Abstract
Full-length cDNA corresponding to Arabidopsis (Arabidopsis thaliana) gene At2g31690, which has been annotated in GenBank as a putative triacylglycerol (TAG) lipase, was obtained by reverse transcription-polymerase chain reaction using RNA from senescing rosette leaves of Arabidopsis as a template. The cognate protein was found to contain the lipase active site sequence, and corresponding recombinant protein proved capable of deesterifying TAG. In vitro chloroplast import assays indicated that the lipase is targeted to chloroplasts. This was confirmed by confocal microscopy of rosette leaf tissue treated with fluorescein isocyanate-labeled, lipase-specific antibody, which revealed that lipase protein colocalizes with plastoglobular neutral lipids. Western-blot analysis indicated that the lipase is expressed in roots, inflorescence stems, flowers, siliques, and leaves and that it is strongly up-regulated in senescing rosette leaf tissue. Transgenic plants with suppressed lipase protein levels were obtained by expressing At2g31690 cDNA in antisense orientation under the regulation of a constitutive promoter. Transgenic plants bolted and flowered at the same time as wild-type plants, but were severely stunted and exhibited delayed rosette senescence. Moreover, the stunted growth phenotype correlated with irregular chloroplast morphology. The chloroplasts of transgenic plants were structurally deformed, had reduced abundance of thylakoids that were abnormally stacked, and contained more plastoglobular neutral lipids than chloroplasts of wild-type plants. These observations collectively indicate that this TAG lipase plays a role in maintaining the structural integrity of chloroplasts, possibly by mobilizing the fatty acids of plastoglobular TAG.
Analysis of the amino acid sequence of fatty acid-deesterifying lipases cloned from animals, bacteria, fungi, and higher plants has shown that they typically contain the 10-amino acid sequence [LIV]-X-[LIVAFY]-[LIAMVST]-G-[HYWV]-S-X-G-[GSTAC] (Derewenda and Derewenda, 1991). The Ser residue of this motif is a key element of the active site of fatty acid-deesterifying lipases (Brick et al., 1995).
Several types of fatty acid-deesterifying lipases, including phospholipase A1 and phospholipase A2, have been identified in plants. Phospholipase A2 catalyzes deesterification of the sn2 fatty acid of phospholipids, yielding free fatty acid and 1-acyl-2-lysophospholipid, and is thought to generate free linolenic acid substrate for the octadecanoid pathway (Grechkin, 1998). The oxylipin products of the octadecanoid pathway, in particular jasmonic acid and methyl jasmonate, regulate a number of cellular processes, including wound and defense responses (Dhondt et al., 2000; Schaller, 2001). Phospholipase A2 is also up-regulated during flower development (Kim et al., 1999; Lee et al., 2003) and, in addition, has been detected in lipid bodies, implicating its involvement in lipid mobilization during seed germination (May et al., 1998). Recently, the DEFECTIVE IN ANTHER DEHISCENCE1 (DAD1) gene in Arabidopsis (Arabidopsis thaliana) has been shown to encode phospholipase A1, an enzyme that catalyzes deesterification of the sn1 fatty acid of phospholipids (Ishiguro et al., 2001). Arabidopsis dad1 mutants show defects in anther dehiscence, pollen maturation, and flower opening. Inasmuch as this mutant phenotype can be rescued by exogenous application of jasmonic acid or linolenic acid, Ishiguro et al. (2001) have proposed that dad1 may catalyze the initial step in the octadecanoid pathway that leads to the synthesis of jasmonic acid. Another Arabidopsis phospholipase that is UV-B inducible also has properties that suggest possible involvement in the octadecanoid pathway (Lo et al., 2004).
Lipolytic acyl hydrolases, another class of fatty acid-deesterifying lipases, are also prevalent in plants. These lipases release fatty acids from a number of different substrates, including phospholipids and wax esters, and, in the case of phospholipids, deesterify at both the sn1 and sn2 positions (Galliard, 1980). A lipolytic acyl hydrolase gene that is up-regulated in senescing petals and is ethylene inducible has been cloned from carnation flowers (Hong et al., 2000). A senescence-inducible lipolytic acyl hydrolase (SAG101) has also been cloned from Arabidopsis, and overexpression of this gene induced precocious leaf senescence (He and Gan, 2002). SAG101 also appears to be involved in signaling that gives rise to pathogen immunity (Feys et al., 2005). A particularly interesting group of lipolytic acyl hydrolases is the patatin family. Patatins are members of a multigene family of vacuolar proteins, which constitute 40% of total soluble potato (Solanum tuberosum) tuber protein and are thought to be storage proteins (Racusen and Foote, 1980; Mignery et al., 1988). However, the functions of patatins are not well established, and it is increasingly apparent that, in addition to being a storage protein, patatin may function as a lipase. Potato patatin, for example, exhibits lipolytic acyl hydrolase activity (Galliard, 1971), and there is some evidence that patatin-like lipases are capable of deesterifying unsaturated fatty acids from membrane lipids that serve as the initial substrate for the octadecanoid pathway (Dhondt et al., 2000; Holk et al., 2002).
Galactolipase functions in the same way as lipolytic acyl hydrolase, with the exception that it specifically cleaves fatty acids from both the sn1 and sn2 positions of galactolipids. Deesterification of galactolipids has been shown to be induced by drought stress, chilling, and senescence (Kaniuga and Gemel, 1984; Kaniuga et al., 1999; Matos et al., 2001). Highly active galactolipase has been partially purified from leaf chloroplasts (Anderson et al., 1974). In addition, a novel patatin-like gene (Vupat1) encoding a protein exhibiting galactolipid acyl hydrolase activity was recently isolated from drought-stressed cowpea (Vigna unguiculata) leaves (Matos et al., 2000, 2001). The cognate protein of Vupat1 exhibited high lipase activity in the presence of monogalactosyl diacylglycerol and digalactosyl diacylglycerol substrates, whereas phosphatidylcholine was not an effective substrate. Also, Vupat1 expression increased during drought stress, suggesting possible involvement in chloroplast membrane degradation induced by water stress (Matos et al., 2000, 2001).
Triacylglycerol (TAG) lipases deesterify fatty acids from TAG, a major storage lipid that, in oil-storing seeds, is localized in oil bodies (Somerville et al., 2000). The enzyme cleaves fatty acids at each of the sn1, sn2, and sn3 positions of TAG. The formation of oil bodies is thought to be triggered by the accumulation of TAG between the monolayers of the endoplasmic reticulum membrane during the final stages of seed development (Huang, 1992; Murphy and Vance, 1999). During seed germination, TAGs are catabolized by TAG lipase and used as a source of energy to support early seedling growth. TAG lipase appears to associate transiently with oil bodies, binding to the oleosin protein coating (Huang, 1992, 1996; Beisson et al., 2001). Indeed, it has been proposed that TAG lipase is able to access its TAG substrate in the interior of the oil body through structural defects in the oil body phospholipid monolayer rendered by phospholipases during postgerminative growth (Noll et al., 2000).
Plastoglobuli, which are lipid bodies localized in chloroplasts, also contain TAG and, increasingly, appear to be structurally analogous to seed oil bodies (Martin and Wilson, 1984; Rey et al., 2000; Austin et al., 2006). For example, there is now ample evidence that plastoglobuli are coated with fibrillin, or plastoglobulins, which are thought to be structural proteins analogous to oleosin present on the surfaces of oil bodies, and that the role of fibrillin, like oleosin, is to prevent coalescence of the particles that they circumscribe (Huang, 1996; Kessler et al., 1999; Rey et al., 2000; Vidi et al., 2006; Ytterberg et al., 2006). Moreover, fibrillin appears to regulate the formation of plastoglobuli from thylakoids in much the same way that oleosin is thought to regulate the formation of seed oil bodies from the endoplasmic reticulum (Huang, 1992; Rey et al., 2000). A role for plastoglobuli in chloroplast senescence has been proposed based on the findings that their size and abundance increase during thylakoid degradation and that they contain both lipid and protein metabolites derived from thylakoid membranes (Sprey and Lichtenthaler, 1966; Lichtenthaler, 1969; Lichtenthaler and Weinert, 1970; Ghosh et al., 1994; Smith et al., 2000; Smith and Ghosh, 2002; Kaup et al., 2002). Moreover, diacylglycerol acyltransferase 1 (DGAT1), which mediates the final acylation step in TAG synthesis, has been shown to be associated with chloroplast membranes in senescing leaves and to be up-regulated coincident with TAG accumulation and increased abundance of plastoglobuli as leaf senescence progresses (Kaup et al., 2002). More recently, two putative TAG-metabolizing enzymes have been identified as plastoglobular constituents by proteomic analyses of plastoglobuli isolated from Arabidopsis (Vidi et al., 2006; Ytterberg et al., 2006). Synthesis of leaf TAG also accompanies the deesterification of polar lipids induced by stress (Sakaki et al., 1990b, 1990c). These observations have prompted the proposal that formation of TAG in chloroplasts may be a means of sequestering free fatty acids that are rapidly deesterified from galactolipids in the event of stress (Sakaki et al., 1990b) and during natural senescence (Kaup et al., 2002). The fatty acid equivalents of this TAG would subsequently be converted to phloem-mobile Suc as senescence progresses or possibly utilized for renewed galactolipid synthesis in the event of recovery from stress (Cohen et al., 2000).
In this study, we have characterized a putative TAG lipase in Arabidopsis that is localized in the plastoglobuli of chloroplasts. Suppression of this lipase in transgenic plants by constitutive expression of a corresponding antisense transgene resulted in severely stunted growth as well as reduced abundance and disorganization of thylakoid membranes. The results are consistent with the view that TAG lipase mobilizes fatty acids from complex plastoglobular lipids.
