Abstract
Nitrogen-fixing symbiosis of legume plants with Rhizobium bacteria is established through complex interactions between two symbiotic partners. Similar to the mutual recognition and interactions at the initial stages of symbiosis, nitrogen fixation activity of rhizobia inside root nodules of the host legume is also controlled by specific interactions during later stages of nodule development. We isolated a novel Fix− mutant, ineffective greenish nodules 1 (ign1), of Lotus japonicus, which forms apparently normal nodules containing endosymbiotic bacteria, but does not develop nitrogen fixation activity. Map-based cloning of the mutated gene allowed us to identify the IGN1 gene, which encodes a novel ankyrin-repeat protein with transmembrane regions. IGN1 expression was detected in all organs of L. japonicus and not enhanced in the nodulation process. Immunoanalysis, together with expression analysis of a green fluorescent protein-IGN1 fusion construct, demonstrated localization of the IGN1 protein in the plasma membrane. The ign1 nodules showed extremely rapid premature senescence. Irregularly enlarged symbiosomes with multiple bacteroids were observed at early stages (8–9 d post inoculation) of nodule formation, followed by disruption of the symbiosomes and disintegration of nodule infected cell cytoplasm with aggregation of the bacteroids. Although the exact biochemical functions of the IGN1 gene are still to be elucidated, these results indicate that IGN1 is required for differentiation and/or persistence of bacteroids and symbiosomes, thus being essential for functional symbiosis.
Legume plants form nitrogen-fixing root nodules in symbiotic associations with soilborne bacteria generally referred to as Rhizobium. The nodulation process of the host legumes is triggered by specific lipochitin-oligosaccharide signal molecules, Nod factors, secreted by rhizobia. Nod factors elicit a series of host responses, such as root hair deformation, preinfection thread (cytoplasmic bridge) formation, and cortical cell division leading to formation of nodule primordia (Spaink and Lugtenberg, 1994; D'Haeze and Holsters, 2002). Subsequently, rhizobia invade the root cortical cells through infection threads and are released into the host cell cytoplasm. The released bacteria reside in the nodule cells as enclosed by peribacteroid membrane (PBM) derived from the host plasma membrane to form a symbiotic organelle symbiosome. Finally, the rhizobia differentiate into the symbiotic form, bacteroids, and induce nitrogen-fixing activity.
These successive developmental stages of symbiotic nodules are established through highly coordinated actions of the genes of both symbiotic partners. Recent comprehensive analyses using expressed sequence tags accumulated from some legume species revealed that the nodulation process involves global and drastic changes in gene expression in the host legumes (Colebatch et al., 2002, 2004; Fedorova et al., 2002; Kouchi et al., 2004; Asamizu et al., 2005). These studies have also allowed us the identification of nodule-specific genes that are activated during the nodule formation process. However, the functions of most of these genes in symbiotic nitrogen fixation are not yet solved.
To identify the host plant genes essential for nodulation process, extensive and systematic efforts have been made during the past decade to isolate various kinds of symbiosis mutants from two model legume species, Lotus japonicus and Medicago truncatula (Schauser et al., 1998; Szczyglowski et al., 1998; Cook, 1999; Kawaguchi et al., 2002; Sandal et al., 2006). These symbiotic mutants have been categorized into three classes, i.e. nonnodulating (Nod−), defect in cooperative histogenesis (Hist−), and formation of ineffective nodules (Fix−) (Kawaguchi et al., 2002). Nod− mutants are attributed to defects in very early steps of symbiotic interactions, and hence bacterial infection and nodule primordium formation both do not occur. Recent progress of the infrastructure for genome analyses of these model legumes has made it possible to identify a number of genes for Nod− loci. They are, for instance, the genes of putative Nod factor receptors (Limpens et al., 2003; Madsen et al., 2003; Radutoiu et al., 2003) and of the following early signaling cascade common for symbiotic interactions with both Rhizobium bacteria and mycorrhizal fungi (Endre et al., 2002; Stracke et al., 2002; Ané et al., 2004; Levy et al., 2004; Imaizumi-Anraku et al., 2005; Tirichine et al., 2006). Most of these cloned genes are primarily involved in Nod factor perception itself or in its immediate downstream signaling pathway(s) that precede the initiation of infection threads and nodule primordia.
