Abstract
Proanthocyanidins (PAs; or condensed tannins) can protect plants against herbivores, contribute to the taste of many fruits, and act as dietary antioxidants beneficial for human health. We have previously shown that in grapevine (Vitis vinifera) PA synthesis involves both leucoanthocyanidin reductase (LAR) and anthocyanidin reductase (ANR). Here we report the characterization of a grapevine MYB transcription factor VvMYBPA1, which controls expression of PA pathway genes including both LAR and ANR. Expression of VvMYBPA1 in grape berries correlated with PA accumulation during early berry development and in seeds. In a transient assay, VvMYBPA1 activated the promoters of LAR and ANR, as well as the promoters of several of the general flavonoid pathway genes. VvMYBPA1 did not activate the promoter of VvUFGT, which encodes the anthocyanin-specific enzyme UDP-glucose:flavonoid-3-O-glucosyltransferase, suggesting VvMYBPA1 is specific to regulation of PA biosynthesis in grapes. The Arabidopsis (Arabidopsis thaliana) MYB transcription factor TRANSPARENT TESTA2 (TT2) regulates PA synthesis in the seed coat of Arabidopsis. By complementing the PA-deficient seed phenotype of the Arabidopsis tt2 mutant with VvMYBPA1, we confirmed the function of VvMYBPA1 as a transcriptional regulator of PA synthesis. In contrast to ectopic expression of TT2 in Arabidopsis, constitutive expression of VvMYBPA1 resulted in accumulation of PAs in cotyledons, vegetative meristems, leaf hairs, and roots in some of the transgenic seedlings. To our knowledge, this is the first report of a MYB factor that controls genes of the PA pathway in fruit, including both LAR and ANR, and this single MYB factor can induce ectopic PA accumulation in Arabidopsis.
Proanthocyanidins (PAs), also known as condensed tannins, are polyphenolic secondary metabolites synthesized via the flavonoid biosynthetic pathway. They are present in many plants and act in defense against plant diseases and in seed dormancy (Debeaujon et al., 2000; Peters and Constabel, 2002). The increase of PAs in important forage crops like alfalfa (Medicago sativa) could protect ruminants against pasture bloat, reduce greenhouse gas, and increase plant disease resistance (Dixon et al., 1996; McMahon et al., 2000). Dietary PAs are present in many fruits and plant products like wine, fruit juices, and teas and contribute to their taste and health benefits. PAs act as potential dietary antioxidants with beneficial effects for human health, including protection against free radical-mediated injury and cardiovascular disease (Bagchi et al., 2000; Middleton et al., 2000; Cos et al., 2004). There is also considerable interest in the PAs found in grapes (Vitis vinifera) because of their importance for the taste and astringency of red and white wine (Glories, 1988). For these reasons, there is a growing interest in metabolic engineering strategies aimed at developing agronomically important food crops and fruits with optimized levels and composition of flavonoids.
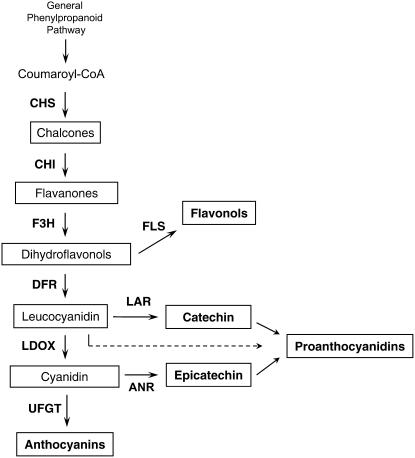
The biosynthesis of PAs, anthocyanins, and flavonols share common steps in the flavonoid pathway, and the genetics and biochemistry of this pathway (Fig. 1) have been characterized in several plant species including Arabidopsis (Arabidopsis thaliana) and grapes (Shirley et al., 1992; Holton and Cornish, 1995; Boss et al., 1996; Winkel-Shirley, 2001).
Figure 1.
Scheme of the flavonoid pathway leading to synthesis of anthocyanins, flavonols, and PAs. The enzymes involved in the pathway are shown as follows: CHS, chalcone synthase; CHI, chalcone isomerase; F3′H, flavonoid-3′-hydroxylase; F3′5′H, flavonoid-3′,5′-hydroxylase; F3H, flavanone-3β-hydroxylase; DFR, dihydroflavonol-4-reductase; LDOX, leucoanthocyanidin dioxygenase; FLS, flavonol synthase; LAR, leucoanthocyanidin reductase; ANR, anthocyanidin reductase; and UFGT, UDP-Glc:flavonoid-3-O-glucosyltransferase. The F3′H and F3′5′H enzymes, which hydroxylate flavanones and dihydroflavonols, were omitted to clarify the scheme of the flavonoid pathway (see Bogs et al., 2006). Note that Arabidopsis lacks LAR.
In Arabidopsis, the biosynthetic pathway leading to PA accumulation has been characterized by using the transparent testa (tt) and tannin-deficient seed (tds) mutants that fail to accumulate PAs in their seed coat (Shirley et al., 1995; Abrahams et al., 2002). Some of the identified TT and TDS loci correspond to enzymes of the general flavonoid pathway and others to enzymes, transporters, and regulators specifically involved in PA accumulation. The structural genes include anthocyanidin reductase (ANR; also called BANYULS) that catalyzes the synthesis of flavan-3-ols such as (−)-epicatechin (Xie et al., 2003), TT19, TT12, and Autoinhibited H+-ATPase isoform 10, which are involved in transport processes of PAs (Debeaujon et al., 2001; Kitamura et al., 2004; Baxter et al., 2005) and TT10 encoding a laccase-type polyphenol oxidase involved in polymerization of flavonoids (Pourcel et al., 2005).
Regulation of flavonoid synthesis occurs mostly via coordinated transcriptional control of the structural genes by the interaction of DNA-binding R2R3 MYB transcription factors, MYC-like basic helix loop helix (bHLH) and WD40 proteins (Mol et al., 1998; Nesi et al., 2000, 2001; Winkel-Shirley, 2001). This has been shown for several regulators of anthocyanin synthesis isolated from maize (Zea mays), Arabidopsis, Antirrhinum, and petunia (Petunia hybrida; Mol et al., 1998).
The Arabidopsis genes TT8, TT2, and TRANSPARENT TESTA GLABRA1 (TTG1), necessary for PA accumulation in the seed coat, were found to encode bHLH, MYB, and WD40 repeat proteins, respectively. By forming a transcription complex, they regulate the expression of several flavonoid structural genes including ANR and TT12 (Walker et al., 1999; Nesi et al., 2000, 2001; Baudry et al., 2004). Although these structural genes were also induced in roots after ectopic expression of TT2 and TT8, the tissue failed to accumulate PAs, suggesting additional factors are required for ectopic PA accumulation in Arabidopsis (Nesi et al., 2001). The transcription factors TT16, TT1, and TTG2 have been shown to influence expression of the PA-specific genes and the organ and cell development important for PA deposition (Johnson et al., 2002; Nesi et al., 2002; Sagasser et al., 2002).
However, the specific regulation of PA synthesis in plant species other than Arabidopsis is not well characterized and until now no MYB factor functionally similar to TT2 has been identified from other plants. Whereas PA synthesis in Arabidopsis is exclusively epicatechin based and limited to the seed coat, many other plants produce both epicatechin- and catechin-based PAs of various amounts and compositions and in a range of different tissues (Dixon et al., 2005). As Arabidopsis lacks a functional leucoanthocyanidin reductase (LAR) gene, it is not known what regulates expression of LAR in PA synthesis in other plants. In grape berries, the first committed steps in PA biosynthesis are catalyzed by LAR and ANR by converting anthocyanidins to flavan-3-ols such as catechin and epicatechin, respectively. Grapevine synthesizes PAs in various compositions in the seeds and skin of the fruit where their accumulation occurs during the early stages of grape berry development, prior to the onset of ripening (termed véraison by viticulturists). At véraison, the flavonoid pathway switches to production of anthocyanins in the skin of red grapes and the grapevine transcription factor VvMYBA, which is only expressed after véraison, has been shown to regulate anthocyanin synthesis in grapes during ripening (Kobayashi et al., 2002; Walker et al., 2007). The tissue and temporal-specific expression of ANR and LAR correlates with PA accumulation prior to véraison in grapes (Bogs et al., 2005), suggesting a separate transcriptional regulator controls PA synthesis in grapevine. Another grapevine transcription factor, VvMYB5a, was recently shown to be expressed in grapes prior to véraison but ectopic expression of this regulator in tobacco (Nicotiana tabacum) affected the metabolism of anthocyanins, flavonols, lignins, and PAs, suggesting it controls a number of different branches of the phenylpropanoid pathway (Deluc et al., 2006). It is not yet clear what transcription factors regulate expression of ANR and LAR in developing grape berries nor how PA synthesis is specifically regulated during fruit development.
In this article, we isolated and characterized a grapevine gene VvMYBPA1 that encodes a MYB transcription factor that is expressed when PAs accumulate during early grape berry development and in seeds. VvMYBPA1 is able to activate the promoters of both of the PA branch genes VvANR and VvLAR1 but also several of the general flavonoid pathway genes of grapevine. VvMYBPA1 did not activate anthocyanin synthesis, suggesting it may be specific for the PA pathway in grapes. Constitutive expression of this transcription factor in Arabidopsis complemented the PA-deficient seed coat phenotype of the Arabidopsis tt2 mutant and also induced ectopic PA accumulation in other tissues including cotyledons, stems, and roots. This grapevine regulator may provide the potential to alter PA synthesis in fruits and crops by metabolic engineering.
