mg−1), whereas a higher AGPase activity was seen in N2-fixing cells (Fig. 2E). A similar expression pattern was noted in polypeptide level and mRNA content (Fig. 2F; data not shown).
Modeling the Suc Network in N2-Fixing Anabaena Filaments
To contribute to an understanding of the interconnection between Suc synthesis and the presence of nucleoside diphosphates, we performed product inhibition assays for the two Anabaena SPSs and modeled C flux under diazotrophic conditions. The formal model was built using the Gepasi framework (Mendes, 1997). The simulation included information from experimental kinetic data obtained in this study from Anabaena sp. in conjunction with values reported in the literature.
Involvement of the two SPSs in Suc production and the relationship between Suc and glycogen metabolism in Anabaena cells diazotrophically grown in the light were investigated through metabolic control analysis (Kacser and Burns, 1973; Heinrich and Rapoport, 1974). Because this study could provide a quantitative depiction of C flow through the two competing metabolic pathways, we included the main reactions of both metabolisms (Fig. 4). The evaluated fluxes are shown in Table I and Figure 4. Fluxes were designated as positive when their net direction was forward (left to right in the equation in Table I) and negative when their direction was backward (right to left in the equation).
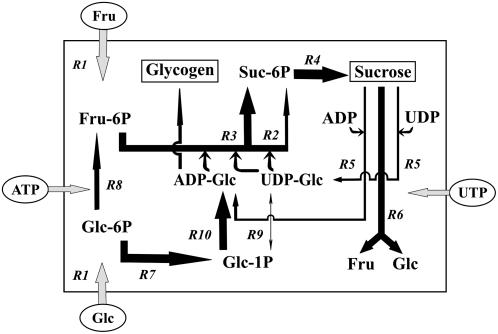
Figure 4.
Metabolic flux map of primary and intermediate reactions involved in Suc and glycogen metabolism in a model of illuminated N2-fixing Anabaena filaments. Arrow widths are proportional to flux values; arrowhead indicates direction of net flux. Each flux is assigned for a number of reactions (R1–R10) indicated in Table I. The intracellular space contained is framed. Metabolites (Fru, Glc, ATP, and UTP) taken up from the extracellular medium are shown in ovals and sink final products (glycogen and Suc) in rectangles.
Table I.
Steady-state properties of the kinetic model of Suc and glycogen metabolism of diazotrophically cultivated Anabaena sp. PCC 7120 cells
| Reaction Name | Vmax | Km or Ki | Net Flux | Comment | Reference | |
|---|---|---|---|---|---|---|
| nmol mg protein−1 min−1 | mm | nmol min−1 mg fresh weight−1 | ||||
| R1: Hexokinase | 8.8 | Km Glc | 0.2 | 1.050 | Clamped concentrations of ATP and ADP | This study; Smith and Moore (1981) |
| Hexose + ATP → hexose-P + ADP | Km Fru | 7.5 | ||||
| Ki ATP | 1.0 | |||||
| R2: SPS-A | 2.8 | Km Fru-6P | 0.4 | 0.090 | Biosynthesis of Suc-6P | This study; Cumino et al. (2002) |
| UDP-Glc + Fru-6P → Suc-6P + UDP | Km UDP-Glc | 1.3 | ||||
| Ki UDP | 2.0 | |||||
| R3: SPS-B | 1.05 | Km Fru-6P | 2.5 | 0.490 | Biosynthesis of Suc-6P | This study; Cumino et al. (2002) |
| XDP-Glc + Fru-6P → Suc-6P + XDP | Km ADP-Glc | 3.0 | ||||
| (X = U or A) | Km UDP-Glc | 4.0 | ||||
| Ki ADP | 4.0 | |||||
| R4: SPP | 4.4 | Km Suc-6P | 0.35 | 0.580 | Net production of Suc | Cumino et al. (2001) |
| Suc-6P → Suc + Pi | Ki Suc | 80 | ||||
| R5: SuS | 3.5 | Km Fru/Km UDP-Glc | 52/2.7 | −0.090 | Flux is in direction of Suc cleavage | Porchia et al. (1999) |
| XDP-Glc + Fru → Suc + XDP | 0.7 | Km Fru/Km ADP-Glc | 4.2/1.3 | |||
| (X = U or A) | 1.8 | Km Suc/Km UDP | 303/1.25 | |||
| 0.7 | Km Suc/Km ADP | 305/1.15 | ||||
| R6: INV | 1.8 | Km Suc | 10 | 0.295 | Irreversible Suc hydrolysis | Vargas et al. (2003) |
| Suc + water → Glc + Fru | ||||||
| R7: Hexose-P mutase | 2.8 | Km Glc-1P | 0.2 | −0.490 | Flux is in direction of Glc-1P production | Pearce and Carr (1969) |
| Glc-1P → Glc-6P | ||||||
| R8: Hexose-P isomerase | 4.2 | Km Fru-6P | 0.25 | 0.190 | Flux is in direction of Fru-6P production | Pearce and Carr (1969) |
| Glc-6P → Fru-6P | Km Glc-6P | 5 | ||||
| R9: UGPase | 52 | Km Glc-1P/Km UTP | 0.1/0.3 | 0.000 | The mass action ratio equals the Keq and the change in free energy is zero at steady state. Under these conditions, the net flux is null. | This study |
| UDP-Glc + PPi → Glc-1P + UTP | 130 | Km UDP-Glc/Km PPi | 0.1/0.2 | |||
| R10: AGPase | 7.8 | Km Glc-1P/Km ATP | 0.07/0.1 | |||
| ADP-Glc + PPi → Glc-1P + ATP | 24 | Km ADP-Glc/Km PPi | 0.4/0.03 | −0.490 | Flux is in direction of ADP-Glc production, involved in glycogen and Suc biosynthesis | Ballicora et al. (2003) |
In all cases, the fluxes and steady-state concentrations of the variable metabolites calculated by the model were in agreement with experimental determinations. Negative fluxes were observed for SuS, hexose-P mutase, and AGPase. This indicated that the reactions catalyzed by these enzymes may operate in vivo in a direction opposite to that outlined in Table I. The flux through SuS should have been toward the cleavage of Suc and the flux through hexose-P mutase toward the formation of Glc-1P, a substrate for the nucleoside-triphosphate-hexose-1P nucleotidyltransferases (AGPase and UGPase). The UGPase reaction operated close to equilibrium, as consistent with previous findings (Roscher et al., 1998; Kleczkowski et al., 2004). The simulation showed the SPS-B flux (0.490 nmol min−1 mg fresh weight−1) to be approximately 5 times higher than the SPS-A flux. The sum of the two fluxes agrees with SPP flux, accounting for the entire Suc biosynthesis capacity.
SPS-A was inhibited by UDP (approximately Ki UDP of 2.0 ± 0.5 mm). This supports the hypothesis that the preferred substrate for this enzyme is UDP-Glc. On the other hand, SPS-B was inhibited only by ADP, whether UDP-Glc or ADP-Glc (approximately Ki ADP of 1.8 ± 0.3 and 3.9 ± 0.5 mm, respectively) served as the glucosyl donor. When the SPS-A reaction was removed from the model, the net flux through SuS was interrupted (−5 × 10−11 nmol min−1 mg fresh weight−1), whereas the fluxes through invertases and UGPase remained unaltered (0.350 and 6 × 10−11 nmol min−1 mg fresh weight−1, respectively).