Hist− mutants represent genetic loci that have an effect on the infection process accompanied by cooperative histogenesis of nodule structures. The mutants of this category are characterized by defects in infection thread formation and/or its growth, as well as incomplete nodule organogenesis with absence of bacterial infected cells. L. japonicus alb1 (Imaizumi-Anraku et al., 1997) and crinkle (Tansengco et al., 2003), and M. truncatula lin (Kuppusamy et al., 2004) are the typical examples of this category.
In contrast, genetic loci in Fix− mutants are attributed to much later stages of symbiotic nodule development, which involves the differentiation of rhizobia to bacteroids followed by induction of nitrogenase, and organization of metabolic functions required for nitrogen fixation in the host nodule cells. Fix− mutants form morphologically normal nodules with infected cells containing endosymbiotic bacteria, but exhibit no or very low nitrogen-fixing activity (Imaizumi-Anraku et al., 1997; Suganuma et al., 2003; Krusell et al., 2005). It is thus clear that genetically controlled specific interactions between legumes and rhizobia are not restricted only to the very early stages of symbiosis, but also exist at rather later stages of nodule development. For example, Rhizobium etli CE3, a compatible microsymbiont of Phaseolus vulgaris, produces Nod factors with the same structures as those from Mesorhizobium loti and can nodulate L. japonicus roots, but the nodules formed by R. etli senesce prematurely with rapid disorganization of the infected cell cytoplasm, and thus fail to further develop nitrogen-fixing symbiosis (Banba et al., 2001). This suggests that specific recognition between rhizobia and host plant cells controlled by unknown mechanisms other than that mediated by Nod factors is required for persistence of symbiosis. Identification of genes for the Fix− loci would provide important clues to elucidate the molecular mechanisms involved in functional symbiosis during the later stages of nodule development. However, identification of genes for the Fix− as well as Hist− loci is still very limited. Only one example is the sst1 mutants of L. japonicus of which genes has been recently identified as a nodule-specific sulfate transporter (Krusell et al., 2005).
We describe here the identification of a novel Fix− mutant designated ineffective greenish nodules 1 (ign1) of the model legume L. japonicus and molecular cloning of the IGN1 gene for the mutated locus by means of a map-based cloning strategy. On the basis of the mutant phenotype and structure of the gene identified, we discuss possible functions of the IGN1 gene in the establishment of functional symbiosis.
RESULTS
The ign1 Mutant Forms Ineffective Nodules
A Fix− mutant, ign1, was isolated by somatic mutation through intensive culture of hypocotyl-derived calli followed by regeneration of the plants (see “Materials and Methods”). Segregation analysis of the F2 progeny after the cross with the wild types Gifu and MG-20 indicated that the ign1 locus is a monogenic recessive (data not shown).