RESULTS
The Grapevine Gene VvMYBPA1 Encodes a MYB Transcription Factor
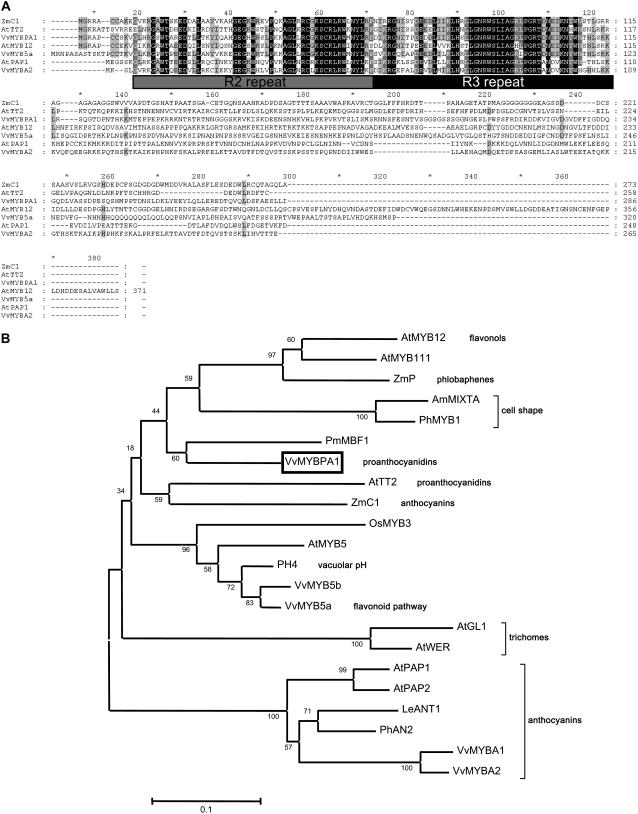
We identified the Tentative Consensus (TC) sequence TC46393 by searching the grape gene index of the The Institute for Genomic Research (TIGR) expressed sequence tag database (Quackenbush et al., 2000; http://www.tigr.org/tdb/tgi/) for MYB transcription factors expressed during early flower and berry development, when PAs are accumulating. The 861-bp open reading frame (ORF) of TC46393 was amplified by PCR from cDNA isolated from developing Shiraz berries sampled 1 week after flowering. The isolated ORF was named VvMYBPA1 (accession no. AM259485) and encoded a protein of 286 amino acid residues with a predicted mass of 32.2 kD and a calculated pI of 9.47. Analyses of the deduced amino acid sequence revealed that VvMYBPA1 contains an N-terminal R2R3 repeat that corresponds to the DNA-binding domain of plant MYB-type proteins (Fig. 2A). Similar to the over 100 members of the MYB protein family in Arabidopsis, the R2R3 repeat region of VvMYBPA1 is highly conserved and contains the motif [D/E]Lx2[R/K]x3Lx6Lx3R for interaction with bHLH proteins, whereas the C-terminal region shows little homology to other MYBs (Fig. 2A; Stracke et al., 2001). Phylogenetic analysis revealed the similarity of VvMYBPA1 to other plant MYB proteins (Fig. 2B). The R2R3 DNA-binding domain of VvMYBPA1 is most closely related to PmMBF1 (AAA82943) of Picea mariana with 81% identical amino acid residues. The PmMBF1 protein has not yet been functionally characterized. The similarity between the R2R3 domain of VvMYBPA1 and AtTT2 (Fig. 2A), which has been shown to regulate PA synthesis in the seed coat of Arabidopsis (Nesi et al., 2001), is clear with 72% amino acid identity. Besides VvMYBPA1 and TT2, the maize MYB factor C1 was also shown to activate the Arabidopsis ANR promoter, which is the first gene leading specifically to PA synthesis (Baudry et al., 2004). Figure 2A shows the comparison of these MYB proteins with the anthocyanin-specific factors VvMYBA2 and AtPAP1 (production of anthocyanin pigment) as well as with AtMYB12, a transcription factor controlling flavonol synthesis (Mehrtens et al., 2005). There are no conserved amino acids in the sequences of VvMYBPA1, TT2, and C1 that are not also present in the other MYB factors (Fig. 2A). Sequence similarity between MYB proteins is generally restricted to the R2R3 domain, but some MYB factors share conserved motifs in their C-terminal domains that may indicate similarities in function (Stracke et al., 2001). However, the C-terminal sequences of VvMYBPA1 (amino acids 116–286) and any other plant MYB factors showed no significant homology and none of the conserved motifs identified by Stracke et al. (2001) were present. The motif Vx2IRTKA[I/L]RC[S/N] conserved between TT2 and OsMYB3 (Nesi et al., 2001) was not found in the sequence of VvMYBPA1 (Fig. 2A).
Figure 2.
A, Alignment of the deduced amino acid sequences of the MYB-type transcriptional regulators ZmC1 (maize), AtTT2, AtPAP1, AtMYB12 (Arabidopsis), and the grapevine regulators VvMYBPA1, VvMYBA2, and VvMYB5a. The R2 and R3 repeats of the MYB domain are indicated below the alignment. Identical amino acids are indicated in black, similar amino acids in gray. Sequences were aligned with the ClustalW program and displayed using the GeneDoc program (Version 2.6.002). B, Phylogenetic tree showing selected plant MYB transcription factors from GenBank or EMBL database. Functions of some of the proteins are given in bold. The ClustalW multiple sequence alignment was formed using the domain of the MYB proteins and the default parameters of the MEGA package (Kumar et al., 2004). The tree was constructed from the ClustalW alignment using the neighbor-joining method by the MEGA program. The scale bar represents 0.1 substitutions per site and the numbers next to the nodes are bootstrap values from 1,000 replicates. The GenBank accession numbers of the MYB proteins are as follows: VvMYBPA1 (AM259485), AtGL1 (P27900), ZmP (P27898), ZmC1 (AAA33482), VvMYBA1 (BAD18977), VvMYBA2 (BAD18978), AtPAP1 (AAG42001), PhAN2 (AAF66727), LeANT1 (AAQ55181), OsMYB4 (T02988), AtMYB5 (U26935), PhMYB1 (Z13996), AmMixta (CAA55725), AtMYB12 (CAB09172), AtMYB111 (AAK97396), PmMBF1 (AAA82943), AtTT2 (Q9FJA2), PH4 (AAY51377), AtPAP1 (AAG42001), AtPAP2 (AAG42002), AtWER (CAC01874), VvMYB5a (AAS68190), and VvMYB5b (Q58QD0).
Taken together, the VvMYBPA1 protein sequence shows the typical features of a plant MYB transcription factor. However, we were not able to detect conserved amino acid homologies between the PA regulators TT2 and VvMYBPA1, which could be used to identify PA-specific MYB regulators from other plant species.
Expression of VvMYBPA1 Correlates with PA Accumulation during Fruit Development
To confirm that VvMYBPA1 is expressed when PAs are accumulating in grape berries, we investigated transcript levels of VvMYBPA1 throughout grape berry (cv Shiraz) development during the season 2000 to 2001 by real-time PCR. VvUbiquitin1 (BN000705) was chosen for normalization of gene expression because it was found to be relatively constant throughout grape berry development (Downey et al., 2003b; Bogs et al., 2005).
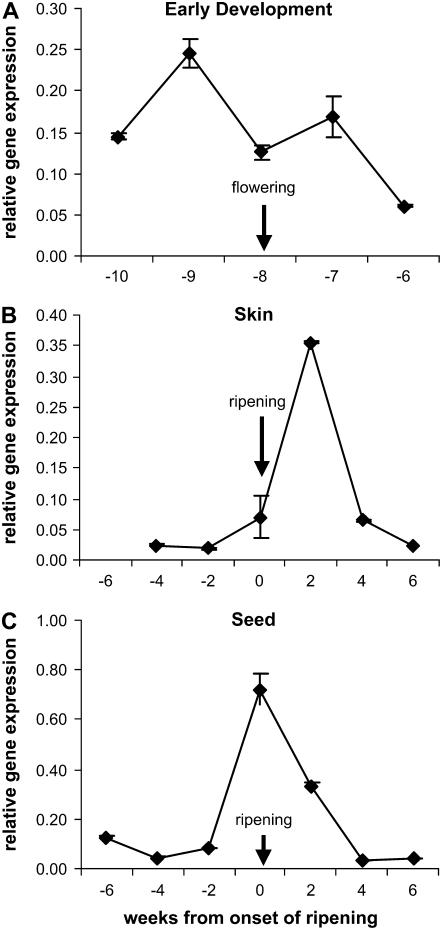
Figure 3 shows VvMYBPA1 is expressed in flowers and grapes early in berry development from 10 to 6 weeks before onset of ripening. This early expression of VvMYBPA1 in developing flowers and grape berries correlates with the accumulation of PAs and the expression of the structural genes leucoanthocyanidin dioxygenase (VvLDOX), VvANR, and VvLAR1 that are involved in PA synthesis in grapevine (Bogs et al., 2005).
Figure 3.
Transcript levels of VvMYBPA1 during grape flower and berry development. Gene expression was determined by real-time PCR and is shown relative to expression of VvUbiquitin1 in each sample. All data is presented as mean of three replicates with error bars indicating ±ses.
In grape berry skins, transcript levels of VvMYBPA1 were relatively low before véraison, which is the onset of ripening, increased to a maximum 2 weeks after véraison, and then declined to a low level (Fig. 3). The concentration of PAs in skins increased from 5 weeks before véraison, reaching a maximum around the time ripening commenced and then declined during ripening (Bogs et al., 2005). In seeds, VvMYBPA1 is expressed before véraison (Fig. 3) when PAs start to accumulate. The expression pattern of VvMYBPA1 in seeds (Fig. 3) correlates with PA synthesis and the expression of VvLAR2 that continued in the seed up until 4 weeks after véraison with a maximum at véraison (Kennedy et al., 2000; Bogs et al., 2005).