RESULTS
Lipase Gene Isolation and Function
Full-length cDNA from Arabidopsis (corresponding to GenBank accession no. At2g31690 and encoding the protein with GenBank accession no. AAD24845) was obtained by reverse transcription (RT)-PCR using RNA isolated from the rosette leaves of 6-week-old plants as a template. The nucleotide and inferred amino acid sequences of the full-length cDNA are shown in Figure 1. A comparison of the genomic and cDNA nucleotide sequences indicated that the gene does not contain introns. Analysis of the inferred amino acid sequence predicts a molecular mass for the full-length protein of approximately 53 kD. The programs TargetP (version 1.0; http://www.cbs.dtu.dk/services/TargetP; Emanuelsson et al., 2000) and Predotar (version 1.03; http://genoplante-info.infobiogen.fr/predotar/predotar.html; Small et al., 2004) predicted the protein is most likely targeted to chloroplasts, and ChloroP (version 1.1; http://www.cbs.dtu.dk/services/ChloroP; Emanuelsson et al., 1999) predicts a plastid transit peptide of 63 amino acids (Fig. 1), cleavage of which would produce a mature protein of approximately 46 kD within chloroplasts. In addition, the cognate protein contains the lipase active site sequence, [LIV]-X-[LIVAFY]-[LIAMVST]-G-[HYWV]-S-X-G-[GSTAC], which is normally a feature of lipases that deesterify fatty acids from complex lipids, as well as predicted phosphorylation sites (Fig. 1). A BLAST search revealed that the next most similar protein (73% identity with an e-value of 2 × 10−171) is another putative Arabidopsis TAG lipase (GenBank accession no. NP_563748), which also contains the lipase active site sequence, [LIV]-X-[LIVAFY]-[LIAMVST]-G-[HYWV]-S-X-G-[GSTAC]. The next 12 closest matches are putative lipases from rice (Oryza sativa cv japonica; namely, GenBank accession nos. ABA92846, ABA92804, NP_915192, BAD82102, NP_915194, and XP_475184) and Arabidopsis (namely, GenBank accession nos. NP_564590, NP_563772, NP_849603, NP_56570, NP_850148, and NP_174326), and these also all contain the lipase motif [LIV]-X-[LIVAFY]-[LIAMVST]-G-[HYWV]-S-X-G-[GSTAC].
Figure 1.
Nucleic acid sequence (GenBank accession no. At2g31690) and corresponding amino acid sequence (protein ID AAD24845) illustrating the lipase active site (black shading), the portion of the sequence utilized for antibody production against a synthetic peptide (gray shading), the position of the ChloroP-predicted transit peptide cleavage site (arrowhead), as well as predicted protein kinase C (underlined) and casein kinase II (bold and italics) phosphorylation sites.
In an effort to confirm that the protein corresponding to accession number AAD24845 corresponds to a lipase, cDNAs corresponding to the full-length and predicted mature (plastid-localized) forms of the protein were overexpressed as recombinant maltose binding protein (MBP) fusion proteins in Escherichia coli. SDS-PAGE and western-blot analysis confirmed the presence of the recombinant proteins in E. coli extracts (data not shown). Purified recombinant fusion proteins were tested for lipase activity in vitro. For this purpose, the MBP fusion proteins were immobilized on amylose resin and tested for their ability to hydrolyze the TAG trilinolein (18:2). In this assay, the full-length AAD24845-MBP fusion protein (MBP-LipF) demonstrated only slightly higher lipase activity than the background registered by MBP on its own (Table I). In contrast, the predicted mature version of the AAD24845 protein (i.e. lacking the predicted transit peptide) fused to MBP (MBP-LipS) exhibited a significantly higher activity compared to both the control MBP and the full-length AAD24845-MBP fusion (Table I). This was true whether the samples were assayed by thin-layer chromatography (TLC) or using the nonesterified fatty acid (NEFA)-C assay kit and suggests that the AAD24845 protein is a bona fide TAG lipase. Furthermore, it appears that cleavage of the predicted transit peptide is required for the protein to attain its fully active form. Lipase activity was undetectable in the presence of phospholipid (soybean [Glycine max] phosphatidylcholine) or galactolipid (monogalactosyl diacylglycerol) substrate.
Table I.
TAG lipase activity of recombinant fusion proteins MBP-LipF and MBP-LipS
Lipase activity was measured by iodine-stained TLC and a NEFA-C assay. Values are means ± se for n = 3. Means identified by different superscripts are significantly different at the 5% level based on Duncan's multiple range test. Percentages relative to the pMal-c2 control are indicated. MBP, Maltose binding protein control produced from the empty pMal-c2 vector; MBP-lipF, full-length lipase fused to MBP; MBP-lipS, truncated lipase (lacking predicted transit peptide) fused to MBP.
| Recombinant Protein | TLC Assay
|
NEFA-C Assay
|
||
|---|---|---|---|---|
| Density Relative to Background × 104 | % Increase | mmol FFA/μg Protein | % Increase | |
| MBP | 8.50a ± 0.30 | – | 23.3a ± 2.04 | – |
| MBP-lipF | 9.17a ± 0.30 | 7.9 | 26.0a,b ± 2.06 | 11.3 |
| MBP-lipS | 11.0b ± 0.05 | 29.5 | 33.3b ± 1.79 | 42.5 |
Expression and Localization Analyses
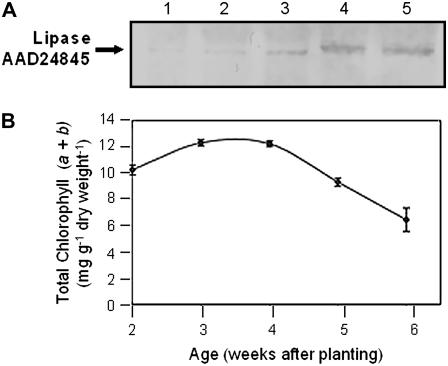
Western blots of total protein extracts from rosette leaves of Arabidopsis plants at different stages of development were probed with antiserum raised against a synthetic peptide corresponding to the putative TAG lipase. An approximately 46-kD polypeptide, which corresponds to the expected size of the mature protein, was barely detectable in leaves from 2- and 3-week-old plants, but showed increased abundance through weeks 5 and 6 (Fig. 2A). The progress of rosette leaf development and senescence was scored by measuring changes in the levels of leaf chlorophyll. Chlorophyll levels proved to be high (10–12 mg g−1 dry weight of leaf tissue) in 2- and 3-week-old leaves, declined by 25% between weeks 4 and 5 as leaf senescence commenced, and by week 6, when the leaves were turning yellow in color, had decreased to approximately 50% of their peak values (Fig. 2B).
Figure 2.
Western blot of total protein isolated from the rosettes of wild-type Arabidopsis plants at various stages of development. A, Protein blot probed with TAG lipase antiserum. Lane 1, Total protein isolated from 2-week-old rosette leaves; lane 2, total protein isolated from 3-week-old rosette leaves; lane 3, total protein isolated from 4-week-old rosette leaves; lane 4, total protein isolated from 5-week-old rosette leaves; lane 5, total protein isolated from 6-week-old rosette leaves. Each lane was loaded with 10 μg of protein. B, Chlorophyll levels in developing and senescing rosette leaves. Age of the leaves (weeks after planting) is indicated.
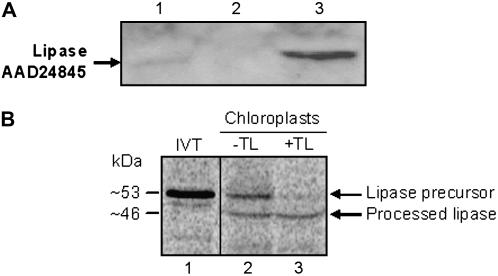
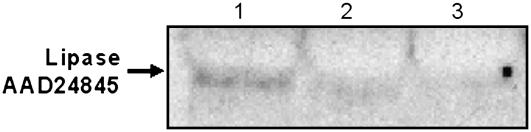
In an effort to confirm that the predicted plastid transit peptide directs the putative TAG lipase to plastids, chloroplasts were purified from the rosette leaves of 4.5-week-old Arabidopsis using a Percoll gradient and analyzed for the presence of the lipase protein by western blotting. Pure intact chloroplasts were subfractionated into total chloroplast membranes and stroma by lysis and centrifugation. The lipase protein was detectable in intact chloroplasts and also in stroma, but not in the chloroplast membrane fraction (Fig. 3A). Indeed, TAG lipase proved to be substantially enriched in the stroma as compared to intact chloroplasts, suggesting that it is primarily a stromal protein (Fig. 3A, compare lanes 1 and 3). In addition, the molecular mass of the stromal TAG lipase proved to be approximately 46 kD, the expected size of the mature protein (Fig. 3A). These data are in agreement with the results of in vitro chloroplast import assays in which the recombinant AAD24845 lipase precursor was incubated with intact chloroplasts isolated from Arabidopsis. These data show that the approximately 53-kD full-length TAG lipase precursor is converted to an approximately 46-kD protein following incubation and reisolation of chloroplasts (Fig. 3B). Furthermore, the processed form of the TAG lipase is resistant to treatment with thermolysin (Fig. 3B, lane 3), which indicates that the protein has been successfully imported into the interior of the chloroplast. In addition, both the western blot and in vitro import data are consistent with the predicted 7-kD size of the transit peptide and confirm that this is a plastid transit peptide capable of directing the protein to chloroplasts.
Figure 3.