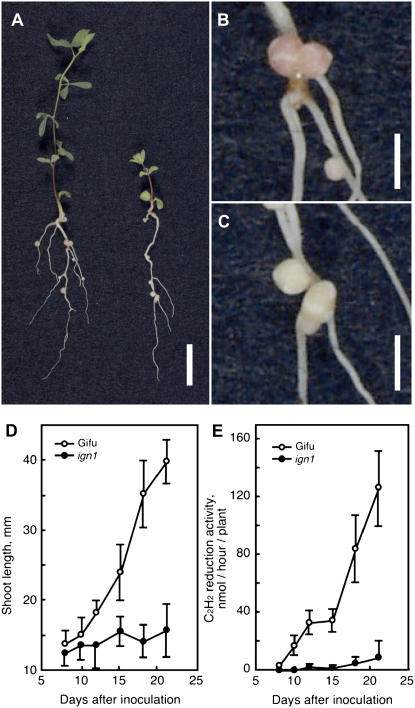
The ign1 mutant displayed typical nitrogen deficiency symptoms, such as chlorotic leaves, pigmented stems, and stunted shoot growth, when grown in nitrogen-free medium following inoculation of M. loti (Fig. 1, A and D). At 18 d after inoculation with M. loti, nodules formed on the ign1 mutant were slightly smaller than those of wild-type Gifu plants and were white to pale green in color, whereas wild-type nodules were red because of the presence of abundant leghemoglobin in the infection zone (Fig. 1, B and C). Nitrogenase activity of the ign1 nodules as measured by acetylene reduction activity was very low as compared with wild-type nodules (Fig. 1E). Immunoblot analysis for nitrogenase proteins from 14-d-old nodules indicated that the component I and II proteins are both present in the bacteroids in ign1 nodules at the level slightly lower than those in wild-type nodules (data not shown).
Figure 1.
Symbiotic phenotypes of the ign1 mutant. Plants were inoculated with M. loti MAFF303099 and grown in vermiculite pots supplied with nitrogen-free medium in an artificially lit growth cabinet. A, Wild-type Gifu (left) and ign1 (right) 18 d after M. loti inoculation. B and C, Root nodules of wild type (B) and of ign1 (C). Bars represent 10 mm in A and 2 mm in B and C. D and E, Shoot growth and acetylene reduction activity, respectively, in the wild-type and ign1 plants after inoculation with M. loti. The data are represented as means of 12 plants with sds indicated by vertical bars.
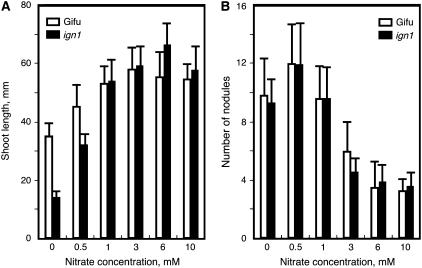
There was no significant difference either in the nodule number per plant (Fig. 2B) or the time course of nodulation between the ign1 mutant and wild-type Gifu plants (data not shown). When the plants were grown with sufficient supply of exogenous nitrogen, the ign1 mutants retained normal growth that is comparable to wild-type plants (Fig. 2A), indicating that the stunted growth of the mutants under symbiotic conditions is simply due to a defect in the nitrogen-fixing ability of the nodules. In addition, nodulation response of the mutants to increasing concentrations of supplemental nitrate was the same as wild-type plants (Fig. 2B). This indicates that nodulation of the ign1 mutants is equally sensitive to exogenous nitrate as that of the wild-type plants. There was no apparent abnormality in the morphology of leaves, stems, and roots of the mutants when grown under nonsymbiotic conditions.
Figure 2.
Effects of nitrate supply on growth and nodule numbers of ign1. The plants were inoculated with M. loti and grown with nutrient solution containing various concentrations of potassium nitrate for 18 d. A, Shoot length. B, Nodule number. The data are means of 12 plants with sds.