Promoter Isolation and Analysis of Grapevine Flavonoid Pathway Genes
To determine which genes of the flavonoid pathway are controlled by VvMYBPA1, we isolated the promoter regions of the genes flavonoid 3′,5′ hydroxylase (VvF3′5′H1; 1,136 bp; accession no. AM259482), chalcone isomerase (VvCHI; 935 bp; accession no. AM259483), VvANR (1,034 bp; accession no. AM259484), and VvLAR1 (1,342 bp; accession no. AM259481) by genome walking (see experimental procedures). These promoter regions were analyzed using the plant DNA cis-elements (PLACE) database (Higo et al., 1999; http://www.dna.affrc.go.jp/htdocs/PLACE/signalscan.html) and contained the consensus sequences of the core DNA-binding sites of MYB (CNGTTR, PLACE accession no. S000176) and bHLH-type (CANNTG, PLACE accession no. S000407) transcription factors. The core MYB site CNGTTR is recognized by the plant transcription factor MYB Ph3 from petunia (Solano et al., 1995), which is involved in regulation of flavonoid biosynthesis and is present in a 86-bp promoter region of the Arabidopsis AtANR promoter necessary for expression in PA-accumulating cells (Debeaujon et al., 2003). Additionally, we cloned the promoter regions of VvLDOX (2,174 bp of accession no. AF290432), UDP-Glc:flavonoid-3-O-glucosyltransferase (VvUFGT; 1,674 bp of accession no. AY955269), and AtANR (also called AtBAN; 1,533 bp of accession no. AT1G61720), which also contain the core DNA-binding sites for MYB and bHLH factors (Gollop et al., 2001; Kobayashi et al., 2001). There are different target recognition sites for different groups of MYB proteins (Jin and Martin, 1999) and in addition to the core MYB DNA-binding site, all promoters contained putative cis-acting regulatory elements for different MYB proteins (data not shown).
VvMYBPA1 Activates Promoters of the Flavonoid Pathway Genes Involved in PA Synthesis
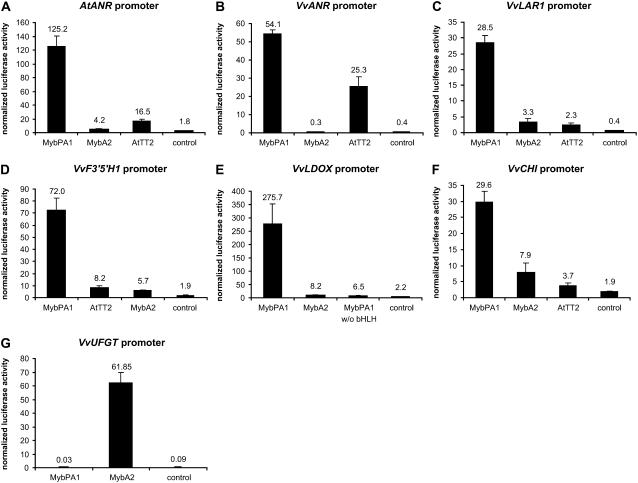
To investigate which structural genes of the flavonoid pathway are activated by VvMYBPA1, we established a transient expression method using grape cells grown in suspension culture and the dual-luciferase assay system. In this system, the cotransfection of effectors (transcription factors) and dual-luciferase reporter plasmids allows quantification of promoter activity by measuring firefly (Photinus pyralis) luciferase activity (promoter of interest inserted into pLuc), which is normalized by measuring Renilla reniformis luciferase activity (pRluc; Horstmann et al., 2004). Therefore, VvMYBPA1 was ligated to pART7 for expression under the control of the 35S promoter of Cauliflower mosaic virus (CaMV). The promoters controlling VvCHI, VvF3′5′H1, VvLDOX, VvANR, AtANR, VvLAR1, and VvUFGT were ligated to pLuc to control the expression of the firefly luciferase reporter gene. The VvCHI, VvF3′5′H1, and VvLDOX promoters were chosen as examples for general flavonoid pathway genes involved in synthesis of flavonols, anthocyanins, and PAs (Fig. 1). In contrast, VvUFGT is specifically required for anthocyanin synthesis, whereas VvANR and VvLAR1 encode the branch point enzymes leading to the synthesis of PAs (Fig. 1). Except for VvLAR1 and VvF3′5′H1, these genes are present as a single copy in the grapevine genome (Sparvoli et al., 1994; Bogs et al., 2005). The grapevine promoters and the Arabidopsis AtANR promoter were then tested as potential targets for VvMYBPA1 (Fig. 4). As controls, we also tested the promoters with the MYB transcription factor VvMYBA2 that specifically activates VvUFGT, controlling anthocyanin synthesis in grapes (Boss et al., 1996; Kobayashi et al., 2002) and TT2 from Arabidopsis that was shown to control AtANR expression and PA synthesis in Arabidopsis seed coat (Nesi et al., 2001). Similar results as for VvMYBA2 were obtained with its isoform VvMYBA1 when transfected with the VvUFGT or VvANR promoter and ENHANCER OF GLABRA3 (EGL3) in the transient expression system (Walker et al., 2007; data not shown).
Figure 4.
VvMYBPA1 activates promoters of flavonoid pathway genes involved in PA synthesis. The MYB transcription factors and promoters used for transfection of grape cell cultures are indicated. Control indicates the activity of the respective promoter transfected without a MYB factor. Each transfection (except E, MybPA1 without bHLH) contained the 35S∷EGL3 construct encoding the bHLH protein EGL3 (GenBank accession no. NM20235) from Arabidopsis and as internal control the Renilla luciferase plasmid pRluc (Horstmann et al., 2004). The normalized luciferase activity was calculated as the ratio between the firefly and the Renilla luciferase activity. Each column represents the mean value of three independent experiments with error bars indicating ±ses.
VvMYBPA1 strongly activated the promoters of the genes VvANR (approximately 135-fold), AtANR (approximately 70-fold), and VvLAR1 (approximately 72-fold), showing its ability to induce the PA-specific branch point genes of Arabidopsis and grapevine (Fig. 4, A–C). VvMYBPA1 also induced the promoters of the general flavonoid pathway genes VvCHI (approximately 16-fold), VvF3′5′H1 (approximately 38-fold), and VvLDOX (approximately 125-fold), suggesting it can activate the whole pathway leading to PA synthesis (Fig. 4, D–F). The anthocyanin-specific promoter of VvUFGT was not affected by VvMYBPA1, whereas VvMYBA2 strongly activated (approximately 600-fold) this promoter (Fig. 4G) but did not activate the other genes. In comparison to VvMYBPA1, the activation of the VvF3′5′H1, VvLDOX, VvCHI, VvANR, VvLAR1, and the AtANR promoter by VvMYBA2 was absent or relatively low (Fig. 4). Similar to VvMYBPA1, the Arabidopsis PA regulator TT2 activated the ANR genes of Arabidopsis and grapevine (Fig. 4, A and B) but TT2 was not able to substantially induce the promoters of VvLAR, VvF3′5′H1, VvLDOX, and VvCHI (Fig. 4, C and F). These results suggest VvMYBPA1 is a specific regulator of PA synthesis, potentially regulating the entire general flavonoid pathway and both of the PA branch genes ANR and LAR, leading to PA formation. Similar to other MYB transcription factors, VvMYBPA1 requires a bHLH protein for promoter activation (Fig. 4E; VvMYBPA1/w/o). Therefore, all standard transfections included a construct expressing EGL3 which encodes a bHLH protein involved in flavonoid pathway regulation in Arabidopsis (Ramsay et al., 2003; Walker et al., 2007). Figure 4 also shows that using VvMYBPA1 without a bHLH protein in our transfection experiments can induce the VvLDOX promoter up to 3-fold compared to the control without the MYB factor, and similar results (3- to 5-fold inductions) were obtained for other promoters and MYB factors (data not shown). However, in comparison to the 125-fold induction of the VvLDOX promoter by VvMYBPA1 and EGL3 we considered a 3-fold induction as insubstantial and possibly provoked by the large amounts of MYB factor and promoter DNA in the transfection assay.
In Arabidopsis, the bHLH protein TT8 has been shown to interact with TT2 and to be required for PA accumulation in the seed coat (Nesi et al., 2000). Therefore, we also tested the ability of TT2 and VvMYBPA1 to interact with TT8 and to activate the ANR promoters of Arabidopsis and grapevine. We found that the ANR promoter activities were not substantially altered when we exchanged EGL3 with TT8 in the transient assays (data not shown). This redundancy of EGL3 and TT8 was also found for their ability to interact with the MYB factors TT2, PAP1, or PAP2 and to activate the dihydroflavonol-4-reductase (AtDFR) promoter of Arabidopsis (Zimmermann et al., 2004).
VvMYBPA1 Complements the Arabidopsis tt2 Mutant PA-Deficient Phenotype
The MYB transcription factor TT2 has been shown to regulate PA synthesis in the seed coat of Arabidopsis and the seeds of tt2 mutants appear yellow due to the lack of PAs (Nesi et al., 2001). To confirm the function of VvMYBPA1 as a regulator of PA synthesis, VvMYBPA1 under the control of the CaMV 35S promoter was introduced into the tt2 mutant. The ORF of VvMYBPA1 was amplified from cDNA by PCR and inserted into the binary vector pART27 to give pART27MYBPA1, which contains kanamycin resistance for selection in planta. This construct was used to transform homozygous tt2 plants by Agrobacterium tumefaciens-mediated transformation.