Differential Expression Patterns of spsA and spsB in Anabaena N2-Fixing Filaments Show That Only SPS-B Is Involved in Heterocyst Metabolic Activity
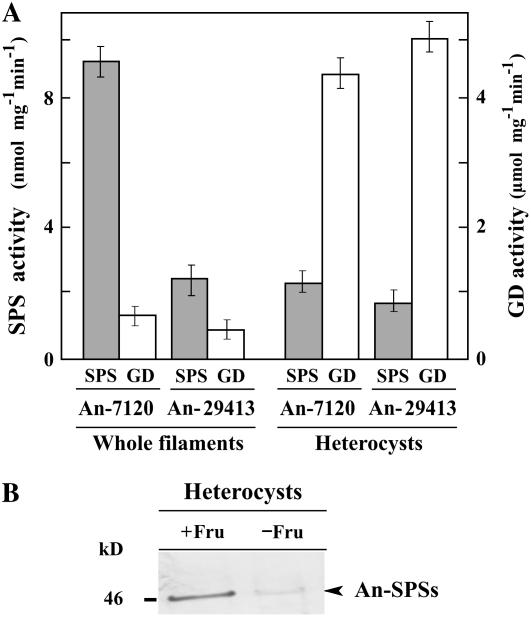
Immunoblot analysis and enzyme activity measurements showed that SPS is present not only in the vegetative cells, but also in the heterocysts of two Anabaena strains (Fig. 5). This was confirmed by incorporation of labeled C into Suc from UDP-[U-14C]Glc (approximately 3,000 cpm min−1 mg fresh weight−1) or ADP-[U-14C]Glc (approximately 2,500 cpm min−1 mg fresh weight−1) into permeabilized heterocysts in Anabaena sp. PCC 7120 in the presence of Fru-6P. Furthermore, only spsB transcripts, which are accompanied by sppA expression, were detected in the heterocysts (Fig. 6A).
Figure 5.
SPS expression in two Anabaena strains grown in N2-fixing conditions. A, SPS activity (gray bars) assayed in whole filaments or heterocyst extracts of Anabaena sp. PCC 7120 (An-7120) and A. variabilis ATCC 29413 (An-29413). Glc-6P dehydrogenase (GD) activity (white bars) was included as a heterocyst marker. Results are the mean ± se (n = 5). B, Immunoblot analysis of SPS expression from heterocyst extracts of Anabaena cells grown in N-deprived conditions with (+Fru) or without (−Fru) 10 mm Fru.
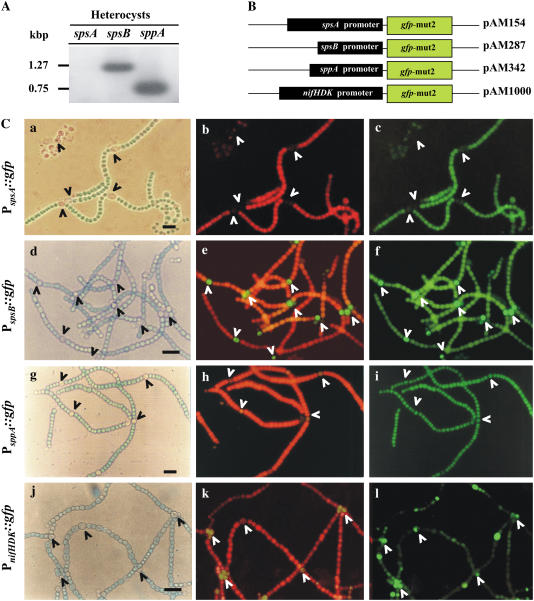
Figure 6.
Cellular localization of spsA, spsB, and sppA expression in N2-fixing filaments of Anabaena sp. PCC 7120. A, RT-PCR analysis from total heterocyst RNA. B, Schematic representation showing a physical map of the GFP fusion constructs. The gfp sequence was fused in frame with upstream sequences of the translational start codon of spsA, spsB, sppA, and nifHDK. C, Cellular expression of PspsA∷gfp, PspsB∷gfp, and PsppA∷gfp reporter transcriptional fusion in Anabaena sp. PCC 7120 filaments subjected to prolonged diazotrophic growth. Each horizontal set of photographs corresponds with the same microscopic field. Contrast photomicrographs were obtained in full-spectrum light (a, d, g, and j). Chlorophyll fluorescence micrographs were taken without the emission filter (b, e, h, and k). GFP fluorescence micrographs were obtained with the corresponding emission filter (c, f, i, and l). Arrowheads indicate heterocysts. All microphotos taken at the same magnification (1,000×). Images photographed and processed with same settings to allow qualitative comparison of fluorescence intensities. Scale bars, 10 μm.
The localization of SPS-A and SPS-B in N2-fixing filament cells was also evidenced with transcriptional fusions of an optimized version of the green fluorescent protein (GFP) gene (gfp-mut2) to putative promoters of spsA, spsB, and sppA (PspsA, PspsB, and PsppA, respectively). DNA fragments of 950, 477, and 559 bp upstream from the translation start site of spsA, spsB, and sppA, respectively, directed the expression of gfp- mut2 in Anabaena sp. PCC 7120 (Fig. 6B). In diazotrophically grown Anabaena cells, spsA expression was detected only in vegetative cells (Fig. 6C, a–c), whereas spsB and sppA expression was localized to both vegetative cells and heterocysts (Fig. 6C, d–i). Anabaena sp. PCC 7120 cells harboring a plasmid (pAM1000) in which a heterocyst cell-specific promoter (PnifHDK) was fused to gfp is shown as control (Fig. 6C, j–l). No fluorescence signal was detected in Anabaena cells containing a control plasmid (pAMpmt−) carrying a promoterless gfp construct (data not shown).
Transcriptional Regulation of spsB under N2 Fixation
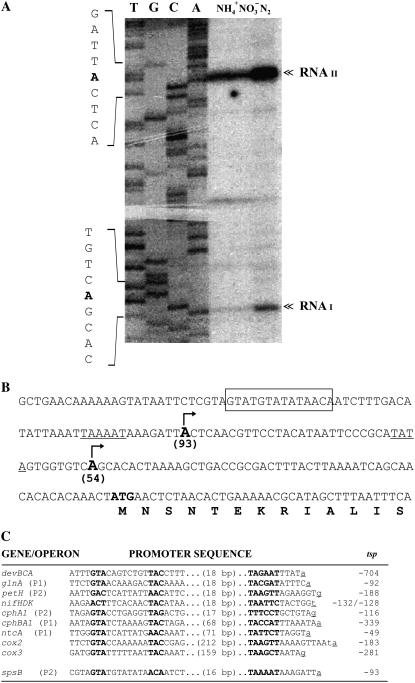
The finding that only spsB is expressed in the heterocysts of N2-fixing filaments led to investigation of possible mechanisms for its transcriptional regulation. This was undertaken through determination of the RNA 5′ ends that correspond to the transcription start points (tsps), mapped by primer extension of RNA obtained from Anabaena sp. PCC 7120 grown under different N conditions. Two RNA sizes (RNAI and RNAII) were observed, starting at the −54 (tspI) and −93 (tspII) nucleotides upstream of the translation initiation site (Fig. 7). Each tsp was confirmed through similar experiments conducted with two different oligonucleotides. RNAI and RNAII were detected under the three N culture conditions assayed, but spsB expression was greater in cells grown in N2 than in combined N (Fig. 7A). The extension product of RNAII was, however, 4 times more abundant than that of RNAI in N2-fixing cells. Analysis of the upstream sequences of the two putative promoters showed a −10 box similar to those of Escherichia coli σ70 promoters (Fig. 7B). Because spsB expression seemed to be regulated by the N source, we went on to investigate in the two putative promoters the possible presence of putative binding sites of N control A (NtcA), a DNA-binding protein involved in the control of transcription of N-regulated genes in cyanobacteria (Herrero et al., 2004). Similar to the consensus sequence motif GTA(N8)TAC described for N2-activated genes (Herrero et al., 2001), the sequence GTA(N8)ACA was found at −39 with respect to tspII (Fig. 7, B and C).