Localization of protein AAD24845 in chloroplasts. A, Western blot of intact chloroplasts, total chloroplast membranes, and stroma isolated from the rosette leaves of 4.5-week-old wild-type Arabidopsis plants probed with TAG lipase antiserum. Lane 1, Intact chloroplasts; lane 2, total chloroplast membranes (thylakoids and envelope membranes); lane 3, stroma isolated from intact chloroplasts. Each lane was loaded with 10 μg of protein. B, Import of the TAG lipase precursor (AAD24845) into isolated chloroplasts. In vitro-translated 35S-Met-labeled TAG lipase precursor was incubated with chloroplasts (50 μg of chlorophyll) in a standard protein import assay for 30 min at 26°C. Following incubation, chloroplasts were reisolated and treated for 30 min on ice without (−TL) or with (+TL) 100 μg/mL thermolysin. Proteins were resolved using SDS-PAGE and radiolabeled proteins were detected in dried gels using a phosphor imager. Lane 1 contains 20% of the 35S-Met-labeled lipase precursor in vitro translation product (IVT) that was added to each reaction. Lanes 2 and 3 contain chloroplasts reisolated following the import assays that were treated without or with thermolysin, respectively. Measured molecular masses (kD) of the precursor and processed, mature forms of the protein are indicated.
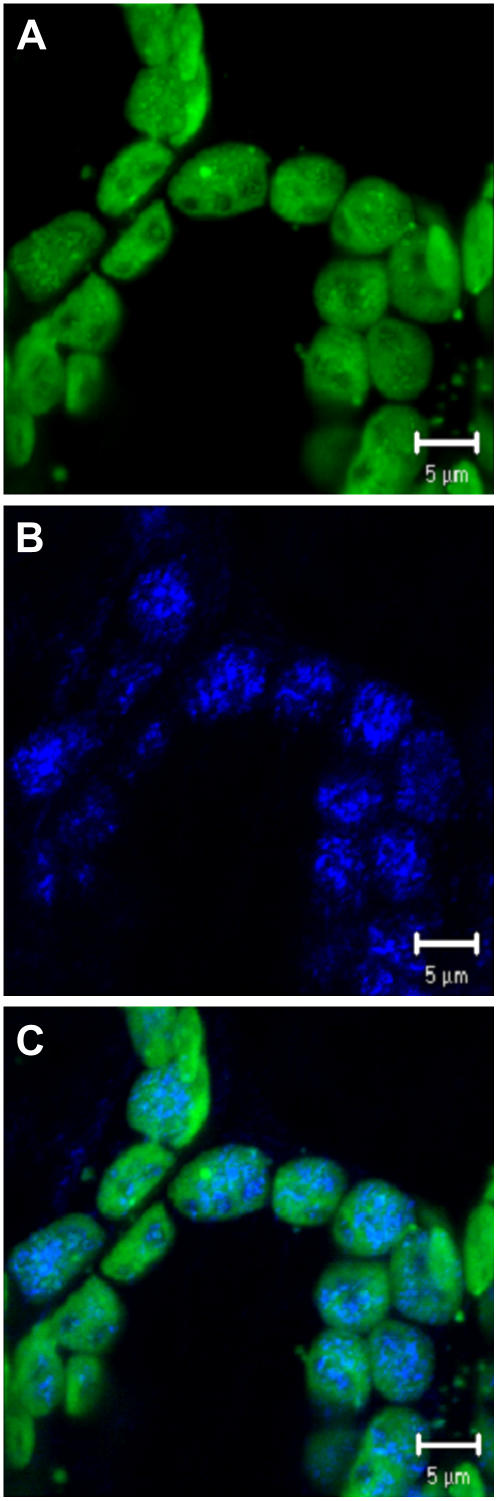
Confocal microscopy of rosette leaves from 4.5-week-old plants provided additional evidence that the putative TAG lipase is localized in chloroplasts. Green pseudocolored autofluorescing chlorophyll reflecting the presence of thylakoids was clearly evident in confocal images of leaf cells, as was blue pseudocolored fluorescence corresponding to FITC antibody-labeled TAG lipase (Fig. 4, A and B). When the chlorophyll fluorescence and antibody fluorescence images were merged, it was clear that the TAG lipase is localized within chloroplasts (Fig. 4C).
Figure 4.
Confocal microscopy of sections from the first leaf pair of 4.5-week-old wild-type plants. The photographs are three-dimensional renderings of five serial optical sections probed with TAG lipase antiserum. A, Autofluorescence of chloroplasts (pseudocolored green). B, Fluorescence of FITC (pseudocolored blue), indicating the presence of the TAG lipase antibody in the chloroplasts. C, Merged image of A and B. Bar = 5 μm.
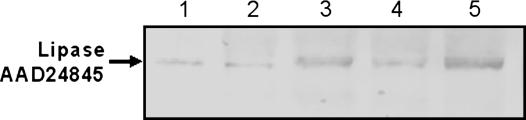
Western blotting of protein isolates from a number of tissues of 6-week-old Arabidopsis plants revealed that the TAG lipase protein is also present in non-green organs, such as roots, suggesting that the transit peptide directs the protein to plastids other than chloroplasts. At 6 weeks of age, inflorescence stems and cauline leaves had not begun to senesce, flowers were a mixture of senescent and nonsenescent, and siliques were developing, but had not yet reached maturity. An approximately 46-kD protein corresponding to the TAG lipase was detected in roots, inflorescence stems, cauline leaves, flowers, and siliques. However, the TAG lipase protein proved to be more abundant in flowers and cauline leaves than in other tissues analyzed (Fig. 5).
Figure 5.
Western blot of total protein extracts from organs of 6-week-old wild-type Arabidopsis plants. The blot was probed with TAG lipase antiserum. Lane 1, Total protein isolated from roots; lane 2, total protein isolated from inflorescence stems; lane 3, total protein isolated from cauline leaves; lane 4, total protein isolated from developing siliques; lane 5, total protein isolated from flowers. Each lane was loaded with 10 μg of protein.
Transgenic Plants with Suppressed Expression of the Putative TAG Lipase
Transgenic Arabidopsis plants expressing antisense full-length cDNA corresponding to the TAG lipase were obtained by vacuum infiltration. T1 seedlings were selected on kanamycin plates, transplanted to soil, and grown to maturity. The presence of the transgene was confirmed by PCR. T1 plants were severely stunted in comparison with wild-type plants, but bolted and flowered at the same time as corresponding wild-type plants. This is illustrated for five transgenic lines (Figure 6, A–D, lines 1–5). That the intensity of the phenotype correlated with the degree of expression of the transgene is illustrated by the phenotypes of segregating T2 plants. For example, for line 2, 75% of the T2 plants exhibited the same degree of stunted growth that was evident for the corresponding T1 plants (Fig. 6, C and E), and a kanamycin screen of the T3 seeds from these plants yielded a 3:1 segregation ratio, indicating that these T2 plants were heterozygous (Fig. 6E). However, 25% of the T2 plants for line 2 exhibited a much more pronounced stunted phenotype and, in the kanamycin screen of the corresponding T3 seeds, 100% of the seedlings survived, indicating that these T2 plants were homozygous (Fig. 6E). There was also an accumulation of anthocyanin in the leaves of homozygous transgenic plants, indicating that they were under stress (Fig. 6F). Although the transgenic plants bolted and flowered at the same time as the wild-type plants, notwithstanding their stunted phenotype (Fig. 6, A–E), rosette senescence was dramatically delayed. Even at 10.5 weeks after planting, by which time the rosettes of wild-type plants were completely dead, the rosettes of transgenic plants were still green. This is illustrated for homozygous line 2 plants in Figure 6G.
Figure 6.
Photographs of transgenic and wild-type plants. A, T1 generation of transgenic plants (lines 1–5), 5.5 weeks after planting. B, Wild-type plants, 5.5 weeks after planting. C, Side profile of line 2 (T1 generation) and wild-type plants, 5.5 weeks after planting. D, Side profile of line 3 (T1 generation) and wild-type plants, 5.5 weeks after planting. E, Wild-type and segregating line 2 (T2 generation) plants 6 weeks after planting, wherein plant 2A is heterozygous for the transgene and plant 2B is homozygous. F, Wild-type and homozygous line 2 (T3 generation) plants, 4.5 weeks after planting. G, Wild-type and homozygous line 2 (T3 generation) plants, 10.5 weeks after planting.
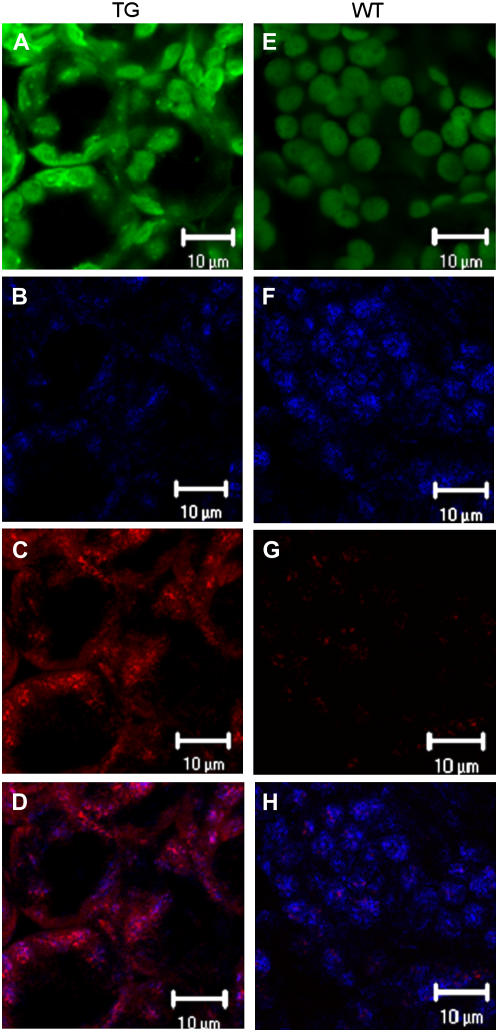
That the phenotype correlated with suppressed expression of the putative TAG lipase gene was confirmed by western blotting. Protein was isolated from rosette leaves of 4.5-week-old wild-type and transgenic plants. At this age, wild-type and transgenic plants were at comparable stages of development, although the transgenic plants were stunted (Fig. 6). The TAG lipase was clearly apparent in rosette leaves from wild-type plants, but was barely detectable in rosette leaf protein from transgenic plants, as is illustrated for lines 2B-1 and 2B-2 in Figure 7. The effect of transgene expression on levels of TAG lipase protein was also assessed by confocal microscopy. Sections from the first leaf pair of 4.5-week-old wild-type and transgenic leaves were treated with FITC-labeled secondary antibody. When these sections were examined by confocal microscopy, it was clear that the TAG lipase protein is localized in chloroplasts in both wild-type and transgenic plants and that the abundance of TAG lipase protein was greatly reduced in transgenic leaf sections in comparison with wild-type leaf sections. This is illustrated for transgenic line 2 in Figure 8, A and B, as compared with wild type in Figure 8, E and F.