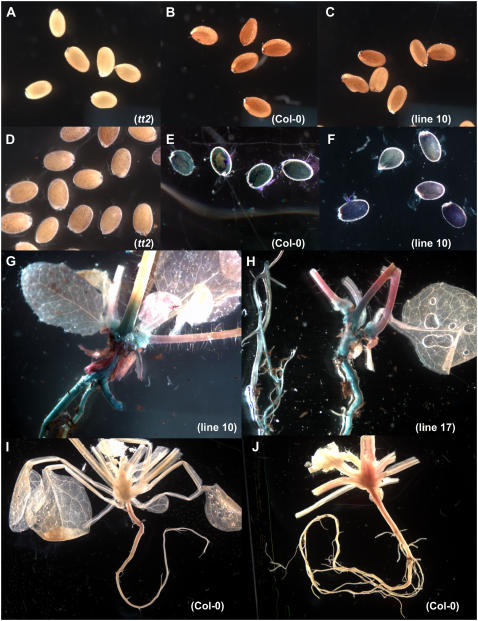
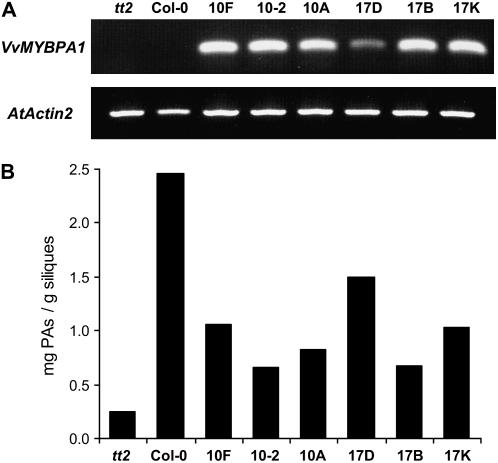
Only around 20% of the kanamycin-resistant T1 seedlings survived on soil and these were grown on to produce seed. Nine kanamycin-resistant tt2 plants independently transformed with pART27MYBPA1 showed wild-type phenotype and developed brown seeds which stained blue for accumulation of PAs when stained with dimethylaminocinnamaldehyde (DMACA; Fig. 5, A–F). DMACA is a useful reagent for detection of molecules of the PA pathway because it reacts with both PA monomers and polymers to form a blue chromophore but does not react with anthocyanidin derivatives (Nagel and Glories, 1991). This demonstrated that constitutive expression of the transgene VvMYBPA1 can complement the tt2 mutant seed phenotype. From the independent tt2 35S∷MYBPA1 lines 10 and 17, T2 plants were generated and analyzed for expression of VvMYBPA1 and PA accumulation. Expression of VvMYBPA1 by the tt2 35S∷MYBPA1 lines 10F, 10-2, 10A, 17D, 17B, and 17 K was confirmed by reverse transcription-PCR (Fig. 6A). Blue PA staining was observed in hypocotyls, roots, seeds, bases of the rosette leaves (Fig. 5), and stipules of leaves (data not shown) after these plants were treated with DMACA, whereas Columbia-0 (Col-0) wild-type plants accumulated PAs exclusively in the developing seeds (Fig. 5). HPLC analysis of the developing siliques revealed that the PA levels of tt2 plants complemented with VvMYBPA1 were 3- to 8-fold higher than in the tt2 background and reached about half of the PA concentration we detected in the Col-0 siliques (Fig. 6B).
Figure 5.
Complementation of the PA-deficient tt2 mutant seed phenotype by expression of VvMYBPA1 and ectopic PA formation of these plants. At least three independent lines showed a similar pattern of PA accumulation. A, Pale Arabidopsis tt2 seeds. B, Brown Col wild-type seeds. C, Brown tt2 35S∷MYBPA1 seeds. D to H, PAs were localized by staining the seeds or plants with DMACA, which specifically stains PAs blue. D, tt2 seeds. E, Col wild-type seeds. F, tt2 35S∷MYBPA1 seeds. G and H, PA formation in the hypocotyls, roots, and bases of the rosette leaves of tt2 35S∷MYBPA1 plants. PA formation in Arabidopsis wild-type plants (Col-0) after DMACA staining was exclusively observed in seeds (I and J).
Figure 6.
VvMYBPA1 expression in Arabidopsis tt2 mutant induced PA accumulation in developing siliques. A, Detection of VvMYBPA1 transcript in tt2 mutant, Col-0 wild-type and tt2 35S∷MYBPA1 lines 10F, 10-2, 10A, 17D, 17B, and 17 K by reverse transcription-PCR. Expression of the Arabidopsis Actin2 gene was used as a positive control. B, Quantification of PAs (total epicatechins) after acid-catalyzed hydrolysis of PA polymers in siliques of Arabidopsis tt2, Col-0 wild-type, and tt2 35S∷MYBPA1 lines 10F, 10-2, 10A, 17D, 17B, and 17 K by HPLC. Technical replicates could not be performed due to the limited amount of silique tissue.
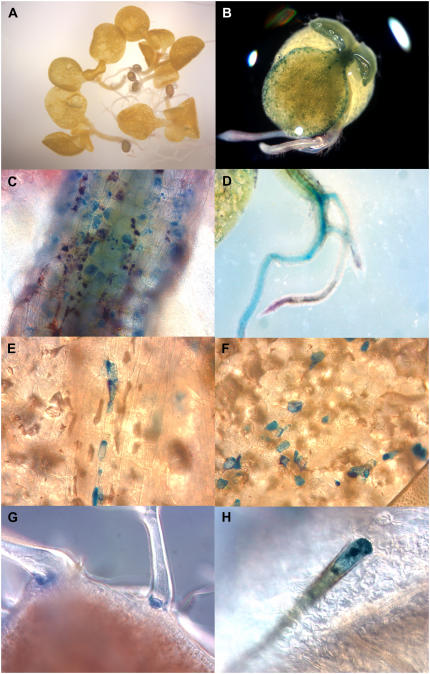
About 80% of the transgenic T1 seedlings showed growth abnormalities with bleaching and necrosis of the first leaves during their development. These plants showed a dwarf phenotype and died about 2 weeks after transferring them to soil. When these seedlings were stained with DMACA we observed accumulation of PAs in cells of cotyledons, hypocotyls, and its apical meristem, roots, trichomes, and basal cells of trichomes indicated by their blue staining (Fig. 7). Control plants (tt2 and Col-0) stained with DMACA did not show any blue staining, indicating their inability to accumulate significant amounts of PAs (Fig. 7, A, I, and J).
Figure 7.
PA accumulation of Arabidopsis tt2 seedlings expressing VvMYBPA1. Plants died before full development of their first leaves. PAs were localized by staining the plants with DMACA, which specifically stains PAs blue. At least five independent lines showed a similar pattern of PA accumulation. A, Arabidopsis tt2 seedlings as negative control. B, Whole tt2 35S∷MYBPA1 seedlings with PAs in the apical meristem and the cotyledons. C, Hypocotyl. D, Root. E and F, Cells of cotyledons. G, Basal cells of trichome. H, Trichome.
DISCUSSION
VvMYBPA1 Encodes a MYB-Type Transcriptional Regulator
In this study, we have identified the transcriptional regulator VvMYBPA1 from grapevine, which has some functional similarity to AtTT2, the only other regulator of PA synthesis isolated so far. Constitutive expression of VvMYBPA1 in Arabidopsis can complement a mutation in tt2 and experimental evidence suggests that VvMYBPA1 regulates PA synthesis during grape berry development. The protein sequence of VvMYBPA1 shows homology to the R2R3 domain of various MYB transcription factors (Fig. 2). However, we were not able to find any of the conserved motifs described by Stracke et al. (2001) in the C-terminal domain of VvMYBPA1 that may indicate similarities in function (Fig. 2). VvMYBPA1 does not display significantly more similarity to TT2 or C1 from maize, which have been shown to activate the ANR promoter (Nesi et al., 2001), than to other functionally unrelated MYB regulators (Fig. 2). Therefore, we were not able to identify conserved amino acid motifs, which could be used to identify PA-specific MYB regulators from other plant species. However, our transient expression assays and complementation experiments indicate that VvMYBPA1 and TT2 share some common functions, suggesting that VvMYBPA1 and TT2 are similar MYB factors. Similar findings were described for AN2 from petunia and C1 from maize, where the function of the proteins in regulating anthocyanin synthesis was not reflected in specific amino acid similarities (Quattrocchio et al., 1998, 1999).
Expression of VvMYBPA1 Correlates with PA Synthesis in Grapes
The functional role of VvMYBPA1 in the regulation of PA synthesis in grape berries is supported by its gene expression pattern during grape berry development (Fig. 3). Development of the grape berry occurs in two successive growth phases and the synthesis of flavonols, anthocyanins, and PAs and the expression of different flavonoid pathway genes is temporally separated during berry development. The first phase, from flowering until véraison (the onset of ripening in grapes), coincides with flavonol and PA synthesis and the second phase, starting with the onset of ripening of the berry, coincides with anthocyanin biosynthesis (Robinson and Davies, 2000; Downey et al., 2003a, 2003b; Bogs et al., 2005). The biosynthesis of PAs, anthocyanins, and flavonols share common steps in the flavonoid pathway, whereas the activities of enzymes specific for PAs, anthocyanins, or flavonols lead exclusively to the synthesis of the respective flavonoid (Fig. 1). Therefore, regulation of this pathway must occur to coordinate synthesis of different flavonoids during grape berry development. Regulation of flavonoid synthesis occurs mostly via coordinated transcriptional control of the structural genes by the interaction of DNA-binding MYB transcription factors and bHLH proteins (Mol et al., 1998). We have demonstrated that the VvMYBPA1 gene, expressed during flower and early berry development and in seeds before ripening, encodes a MYB transcription factor (Fig. 3). The expression pattern correlates with PA accumulation and expression of the PA-specific biosynthetic genes VvLAR1, VvLAR2, and VvANR (Bogs et al., 2005). PA synthesis appears to continue in the seed up until 2 to 4 weeks after véraison (Kennedy et al., 2000), which coincides with the expression pattern of VvLAR2 (Bogs et al., 2005) and VvMYBPA1 in seeds (Fig. 3). Both VvLAR2 and VvMYBPA1 expression reached their maximum in seeds at véraison and this corresponds to the peak of PA monomer accumulation at this time in seeds (Bogs et al., 2005). In grape skins, PA accumulation appeared to be complete by véraison and maximum transcript levels of VvANR and VvLAR2 were detected 4 weeks before véraison (Bogs et al., 2005). The transcript level of VvMYBPA1 in skins was relatively low before véraison, increased to a maximum 2 weeks after véraison, and then declined to a low level (Fig. 3). It is unclear if VvMYBPA1 expression before véraison is sufficient to induce PA synthesis in skins or whether another regulator activates PA synthesis in this tissue. However, a maximum of VvMYBPA1 expression in skins could take place earlier than 4 weeks before véraison, where we have no expression data because it was not possible to separate seeds and skins from very small berries. This would explain VvMYBPA1 expression in skins before véraison, but the reason for its relatively high transcript level 2 weeks after véraison remains unclear. It is possible that in the skin of ripening grapes VvMYBPA1 activates the shared genes of the flavonoid pathway required for anthocyanin synthesis, as the grape color regulator VvMYBA appears to be relatively specific for UFGT (Fig. 4). Taken together, the expression pattern of VvMYBPA1 suggests that the encoded protein is involved in regulation of PA biosynthesis in grapevine at least during early fruit development and in seeds.