Figure 7.
Origins of spsB transcription and analysis of the putative promoter regions. A, Primer extension mapping of the tsps of spsB carried out with total RNA (30 μg) from Anabaena cells grown in different N sources and the oligonucleotides B-ol-tsp3. A similar result was obtained with B-ol-tsp4 (data not shown). The sequencing ladders presented (lanes T, G, C, and A) were generated with the same primers used in the primer extension reactions. Arrowhead points to the extension product identifying the putative tsps. B, Nucleotide sequence of the spsB upstream region. The tsps (arrows) and start translation codon are in bold. Numbers in parentheses indicate the relative position from the translation start. A sequence similar to the consensus NtcA-binding sequence is boxed. The −10 box regions are underlined. C, Sequence alignments of NtcA-activated promoters (P) whose products act during differentiation and in the mature Anabaena heterocyst (devBCA, ATP-binding cassette transporter; glnA, Gln synthetase; petH, ferredoxin-NADP reductase; nifHDK, nitrogenase complex; cphA1, cyanophycin synthetase; cphBA1, cyanophycinase; ntcA, N regulator; cox2 and cox3, terminal respiratory oxidases). Location of the tsps with respect to the tsp of the corresponding gene is indicated. The consensus for NtcA-binding site and −10 hexamers is in bold.
DISCUSSION
A close relationship between Suc metabolism and the N2 fixation process has been described in legume-Rhizobium symbiosis. Degradation of Suc imported from the plant by SuS is a key initial step in the development and normal function of the nodule and maintenance of nitrogenase activity (Gordon et al., 1999). Suc biosynthesis, however, is unlikely to take place in the heterotrophic nodule. A different scenario of N2 fixation is the filament of diazotrophic cyanobacteria, where Suc metabolism has been described (Curatti et al., 2002) and Suc cleavage takes place only in photosynthetic vegetative cells (Curatti et al., 2006). This study focused on the fact that Suc biosynthesis occurs in N2-fixing filaments of Anabaena sp. PCC 7120, modeling the C flux between Suc and glycogen. The presence of an active SPS in the heterocysts (SPS-B) was uncovered in addition to a specific isoform in the vegetative cells (SPS-A), both of which relate to the dynamic mechanism underlying N2 fixation.
Although Suc biosynthesis is higher in diazotrophically grown filaments than in cells grown under combined N conditions (Figs. 2 and 3), a very small amount of Suc accumulation is detectable due to a high rate of disaccharide degradation (Curatti et al., 2002; Vargas et al., 2003; Table I). This notable finding of Suc turnover may provide additional support for the role of Suc as an intermediate in the flux of reduced C in the N2-fixing filament (Wolk et al., 1994; Curatti et al., 2002). However, contrary to previous reports (Schilling and Ehrnsperger, 1985), we found that Suc biosynthesis can take place not only in vegetative cells, but also in heterocysts. Unlike plant root nodules, heterocysts are able to synthesize the disaccharide (Figs. 5 and 6), as well as import C from adjacent cells, as was similarly described to occur in heterotrophic plant tissues (Roscher et al., 1998; Babb and Haigler, 2001; Im, 2004).
Whereas the presence of one SPS gene is likely to be characteristic of unicellular cyanobacterium strains, filamentous N2-fixing cyanobacteria were reported to have two genes (spsA and spsB) present (Curatti et al., 1998; Cumino et al., 2002; Salerno and Curatti, 2003). Each gene, however, is transcribed in Anabaena independently of the N2 fixation process. The two genes are simultaneously expressed in diazotrophically and combined N-grown vegetative cells (Figs. 3 and 6). The possibility that the two SPS isoforms fulfill distinct metabolic functions was explored by examining the distribution of Suc biosynthesis genes in the Anabaena sp. PCC 7120 genome. Interestingly, spsA and sppA are located in the same genome half (the 265°–85° region); meanwhile, spsB is found in the other genome half (85°–265° region), reported by Sugaya et al. (2004) to contain most of the protein genes performing housekeeping functions essential to cyanobacteria and genes that code for proteins with functions needed in a particular environment (such as for N2 fixation and metabolism under N deprivation conditions, respectively).
Because control of spsB expression may occur at transcriptional initiation (Fig. 7), primer extension experiments were conducted. These indicated that the putative spsB promoter inferred from tspII shares a sequence similar to that of promoters activated by NtcA. The latter is a cAMP receptor protein family transcription factor, which, in the absence of ammonium, promotes the expression of alternative N source assimilation genes (Herrero et al., 2001). Suc biosynthesis may thus be coordinated with N assimilation by NtcA. SPS-B, similarly to the products of NtcA-activated genes, may be involved in heterocyst differentiation, development, and function (Herrero et al., 2004). Transcriptional regulation and the absence of allosteric modulation by Glc-6P and Pi are likely to be a particular feature of cyanobacterial SPSs (Porchia and Salerno, 1996; Curatti et al., 1998; Desplats et al., 2005).
Experimental data from this study point to the interconnection between glycogen and Suc metabolism in Anabaena N2-fixing filaments. Glycogen is produced and stored only during the day, serving as the predominant metabolic fuel at night (Stal and Moezelaar, 1997), but Suc is biosynthesized in both light or darkness (Fig. 2, A and B). Glycogen may, therefore, be the C source for disaccharide formation at night. Chen et al. (2005) recently reported that SPS performs a crucial function in plants in synthesizing Suc during starch mobilization at night. On the other hand, the considerable accumulation of glycogen in Anabaena during N2 fixation in the light (Ernst and Böger, 1985) is in accordance with a high level of AGPase activity (Fig. 2, E and F). This study, however, found that AGPase flux alone, as calculated by metabolic simulation, is insufficient to supply the ADP-Glc needed for glycogen synthesis and Suc production through SPS-B (Table I; Fig. 4). A concomitant production of ADP-Glc might also be ascribed to Suc cleavage by SuS in the vegetative cells (Table I) in keeping with this enzyme's reported role (Porchia et al., 1999; Curatti et al., 2002, 2006). A null flux through SuS when the SPS-A reaction was removed from the metabolic model was also seen, leading us to speculate that the activity of the two enzymes may be connected in the vegetative cells of the N2-fixing filament. Building on the recent report by Curatti et al. (2006), our findings suggest that susA and spsA may be similarly down-regulated in the developmental heterocyst differentiation program.