Infected Cells in the ign1 Nodules Collapse Prematurely
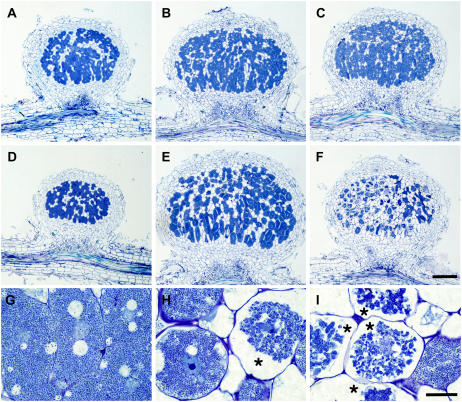
In the wild-type L. japonicus Gifu, nodules are first visible as small bumps around 4 d post inoculation (dpi) and nitrogen fixation is initiated at 10 to 12 d (Kouchi et al., 2004). This is also the case for the ign1 mutants. The appearance of the ign1 nodules were indistinguishable from wild-type nodules until the time of the onset of nitrogen fixation, but thereafter became pale green rapidly, indicating premature senescence. Figure 3 shows light micrographs of nodule sections. At 14 dpi, the nodule structure of the ign1 mutants was almost the same as that of wild-type plants (Fig. 3, A and D). As nodules developed, however, the infected cells of the ign1 nodules disintegrated rapidly and the intact infected cell with dense cytoplasm was hardly seen in the nodules at 21 dpi (Fig. 3, E and F). At initial stages of nodulation, infected cells were developed in the central zone of the ign1 nodules similarly to the wild-type nodules, and contained as many bacteroids as those of wild-type nodules. Thereafter, infected cells of the mutant nodules exhibited obvious disorganization of cytoplasm; the cytoplasm shrank and was broken away from the cell wall and the bacteroids appeared to aggregate (Fig. 3, H and I). At 21 dpi, most of the infected cells of the mutant nodules displayed severe disintegration of the intracellular structures and coarse-grained bacteroids were found to be localized around the cell nucleus (Fig. 3H).
Figure 3.
Light micrographs of nodule structure of wild-type and ign1 plants. A to F, Comparisons of nodule structures of wild type (A–C) and ign1 mutant (D–F) at 14 (A and D), 17 (B and E), and 21 dpi (C and F). Bar = 200 μm. G to I, Infected cells of 17 dpi wild-type nodule (G) and of ign1 nodules at 17 (H) and 21 dpi (I). Disintegrated infected cells of ign1 nodules are indicated by asterisks. Bar = 20 μm.
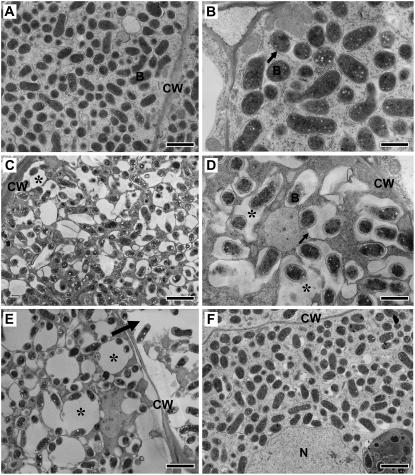
To examine the mutant nodule phenotype in more detail, ultrathin sections of the nodules at 8 to 13 dpi were observed by transmission electron microscopy (Fig. 4). Distinctive features of the infected cells of the ign1 nodules were evident at the ultrastructural level even at such early developmental stages. In wild-type Gifu nodules, infected cells were filled with PBM-enclosed bacteroids (symbiosomes; Fig. 4, A and B). Most of the symbiosomes were spherical and the PBM was adjacent to bacteroids. Some symbiosomes appeared to fuse with each other to form larger symbiosomes containing more than two bacteroids. The most striking difference in infected cells of the ign1 nodules as compared with those of wild-type nodules appeared on the symbiosome structures. Symbiosomes in the infected cells of ign1 nodules were irregularly shaped, enlarged relative to the bacteroids inside, and as a consequence the symbiosome space between the PBM and bacteroids increased in size. The abnormal symbiosome structures were clearly observed in 8 dpi nodules (Fig. 4C) and became more severe along the nodule age (Fig. 4E). Occasionally these structures seemed to be similar to lytic vacuoles as described recently for the prematurely senesced nodules of the sst1 (sulfate transporter) mutant of L. japonicus (Krusell et al., 2005). Bacteroids in the ign1 nodules appeared to be smaller in size and less dense than those of the wild type, suggesting that the bacteroid differentiation is inhibited and/or at least delayed in the ign1 nodules. Premature senescence is more or less a common feature of ineffective nodules formed on Fix− mutants. Abnormal enlargement of symbiosomes has been described as an early symptom of the premature senescence in the nodules of some Fix− mutants (Suganuma et al., 2003; Krusell et al., 2005; Hossain et al., 2006). To examine the possibility if the ineffectiveness (lack of nitrogen fixation) causes such abnormal symbiosome structures, we observed ultrastructures of the infected cells of the ineffective nodules formed by a nifH mutant (ΔnifH) of M. loti that lacks completely nitrogenase expression (Fig. 4F). The infected cell structures of ΔnifH nodules are almost comparable to those of wild-type nodules at least during early stages of nodule development.