Figure 7.
Western blot of total protein extracts from the rosette leaves of 4.5-week-old wild-type and transgenic Arabidopsis plants. The blot was probed with TAG lipase antiserum. Lane 1, Rosette leaf protein from wild-type plants; lane 2, rosette leaf protein from transgenic line 2B-1; lane 3, rosette leaf protein from transgenic line 2B-2. Each lane was loaded with 15 μg of protein.
Figure 8.
Confocal microscopy of sections from the first leaf pair of 4.5-week-old transgenic (T3 generation) homozygous line 2 plants (A–D) and of wild-type plants (E–H). A and E, Autofluorescence of chloroplasts pseudocolored green. B and F, Fluorescence of FITC pseudocolored blue, indicating antibody bound to TAG lipase protein. C and G, Fluorescence of Nile Red associated with neutral lipid (pseudocolored red). D, Merged image of B and C. H, Merged image of F and G. Bar = 10 μm.
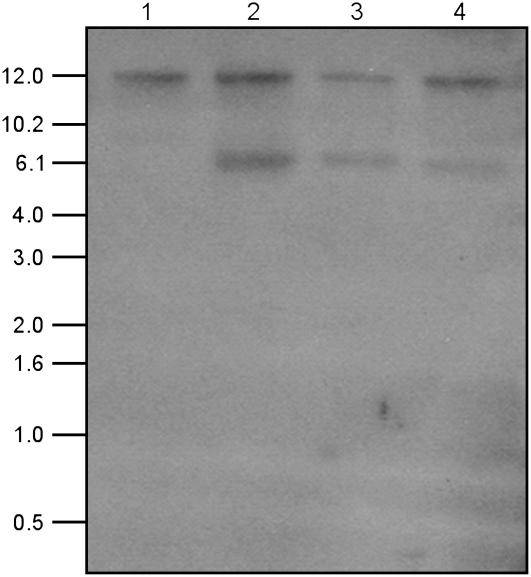
Southern-blot analyses confirmed GenBank data indicating that the Arabidopsis genome contains only one copy of the At2g31690 gene. When rosette leaf genomic DNA was digested with the restriction enzyme HindIII, for which there is no site in the open reading frame of the lipase gene, fractionated on an agarose gel, blotted, and probed with full-length At2g31690 cDNA corresponding to the TAG lipase, only one restriction fragment was visible for wild-type plants (Fig. 9). In corresponding blots for transgenic line 2, one additional restriction fragment corresponding to a single copy of the transgene was also discernible (Fig. 9). Transgenic lines 2B and 2C were homozygous with respect to the transgene and, as expected, the restriction fragments corresponding to the endogenous gene and the single transgene were of comparable intensity (Fig. 9). However, transgenic line 2A was heterozygous with respect to the transgene and, accordingly, the intensity of the transgene restriction fragment was approximately one-half that of the endogenous gene (Fig. 9).
Figure 9.
Southern blot of genomic DNA isolated from rosettes of 4-week-old wild-type and transgenic (T2 generation) line 2 Arabidopsis plants. DNA was cut with HindIII and the blot was probed with full-length cDNA of Arabidopsis TAG lipase, At2g31690. Lane 1, DNA from wild-type plant; lane 2, DNA from homozygous 2B plant; lane 3, DNA from homozygous 2C plant; lane 4, DNA from heterozygous 2A plant. Molecular mass markers (kb) are indicated.
Colocalization of Lipase with Chloroplast Neutral Lipids
Confocal microscopy of sections from the first leaf pair of 4.5-week-old wild-type and transgenic plants indicated that the TAG lipase colocalizes with neutral lipids in chloroplasts. Lipase protein was visualized using FITC-labeled antibody against the TAG lipase primary antibody and neutral lipid (e.g. TAG) was stained with the fluorescent dye, Nile Red. For wild-type leaves, TAG lipase protein was abundant and clearly localized in the chloroplasts (Fig. 8, E and F). Levels of chloroplast neutral lipids, which are localized in plastoglobuli, were low, but detectable (Fig. 8G). However, because the neutral lipid levels were so low in wild-type leaves, merging of the Nile Red-stained image and the TAG lipase image did not provide clear evidence for colocalization of neutral lipid and TAG lipase (Fig. 8H). As in wild-type leaves, the TAG lipase of transgenic leaves was also localized in chloroplasts (Fig. 8, A and B). However, levels of lipase were greatly reduced in transgenic as compared to wild-type plants (Fig. 8, B and F), whereas neutral lipid levels were enhanced in transgenic plants (Fig. 8, C and G). In addition, merging of the TAG lipase image and the Nile Red-stained image for transgenic leaf sections provided clear evidence for colocalization of TAG lipase protein and neutral lipids likely contained within plastoglobuli (Fig. 8D). Of particular note is the fact that the high levels of TAG lipase protein in wild-type leaf segments are accompanied by low levels of neutral lipids (Fig. 8, F and G), whereas in transgenic leaf segments high levels of neutral lipids are accompanied by low levels of TAG lipase protein (Fig. 8, B and C). Inasmuch as the neutral lipids visualized by Nile Red staining would include TAG, this is consistent with the notion that TAG lipase hydrolyzes chloroplast TAG. Moreover, in both wild-type and transgenic leaf sections, lipase protein and neutral lipids appear as clusters, suggesting that they are colocalized in plastoglobuli (Fig. 8), which are known to contain neutral lipids, such as TAG (Kaup et al., 2002). These findings are thus consistent with the notion that TAG lipase hydrolyzes chloroplast TAG contained in plastoglobuli.
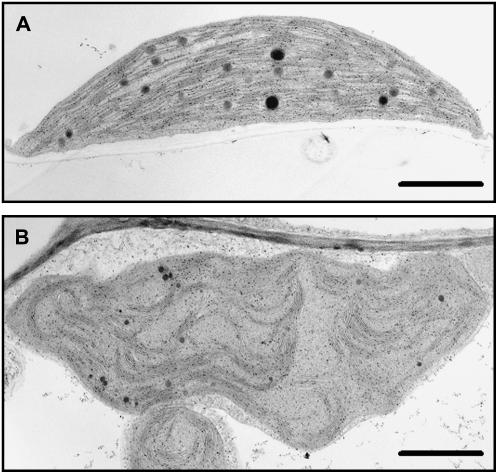
Transmission electron microscopy of thin sections from the first leaf pair of 3.5-week-old wild-type and transgenic plants revealed that suppression of this putative TAG lipase also resulted in chloroplast structural aberrations. Chloroplasts in wild-type leaves proved to be compact with tightly stacked thylakoids and several large plastoglobuli (Fig. 10A). By contrast, chloroplasts of transgenic plants were deformed with wavy outer envelopes and reduced levels of thylakoid membranes that were loosely stacked. Distinct plastoglobuli that were smaller than those evident in wild-type chloroplasts were also discernible in the chloroplasts of transgenic leaves (Fig. 10, A and B). However, in keeping with the greater abundance of neutral lipid in the chloroplasts of transgenic plants depicted by Nile Red staining (Fig. 8, C and G), there were also large numbers of small osmium-stained stromal particles evident in thin-sectioned transgenic chloroplasts that are likely plastoglobuli (Fig. 10B).
Figure 10.
Transmission electron micrographs of chloroplasts in the first leaf pair of 3.5-week-old wild-type and transgenic (T3 generation) homozygous line 2 plants. A, Chloroplast in a wild-type leaf. B, Chloroplast in a transgenic leaf. Bar = 1 μm.
DISCUSSION
The Arabidopsis gene characterized in this study is identified as a putative TAG lipase in GenBank (At2g31690) based on its degree of sequence identity to known TAG lipases. Western-blot data indicated that this putative TAG lipase (protein accession no. AAD24845) is present in flowers, inflorescence stems, cauline and rosette leaves, siliques, and roots of Arabidopsis plants. Moreover, several lines of evidence indicate that it is targeted to plastids. Examination of the deduced protein sequence using the programs TargetP (version 1.01), Predator (version 1.03), and ChloroP (version 1.1) predicted that it is targeted to chloroplasts and possesses a 63-amino acid (approximately 7 kD) transit peptide (Fig. 1). Western-blot analysis of leaf subcellular fractions indicated that the lipase is localized in chloroplasts (Fig. 3A). This contention was further supported by confocal microscopy, which showed that FITC-labeled antibody against the AAD24845-specific antibody recognized a protein that was localized to chloroplasts (Fig. 4) and by in vitro chloroplast import assays, which showed that the protein is imported and processed to generate a mature protease-protected protein that is approximately 7 kD shorter than the original full-length (precursor) protein (Fig. 3B). Of note is that the TAG lipase was also detectable by western blotting in roots that do not contain chloroplasts, suggesting that it is also associated with plastid types other than chloroplasts. Storage plastids that contain neutral lipids, including TAG, are known to be present in tissues that are nonphotosynthetic (Xue et al., 1997; Staehelin and Newcomb, 2000; Murphy, 2001), and it is therefore to be expected that one or more lipases are targeted to such plastids.