Suc synthesis in the heterocyst could be paralleled to Suc synthesis in heterotrophic plant tissues. The heterocyst, however, is very distinct from cotton (Gossypium hirsutum) fibers, etiolated hypocotyls, or germinating seeds, for example, in which Suc pool regulation is modulated by simultaneous degradation and resynthesis and Suc cleavage by SuS plays a central role (Geigenberger and Stitt, 1991; Geigenberger et al., 1999; Nguyen-Quoc and Foyer, 2001). As Anabaena heterocysts lack SuS activity (Curatti et al., 2006), Suc degradation occurs solely via A/N-Inv, which Böhme (1998) proposed necessary for C supply, maintenance of a high respiration rate, and provision of the C acceptors for ammonia required for N2 fixation.
Substrate cycling involving carbohydrate turnover in plant tissues has been widely reported, but its mechanisms and functions remain poorly understood (Alonso et al., 2005). Our results suggest that a Suc-cycling mechanism may be operating in heterocysts, allowing cell metabolism to shift easily from Suc production to degradation, through A/N-Inv, hexokinase, hexose-P mutase, hexose-P isomerase, AGPase, SPS-B, and SPP activity. Although the precise metabolic role of the Suc cycles involving SPS/Inv has not yet been determined, several functions in plants have been proposed (Roscher et al., 1998; Geigenberger et al., 1999). It has been suggested that Suc cycles allow a pathway's net flux a high degree of sensitivity to respond to factors modulating rates of synthesis and degradation, controlling respiration, maintaining osmotic potential, controlling sugar accumulation, and promoting sugar signaling (Rohwer and Botha, 2001; Roby et al., 2002). Whereas we have been able to demonstrate the operational feasibility of Suc cycles, further work will be needed to elucidate Suc cycle function in the heterocyst.
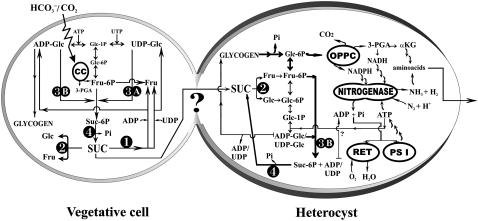
Our findings are depicted in the model in Figure 8. Suc translocated from the vegetative cells into the heterocysts may be the respiratory substrate (Wolk et al., 1994; Herrero et al., 2004), after hydrolysis to Glc and Fru by a high level of invertase activity (Schilling and Ehrnsperger, 1985; Curatti et al., 2002). The resulting hexoses, after phosphorylation by hexokinases, can enter the oxidative pentose-P cycle and respiration to provide NADPH and C skeletons for ammonium assimilation and act as glycogen synthesis precursors. Suc resynthesis by SPS-B may take place in the heterocysts, likely from glycogen degradation products, contributing to an operational Suc-cycling mechanism. It may be that SPS-B is contributing to the ADP pool and thus to the high ADP-ATP ratio reported in Anabaena heterocysts (Privalle and Burris, 1983) and to modulate nitrogenase activity through the inhibition of proteolytic degradation of the dinitrogenase reductase subunit (Durner et al., 1996).
Figure 8.
Schematic diagram of Suc pathway in N2-fixing Anabaena filament cells. Suc enzyme localization in Anabaena sp. PCC 7120 illuminated filaments based on this study and Curatti et al. (2006). Photosynthetic C fixation through the Calvin cycle (CC) occurs in the vegetative cells and could lead to Suc and glycogen biosynthesis. Heterocysts act as an important sink for carbohydrates from vegetative cells and as a source of fixed N (Wolk et al., 1994). In heterocysts, which could also synthesize glycogen and Suc, the reductants for N2 and O2 reduction are generated by the activity of the oxidative pentose-P cycle (OPPC), the NADPH heterocyst-specific ferrodoxin, and respiratory electron transport (RET), as well as the ATP synthesis by cyclic phosphorylation (PSI). Suc enzymes are indicated as (1) SuS; (2) A/N-Inv; (3A) SPS-A; (3B) SPS-B; and (4) SPP. αKG, α-Ketoglutarate.
Ongoing work is focusing on creating spsA and spsB mutant strains to further pinpoint the role of Suc biosynthesis in the N2-fixing filaments. Future studies on the role of Suc during the early and later stages of heterocyst differentiation should help to bring some light on the physiological role of Suc in the N source-dependent development of these specialized cells.
MATERIALS AND METHODS
Biological Material and Culture Conditions
Axenic cultures of Anabaena sp. strain PCC 7120 and Anabaena variabilis ATCC 29413 were grown photoautotrophically under a 12-h light/12-h dark cycle with white fluorescent light at 30°C with orbital shaking. Media used for growth were BG11 (prepared with KNO3 instead of NaNO3), BG110 (BG11 medium lacking nitrate), and BG110NH4+/TES [BG11 medium lacking nitrate and supplemented with 2.5 mm NH4Cl and 5 mm N-tris(hydroxymethyl) methyl-2-aminoethane sulfonic acid (TES-KOH buffer, pH 7.5); Curatti et al., 2002]. Cells were harvested at the late-exponential phase by centrifugation during the midlight or middark period, washed, and either stored at −80°C or toluene permeabilized. For strains containing the PspsA∷gfp, PspsB∷gfp, and PnifHDK∷gfp transcriptional reporters (plasmids pAM154, pAM287, and pAM1000, respectively), neomycin at 25 and 100 μg mL−1 was added to liquid and solid media, respectively (Yoon and Golden, 2001).
Escherichia coli strains were grown at 37°C in Luria-Bertani liquid or agar medium supplemented with appropriate antibiotics according to standard protocols (Sambrook and Russell, 2001). E. coli BL21(λDE3):pLysS strain (Novagen) was used for the overproduction of SPS-A and SPP proteins of Anabaena sp. PCC 7120, as previously reported by Cumino et al. (2001, 2002).
Cell Fractionation and Heterocyst Isolation
Membrane and periplasm proteins were purified from 2 L of Anabaena sp. PCC 7120 cultures, according to Norling et al. (1998) and Zhu et al. (1999), respectively. Heterocysts were isolated from diazotrophically grown cells of A. variabilis sp. ATCC 29413 and Anabaena sp. PCC 7120, according to Zhou et al. (1998), with or without Fru in the medium (Jensen et al., 1986). Heterocyst preparations were monitored by light microscopy and used once heterocyst purity was approximately greater than 95% of the total cell number. Quantification of proteins was done using Bradford's dye-binding assay.
Protein Extraction and Immunoblot Analysis
Total cell extracts from Anabaena vegetative cells and heterocysts were prepared as previously reported (Cumino et al., 2001) and desalted through Sephadex G-50 columns. Proteins were separated by SDS-PAGE using a MiniProtean system (Bio-Rad) and five-well combs (0.75 mm) on 12% or 15% polyacrylamide gels for SPS/AGPase and SPP analyses, respectively (Cumino et al., 2001). Polypeptides were visualized with Coomassie Blue or electroblotted onto nitrocellulose membranes (HyBond C; Amersham), which were probed with the polyclonal antibodies anti-An-SPS, anti-An-AGPase, or anti-An-SPP and raised in rabbits against His6∷SPS-A (Cumino et al., 2002), His6∷AGPase (this study), or native SPP purified from Anabaena cells (Cumino et al., 2001), respectively.