VvMYBPA1 Activates the Promoters of General Flavonoid Pathway and PA Branch Genes
As well as activating the promoters of the two PA-specific genes, VvANR and VvLAR, VvMYBPA1 also activated the promoters of general flavonoid pathway genes, whereas the Arabidopsis TT2 regulator appeared to be more specific for ANR in this transient assay (Fig. 4). Anthocyanin and PA biosynthesis share the general flavonoid pathway enzymes until LDOX catalyzes the synthesis of anthocyanidins (Abrahams et al., 2003), which are substrates for both anthocyanin synthesis via UFGT and PA synthesis catalyzed by ANR and LAR (Fig. 1). Therefore, UFGT and ANR/LAR are branch enzymes leading to anthocyanin or PA accumulation, respectively, and represent possible control points between the two flavonoid branches. Our results obtained with the transient promoter assay revealed the ability of VvMYBPA1 to activate the PA-specific branch genes VvANR and VvLAR1, showing its capacity to control PA synthesis in grapevine. Furthermore, VvMYBPA1 was not able to induce the VvUFGT promoter, suggesting it specifically regulates PA biosynthesis. In addition, we determined that VvMYBA2, which was shown to induce anthocyanin accumulation in grapes (Kobayashi et al., 2002; Walker et al., 2007), activates the VvUFGT promoter but not the VvANR or VvLAR1 promoters, showing it specifically controls anthocyanin synthesis. These results suggest that in grapes the transcription factors VvMYBA2/VvMYBA1 and VvMYBPA1 control whether anthocyanins or PAs are synthesized by regulating expression of VvUFGT and VvANR/VvLAR, respectively. However, we cannot exclude that there are additional MYB transcription factors involved in regulation of anthocyanin and/or PA synthesis in grapevine.
In maize, most of the genes involved in anthocyanin synthesis are controlled by the MYB factor C1 interacting with the bHLH protein R (Goff et al., 1992), whereas the synthesis of phlobaphene, a pigmented polymeric flavonoid compound, is controlled by the MYB factor P1, independent of a bHLH protein, and MYB factor P1 activates only a subset of flavonoid biosynthetic genes (Grotewold et al., 1994). Interestingly, the change of six amino acids in the MYB domain of P1 allows the protein to interact with R and activate all the anthocyanin genes, resulting in anthocyanin formation (Grotewold et al., 2000). These results emphasize the importance of the interaction between MYB and bHLH proteins for the regulatory specificity of transcription factors with very similar DNA-binding domains.
For the grapevine MYB factors VvMYBA and VvMYBPA1 the interaction with a bHLH protein (EGL3) was essential for their ability to activate gene expression (Fig. 4E). However, as we used EGL3 as bHLH protein in all our promoter experiments, their specificity to control either the anthocyanin-specific gene UFGT or the PA-specific genes ANR and LAR seems not to depend on the identity of the interacting bHLH protein (Fig. 4). The difference in specificity of VvMYBA, VvMYBPA1, and TT2 is likely to be due to their ability to bind to different sites in the promoters of grapevine and Arabidopsis genes. For example, differences of the MYB factors would change their ability to bind the promoters and/or other unidentified factors which are present in the grapevine cell cultures.
Another grapevine MYB, VvMYB5a, recently identified by Deluc et al. (2006) is also expressed early in berry development when PAs are synthesized. Ectopic expression of VvMYB5a in tobacco increased expression of a number of structural genes in the phenylpropanoid pathway and modified the levels of anthocyanins, PAs, flavonols, and lignins in the flowers. The transgenic tobacco flowers had increased levels of epicatechin but not catechin but it is not yet clear whether VvMYB5a can activate the genes specifically involved in PA synthesis, such as ANR and LAR, or whether it controls the pathway generally, leading to an increase in all flavonoids.
Control of the pathway in grapevine is thus analogous to that in Arabidopsis, where the MYB transcription factor TT2 controls PA synthesis, whereas PAP1/PAP2 control anthocyanin biosynthesis. However, the Arabidopsis flavonoid MYB factors seem to control different steps in the flavonoid pathway than the similar factors from grapevine. In previous studies it was shown that TT2 controls expression of the flavonoid late biosynthetic genes including DFR, LDOX, AtANR (BAN), and TT12, whereas transcript levels of the flavonoid early biosynthetic genes like CHS, CHI, F3′H, or F3H were not affected by TT2 (Nesi et al., 2001). In contrast, our quantitative analysis of VvCHI, VvF3′5′H1, VvLDOX, VvANR, and VvLAR1 promoter activities revealed that VvMYBPA1 is able to induce promoters of both early and late flavonoid biosynthetic genes (Fig. 4). Thus, regulation of the flavonoid pathway in developing and ripening grape berries appears to differ from that in the Arabidopsis seed coat. The grapevine regulator of anthocyanin synthesis, VvMYBA2, appears to substantially activate only the promoter of UFGT and other factors appear to be required to activate the other structural genes of the pathway to synthesize anthocyanins. In contrast, VvMYBPA1 appears capable of activating the shared genes of the pathway as well as both of the PA-specific steps (ANR and LAR) and may be sufficient to induce PA synthesis in both the seeds and skin of the fruit. These different transcription factors appear to contribute to the temporal and spatial regulation of the flavonoid pathway in grapes to provide for PA synthesis in the skin and seeds of the developing berry and only anthocyanin synthesis in the skin of the ripening fruit.
VvMYBPA1 Complements the tt2 Seed Phenotype and Can Induce Ectopic PA Accumulation in Arabidopsis
The complementation of the PA-deficient seed phenotype of the tt2 mutant by VvMYBPA1 indicates that this grapevine transcription factor is capable of substituting for the function of the Arabidopsis PA regulator TT2 (Fig. 5). Although TT2 controls PA synthesis in the Arabidopsis seed coat and its ectopic expression was shown to induce expression of the AtANR promoter, Arabidopsis 70S∷TT2 plants failed to accumulate PAs in any tissue other than seeds (Nesi et al., 2001). In contrast, Arabidopsis 35S∷MYBPA1 plants accumulated PAs in their cotyledons, hypocotyls, roots, trichomes, and the bases of rosette leaves as well as in seed coats (Figs. 5 and 7). A similar organ- and cell-specific pattern of PA accumulation was observed in Arabidopsis plants simultaneously overexpressing TT2 and PAP1, which reflected the ANR promoter activity in Arabidopsis (Sharma and Dixon, 2005). Our promoter studies (Fig. 4) and analysis of the 35S∷MYBPA1 Arabidopsis plants (Figs. 5 and 7) suggest that in grapevine VvMYBPA1 regulates the whole flavonoid pathway branch leading to PA synthesis (early and late flavonoid biosynthetic genes), whereas in Arabidopsis TT2 controls only the late flavonoid biosynthetic genes (Nesi et al., 2001). Therefore, coexpression of TT2 and PAP1 was needed for induction of the early and late flavonoid biosynthetic genes and ectopic formation of PAs in Arabidopsis (Sharma and Dixon, 2005), whereas the grapevine MYBPA1 appears capable of activating PA synthesis without additional MYB genes. The majority of our 35S∷MYBPA1 transgenic seedlings developed growth abnormalities and accumulated relatively high levels of PAs in roots, hypocotyl, and apical meristem, which could have lead to the death of these plants before they fully developed their first leaves. Presumably, these plants expressed much higher levels of VvMYBPA1 transcript than those lines (e.g. lines 10 and 17) that grew past this stage. Similar toxic effects for Arabidopsis were observed by constitutive coexpression of TT2, PAP1, and the maize regulatory gene leaf color, which resulted in PA formation in roots and leaves and death of the plants (Sharma and Dixon, 2005). As Arabidopsis wild-type plants accumulate PAs only in the seed coat, it may be that only these specific cells are equipped to accumulate PAs. Transgenic plants ectopically synthesizing PAs in other cells may not be able to process the PA or compartmentalize it into specific cell types or vacuoles so the PAs or their precursors could become toxic in these cells, resulting in death of the plants. Alternatively, overexpression of VvMYBPA1 may induce formation of other cytotoxic metabolites or could lead to other dysfunction in these plants.
It would be interesting to determine whether ectopic expression of VvMYBPA1 in plants like alfalfa and clover (Trifolium repens) can induce PA formation in leaves or other tissue, because it is of great interest to engineer PAs in forage crops to reduce the risk of pasture bloat for ruminants (Dixon et al., 2005). However, our present results showing that PA formation after overexpression of VvMYBPA1 is limited to certain cell types suggest that additional factor(s) may be needed for ectopic accumulation of PAs, at least in Arabidopsis. Recently, Xie et al. (2006) have shown that expression of ANR in alfalfa or coexpression of ANR and PAP1 in tobacco can induce ectopic PA accumulation. It would be interesting to test whether VvMYBPA1 is also able to induce PA accumulation in these plants.