The findings that the putative TAG lipase has a plastid transit peptide, is localized in chloroplasts, and is found as well in tissues that contain storage plastids are all consistent with the contention that it is a plastid lipase. Moreover, there are several lines of evidence indicating that the protein is in fact a lipase. First, the amino acid sequence contains the 10-residue lipase active site sequence, [LIV]-X-[LIVAFY]-[LIAMVST]-G-[HYWV]-S-X-G-[GSTAC]. This motif is normally a feature of lipases that deesterify fatty acids from complex lipids (Derewenda and Derewenda, 1991; Hong et al., 2000; Ishiguro et al., 2001; He and Gan, 2002; Lo et al., 2004) and, accordingly, its presence in the inferred amino acid sequence indicates that AAD24845 is a lipase. Second, it is evident from confocal micrographs that the protein colocalizes with plastoglobular neutral lipids, including TAGs, in chloroplasts, which is consistent with its annotation as a putative TAG lipase in GenBank. Third, chloroplastic neutral lipids visualized by Nile Red staining, which include TAG, proved to be much more abundant in the leaves of AAD24845-suppressed transgenic plants than in leaves of corresponding wild-type plants containing higher levels of the protein, which is again consistent with the contention that the AAD24845 protein deesterifies TAG. Last, in vitro biochemical analysis of recombinant versions of the protein fused to MBP demonstrated that AAD24845 possesses TAG lipase activity and that activity is highest when the putative transit peptide is removed (Table I). These data collectively indicate that this lipase is capable of deesterifying TAG and associates with neutral lipids in the chloroplast.
It is apparent from GenBank that lipase AAD24845 is encoded by a single-copy gene and this was confirmed by Southern blotting in this study. That down-regulation of expression of lipase gene At2g31690 had been achieved in antisense At2g31690 transgenic plants was confirmed by western-blot analysis of levels of the lipase protein AAD24845 in leaves at comparable stages of development and also by confocal microscopy. The antibodies used for detection of the putative TAG lipase protein were raised against a synthetic peptide deemed to be unique to AAD24845 based on BLAST interrogation of its sequence. The severely stunted nature of transgenic plants with suppressed AAD24845 indicates that this protein plays a seminal role in plant growth. It appears to be constitutively expressed at low levels, but, judging from western-blot analysis, there is clear up-regulation of expression in senescing leaves. Senescing chloroplasts are known to be enriched in plastoglobuli, which form coincident with the breakdown of thylakoid membranes (Harwood, 1980; Steinmüller and Tevini, 1985; Murphy, 2001; Lee and Chen, 2002). Although the precise way in which plastoglobuli function has not been fully elucidated, their accumulation and increase in size during thylakoid degradation suggest that they temporarily store TAG formed from galactolipid fatty acids deesterified during leaf senescence (Sprey and Lichtenthaler, 1966; Lichtenthaler, 1969; Lichtenthaler and Weinert, 1970; Kaup et al., 2002). Accordingly, western-blot and confocal microscopy data indicating that the AAD24845 protein not only is abundant in senescing leaves, but also is colocalized with neutral lipids in chloroplasts raise the possibility that it may participate in phloem mobilization of lipids characteristic of senescing leaves by deesterifying fatty acids from plastoglobular TAG. That AAD24845 plays a critical role in leaf senescence is supported by the finding that suppression of its expression delayed the onset of rosette senescence. Although the AAD24845-suppressed plants were severely stunted, bolting and flowering were temporally coincident in wild-type and transgenic plants. However, rosettes of transgenic plants stayed green for at least 2 weeks longer than those of corresponding wild-type plants. For example, rosettes of wild-type plants were almost completely dead at 8.5 weeks after planting, whereas rosettes of 10.5-week-old transgenic plants were still green.
Plastoglobuli are present in chloroplasts of both senescing and nonsenescing leaves, but are much more distinct and abundant in senescing leaves. Moreover, their increased size and abundance in senescing leaves has been shown to be paralleled by an increase in TAG originating from galactolipid and up-regulation of DGAT1, which mediates the final acylation step in the formation of TAG (Kaup et al., 2002). These observations were interpreted as indicating that DGAT1 plays a role in senescence by sequestering fatty acids deesterified from galactolipids into TAG and that this is an intermediate step in the conversion of thylakoid fatty acids to phloem-mobile Suc (Kaup et al., 2002). It is interesting to note that other lipid-metabolizing enzymes, including two other putative TAG-synthesizing proteins, were detected in recent proteomic analyses of plastoglobuli from Arabidopsis (Vidi et al., 2006; Ytterberg et al., 2006). These observations are consistent with the proposed role of plastoglobule-associated TAG in lipid mobilization and thylakoid remodeling. It is apparent from this study that the putative TAG lipase corresponding to AAD24845 is also associated with plastoglobuli, is strongly up-regulated in senescing leaf tissue, and thus could be the lipase that deesterifies, and hence remobilizes, the fatty acids of TAG formed from thylakoids. It is also noteworthy that Vidi et al. (2006) did not detect any lipases in their proteomic analysis of plastoglobuli. However, their study only examined one of two plastoglobule-containing fractions with different densities; indeed, plastoglobuli with distinct densities have been reported previously (Bailey and Whyborn, 1963; Smith et al., 2000). Vidi et al. (2006) examined the plastoglobuli-containing fraction with the lowest density, which would presumably have the highest TAG content. In contrast, the higher density particles in the fraction not examined by Vidi et al. (2006) would presumably have a lower TAG content. Therefore, it is possible that generation of plastoglobuli with different densities may be due to hydrolysis of plastoglobule TAG by lipases such as that described herein, and it remains possible that lipases would only be detected in association with higher density plastoglobuli. The manner by which the AAD24845 lipase might associate with plastoglobuli remains unknown. However, it is possible that it occurs via interaction with other plastoglobule-specific proteins, such as plastoglobulins (Pozueta-Romero et al., 1997; Kessler et al., 1999; Vidi et al., 2006; Ytterberg et al., 2006). It is also possible that this lipase resides in the stroma until such time as catalysis of TAG is needed, at which time it might transition to a plastoglobule-associated form.
Of particular interest is the finding that the chloroplasts of AAD24845-suppressed transgenic plants appeared deformed and had fewer thylakoids than the chloroplasts of wild-type plants. Moreover, thylakoids of transgenic plants were loosely stacked in comparison with grana of wild-type plants. These observations suggest that putative TAG lipase AAD24845 plays a role in maintaining the structural integrity of thylakoids and that lack of grana structural integrity in transgenic plants accounts for their stunted growth. Transgenic plants were clearly under stress throughout growth and development as indicated by the consistent presence of anthocyanin in the leaves. Previous studies have demonstrated activation of TAG synthesis and a concurrent decrease in galactolipids in ozone-fumigated leaves (Sakaki et al., 1990a, 1990b, 1990c). Activation of TAG synthesis in the ozone-treated leaves is thought to reflect sequestration of fatty acids deesterified from galactolipids in response to ozone stress, which would otherwise disrupt the structure of thylakoid membranes (Sakaki et al., 1990b). Similar findings, a decline in polar lipid paralleled by an increase in TAG, have been reported for drought-stressed cotton leaves (El-Hafid et al. 1989). These observations raise the possibility that renewed growth upon abatement of stress, or even continued growth in the event of chronic sublethal stress, are contingent upon mobilization of the fatty acid equivalents of this TAG. This could be achieved by direct reutilization of deesterified TAG fatty acids. However, the possibility that deesterified TAG fatty acids are degraded and their carbon equivalents used for the synthesis of new galactolipid fatty acids is not precluded. The findings in this study that lipase AAD24845 colocalizes with chloroplast neutral lipids, including TAG, and that its suppression curtails growth suggest that it could play a role in this remobilization.
MATERIALS AND METHODS
Plant Material
Arabidopsis (Arabidopsis thaliana ecotype Columbia) was grown on Promix BX (Premier Brands). Seeds were sown in 6-inch pots, covered with plastic wrap, and stratified at 4°C for 2 d to achieve uniform germination. Plants were transferred to a growth chamber operating at 16-h-light/8-h-dark cycles, 23°C ± 3°C, and 150 μmol m−2 s−1 photosynthetically active radiation. Each pot contained eight to 10 plants, which were grown for a period of 7 to 8 weeks after transfer to the growth chamber with senescence starting after 4 weeks and seed production finished by 8 weeks. Rosette leaves were harvested at 2, 3, 4, 5, and 6 weeks of age. In some cases, additional organs were harvested from 6-week-old plants, specifically roots, inflorescence stems, cauline leaves, flowers, and siliques.
Isolation of At2g31690 cDNA and Production of Transgenic Plants
Full-length cDNA corresponding to GenBank sequence At2g31690 (encoding protein AAD24845) was obtained by RT-PCR using RNA isolated from 6-week-old Arabidopsis rosette leaves as the template. The upstream primer (5′-CGTGTCGACTTCCATCAATGGCCTTGATCC-3′) contained the restriction enzyme site for SalI and the downstream primer (5′-GCCAAGCTTACGCTGACGTTGTACGAATTATAG-3′) contained the restriction enzyme site for HindIII. The PCR product was subcloned into pBluescript KS that had been digested with SalI and HindIII, amplified in Escherichia coli DH5-α, and sequenced.