Enzyme Assays
Because SPS-A and SPS-B show different substrate specificity (both SPSs accept UDP-Glc as the glucosyl donor, but only SPS-B uses ADP-Glc), total SPS activity was assayed in a reaction mixture containing Fru-6P and UDP-Glc, and SPS-B activity was carried out in the presence of ADP-Glc (Porchia and Salerno,1996). SPS-B catalytic efficiency with ADP-Glc is twice that with UDP-Glc (Porchia and Salerno, 1996). SuS, SPP, and A/N-Inv activity were measured as previously reported (Porchia et al., 1999; Cumino et al., 2001; Vargas et al., 2003). SPP activity could not be measured in crude extracts, as unspecific phosphatases interfered with the assay. Glc-6P dehydrogenase activity, used as an enrichment marker in heterocyst preparation (30- to 60-fold higher specific activity in heterocysts versus vegetative cells), was estimated by monitoring the Glc-6P-dependent increase in NADPH absorbance (Summers et al., 1995). Malate dehydrogenase activity, used as a cytoplasmic marker, was assayed in the direction of oxalacelate reduction (Reng et al., 1993). UGPase and AGPase activity were determined in either the pyrophosphorolysis or the synthesis direction, as described in Sowokinos et al. (1993) and Ballicora et al. (2003). Hexokinase, hexose-P isomerase, and hexose-P mutase were assayed according to Pearce and Carr (1969).
The Ki values for UDP and ADP (the enzyme-inhibitor dissociation constant for product-competitive inhibition) for SPS-A and SPS-B were determined with homogeneous preparations of each isoenzyme (Porchia and Salerno, 1996; Cumino et al., 2002). Anabaena hexokinase and UGPase kinetic parameters were determined in partially purified enzyme fractions, following a purification procedure similar to that described by Porchia and Salerno (1996).
Permeabilization of Anabaena Cells and in Situ Suc Synthesis
Filaments of Anabaena sp. PCC 7120 grown autotrophically in BG110 or BG11 medium were permeabilized as previously described (Salerno et al., 2004). The permeabilization of heterocysts was performed according to Bottomley et al. (1980). In situ Suc synthesis was determined in the assay mixture (50 μL total volume) containing 5 mm UDP-[U-14C]Glc or ADP-[U-14C]Glc (specific activity 2 × 105 cpm μmol−1 or 4.4 × 105 cpm μmol−1, respectively), 10 mm Fru-6P, 100 mm HEPES-NaOH (pH 7.5), 10 mm MgCl2, and an aliquot of toluene-treated cells. Labeled Suc was separated by paper chromatography and quantified as previously described (Porchia and Salerno, 1996).
Carbohydrate and Metabolite Determination
Suc and glycogen were determined in ethanol extracts as described by Geigenberger et al. (1996). Suc content was quantified by measuring Fru and Glc after hydrolysis with acid invertase using a coupled-enzyme method (Puebla et al., 1997). For glycogen determination, pellets of the ethanol extraction were solubilized by heating to 95°C in 0.1 n NaOH for 30 min. After acidification to pH 4.9 with an HCl/sodium-acetate mixture (pH 4.9), part of the suspension was digested overnight with amyloglucosidase and amylase. The intermediate metabolites, Fru-6P, Glc-6P, Glc-1P, UDP-Glc, and Suc-6P, were determined in perchloric acid extracts using an enzymatic cycling assay (Salerno et al., 1996). Enzymes and chemicals for biochemical analyses were obtained from Boehringer and Sigma-Aldrich.
Metabolic System Delineation
Control of the steady-state behavior of the Suc-glycogen metabolism relationship in Anabaena N2-fixing filaments was determined through metabolic control analysis, as developed by Kacser and Burns (1973) and Heinrich and Rapoport (1974). The capabilities of this mathematical model are not limited to enzymes obeying the Michaelis-Menten formulation, but can also be extended to complex rate expressions. The enzymatic reactions simulated in this model respond to kinetic mechanisms different from those described in a previous maize (Zea mays) leaf model (Barreiro, 1999). This method also allows introduction of flux inhibition and activation mechanisms into the model.
To restrict the model to reactions directly involved in Suc synthesis and degradation, we omitted the Calvin cycle and pentose-P pathway, the major pathway of cyanobacteria Glc catabolism (Summers et al., 1995). Elementary flux modes were calculated using METATOOL software (Pfeiffer et al., 1999). Steady-state calculation of the kinetic models was performed using Gepasi version 3.30 (Mendes, 1997).
The kinetic parameters and thermodynamic data for all enzymes used in the model are summarized in Table I. Kinetic constant values were either taken from the literature, where available, or estimated, as indicated in Table I. The reactions in this model were assumed reversible unless information on irreversibility in vivo had been reported (Rohwer and Botha, 2001). The fixed extracellular metabolite concentrations were 10 mm Fru, 10 mm Glc, 5 mm ATP, 5 mm UTP, and 2 mm 3-phosphoglycerate. The model was validated and its performance assessed against independent experimental observations of the determination of final products (Suc and glycogen) and intermediate metabolites (Fru-6P, Glc-6P, Glc-1P, UDP-Glc, and Suc-6P). Maximal enzyme activity, metabolite concentration, and flux value were determined. For metabolic flux measurements, Anabaena sp. PCC 7120 cells were grown in BG110 medium and harvested by centrifugation in the midlight phase. Data were obtained from independent experiments conducted in triplicate. Cells were disrupted by sonication and incubated with extracellular fixed metabolites. Metabolites and enzyme activities were measured in aliquots of the same sample taken at six different time points.
Isolation, Manipulation, and Analysis of Nucleic Acids
Plasmids were isolated and modified according to standard protocols (Sambrook and Russell, 2001). Genomic DNA was isolated from cyanobacteria as previously described (Cai and Wolk, 1990). RNA from Anabaena was obtained according to Vargas et al. (2003). RNA quality and PCR products were analyzed by electrophoresis in 1% agarose gels.
RT-PCR
For RT-PCR analysis, total RNA treated with DNase (RQ1 RNase-free DNase; Promega) was reverse transcribed using Moloney murine leukemia virus reverse transcriptase (Promega) and specific primers for spsA, spsB, and sppA (Cumino et al., 2001, 2002). The primers designed for AGPase gene detection were agp-f (5′-CGGGATCCATGAAAAAAGTCTTAGCAATTATTCT-3′) and agp-r (5′-CGGAATTCGCTAAATGATTGTGCCATCTGTAATAAC-3′). PCR reactions were run on a PTC-100 thermal cycler (model-96V; MJ Research) for 25 cycles of 94°C (1 min), 50°C (1 min), and 72°C (1 min) plus a single step at 72°C for 5 min. To determine the optimal amount of input RNA, the 3-fold diluted template RNA was amplified in RT-PCR assays under identical reaction conditions to construct a standard curve for each gene product. Once the optimal amount of input RNA was determined for each gene product, RT-PCR was carried out under identical reaction conditions to detect differential transcript levels of genes in the distinct N source cultures. Under these conditions, PCR amplification occurs in the linear range. Amplification of the encoding gene of the subunit of ribonuclease P (rpnB) was used as an internal control in RT-PCR determinations, as described by Zhu et al. (2001). Control reactions were incubated at 30°C for 30 min in the presence of RNaseA to ascertain RNA dependence on the RT-PCR signals. Control reactions were incubated at 30°C for 30 min in the presence of RNaseA to ascertain RNA dependence on the RT-PCR signals. Blots were quantified using an imaging analyzer (Fotodyne model express zoom lens system) and its dedicated software (TotalLab image analysis software).