Figure 4.
Ultrastructure of infected cells of wild-type and ign1 nodules. A and B, Wild-type nodules at 10 (A) and 13 dpi (B). C to E, ign1 nodule infected cells at 8 (C), 10 (D), and 13 dpi (E). Irregularly shaped, enlarged symbiosomes are indicated by asterisks. An arrow in E indicates a collapsed infected cell. F, Infected cells of 10 dpi nodules formed by a ΔnifH mutant strain of M. loti. CW, Cell wall; B, bacteroid; N, nucleus. PBM is indicated by arrows in B and D. Bars indicate 2 μm (A, C, E, and F) and 1 μm (B and D).
Map-Based Cloning of the IGN1 Gene
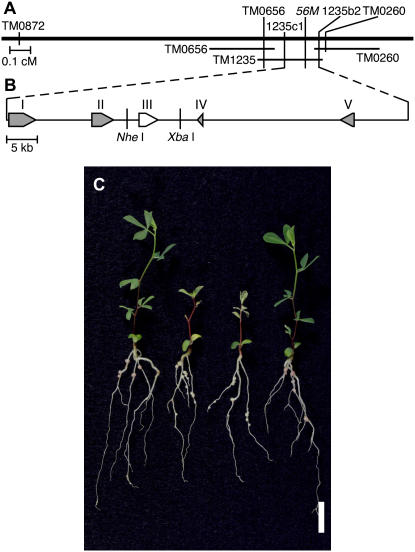
To identify the gene affected in the ign1 mutant, we employed a map-based cloning strategy using L. japonicus accession MG-20 Miyakojima as a crossing partner (Kawaguchi et al., 2001). The IGN1 locus was positioned on the short arm of chromosome 5 between the microsatellite (SSR) markers TM0260 (Asamizu et al., 2003) and TM0656 (http://www.kazusa.or.jp/lotus; Fig. 5A; see also Sandal et al., 2006). This region was mostly covered by two transformation-competent artificial chromosome clones (Liu et al., 1999), TM0260 and TM1235, derived from the MG-20 genome. Derived-cleaved-amplified polymorphic sequence (dCAPS) markers were designed from the sequences of these transformation-competent artificial chromosome clones and used to further delimit the IGN1 locus. Finally, we positioned the IGN1 locus within a region of 75-kb length between dCAPS markers 1235b2 and 1235c1 on TM1235. Five putative open reading frames (ORFs) were predicted in this region (Fig. 5B). Direct sequencing of deduced exons and introns of these ORFs in both the ign1 and wild-type Gifu genome revealed a one-base deletion in a predicted exon of the third ORF that encodes an ankyrin-repeat protein. We further performed RNA interference experiments for each ORF by a hairy root transformation procedure for wild-type Gifu plants (Kumagai and Kouchi, 2003). As a consequence, only the RNA interference for the third ORF reproduced the ign1 mutant phenotype in the nodules on transformed roots (data not shown). Thus, we suspected this ORF to be the prime candidate IGN1 gene.
Figure 5.