Suppression of the endogenous At2g31690 gene was achieved by expressing full-length At2g31690 cDNA in the antisense orientation under the regulation of a constitutive promoter in transgenic plants. At2g31690 cDNA was excised from pBluescript KS by double digestion with SalI/HindIII and subcloned into the binary vector, pKYLX71 (Schardl et al., 1987), in the antisense orientation under the regulation of two copies of the constitutive cauliflower mosaic virus promoter (CaMV 35S). The pKYLX71 vector contains a tetracycline resistance gene in the bacterial replication region as well as a kanamycin resistance gene (NPT II) within the T-DNA region. The recombinant binary vector was introduced into Agrobacterium tumefaciens, strain GV3101, by electroporation. The resulting transformed bacteria were used to infect the flowers of 4-week-old Arabidopsis plants by vacuum infiltration (Bechtold et al., 1993). Transgenic plants were selected by germinating seeds from the vacuum-infiltrated plants on medium containing kanamycin. Seeds were surface sterilized in a solution of 1% sodium hypochlorite and 0.1% Tween 80, rinsed three times in sterile water, and plated on 0.7% agar containing 0.5× Murashige and Skoog salt (Sigma-Aldrich) and 50 μg mL−1 kanamycin. Plates were maintained at 4°C for 2 d and then transferred to a tissue culture chamber operating at 22°C ± 3°C with 16-h-light (150 μmol m−2 s−1 photosynthetically active radiation)/8-h-dark cycles. After 12 d, surviving seedlings were transferred to soil (Promix BX) and grown to maturity in a growth chamber under conditions specified above. Seeds were harvested and the selection process was repeated with successive generations until homozygous lines exhibiting 100% germination on kanamycin-containing medium were obtained. The presence of the transgene in the transgenic plants was confirmed by PCR using genomic DNA as a template. DNA was isolated from the rosette leaves of 4-week-old plants using the Wizard genomic DNA extraction kit (Promega). Genomic DNA from wild-type plants was used as a control.
Molecular Analyses
Leaf genomic DNA for Southern-blot analysis was isolated according to Wang et al. (2001) and digested with HindIII. The digested products (10 μg DNA) were fractionated on 1.0% agarose gel, immobilized on a nylon membrane, and hybridized with full-length lipase cDNA as described by Wang and Arteca (1995).
For western-blot analysis, total protein was extracted from tissues at specified ages. The tissue (200 μg) was homogenized on ice with a mortar and pestle in 500 μL of extraction buffer (50 mm HEPES, pH 7.4). Homogenate protein was quantified according to Ghosh et al. (1988), fractionated on 10% SDS-polyacrylamide gels, and the separated proteins transferred to Hybond-P membranes (Amersham-Pharmacia). Immunoblotting was performed according to Wang et al. (2001) using a rabbit polyclonal antiserum specific for the AAD24845 protein (see below) as the primary antibody and alkaline phosphatase-conjugated goat anti-rabbit IgG (BioShop Canada) as the secondary antibody. Antibodies against AAD24845 were raised in rabbits using a synthetic peptide (CTNSRSVDSDEDEDSDN; Fig. 1) as antigen according to Kaup et al. (2002). Equal loading was confirmed by densitometry of corresponding SDS-PAGE gels stained with Coomassie Blue.
Chloroplast Isolation and Fractionation
For the purpose of western-blot analyses, chloroplasts were isolated from the rosette leaves of 4.5-week-old, soil-grown Arabidopsis rosettes using a Percoll gradient as described by Kunst (1998). The upper, large diffuse band containing broken chloroplasts was removed and the lower band containing intact chloroplasts was collected and used for chloroplast subfractionation. Chloroplastic membranes (including thylakoids and envelope membranes) and stroma were isolated from this fraction of intact chloroplasts according to Kunst (1998).
Chlorophyll was extracted and quantified as described by Porra et al. (1989).
Confocal Microscopy
Discs (4-mm diameter) were cut from the center portion of the first leaf pairs. Leaf discs were vacuum infiltrated for 10 min in 4% paraformaldehyde in phosphate-buffered saline (PBS) and stored at 4°C until stained. Discs were washed with PBS twice (30 min each) at room temperature to remove the fixative and once with 1% Triton X-100 in PBS to permeabilize the tissue. Discs were then gently shaken overnight at room temperature with purified lipase antibody (1:50) in PBS containing 1% bovine serum albumin (BSA), washed twice in PBS, and probed with goat anti-rabbit antibody conjugated to FITC (Sigma; 1:100 in PBS plus 1% BSA) for 2 h in the dark. Discs were then washed twice with PBS and mounted on slides in 70% glycerol. Nile Red staining was performed according to Fowler and Greenspan (1985). Briefly, a stock solution of 0.05% (w/v) Nile Red in acetone was diluted to 2.5 μg/mL in 75% glycerol, vortexed, and added directly to the tissue section before the coverslip was put in place.
Samples were observed using a Zeiss LSM 510 confocal laser-scanning microscope attached to an Axiovert inverted microscope.
Electron Microscopy
Segments of tissue (approximately 2 mm2) were cut from the center of first leaf pairs, vacuum infiltrated in 20 mm sodium phosphate buffer (pH 7.2), and fixed overnight at 4°C in 4% gluteraldehyde in 20 mm sodium phosphate buffer (pH 7.2). Samples were then washed four times in 20 mm phosphate buffer (pH 7.2), postfixed in 1% osmium tetroxide in 20 mm phosphate buffer (pH 7.2) for 2 h at 4°C, and washed four times in water. They were then dehydrated in a graded series of acetone, washed four times in 100% acetone, and embedded in Epon-Araldite. Ultrathin sections (70–90 nm) were stained in lead citrate and uranyl acetate, and examined with a Philips CM 10 electron microscope operating at 60 kV.
Preparation of Recombinant Fusion Proteins and Lipase Activity Assay
To obtain recombinant AAD24845 protein, two versions of At2g31690 cDNA were subcloned into the expression vector pMal-c2X (New England Biolabs), which produces MBP fusion proteins. Full-length cDNA (corresponding to the full-length putative precursor protein) was amplified by PCR using two primer adapters (U1, TCTTGTCGACATGGCCTTGATCCAAAACCC, SalI site underlined; D, TAGTAAGCTTTACGCTGACGTTGTACGAAT, HindIII site underlined) that incorporated restriction sites to facilitate subcloning. The PCR product was subcloned into pMal-c2X to form pMal-lipF, which encodes the recombinant fusion protein MBP-LipF. The truncated cDNA corresponding to the predicted mature form of the protein lacking the predicted transit peptide was obtained by eliminating the first 162 bp of the full-length cDNA. This was accomplished by amplification using PCR with primer-adapters (U2, TCTGTCGACGCACCAGTGATTCTAAATTCTCCGG, SalI site underlined; D, same as above). The PCR product was subcloned into pMal-c2X to form pMal-lipS, which encodes the recombinant fusion protein MBP-LipS. Both constructs were expressed in E. coli and purified according to the manufacturer's recommendations.
TAG lipase activity of the recombinant MBP fusion proteins was measured in vitro. To prepare the recombinant proteins, overnight cultures of each of the E. coli clones containing pMal-lipF, pMal-lipS, or pMal (empty control vector) were prepared in 2× yeast extract tryptone (YT) containing 50 μg/mL ampicillin and 0.2% (w/v) Glc. The next day, these cultures were used to inoculate 35 mL of fresh 2× YT containing 0.2% Glc and incubated at 37°C for 1.5 h. To induce expression of the recombinant fusion proteins, isopropylthio-β-galactoside was added to a final concentration of 0.3 mm and incubation continued at 30°C for 3.5 h. Cells were collected by centrifugation, resuspended in 1 mL of lysis buffer (100 mm Tris, pH 8.0, 150 mm NaCl, and 0.1% Triton X-100), and sonicated. The lysates were cleared by centrifugation and the soluble extracts were added to 200 μL of amylose resin slurry (New England Biolabs), which had been equilibrated with lysis buffer. After allowing the MBP fusion proteins to bind for at least 15 min at 4°C, the amylose resin was washed three times with lysis buffer. Another 1 mL of lysis buffer was added to the resin and the suspension was divided into four equal parts. Resin-bound protein was eluted from one aliquot with 20 mm maltose in lysis buffer for estimating the protein content (Bradford, 1976). Each tube contained approximately 1 μg protein. The other three aliquots were briefly centrifuged to pellet the resin, the supernatants were withdrawn, and the resin-bound protein was dried under vacuum. Ten microliters of trilinolein (18:2) were added to the resin-bound protein and the suspension was incubated at room temperature for 1 h. Forty microliters of 100 mm Tris-HCl (pH 7.5) plus 2 mm dithiothreitol were added, the suspension was vortexed for 15 s, and the reaction incubated overnight at room temperature.
Following overnight incubation, lipids were extracted essentially as described by Bligh and Dyer (1959). Briefly, 3 volumes of methanol:chloroform (2:1) were added, and the suspension was mixed well and incubated for 10 min at room temperature. One volume of chloroform and 0.8 volumes of 0.73% (w/v) NaCl were added, and the suspensions were mixed well and centrifuged briefly. The bottom organic layer was carefully withdrawn, transferred to a clean tube, and dried under nitrogen. Four hundred microliters of methanol:chloroform (6:1) were added to redissolve the dry sample. Twenty microliters of the sample were loaded onto a TLC plate (SIL-G25, 0.25-mm silica gel layer; Macherey-Nagel), the samples were fractionated by running in a nonpolar solvent (petroleum ether:diethyl ether:acetic acid; 70:30:1), and lipids were visualized with iodine. The iodine-stained TLC plate was then scanned with a densitometer. Alternatively, free fatty acids present in the samples following the overnight reactions were analyzed using a NEFA-C ACS-ACOD assay kit (Wako Chemicals), according to the manufacturer's recommendations. One-hundred-fifty microliters of each sample were used for the NEFA-C reaction.