Unlike Arabidopsis, grapevine synthesizes PAs of different polymer lengths and composition in leaves, flowers, and in the skin and seeds of the developing fruit (Kennedy et al., 2001; Bogs et al., 2005). It appears that regulation of PA synthesis in grapes differs from the regulation of PA in Arabidopsis, which is restricted to the developing seed coat. In grapes, VvMYBPA appears capable of activating the both the early and late shared genes of the flavonoid pathway as well as the PA-specific genes encoding both ANR and LAR. There is considerable interest in grape PAs because of their importance for the color and taste of wine and their antioxidant capacity promoting health benefits in a number of model cell and animal systems (Bagchi et al., 2000). Our data suggest that VvMYBPA1 regulates PA formation in grapes and alteration of VvMYBPA1 expression may have the potential to manipulate the amount and composition of PAs in grape berries and possibly in other fruits.
MATERIALS AND METHODS
Plant Material
Grapevine (Vitis vinifera) tissues of cultivar Shiraz were collected from a commercial vineyard during the 2000 to 2001 season. Approximately 100 berries from at least 20 bunches were collected at weekly intervals throughout berry development from floral initiation until harvest, as described in Downey et al. (2003a). All samples were frozen in liquid nitrogen upon collection in the field and stored at −80°C until analyzed.
Arabidopsis (Arabidopsis thaliana) Col-0 and tt2 (SALK_005260) seeds were provided by The Arabidopsis Biological Resource Center.
Preparation of cDNAs
Total grapevine RNA was isolated from the various plant tissues as described in Downey et al. (2003b). Arabidopsis RNA was isolated from leaves with RNeasy kit (Qiagen) following the suppliers protocol. The quality of RNA was verified by demonstration of intact ribosomal bands following agarose gel electrophoresis in addition to the absorbance ratios (A260/280) of 1.8 to 2.0. For cDNA synthesis, 4 μg of grapevine or 1 μg of Arabidopsis total RNA were reverse transcribed using oligo d(T)18 and SuperScript III reverse transcriptase (Invitrogen Life Technologies) following the protocol of the supplier.
Cloning of VvMYBPA1 and Plant Transformation
The ORF of VvMYBPA1 was inserted into the binary vector pART27 for expression of the gene in Arabidopsis. Therefore, the VvMYBPA1 ORF was amplified by PCR from grapevine (cv Shiraz) cDNA (from RNA isolated 10 weeks before véraison) using PfuTurbo polymerase (Stratagene) and the primers MybPAartF (5′-TGAGGTACCGAGAGAGATATGGGCAGAGCAC-3′) and MybPAartR (5′-TGAGGATCCTGATCTTTTGGTCTCTCTGCAA-3′). The generated PCR fragment was purified, digested with BamHI and KpnI, and cloned in the vector pART7 (Gleave, 1992) to give pART7MYBPA1 where VvMYBPA1 is under the control of the CaMV 35S promoter. The nucleotide sequence of the VvMYBPA1 ORF (accession no. AM259485) in pART7MYBPA1 was determined by DNA sequencing. The expression cassette present in pART7MYBPA1 was isolated as NotI fragment, cloned into the NotI site of the binary vector pART27 (Gleave, 1992), and transferred into Agrobacterium tumefaciens (AGLI) by electroporation. Arabidopsis tt2 (SALK005260) ecotype Col was transformed using the floral-dip method (Clough and Bent, 1998). T1 transgenic plants were selected on one-half-strength Murashige and Skoog media containing 8 g/L agar and 35 mg/L kanamycin. Kanamycin-resistant T1 seedlings were transferred to soil and grown at 20°C in a growth chamber (Phoenix Biosystems) with a 16-h day length and a light intensity of 150 μmol m−2 s−1. Seeds of individual self-fertilized T2 lines were collected and single-copy insertion lines were selected based on a Mendelian segregation ratio.
HPLC Analysis and DMACA Staining of PAs
Immature Arabidopsis siliques were finely ground and 80 mg were extracted in 400 μL 70% (v/v) acetone containing 0.1% (w/v) ascorbate for 18 h at room temperature on a rotating wheel in darkness. Samples were centrifuged and 300 μL aliquots of the supernatant were transferred to fresh tubes and vaccum dried at 35°C for 60 min. The pellet was resuspended in 100 μL phloroglucinol buffer (0.25 g ascorbate, 1.25 g phloroglucinol, 215 μL concentrated HCl, 25 mL methanol) and incubated at 50°C for 20 min, then neutralized with 100 μL sodium acetate (200 mm, pH 7.5) for the analysis of PAs. Reverse-phase HPLC was used for analysis of PAs as described by Downey et al. (2003a).
The presence of PAs in plant tissue was detected by staining the tissues with DMACA solution (1% DMACA, 1% 6 n HCl in methanol). Dried seeds were stained for 6 to 14 h and seedlings for 10 to 30 min. The tissues were then transferred to distilled water and blue staining of the tissue was visualized with a microscope and documented using a digital camera.
Cloning of the Reporter and Effector Constructs for the Transient Promoter Assays
The Universal GenomeWalker kit (CLONTECH) was used to isolate promoter fragments of VvCHI, VvF3′5′H1, VvANR, and VvLAR1. Four libraries of adaptor-ligated genomic fragments were constructed from grapevine (Shiraz) genomic DNA restricted by DraI, EcoRV, PvuII, or StuI endonucleases and generated according to the GenomeWalker protocol. These genomic DNA libraries served as templates for the promoter isolation. Outer and nested gene-specific primers were designed to the 5′ ends of the cDNA sequences of VvCHI (accession no. X75963), VvF3′5′H1 (accession no. AJ880356), VvANR (accession no. CAD91911), and VvLAR1 (accession no. AJ865336) and primary and secondary PCRs were performed with the outer adapter primer AP1 and the nested adapter primer AP2, respectively. Primer design and PCR conditions for genome walking were performed according to the manufacturer's instructions. The amplified promoter fragments of the nested PCRs were cloned into pDrive (Qiagen) and sequenced. These DNA sequences were then used to design specific primers for the amplification of the respective promoter from grapevine (Shiraz) genomic DNA using PfuTurbo polymerase (Stratagene). The primers used for these PCR reactions contained restriction sites (in bold) for cloning the promoters into the luciferase reporter vector pLuc (Horstmann et al., 2004) as a BamHI/XhoI fragment. Their DNA sequences were as follows: CHIf (5′-ATAGGATCCTGGAATTATGGAAGACAAATAGTCAA-3′), CHIr (5′-TTACTCGAGGATATGGCTGCAGAGAAACGA-3′), ANRf (5′-CGAGGATCCCATTCATAGTCAAATTACAAAAATCAA-3′), ANRr (5′-ATACTCGAGATATGCCCTCACTTCCAAATTC-3′), F35Hf (5′-CGAGGATCCCAAAAAGAGTTGGAAATACAACGA-3′), F35Hr (5′-ATACTCGAGTGAGTATAGGATAGTGAAGGTGGCTAT-3′), LARf (5′-CGAGGATCCTCGGAATAATTTCATAGGGCTTT-3′), and LARr (5′-ATACTCGAGTCTGATGATGCTTCTTCTCTACTACTC-3′).
A 1,674-bp DNA fragment of the VvUFGT promoter (accession no. AY955269) was amplified by PCR from the plasmid pART7UFGT:GFP (gift from Paul Boss, Commonwealth Scientific and Industrial Research Organization, Australia) using Pfx polymerase (Invitrogen) with the primers UFGTp2F (5′-ACGGGATCCTCATGCGTCCACCTATTATCAA-3′) and UFGTpR (5′-GTACTCGAGGGTTGGAATGGGGGATGTTA-3′). A 1,533-bp DNA fragment of the AtANR promoter was amplified with PfuTurbo polymerase using the primers AtANRf (5′-CGAGGATCCCTGGGAAGACAATCGCTTTA-3′) and AtANRr (5′-ATCTCGAGTTGAAATTACAGAGATAGAGATTTAGTTG-3′). A 2,174-bp DNA fragment of the VvLDOX promoter was amplified with PfuTurbo polymerase using the primers LDOXf (5′-CGAGGATCCGTTTGCTTCCATCCCAATCTCACT-3′) and LDOXr (5′-TGTCTCGAGAAATATCACTGATCTACTTGTTTTCC-3′). These PCR fragments were gel purified, digested with BamHI and XhoI, and cloned between the respective sites of the vector pLuc. All described grapevine promoters were amplified from grapevine (Shiraz) genomic DNA.
For transient expression of TT2 and VvMYBA2 their ORFs were amplified from cDNA by PCR using PfuTurbo polymerase and cloned into the vector pART7, which contains the CaMV 35S constitutive promoter. Therefore, TT2 was amplified using the primers TT2F (5′-AGGTCGACATGGGAAAGAGAGCAACTACTAGTG-3′) and TT2R (5′-TACTCGAGTCAACAAGTGAAGTCTCGGAGC-3′) from cDNA of Col-0 siliques. The PCR fragment was digested with SalI and XhoI and ligated into pART7, digested with the same enzymes. The ORF of VvMYBA2 was amplified from grapevine postvéraison berry skin cDNA using the primers MybAF (5′-CGCCTCGAGCTCGATGGAGAGCTTAGGAGTTAG-3′) and MybAR (5′-CGCTCTAGATAAATCAGATCAAATGATTTACTT-3′). The PCR fragment was digested with XhoI and XbaI and ligated into pART7, digested with the same enzymes. All described PCR fragments were subjected to DNA sequencing before analysis in the transient assay system.