Construction of GFP Fusion Reporters
To construct the spsA promoter∷gfp (PspsA∷gfp), spsB promoter∷gfp (PspsB∷gfp), sppA promoter∷gfp (PsppA∷gfp), and nifHDK promoter∷gfp (PnifHDK∷gfp) transcriptional reporters, three DNA fragments containing upstream sequences of the translational start codons of spsA, spsB, sppA, and nifHDK were amplified from Anabaena sp. PCC 7120 genomic DNA by PCR using the primers spsA-f (5′-GTCTTTTGATATATACTTTCTGCATAGCTT-3′) and spsA-r (5′-ACCACCGGCTTCTTCTTGACCTATTTC-3′); spsB-f (5′-GCTATAGTTGTGATTTCTTACGTATAT-3′) and spsB-r (5′-TGCCCTCCAGCTTCTTCTTTGCCAATTT-3′); sppA-f (5′-CCGTCGGGAATCTGAAATAAAGCGTATA-3′) and sppA-r (5′-TACTCTGGTAGGTAATGATGCAGCCCTGGC-3′); and nif-f (5′-CTGAGACTGCACATCAAGGTAGAAG-3′) and nif-r (5′-TGTTCTCTTTTCCTGCAATTGGTTG-3′).
Amplification products were of 950, 477, 559, and 1,000 bp, respectively. Each DNA fragment was ligated to pGEM-T Easy (Promega). After digestion with EcoRI, each resulting DNA fragment was ligated into a shuttle vector derived from pAM505 and pKEN2-GFPmut2 (source of the gfp-mut2 sequence) in frame with the GFP coding sequence (Yoon and Golden, 2001). The resulting conjugal plasmids, pAM154, pAM287, pAM342, and pAM1000, containing spsA-gfp, spsB-gfp, sppA-gfp, and nifHDK-gfp transcriptional fusions, respectively, were confirmed by restriction digestions and DNA sequencing. Plasmid pAM1000 was used as a control for localization of GFP in heterocysts and plasmid pAMC486, containing the large subunit of Rubisco gene promoter (PrbcL), was used to detect GFP expression in vegetative cells (Yoon and Golden, 2001; Curatti et al., 2006). Strains carrying the promoterless gfp construct pAMpmt− were used as negative controls. Plasmids were transferred by conjugation into Anabaena sp. PCC 7120 based on the method described by Elhai and Wolk (1988). Plasmids pAM505, pKEN2-GFPmut2, and pAMC486 were kindly provided by Dr. James W. Golden (Texas A&M University).
Fluorescence Analysis and Microscopy
Cells grown in BG110 were used for photography. Fluorescence and phase contrast micrographs were taken on a Nikon microscope (model E600) with an X100 objective using specific emission filter sets. GFP fluorescence images were taken by illumination with light (450- to 490-nm wavelength) and photographing emission through a filter of 510-nm wavelength narrow band pass with a 3-s exposure. Red emission from photosynthetic pigments was photographed without the 510-nm filter. Images were captured with a Nikon digital camera (model E995) attached to the microscope and processed with Adobe Photoshop version 4.0 software. The presence of a thick cell envelope, changes in cytoplasm granularity, cyanophycin granulate formation at the cell poles, and absence of chlorophyll fluorescence distinguished heterocysts from vegetative cells (Elhai and Wolk, 1990).
Primer Extension Mapping of tsps
Primer extension experiments were performed according to Sambrook and Russell (2001) from total RNA (30 μg) and using SuperScript II RNase H− reverse transcriptase (Invitrogen) and oligonucleotides B-ol-tsp3 (5′-TTTGTTCAGCATTTACATTTAATCTCTTTA-3′), B-ol-tsp4 (5′-TGCCCTCCAGCTTCTTCTTTGCCAATTT-3′), covering positions −185 to −155 and +61 to +89, with respect to the spsB translational start, respectively. Primers were end labeled with [γ-32P]ATP using T4 polynucleotide kinase (Invitrogen). cDNA products were purified and resolved on a sequencing gel along sequencing ladders generated using the same primers as those used in the primer extension experiments. Nucleotide sequencing was carried out using the dideoxy-chain termination method with a Sequenase quick-denature plasmid sequencing kit (USB Corporation).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers AJ302071 and AJ302073.
Acknowledgments
We are grateful to Dr. James W. Golden at Texas A&M University, College Station, Texas, for facilitating our use of the vectors pAM505 and pKEN-GFPmut2; Dr. Horacio G. Pontis and our colleagues at the Centro de Investigaciones Biológicas, Fundación para Investigaciones Biológicas Aplicadas, Mar del Plata, Argentina, for many helpful discussions; and C. Fernández for technical assistance.
This work was supported by the Agencia Nacional de Promoción Científica y Tecnológica, Consejo Nacional de Investigaciones Científicas y Tecnológicas, Fundación para Investigaciones Biológicas Aplicadas, and Universidad Nacional de Mar del Plata, Argentina.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Graciela L. Salerno (gsalerno@fiba.org.ar).
Open Access articles can be viewed online without a subscription.