In Vitro Translation and Chloroplast Protein Import Assays
For the purpose of preparing protein to be used for in vitro chloroplast import assays, cDNA encoding the putative lipase precursor protein was subcloned into the NheI and SalI recognition sites of pET21B (Novagen) following PCR using primer adapters, such that the corresponding protein contained its naturally occurring N terminus. [35S]Met-labeled protein was generated in vitro using a coupled transcription-translation system containing rabbit reticulocyte lysate according to the manufacturer's recommendations (Promega).
Arabidopsis (ecotype Columbia) seedlings were grown on agar plates supplemented with 0.5× Murashige and Skoog growth medium and 1% (w/v) Suc as described previously (Smith et al., 2002), and intact chloroplasts were isolated using a protocol described by Schulz et al. (2004) with minor modifications. Briefly, 20 to 30 g of tissue were harvested from 2- to 3-week-old seedlings, homogenized in ice-cold grinding buffer (50 mm HEPES-KOH, pH 7.5, 330 mm sorbitol, 2 mm EDTA, 1 mm MgCl2, 1 mm MnCl2, 50 mm ascorbic acid, 0.25% [w/v] BSA) using a PowerGen homogenizer (Fisher Scientific), and filtered through two layers of Miracloth. The filtrate was centrifuged at 1,000g for 8 min (JLA 10.5 rotor; Beckman-Coulter). The pellet was resuspended in 8 mL of grinding buffer and layered onto two Percoll step gradients as described (Smith et al., 2002). The gradients were centrifuged in a swinging-bucket rotor (JS13.1; Beckman-Coulter) at 7,700g for 15 min with slow acceleration and deceleration. Intact chloroplasts collected from the 85%-40% Percoll interface were washed in high-salt (HS) buffer (50 mm HEPES-KOH, pH 7.5, 330 mm sorbitol) and pelleted by centrifugation.
In vitro chloroplast import assays were performed essentially as described in Smith et al. (2002). Reaction mixtures contained freshly prepared, intact chloroplasts equivalent to 50 μg of chlorophyll, 50 mm HEPES-KOH, pH 7.5, 330 mm sorbitol, 5 mm magnesium acetate, 25 mm potassium acetate, 1 mm dithiothreitol, 1 mm GTP, 5 mm ATP, 10 mm Met, 4 μL of [35S]Met-labeled lipase precursor protein, and HS buffer to a final volume of 100 μL. Import reactions were allowed to proceed for 30 min at 26°C, reactions were stopped with the addition of 400 μL of ice-cold HS buffer, and chloroplasts were reisolated through a 40% Percoll cushion. The recovered chloroplasts were resuspended in 100 μL of HS buffer containing 2 mm CaCl2 and were treated without or with 100 μg/mL thermolysin for 30 min on ice. The reaction was stopped with the addition of EDTA to a final concentration of 20 mm and the chloroplasts were recovered by centrifugation of the reaction mixture for 5 min at 2,000g. Chloroplast proteins were resolved by SDS-PAGE and radiolabeled proteins were detected in dried gels using a phosphor imager (Typhoon; Amersham Biosciences).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers NP_180727, AAY78709, and AAD24845.
Acknowledgments
We are grateful to Mr. Dale Weber for expert technical assistance with microscopy and to Dr. Alexandre Joyeux for technical support.
This work was supported by the Natural Sciences and Engineering Research Council of Canada (Discovery grants to J.E.T. and M.D.S.), and Wilfrid Laurier University is gratefully acknowledged (by M.D.S.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: John E. Thompson (jet@sciborg.uwaterloo.ca).
Open Access articles can be viewed online without a subscription.
References
- Anderson MM, McCarty RE, Zimmer EA (1974) The role of galactolipids in spinach chloroplast lamellar membranes. I. Partial purification of bean leaf galactolipid lipase and its action on sub-chloroplast particles. Plant Physiol 53 699–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin II Jr, Frost E, Vidi P-A, Kessler F, Staehelin A (2006) Plastoglobules are lipoprotein subcompartments of the chloroplast that are permanently coupled to thylakoid membranes and contain biosynthetic enzymes. Plant Cell 18 1693–1703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey JL, Whyborn AG (1963) The osmiophillic globules of chloroplasts II. Globules of the spinach-beet chloroplasts. Biochim Biophys Acta 1143 113–134 [Google Scholar]
- Bechtold N, Ellis J, Pelletier G (1993) In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C R Acad Sci III Sci Vie 316 1194–1199 [Google Scholar]
- Beisson F, Ferte N, Bruley S, Voultoury R, Verger R, Arondel V (2001) Oil-bodies as substrates for lipolytic enzymes. Biochim Biophys Acta 1531 47–58 [DOI] [PubMed] [Google Scholar]
- Bligh EG, Dyer WJ (1959) A rapid method of total lipid extraction and purification. Can J Bot 37 911–917 [DOI] [PubMed] [Google Scholar]
- Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72 248–254 [DOI] [PubMed] [Google Scholar]
- Brick DJ, Brumlik MJ, Buckley JT, Cao J-X, Davies PC, Misra S, Tranbarger TJ, Upton C (1995) A new family of lipolytic plant enzymes with members in rice, Arabidopsis and maize. FEBS Lett 377 475–480 [DOI] [PubMed] [Google Scholar]
- Cohen A, Khozin-Goldberg I, Adlerstein D, Bigogno C (2000) The role of triacylglycerol as a reservoir of polyunsaturated fatty acids for the rapid production of chloroplastic lipids in certain microalgae. Biochem Soc Trans 28 740–743 [PubMed] [Google Scholar]
- Derewenda ZS, Derewenda U (1991) Relationships among serine hydrolases: evidence for a common structural motif in triacylglyceride lipases and esterases. Biochem Cell Biol 69 842–851 [DOI] [PubMed] [Google Scholar]
- Dhondt S, Geoffroy P, Stelmach BA, Legrand M, Heitz T (2000) Soluble phospholipase A2 activity is induced before oxylipin accumulation in tobacco mosaic virus-infected tobacco leaves and is contributed by patatin-like enzymes. Plant J 23 431–440 [DOI] [PubMed] [Google Scholar]
- El-Hafid L, Pham AT, Zuily-fodil Y, da Silva JV (1989) Enzymatic breakdown of polar lipids in cotton leaves under water stress. I. Degradation of monogalactosyl-diacylglycerol. Plant Physiol Biochem 27 495–502 [Google Scholar]
- Emanuelsson O, Nielsen H, Brunak S, von Heijne G (2000) Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol 300 1005–1016 [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, von Heijne G (1999) ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci 8 978–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feys BJ, Wiermer M, Bhat RA, Moisan LJ, Medina-Escobar N, Neu C, Cabral A, Parker JE (2005) Arabidopsis SENESCENCE-ASSOCIATED GENE 101 stabilizes the signals within an ENHANCED DISEASE SUSCEPTIBILITY1 complex in plant innate immunity. Plant Cell 17 2601–2613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler SD, Greenspan P (1985) Application of Nile red, a fluorescent hydrophobic probe, for the detection of neutral lipid deposits in tissue sections: comparison with oil red O. J Histochem Cytochem 33 833–836 [DOI] [PubMed] [Google Scholar]
- Galliard T (1971) The enzymatic deacylation of phospholipids and galactolipids in plants. Purification and properties of a lipolytic acyl hydrolase from potato tubers. Biochem J 121 379–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galliard T (1980) Degradation of acyl lipids: hydrolytic and oxidative enzymes. In PK Stumpf, ed, The Biochemistry of Plants, Vol IV. Academic Press, New York, pp 85–114
- Ghosh S, Gepstein S, Heikkila JJ, Dumbroff EB (1988) Use of a scanning densitometer or an ELISA plate reader for measurement of nanogram amounts of protein in crude extracts from biological tissues. Anal Biochem 169 227–233 [DOI] [PubMed] [Google Scholar]
- Ghosh S, Hudak KA, Dumbroff EB, Thompson JE (1994) Release of photosynthetic protein catabolites by blebbing from thylakoids. Plant Physiol 106 1547–1553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grechkin A (1998) Recent developments in biochemistry of the plant lipoxygenase pathway. Prog Lipid Res 37 317–352 [DOI] [PubMed] [Google Scholar]
- Harwood JL (1980) Plant acyl lipids: structure, distribution and analysis. In PK Stumpf, ed, Biochemistry of Plants, Vol 4. Academic Press, New York, pp 2–56
- He Y, Gan S (2002) A gene encoding an acyl hydrolase is involved in leaf senescence in Arabidopsis. Plant Cell 14 805–815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holk A, Rietz S, Zahn M, Quader H, Scherer GFE (2002) Molecular identification of cytosolic, patatin-related phospholipases A from Arabidopsis with potential functions in plant signal transduction. Plant Physiol 130 90–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Y, Wang TW, Hudak KA, Schade F, Froese CD, Thompson JE (2000) An ethylene-induced cDNA encoding a lipase expressed at the onset of senescence. Proc Natl Acad Sci USA 97 8717–8722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang AHC (1992) Oil bodies and oleosins in seeds. Annu Rev Plant Physiol Plant Mol Biol 43 177–200 [Google Scholar]