Transient Transfection Experiments and Dual Luciferase Assay
A transient assay was developed using a cell suspension of a Chardonnay petiole callus culture, using the methods detailed in Takos et al. (2006), modified from Torregrosa et al. (2002), and utilizing a dual luciferase system (Horstmann et al., 2004). In brief, gold particles were coated with a mixture of DNA constructs (150 ng of the respective plasmid, giving a total plasmid concentration of 750 ng/shot) and used to bombard immobilized grape cells. After incubation in the dark at 27°C for 48 h, the harvested cells were assayed for the luciferase activities using the dual-luciferase reporter assay system (Promega) measured with TD-20/20 Luminometer (Turner Design). The relative luciferase activity was calculated as the ratio between the firefly (Photinus pyralis) and the Renilla (control) luciferase activity. All transfection experiments were performed in triplicate and each set of promoter experiments was repeated with similar relative ratios to the respective control.
Expression Analysis of VvMYBPA1
Transcript levels of VvMYBPA1 in grapevine were measured by real-time PCR, using SYBR green method on a Rotor-Gene 2000 (version 4.2) real-time cycler (Corbett Research). Each PCR reaction (15 μL) contained: 266 nm primer (each), cDNA (diluted 1:60), and 1× ABsolute QPCR SYBR Green ROX mix (ABgene House). The thermal cycling conditions were 95°C for 15 min followed by 95°C for 30 s, 58°C for 25 s, and 72°C for 25 s for 30 or 35 cycles, followed by a melt cycle from 50°C to 96°C.
The expressed sequence tag clone TC46393 (TIGR database) was used to design the primers MYBPA1F (5′-AGATCAACTGGTTATGCTTGCT-3′) and MYBPA1R (5′-AACACAAATGTACATCGCACAC-3′) that were used to detect the transcript level of VvMYBPA1 in grapevine and amplified a 190-bp PCR fragment from the 3′ untranslated region of the gene. With all cDNAs used the primer set gave a single PCR product that was verified by determining the melt curves for the product at the end of each run, by analysis of the product using gel electrophoresis, and by comparing the DNA sequence of the PCR product with the gene sequence. The efficiency of the primers was tested in preliminary experiments with dilutions of the purified PCR product and maintained an r2 value ≥ 0.98. The expression of genes was normalized to VvUbiquitin1 (TC32075, TIGR database), which transcripts were detected by amplifying a 182-bp product with the primers VvUbiquitin Forward (5′-GTGGTATTATTGAGCCATCCTT-3′) and VvUbiquitin Reverse (5′-AACCTCCAATCCAGTCATCTAC-3′). All samples were measured in triplicate. The difference between the cycle threshold (Ct) of the target gene and the Ct of Ubiquitin, ΔCt = CtTarget − CtUbiquitin, was used to obtain the normalized expression of target genes, which corresponds to 2−ΔCt. The Rotor Gene 2000 software (Corbett Research) and the Q-Gene software (Muller et al., 2002) were used to calculate the mean normalized expression of the genes.
For detection of the VvMYBPA1 transcript in Arabidopsis (Fig. 7), PCR reactions were performed as described above and analyzed on a 1.5% agarose gel containing ethidium bromide. The primers MYBf (5′-CAACTGACAACTCTCTGGACAA-3′) and MYBr (5′-GATCTTTTGGTCTCTCTGCAAC-3′) were used to amplify a 146-bp PCR from the 3′ translated region of the gene. To determine whether the similar amounts of cDNA were applied to all samples, a 268-bp PCR fragment from the Arabidopsis Actin2 gene (accession no. NM_112764) was amplified with the primers ACT2F (5′-ATTCAGATGCCCAGAAGTCTTGTTCC-3′) and ACT2R (5′-ACCACCGATCCAGACACTGTACTTCC-3′).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number AM259485.
Acknowledgments
We wish to thank to Andrew Turner for help with the luciferase analysis, Eva Decker for the pluc and pRluc vectors, Nicole Cordon for help with HPLC analysis, and Karin Sefton and Debra McDavid for excellent technical assistance.
This work was supported by the Australian Government's Cooperative Research Centres Program and the Grape and Wine Research and Development Corporation.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Simon P. Robinson (simon.robinson@csiro.au).
Open Access articles can be viewed online without a subscription.
References
- Abrahams S, Lee E, Walker AR, Tanner GJ, Larkin PJ, Ashton AR (2003) The Arabidopsis TDS4 gene encodes leucoanthocyanidin dioxygenase (LDOX) and is essential for proanthocyanidin synthesis and vacuole development. Plant J 35 624–636 [DOI] [PubMed] [Google Scholar]
- Abrahams S, Tanner GJ, Larkin PJ, Ashton AR (2002) Identification and biochemical characterization of mutants in the proanthocyanidin pathway in Arabidopsis. Plant Physiol 130 561–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagchi D, Bagchi M, Stohs SJ, Das DK, Ray SD, Kuszynski CA, Joshi SS, Pruess HG (2000) Free radicals and grape seed proanthocyanidin extract: importance in human health and disease prevention. Toxicology 148 187–197 [DOI] [PubMed] [Google Scholar]
- Baudry A, Heim MA, Dubreucq B, Caboche M, Weisshaar B, Lepiniec L (2004) TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana. Plant J 39 366–380 [DOI] [PubMed] [Google Scholar]
- Baxter IR, Young JC, Armstrong G, Foster N, Bogenschutz N, Cordova T, Peer WA, Hazen SP, Murphy AS, Harper JF (2005) A plasma membrane H+-ATPase is required for the formation of proanthocyanidins in the seed coat endothelium of Arabidopsis thaliana. Proc Natl Acad Sci USA 102 2649–2654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogs J, Downey M, Harvey JS, Ashton AR, Tanner GJ, Robinson SP (2005) Proanthocyanidin synthesis and expression of genes encoding leucoanthocyanidin reductase and anthocyanidin reductase in developing grape berries and grapevine leaves. Plant Physiol 139 652–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogs J, Ebadi A, McDavid D, Robinson SP (2006) Identification of the flavonoid hydroxylases from grapevine and their regulation during fruit development. Plant Physiol 140 279–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boss PK, Davies C, Robinson SP (1996) Analysis of the expression of anthocyanin pathway genes in developing Vitis vinifera L. cv Shiraz grape berries and the implications for pathway regulation. Plant Physiol 111 1059–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16 735–743 [DOI] [PubMed] [Google Scholar]
- Cos P, De Bruyne T, Hermans N, Apers S, Berghe DV, Vlietinck AJ (2004) Proanthocyanidins in health care: current and new trends. Curr Med Chem 11 1345–1359 [DOI] [PubMed] [Google Scholar]
- Debeaujon I, Léon-Kloosterziel KM, Koornneef M (2000) Influence of the testa on seed dormancy, germination, and longevity in Arabidopsis. Plant Physiol 122 403–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debeaujon I, Nesi N, Perez P, Devic M, Grandjean O, Caboche M, Lepiniec L (2003) Proanthocyanidin-accumulating cells in Arabidopsis testa: regulation of differentiation and role in seed development. Plant Cell 15 2514–2531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debeaujon I, Peeters AJM, Léon-Kloosterziel KM, Koornneef M (2001) The TRANSPARENT TESTA12 gene of Arabidopsis encodes a multidrug secondary transporter-like protein required for flavonoid sequestration in vacuoles of the seed coat endothelium. Plant Cell 13 853–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deluc L, Barrieu F, Marchive C, Lauvergeat V, Decendit A, Richard T, Carde JP, Merillon JM, Hamdi S (2006) Characterization of a grapevine R2R3-MYB transcription factor that regulates the phenylpropanoid pathway. Plant Physiol 140 499–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon RA, Lamb CJ, Masoud S, Sewalt VJH, Paiva NL (1996) Metabolic engineering: prospects for crop improvement through the genetic manipulation of phenylpropanoid biosynthesis and defense responses—a review. Gene 179 61–71 [DOI] [PubMed] [Google Scholar]
- Dixon RA, Xie DY, Sharma SB (2005) Proanthocyanidins—a final frontier in flavonoid research? New Phytol 165 9–28 [DOI] [PubMed] [Google Scholar]
- Downey MO, Harvey JS, Robinson SP (2003. a) Analysis of tannins in seeds and skins of Shiraz grapes throughout berry development. Aust J Grape Wine Res 9 15–27 [Google Scholar]
- Downey MO, Harvey JS, Robinson SP (2003. b) Synthesis of flavonols and expression of flavonol synthase genes in developing grape berries of Shiraz and Chardonnay (Vitis vinifera L.). Aust J Grape Wine Res 9 110–121 [Google Scholar]
- Gleave AP (1992) A versatile binary vector system with a T-DNA organisational structure conducive to efficient integration of cloned DNA into the plant genome. Plant Mol Biol 20 1203–1207 [DOI] [PubMed] [Google Scholar]
- Glories Y (1988) Anthocyanins and tannins from wine: organoleptic properties. In V Cody, E Middleton, JB Harborne, A Beretz, eds, Plant Flavonoids in Biology and Medicine II: Biochemical, Cellular, and Medicinal Properties. Alan R. Liss, Inc., New York, pp 123–134 [PubMed]
- Goff SA, Cone KC, Chandler VL (1992) Functional analysis of the transcriptional activator encoded by the maize B gene: evidence for a direct functional interaction between two classes of regulatory proteins. Genes Dev 6 864–875 [DOI] [PubMed] [Google Scholar]
- Gollop R, Farhi S, Peri A (2001) Regulation of the leucoanthocyanidin dioxygenase gene expression in Vitis vinifera. Plant Sci 161 579–588 [Google Scholar]