References
- Alonso AP, Vigeolas H, Raymond P, Rolin D, Dieuaide-Noubhani M (2005) A new substrate cycle in plants: evidence for a high glucose-phosphate-to-glucose turnover from in vivo steady-state and pulse-labeling experiments with [13C]glucose and [14C]glucose. Plant Physiol 138 2220–2232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babb VM, Haigler CH (2001) Sucrose phosphate synthase activity rises in correlation with high-rate cellulose synthesis in three heterotrophic systems. Plant Physiol 127 1234–1242 [PMC free article] [PubMed] [Google Scholar]
- Ballicora MA, Iglesias AA, Preiss J (2003) ADP-glucose pyrophosphorylase, a regulatory enzyme for bacterial glycogen synthesis. Microbiol Mol Biol Rev 67 213–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barreiro R (1999) Metabolic control analysis of carbon pathways. PhD thesis. UMI Dissertation Services, Ann Arbor, MI
- Berman-Frank I, Lundgren P, Falkowski P (2003) Nitrogen fixation and photosynthetic oxygen evolution in cyanobacteria. Res Microbiol 154 157–164 [DOI] [PubMed] [Google Scholar]
- Böhme H (1998) Regulation of nitrogen fixation in heterocyst-forming cyanobacteria. Trends Plant Sci 3 346–351 [Google Scholar]
- Bottomley PJ, van Baalen C, Tabita FR (1980) Heterocyst differentiation and tryptophan metabolism in the cyanobacterium Anabaena sp CA. Arch Biochem Biophys 203 204–213 [DOI] [PubMed] [Google Scholar]
- Buikema WJ, Haselkorn R (1991) Isolation and complementation of nitrogen fixation mutants of the cyanobacterium Anabaena sp. strain PCC 7120. J Bacteriol 173 1879–1885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y, Wolk CP (1990) Use of a conditional lethal gene in Anabaena sp. PCC 7120 to select for double recombinants and to entrap insertion sequences. J Bacteriol 172 3138–3145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Hajirezaei M, Börnke F (2005) Differential expression of sucrose-phosphate synthase isoenzymes in tobacco reflects their functional specialization during dark-governed starch mobilization in source leaves. Plant Physiol 139 1163–1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford NM, Kahn ML, Leustek T, Long SR (2000) Nitrogen and sulfur. In BB Buchanan, W Gruissem, RL Jones, eds, Biochemistry and Molecular Biology of Plants. American Society of Plant Physiologists, Rockville, MD, pp 786–849
- Cumino A, Curatti L, Giarrocco L, Salerno GL (2002) Sucrose metabolism: Anabaena sucrose-phosphate synthase and sucrose-phosphate phosphatase define minimal functional domains shuffled during evolution. FEBS Lett 517 19–23 [DOI] [PubMed] [Google Scholar]
- Cumino A, Ekeroth C, Salerno GL (2001) Sucrose-phosphate phosphatase from Anabaena sp. strain PCC 7120: isolation of the protein and gene revealed significant structural differences from the higher-plant enzyme. Planta 214 250–256 [DOI] [PubMed] [Google Scholar]
- Curatti L, Flores E, Salerno G (2002) Sucrose is involved in the diazotrophic metabolism of the heterocyst-forming cyanobacterium Anabaena sp. FEBS Lett 513 175–178 [DOI] [PubMed] [Google Scholar]
- Curatti L, Folco E, Desplats P, Abratti G, Limones V, Herrera-Estrella L, Salerno GL (1998) Sucrose-phosphate synthase from Synechocystis sp. PCC 6803: identification of the spsA gene and characterization of the enzyme expressed in E. coli. J Bacteriol 180 6776–6779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curatti L, Giarrocco L, Salerno GL (2006) Sucrose synthase and RuBisCo expression is similarly regulated by the nitrogen source in the nitrogen-fixing cyanobacterium Anabaena sp. Planta 223 891–900 [DOI] [PubMed] [Google Scholar]
- Desplats P, Folco E, Salerno GL (2005) Sucrose may play an additional role to that of an osmolyte in Synechocystis sp. PCC 6803 salt-shocked cells. Plant Physiol Biochem 43 133–138 [DOI] [PubMed] [Google Scholar]
- Durner J, Böhm I, Knörzer OC, Böger P (1996) Proteolytic degradation of dinitrogenase reductase from Anabaena variabilis (ATCC 29413) as a consequence of ATP depletion and impact of oxygen. J Bacteriol 178 606–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elhai J, Wolk CP (1988) Conjugal transfer of DNA to cyanobacteria. Methods Enzymol 167 747–754 [DOI] [PubMed] [Google Scholar]
- Elhai J, Wolk CP (1990) Developmental regulation and spatial pattern of expression of the structural genes for nitrogenase in the cyanobacterium Anabaena. EMBO J 9 3379–3388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst A, Böger P (1985) Glycogen accumulation and the induction of nitrogenase activity in the heterocyst-forming cyanobacterium Anabaena variabilis. J Gen Microbiol 131 3147–3153 [Google Scholar]
- Ernst A, Reich S, Böger P (1990) Modification of dinitrogenase reductase in the cyanobacterium Anabaena variabilis due to C starvation and ammonia. J Bacteriol 172 748–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay P (1976) Factors influencing dark nitrogen fixation in a blue-green alga. Appl Environ Microbiol 31 376–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geigenberger P, Lerchl J, Stitt M, Sonnewald U (1996) Phloem-specific expression of pyrophosphatase inhibits long distance transport of carbohydrates and amino acids in tobacco plants. Plant Cell Environ 19 43–55 [Google Scholar]
- Geigenberger P, Reimholz R, Deiting U, Sonnewald U, Stitt M (1999) Decreased expression of sucrose phosphate synthase strongly inhibits the water stress-induced synthesis of sucrose in growing potato tubers. Plant J 19 119–129 [DOI] [PubMed] [Google Scholar]
- Geigenberger P, Stitt M (1991) Regulation of carbon partitioning between sucrose and nitrogen assimilation in cotyledons of germinating Ricinus communis. Planta 185 563–568 [DOI] [PubMed] [Google Scholar]
- Gordon AJ, Minchin FR, James CL, Komina O (1999) Sucrose synthase in legume nodules is essential for nitrogen fixation. Plant Physiol 120 867–878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrich R, Rapoport TA (1974) A linear steady-state treatment of enzymatic chains: critique of the crossover theorem and a general procedure to identify interaction sites with an effector. Eur J Biochem 42 97–105 [DOI] [PubMed] [Google Scholar]
- Herrero A, Muro-Pastor AM, Flores E (2001) Nitrogen control in cyanobacteria. J Bacteriol 183 411–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrero A, Muro-Pastor AM, Valladares A, Flores E (2004) Cellular differentiation and the NtcA transcription factor in filamentous cyanobacteria. FEMS Microbiol Rev 28 469–487 [DOI] [PubMed] [Google Scholar]
- Im KH (2004) Expression of sucrose-phosphate synthase (SPS) in non-photosynthetic tissues of maize. Mol Cells 17 404–409 [PubMed] [Google Scholar]
- Jensen BB, Cox RP, Burris RH (1986) Isolation of cyanobacterial heterocysts with high and sustained dinitrogen-fixation capacity supported by endogenous reductants. Arch Microbiol 145 241–247 [DOI] [PubMed] [Google Scholar]
- Kacser H, Burns JA (1973) The control of flux. Symp Soc Exp Biol 27 65–104 [PubMed] [Google Scholar]
- Kleczkowski LA, Geisler M, Ciereszko I, Johansson H (2004) UDP-glucose pyrophosphorylase an old protein with new tricks. Plant Physiol 134 912–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockau W, Peterson RB, Wolk CP, Burris RH (1978) Modes of reduction of nitrogen in heterocysts isolated from Anabaena species. Biochim Biophys Acta 502 298–308 [DOI] [PubMed] [Google Scholar]