Map-based cloning of IGN1 and complementation test. A, Genetic linkage map of the IGN1 region on a part of the short arm of L. japonicus chromosome 5. The rough map position of IGN1 is also given in the previous article (Sandal et al., 2006). B, Candidate genes in the region between dCAPS markers, 1235c1 and 1235b2. I, β-Ketoacyl-CoA synthase; II, protein phosphatase 2C; III, IGN1; IV, hypothetical protein; V, pentatricopeptide repeat-containing protein. C, Complementation of the ign1 mutant with an NheI-XbaI fragment containing the complete wild-type IGN1 gene. From left to right: wild type (Gifu), ign1, ign1 transformed with empty vector, ign1 transformed with IGN1 gene. Plants were grown under nitrogen-free conditions and harvested 19 d after inoculation with M. loti. Bar represents 10 mm.
Since we isolated only a single allele of the ign1 locus, it was imperative to test the complementation of the mutant phenotype with the candidate gene. A DNA fragment of 8.6-kb length was excised from a genomic library of L. japonicus Gifu plants, which contains the entire ORF of the candidate gene together with the 1.9-kb upstream and 3.5-kb downstream sequences (Fig. 5B), and transformed into the ign1 plants according to a transformation procedure mediated by Agrobacterium tumefaciens (see “Materials and Methods”). The resultant transgenic plants showed normal growth comparable to wild-type plants under symbiotic conditions and nitrogenase activity of the nodules formed on the transgenic plants was fully recovered to the level of wild-type nodules (Fig. 5C; Table I). Taking the overall results together, we concluded that the gene encoding an ankyrin-repeat protein is the IGN1 gene.
Table I.
Complementation of ign1 mutant with an NheI-XbaI fragment containing the complete IGN1 gene
Plants were grown with nitrogen-free medium and harvested 19 d after inoculation with M. loti. Data are the means of 12 plants each.
| Line | Shoot Length | C2H2 Reduction Activity |
|---|---|---|
| mm ± sd | nmol/h/plant ± sd | |
| Gifu | 32.46 ± 4.03 | 70.51 ± 18.4 |
| ign1 | 16.92 ± 3.95 | 02.51 ± 4.54 |
| ign1/empty vector | 14.33 ± 1.61 | 01.62 ± 2.17 |
| ign1/NheI-XbaI fragment | 34.31 ± 4.35 | 73.51 ± 18.38 |
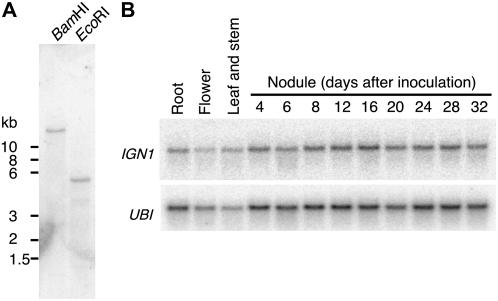
Genomic Southern hybridization suggested that IGN1 is present as a single copy in the L. japonicus genome (Fig. 6A). Its transcripts (about 2.3 kb) were detected in flowers, shoots, roots, and nodules at low levels, and the expression levels did not change through nodule development (Fig. 6B).
Figure 6.
DNA and RNA blot analyses of the IGN1 gene. A, Southern-blot analysis of genomic DNA from L. japonicus. DNA was digested by BamHI or EcoRI. B, Expression analysis of IGN1 in various organs and during nodule development of L. japonicus by northern hybridization. UBI, Polyubiquitin transcripts.
Structure of the IGN1 Protein
A full-length cDNA for the IGN1 mRNA was amplified by a combination of 5′ and 3′ RACE procedures from total RNA of nodules (see “Materials and Methods”). Alignment of the cDNA sequence with the genomic sequence defined the structure of the IGN1 gene with three introns (Fig. 7A). The predicted ORF is 1,788 nucleotides long, which is preceded by a leader sequence of 129 nucleotides and followed by a 3′ untranslated region of 320 nucleotides. The conceptual IGN1 protein is 596 amino acids long (MW = 64,510) and contains ankyrin repeats with transmembrane regions (Fig. 7B). The amino terminus is rich in Pro, Glu, Ser, and Thr, as has been described for the PEST sequence that serve as a signal module for conditional or constitutive proteolysis (Rechsteiner and Rogers, 1996). The PEST sequence region is followed by seven repeats of ankyrin sequences. Stacking of repeated ankyrin sequences serves as a domain for protein-protein interactions (Sedgwick and Smerdon, 1999). Secondary structure prediction indicated that all of seven ankyrin sequences in the IGN1 have a pair of helices to constitute the architecture of the ankyrin-repeat domain (Fig. 7C). In the carboxy-terminal region of the protein, four transmembrane domains were predicted. The mutation in ign1 was a one-base deletion immediately before the second transmembrane domain (Fig. 7D), thus resulting in a frame shift followed by an abrupt stop codon. We could not find any signal sequence to localize IGN1 into specific organelles by computer analyses using the TargetP program.