- Grotewold E, Drummond BJ, Bowen B, Peterson T (1994) The myb-homologous P gene controls phlobaphene pigmentation in maize floral organs by directly activating a flavonoid biosynthetic gene subset. Cell 76 543–553 [DOI] [PubMed] [Google Scholar]
- Grotewold E, Sainz MB, Tagliani L, Hernandez JM, Bowen B, Chandler VL (2000) Identification of the residues in the Myb domain of maize C1 that specify the interaction with the bHLH cofactor R. Proc Natl Acad Sci USA 97 13579–13584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higo KY, Ugawa Iwamoto M, Korenaga T (1999) Plant cis-acting regulatory DNA elements (PLACE) database. Nucleic Acids Res 27 297–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holton TA, Cornish EC (1995) Genetics and biochemistry of anthocyanin biosynthesis. Plant Cell 7 1071–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horstmann V, Huether CM, Jost W, Reski R, Decker EL (2004) Quantitative promoter analysis in Physcomitrella patens: a set of plant vectors activating gene expression within three orders of magnitude. BMC Biotechnol 4 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H, Martin C (1999) Multifunctionality and diversity within the plant MYB-gene family. Plant Mol Biol 41 577–585 [DOI] [PubMed] [Google Scholar]
- Johnson CS, Kolevski B, Smyth DR (2002) TRANSPARENT TESTA GLABRA2, a trichome and seed coat development gene of Arabidopsis, encodes a WRKY transcription factor. Plant Cell 14 1359–1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy JA, Hayasaka Y, Vidal S, Waters EJ, Jones GP (2001) Composition of grape skin proanthocyanidins at different stages of berry development. J Agric Food Chem 49 5348–5355 [DOI] [PubMed] [Google Scholar]
- Kennedy JA, Matthews MA, Waterhouse AL (2000) Changes in grape seed polyphenols during ripening. Phytochemistry 55 77–85 [DOI] [PubMed] [Google Scholar]
- Kitamura S, Shikazono N, Tanaka A (2004) TRANSPARENT TESTA 19 is involved in the accumulation of both anthocyanins and proanthocyanidins in Arabidopsis. Plant J 37 104–114 [DOI] [PubMed] [Google Scholar]
- Kobayashi S, Ishimaru M, Ding CK, Yakushiji H, Goto N (2001) Comparison of UDP-glucose:flavonoid 3-O-glucosyltransferase (UFGT) gene sequences between white grapes (Vitis vinifera) and their sports with red skin. Plant Sci 160 543–550 [DOI] [PubMed] [Google Scholar]
- Kobayashi S, Ishimaru M, Hiraoka K, Honda C (2002) Myb-related genes of the Kyoho grape (Vitis labruscana) regulate anthocyanin biosynthesis. Planta 215 924–933 [DOI] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Nei M (2004) MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform 5 150–163 [DOI] [PubMed] [Google Scholar]
- McMahon LR, McAllister TA, Berg BP, Majak W, Acharya SN, Popp JD, Coulman BE, Wang Y, Cheng KJ (2000) A review of the effects of forage condensed tannins on ruminal fermentation and bloat in grazing cattle. Can J Plant Sci 80 469–485 [Google Scholar]
- Mehrtens F, Kranz H, Bednarek P, Weisshaar B (2005) The Arabidopsis transcription factor MYB12 is a flavonol-specific regulator of phenylpropanoid biosynthesis. Plant Physiol 138 1083–1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middleton EJR, Kandaswami C, Theoharidis TC (2000) The effects of plant flavonoids on mammalian cells: implications for inflammation, heart disease and cancer. Pharmacol Rev 52 673–751 [PubMed] [Google Scholar]
- Mol J, Grotewold E, Koes R (1998) How genes paint flowers and seeds. Trends Plant Sci 3 212–217 [Google Scholar]
- Muller PY, Janovjak H, Miserez AR, Dobbie Z (2002) Processing of gene expression data generated by quantitative real-time RT-PCR. Biotechniques 32 1372–1379 [PubMed] [Google Scholar]
- Nagel CW, Glories Y (1991) Use of a modified dimethylaminocinnamaldehyde reagent for analysis of flavonols. Am J Enol Vitic 42 364–366 [Google Scholar]
- Nesi N, Debeaujon I, Jond C, Pelletier G, Caboche M, Lepiniec L (2000) The TT8 gene encodes a basic helix-loop-helix domain protein required for expression of DFR and BAN genes in Arabidopsis siliques. Plant Cell 12 1863–1878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesi N, Debeaujon I, Jond C, Stewart AJ, Jenkins GI, Caboche M, Lepiniec L (2002) The TRANSPARENT TESTA16 locus encodes the ARABIDOPSIS BSISTER MADS domain protein and is required for proper development and pigmentation of the seed coat. Plant Cell 14 2463–2479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesi N, Jond C, Debeaujon I, Caboche M, Lepiniec L (2001) The Arabidopsis TT2 gene encodes an R2R3 MYB domain protein that acts as a key determinant for proanthocyanidin accumulation in developing seed. Plant Cell 13 2099–2114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters DJ, Constabel CP (2002) Molecular analysis of herbivore-induced condensed tannin synthesis: cloning and expression of dihydroflavonol reductase from trembling aspen (Populus tremuloides). Plant J 32 701–712 [DOI] [PubMed] [Google Scholar]
- Pourcel L, Routaboul JM, Kerhoas L, Caboche M, Lepiniec L, Debeaujon I (2005) TRANSPARENT TESTA10 encodes a laccase-like enzyme involved in oxidative polymerization of flavonoids in Arabidopsis seed coat. Plant Cell 17 2966–2980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quackenbush J, Liang F, Holt I, Pertea G, Upton J (2000) The TIGR gene indices: reconstruction and representation of expressed gene sequences. Nucleic Acids Res 28 141–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quattrocchio F, Wing J, van der Woude K, Souer E, de Vetten N, Mol J, Koes R (1999) Molecular analysis of the anthocyanin2 gene of petunia and its role in the evolution of flower color. Plant Cell 11 1433–1444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quattrocchio F, Wing JF, van der Woude K, Mol JNM, Koes R (1998) Analysis of bHLH and MYB domain proteins: species specific regulatory differences are caused by divergent evolution of target anthocyanin genes. Plant J 13 475–488 [DOI] [PubMed] [Google Scholar]
- Ramsay NA, Walker AR, Mooney M, Gray JC (2003) Two basic-helix-loop-helix genes (MYC-146 and GL3) from Arabidopsis can activate anthocyanin biosynthesis in a white-flowered Matthiola incana mutant. Plant Mol Biol 52 679–688 [DOI] [PubMed] [Google Scholar]
- Robinson SP, Davies C (2000) Molecular biology of grape berry ripening. Aust J Grape Wine Res 6 175–188 [Google Scholar]
- Sagasser M, Lu GH, Hahlbrock K, Weisshaar B (2002) A. thaliana TRANSPARENT TESTA 1 is involved in seed coat development and defines the WIP subfamily of plant zinc finger proteins. Genes Dev 16 138–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma SB, Dixon RA (2005) Metabolic engineering of proanthocyanidins by ectopic expression of transcription factors in Arabidopsis thaliana. Plant J 44 62–75 [DOI] [PubMed] [Google Scholar]
- Shirley BW, Hanley S, Goodman HM (1992) Effects of ionizing radiation on a plant genome: analysis of two Arabidopsis transparent testa mutations. Plant Cell 4 333–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirley BW, Kubasek WL, Storz G, Bruggemann E, Koornneef M, Ausubel FM, Goodman HM (1995) Analysis of Arabidopsis mutants deficient in flavonoid biosynthesis. Plant J 8 659–671 [DOI] [PubMed] [Google Scholar]
- Solano R, Nieto C, Pazares J (1995) Myb.Ph3 transcription factor from Petunia hybrida induces similar DNA-bending/distortions on its 2 types of binding-site. Plant J 8 673–682 [DOI] [PubMed] [Google Scholar]
- Sparvoli F, Martin C, Scienza A, Gavazzi G, Tonelli C (1994) Cloning and molecular analysis of structural genes involved in flavonoid and stilbene biosynthesis in grape (Vitis vinifera L.). Plant Mol Biol 24 743–755 [DOI] [PubMed] [Google Scholar]
- Stracke R, Werber M, Weisshaar B (2001) The R2R3-MYB gene family in Arabidopsis thaliana. Curr Opin Plant Biol 4 447–456 [DOI] [PubMed] [Google Scholar]
- Takos AM, Jaffe FW, Jacob SR, Bogs J, Robinson SP, Walker AR (2006) Light-induced expression of a MYB gene regulates anthocyanin biosynthesis in red apples. Plant Physiol 142 1216–1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torregrosa L, Verries C, Tesniere C (2002) Grapevine (Vitis vinifera L.) promoter analysis by biolistic-mediated transient transformation of cell suspensions. Vitis 41 27–32 [Google Scholar]
- Walker AR, Davison PA, Bolognesi-Winfield AC, James CM, Srinivasan N, Blundel TL, Esch JJ, Marks MD, Gray JC (1999) The TRANSPARENT TESTA GLABRA1 locus, which regulates trichome differentiation and anthocyanin biosynthesis in Arabidopsis, encodes a WD40 repeat protein. Plant Cell 11 1337–1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker AR, Lee E, Bogs J, McDavid DAJ, Thomas MR, Robinson SP (2007) White grapes arose through mutation of two similar and adjacent regulatory genes. Plant J (in press) [DOI] [PubMed]
- Winkel-Shirley B (2001) Flavonoid biosynthesis: a colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol 126 485–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie DY, Sharma SB, Paiva NL, Ferreira D, Dixon RA (2003) Role of anthocyanidin reductase, encoded by BANYULS in plant flavonoid biosynthesis. Science 299 396–399 [DOI] [PubMed] [Google Scholar]
- Xie DY, Sharma SB, Wright E, Wang ZY, Dixon RA (2006) Metabolic engineering of proanthocyanidins through co-expression of anthocyanidin reductase and the PAP1 MYB transcription factor. Plant J 45 895–907 [DOI] [PubMed] [Google Scholar]
- Zimmermann IM, Heim MA, Weisshaar B, Uhrig JF (2004) Comprehensive identification of Arabidopsis thaliana MYB transcription factors interacting with R/B-like BHLH proteins. Plant J 40 22–34 [DOI] [PubMed] [Google Scholar]