- Ludden P, Barris RH (1986) Biochemical basis of plant breeding. In CA Neyra, ed, Nitrogen Metabolism, Vol 2. CRC Press, Boca Raton, FL, pp 41–59
- Mendes P (1997) Biochemistry by numbers: simulation of biochemical pathways with Gepasi 3. Trends Biochem Sci 22 361–363 [DOI] [PubMed] [Google Scholar]
- Nguyen-Quoc B, Foyer CH (2001) A role for “futile cycles” involving invertase and sucrose synthase in sucrose metabolism of tomato fruit. J Exp Bot 52 881–889 [DOI] [PubMed] [Google Scholar]
- Norling B, Zak E, Andersson B, Pakrasi H (1998) 2D-isolation of pure plasma and thylakoid membranes from the cyanobacterium Synechocystis sp. PCC 6803. FEBS Lett 436 189–192 [DOI] [PubMed] [Google Scholar]
- Pearce J, Carr N (1969) The incorporation and metabolism of glucose by Anabaena variabilis. J Gen Microbiol 54 451–462 [DOI] [PubMed] [Google Scholar]
- Peters GA, Meeks JC (1989) The Azolla-Anabaena symbiosis: basic biology. Annu Rev Plant Physiol Plant Mol Biol 40 193–210 [Google Scholar]
- Pfeiffer T, Sanchez-Valdenebro I, Nuno JC, Montero F, Schuster S (1999) METATOOL: for studying metabolic networks. Bioinformatics 15 251–257 [DOI] [PubMed] [Google Scholar]
- Porchia AC, Curatti L, Salerno GL (1999) Sucrose metabolism in cyanobacteria: sucrose synthase form Anabaena sp. strain PCC 7119 is remarkably different from the plant enzymes with respect to substrate affinity and amino-terminal sequence. Planta 210 34–40 [DOI] [PubMed] [Google Scholar]
- Porchia AC, Salerno GL (1996) Sucrose biosynthesis in a prokaryotic organism: presence of two sucrose-phosphate synthases in Anabaena with remarkable differences compared with the plant enzymes. Proc Natl Acad Sci USA 93 13600–13604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Privalle LS, Burris RH (1983) Adenine nucleotide levels in and nitrogen fixation by the cyanobacterium Anabaena sp. strain 7120. J Bacteriol 154 351–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Privalle LS, Burris RH (1984) D-erythrose supports nitrogenase activity in isolated Anabaena sp. strain 7120 heterocysts. J Bacteriol 157 350–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puebla AF, Salerno GL, Pontis HG (1997) Fructan metabolism in two species of Bromus subjected to chilling and water stress. New Phytol 136 123–129 [Google Scholar]
- Reng W, Riessland R, Scheibe R, Jaenicke R (1993) Cloning, site-specific mutagenesis, expression and characterization of full-length chloroplast NADP-malate dehydrogenase from Pisum sativum. Eur J Biochem 217 189–197 [DOI] [PubMed] [Google Scholar]
- Roby C, Cortes S, Gromova M, Le Bail JL, Roberts JK (2002) Sucrose cycling in heterotrophic plant cell metabolism: first step towards an experimental model. Mol Biol Rep 29 145–149 [DOI] [PubMed] [Google Scholar]
- Rohwer JM, Botha FC (2001) Analysis of sucrose accumulation in the sugar cane culm on the basis of in vitro kinetic data. Biochem J 358 437–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roscher A, Emsley L, Raymond P, Roby C (1998) Unidirectional steady state rates of central metabolism enzymes measured simultaneously in a living plant tissue. J Biol Chem 273 25053–25061 [DOI] [PubMed] [Google Scholar]
- Salerno G, Echeverria M, Pontis H (1996) Activation of sucrose-phosphate synthase by a protein factor/sucrose-phosphate phosphatase. Cell Mol Biol 42 665–672 [PubMed] [Google Scholar]
- Salerno GL, Curatti L (2003) Origin of sucrose metabolism in higher plants: when, how and why? Trends Plant Sci 8 63–69 [DOI] [PubMed] [Google Scholar]
- Salerno GL, Porchia AC, Vargas WA, Abdian PL (2004) Fructose-containing oligosaccharides: novel compatible solutes in Anabaena cells exposed salt stress. Plant Sci 167 1003–1008 [Google Scholar]
- Sambrook J, Russell DW (2001) Molecular Cloning: A Laboratory Manual, Ed 3. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Schilling N, Ehrnsperger K (1985) Cellular differentiation of sucrose metabolism in Anabaena variabilis. Z Naturforsch 40 776–779 [Google Scholar]
- Smith A, Moore J (1981) Transport of carbohydrates into cyanobacteria. Methods Enzymol 167 550–559 [Google Scholar]
- Sowokinos JR, Spychalla JP, Desborough SL (1993) Pyrophosphorylases in Solanum tuberosum (IV. Purification, tissue localization, and physicochemical properties of UDP-glucose pyrophosphorylase). Plant Physiol 101 1073–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stal L (2003) Smart modeling of unusual cyanobacteria—an enigma solved? New Phytol 160 455–462 [DOI] [PubMed] [Google Scholar]
- Stal LJ, Moezelaar R (1997) Fermentation in cyanobacterial. FEMS Microbiol Rev 21 179–211 [Google Scholar]
- Sugaya N, Sato M, Murakami H, Imaizumi A, Aburatani S, Horimoto K (2004) Causes for the large genome size in a cyanobacterium Anabaena sp. PCC7120. Genome Inform 15 229–238 [PubMed] [Google Scholar]
- Summers ML, Wallis JG, Campbell EL, Meeks JC (1995) Genetic evidence of a major role for glucose-6-phosphate dehydrogenase in nitrogen fixation and dark growth of the cyanobacterium Nostoc sp. strain ATCC 29133. J Bacteriol 177 6184–6194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargas W, Cumino A, Salerno GL (2003) Cyanobacterial alkaline/neutral invertases: origin of sucrose hydrolysis in the plant cytosol? Planta 216 951–960 [DOI] [PubMed] [Google Scholar]
- Winter H, Huber S (2000) Regulation of sucrose metabolism in higher plants: localization and regulation of activity of key enzymes. CRC Crit Rev Plant Sci 19 31–67 [DOI] [PubMed] [Google Scholar]
- Wolk CP (1968) Movement of carbon from vegetative cells to heterocysts in Anabaena cylindrica. J Bacteriol 96 2138–2143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolk CP, Ernst A, Elhai J (1994) Heterocyst metabolism and development. In DA Bryant, ed, Molecular Biology of Cyanobacteria. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 769–823
- Wolk PC (2000) Heterocyst formation in Anabaena. In YV Brun, LJ Shimkets, eds, Prokaryotic Development. American Society for Microbiology, Washington, DC, pp 83–104
- Wong FC, Meeks JC (2001) The hetF gene product is essential to heterocyst differentiation and affects HetR function in the cyanobacterium Nostoc punctiforme. J Bacteriol 183 2654–2661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon HS, Golden JW (2001) PatS and products of nitrogen fixation control heterocyst pattern. J Bacteriol 183 2605–2613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang CC, Laurent S, Sakr S, Peng L, Bedu S (2006) Heterocyst differentiation and pattern formation in cyanobacteria: a chorus of signals. Mol Microbiol 59 367–375 [DOI] [PubMed] [Google Scholar]
- Zhou R, Cao Z, Zhao J (1998) Characterization of HetR protein turnover in Anabaena sp. PCC 7120. Arch Microbiol 169 417–423 [DOI] [PubMed] [Google Scholar]
- Zhu J, Jäeger K, Black T, Zarka K, Koksharova O, Wolk CP (2001) HcwA, an autolysin, is required for heterocyst maturation in Anabaena sp. strain PCC 7120. J Bacteriol 183 6841–6851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z, Sun D, Davidson VL (1999) Localization of periplasmic redox proteins of Alcaligenes faecalis by a modified general method for fractionating gram-negative bacteria. J Bacteriol 181 6540–6542 [DOI] [PMC free article] [PubMed] [Google Scholar]