- Huang AHC (1996) Oleosin and oil bodies in seeds and other organs. Plant Physiol 110 1055–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiguro S, Kawai-Oda A, Ueda J, Nishida I, Okada K (2001) The DEFECTIVE IN ANTHER DEHISCENCE1 gene encodes a novel phospholipase A1 catalyzing the initial step of jasmonic acid biosynthesis, which synchronizes pollen maturation, anther dehiscence, and flower opening in Arabidopsis. Plant Cell 13 2191–2209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaniuga Z, Gemel J (1984) Galactolipase activity and free fatty acid levels in chloroplasts—novel approach to characteristics of chilling sensitivity of plants. FEBS Lett 171 55–58 [Google Scholar]
- Kaniuga Z, Saczynska V, Miskiewicz E, Garstka M (1999) Degradation of leaf polar lipids during chilling and post-chilling rewarming of Zea mays genotypes reflects differences in their response to chilling stress. The role of galactolipase. Acta Physiol Plant 21 45–56 [Google Scholar]
- Kaup MT, Froese CD, Thompson JE (2002) A role for diacylglycerol acyltransferase during leaf senescence. Plant Physiol 129 1616–1626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler F, Schnell D, Blobel G (1999) Identification of proteins associated with plastoglobules isolated from pea (Pisum sativum L.) chloroplasts. Planta 208 107–113 [DOI] [PubMed] [Google Scholar]
- Kim JY, Chung YS, Ok SH, Lee SG, Chung WI, Kim IY, Shin JS (1999) Characterization of the full-length sequences of phospholipase A2 induced during flower development. Biochim Biophys Acta 1489 389–392 [DOI] [PubMed] [Google Scholar]
- Kunst L (1998) Preparation of physiologically active chloroplasts from Arabidopsis. In JM Martínez-Zapater, J Salinas, eds, Methods in Molecular Biology: Arabidopsis Protocols, Vol 82. Humana Press, Totowa, NJ, pp 43–48 [DOI] [PubMed]
- Lee HY, Bahn SC, Kang YM, Lee KH, Kim HJ, Noh EK, Palta JP, Shin JS, Ryu SB (2003) Secretory low molecular weight phospholipase A2 plays important roles in cell elongation and shoot gravitropism in Arabidopsis. Plant Cell 15 1990–2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee R-H, Chen SCG (2002) Programmed cell death during rice leaf senescence is nonapoptotic. New Phytol 155 25–30 [DOI] [PubMed] [Google Scholar]
- Lichtenthaler HK (1969) The plastoglobuli of spinach: their size, isolation and lipoquinone composition. Protoplasma 68 65–66 [Google Scholar]
- Lichtenthaler HK, Weinert H (1970) The correlation between lipoquinone accumulation and plastoglobuli formation in the chloroplasts of Ficus elastica Roxb. Z Naturforsch 25b 619–623 [Google Scholar]
- Lo M, Taylor C, Wang L, Nowack L, Wang T-W, Thompson JE (2004) Characterization of an ultraviolet B-induced lipase in Arabidopsis. Plant Physiol 135 947–958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin BA, Wilson RF (1984) Subcellular localization of TAG synthesis in spinach leaves. Lipids 19 117–121 [DOI] [PubMed] [Google Scholar]
- Matos AR, d'Arcy-Lamet A, Franca M, Zuily-Fodil Y, Pham-Thi AT (2000) A patatin-like protein with galactolipase activity is induced by drought stress in Vigna unguiculata leaves. Biochem Soc Trans 28 779–781 [PubMed] [Google Scholar]
- Matos AR, d'Arcy-Lameta A, França M, Petres S, Edelman L, Kader J, Zuily-Fodil Y, Pham-Thi AT (2001) A novel patatin-like gene stimulated by drought stress encodes a galactolipid acyl hydrolase. FEBS Lett 491 188–192 [DOI] [PubMed] [Google Scholar]
- May C, Preisig-Müller R, Höhne M, Gnau P, Kindl H (1998) A phospholipase A2 is transiently synthesized during seed germination and localized to lipid bodies. Biochim Biophys Acta 1393 267–276 [DOI] [PubMed] [Google Scholar]
- Mignery GA, Pikaard CS, Park WD (1988) Molecular characterization of the patatin multigene family of potato. Gene 62 27–44 [DOI] [PubMed] [Google Scholar]
- Murphy DJ (2001) Biogenesis and functions of lipid bodies in animals, plants and microorganisms. Prog Lipid Res 40 325–438 [DOI] [PubMed] [Google Scholar]
- Murphy DJ, Vance J (1999) Mechanisms of lipid-body formation. Trends Biochem Sci 24 109–115 [DOI] [PubMed] [Google Scholar]
- Noll F, May C, Kindl H (2000) Phospholipid monolayer of plant lipid bodies attacked by phospholipase A2 shows 80 nm holes analyzed by atomic force microscopy. Biophys Chem 86 29–35 [DOI] [PubMed] [Google Scholar]
- Porra RJ, Thompson WA, Kriedemann PE (1989) Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: verification of the concentration of chlorophyll standards by atomic spectroscopy. Biochim Biophys Acta 975 384–394 [Google Scholar]
- Pozueta-Romero J, Rafia F, Houlne G, Cheniclet C, Carde JP, Schantz ML, Schantz R (1997) A ubiquitous plant housekeeping gene, PAP, encodes a major protein component of bell pepper chromoplasts. Plant Physiol 115 1185–1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racusen D, Foote M (1980) A major soluble glycoprotein of potato tuber. J Food Biochem 4 43–52 [Google Scholar]
- Rey P, Gillet B, Römer S, Eymery F, Massimino J, Peltier G, Kuntz M (2000) Over-expression of a pepper plastid lipid-associated protein in tobacco leads to changes in plastid ultrastructure and plant development upon stress. Plant J 21 483–494 [DOI] [PubMed] [Google Scholar]
- Sakaki T, Kondo N, Yamada M (1990. a) Free fatty acids regulate two galactosyltransferases in chloroplast envelope membranes isolated from spinach leaves. Plant Physiol 94 781–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaki T, Kondo N, Yamada M (1990. b) Pathway for the synthesis of triacylglycerols from monogalactosyl diacylglycerols in ozone-fumigated spinach leaves. Plant Physiol 94 773–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaki T, Saito K, Kawaguchi A, Kondo N, Yamada M (1990. c) Conversion of monogalactosyl diacylglycerols to triacylglycerols in ozone-fumigated spinach leaves. Plant Physiol 94 766–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller F (2001) Enzymes of the biosynthesis of octadecanoid-derived signaling molecules. J Exp Bot 52 11–23 [PubMed] [Google Scholar]
- Schardl CL, Byrd AD, Benzio G, Altschuler MA, Hildebrand DF, Hunt AG (1987) Design and construction of a versatile system for the expression of foreign genes in plants. Gene 61 1–11 [DOI] [PubMed] [Google Scholar]
- Schulz A, Knoetzel J, Scheller HV, Mant A (2004) Uptake of a fluorescent dye as a swift and simple indicator of organelle intactness: import-competent chloroplasts from soil-grown Arabidopsis. J Histochem Cytochem 52 701–704 [DOI] [PubMed] [Google Scholar]
- Small I, Peeters N, Legeai F, Lurin C (2004) Predotar: a tool for rapidly screening proteomes for N-terminal targeting sequences. Proteomics 4 1581–1590 [DOI] [PubMed] [Google Scholar]
- Smith MD, Fitzpatrick L, Keegstra K, Schnell DJ (2002) In vitro analysis of chloroplast protein import. In JS Bonifacino, M Dasso, JB Harford, J Lippinicott-Schwartz, KM Yamada, eds, Current Protocols in Cell Biology, Supplement 14. Wiley and Sons, New York, pp 11.16.1–11.16.21
- Smith MD, Ghosh S (2002) Lipid bodies of plastids. In A Goyal, SL Mehta, ML Lodha, eds, Reviews in Plant Biochemistry and Biotechnology, Vol 1. Indian Agricultural Research Institute, New Delhi, India, pp 209–216
- Smith MD, Licatalosi DD, Thompson JE (2000) Co-association of cytochrome f catabolites and plastid-lipid-associated protein with chloroplast lipid particles. Plant Physiol 124 211–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somerville C, Browse J, Jaworski JG, Ohlrogge JB (2000). Lipids. In RB Buchanan, W Gruissem, RL Jones, eds, Biochemistry and Molecular Biology of Plants. American Society of Plant Biologists, Rockville, MD, pp 456–527
- Sprey B, Lichtenthaler HK (1966) Zur frage der beziehungen zwischen plastoglobuli und thylakoidgense in gertenkeimlingen. Z Naturforsch 21b 697–699 [Google Scholar]
- Staehelin LA, Newcomb EH (2000) Membrane structure and membranous organelles. In RB Buchanan, W Gruissem, RL Jones, eds, Biochemistry and Molecular Biology of Plants. American Society of Plant Biologists, Rockville, MD, pp 2–50
- Steinmüller D, Tevini M (1985) Composition and function of plastoglobuli I. Isolation and purification from chloroplasts and chromoplasts. Planta 163 201–207 [DOI] [PubMed] [Google Scholar]
- Vidi P-A, Kanwisher A, Baginsky S, Austin JR, Csucs G, Dörmann P, Kessler F, Bréhélin C (2006) Tocopherol cyclase (VTE1) localization and vitamin E accumulation in chloroplast plastoglobule lipoprotein particles. J Biol Chem 281 11225–11234 [DOI] [PubMed] [Google Scholar]
- Wang TW, Arteca RN (1995) Identification and characterization of cDNAs encoding ethylene biosynthetic enzymes from Pelargonium × hortorum cv Snow Mass leaves. Plant Physiol 109 627–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang TW, Lu L, Wang D, Thompson JE (2001) Isolation and characterization of senescence-induced cDNAs encoding deoxyhypusine synthase and eukaryotic translation initiation factor 5A from tomato. J Biol Chem 276 17541–17549 [DOI] [PubMed] [Google Scholar]
- Xue L, McCune LM, Kleppinger-Sparace KF, Brown MJ, Pomeroy MK, Sparace SA (1997) Characterization of the glycerolipid composition and biosynthetic capacity of pea root plastids. Plant Physiol 113 549–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ytterberg AJ, Peltier J-B, Van Wijk KJ (2006) Protein profiling of plastoglobules in chloroplasts and chromoplasts. A surprising site for differential accumulation of metabolic enzymes. Plant Physiol 140 984–997 [DOI] [PMC free article] [PubMed] [Google Scholar]