Figure 7.
Genomic organization of IGN1 and structure of the translation product. A, Intron-exon structure of IGN1 gene. Exons are shown as boxes. B, Predicted amino acid sequence of IGN1 protein. The PEST sequence is boxed. ANKR, Ankyrin repeat (bold and underlined); TM, transmembrane domain (double underlined). C, Alignment of the seven ankyrin repeats of IGN1 with the ankyrin consensus. Amino acids identical to the consensus are shown as bold. Helices predicted by Predict-Protein are shown as white boxes. D, Mutation site in the ign1 genome.
A number of expressed sequence tags from soybean (Glycine max) and M. truncatula were found to share significant similarity with IGN1 at the nucleotide level, suggesting that IGN1 is well conserved in legume plants. A database search using the deduced amino acid sequence of the IGN1 resulted in detection of cDNA clones from Arabidopsis (Arabidopsis thaliana; At3g12360) and Oryza sativa (AK100495), of which the encoded proteins exhibit significant homology with the IGN1 protein. Similarities of their deduced amino acid sequences with the IGN1 protein are 77.2% and 68.3%, respectively. However, physiological functions of these putative IGN1 orthologs are yet to be characterized.
IGN1 Protein Localizes on the Plasma Membrane
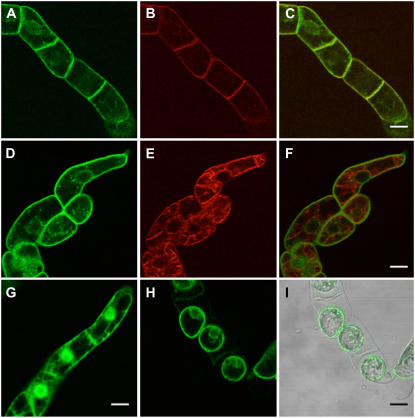
To investigate subcellular localization of IGN1 protein in plant cells we constructed a translational fusion of the IGN1 gene to the green fluorescent protein (GFP) gene. GFP was fused in frame at the N-terminal end of IGN1. The fusion gene was situated downstream of the 35S promoter from Cauliflower mosaic virus, and introduced into tobacco (Nicotiana tabacum) Bright-Yellow 2 (BY2) cells by Agrobacterium-mediated transformation (see “Materials and Methods”). Figure 8 shows GFP fluorescence in the transgenic BY2 cells under a confocal laser-scanning microscope. The fluorescence from the GFP-IGN1 fusion was found only in cell peripherals lining the plasma membrane (Fig. 8, A and D), whereas fluorescence from free GFP was also found in cytoplasm and in the nucleus (Fig. 8G). To verify that the IGN1 fused with GFP resides on the plasma membrane, the transgenic BY2 cells were pulse labeled with FM4-64, which is known as an endocytic tracer translocated from plasma membrane to vacuolar membranes in plant cells (Bolte et al., 2004). As shown in Figure 8B, FM4-64 first labeled the periphery of the cells, which overlap with the fluorescence of GFP-IGN1 (Fig. 8C). Ten hours after labeling, FM4-64 was transported to the tonoplasts by endocytosis (Fig. 8E) and did not overlap with the fluorescence of IGN1-GFP (Fig. 8F). We also observed GFP-IGN1 fluorescence from plasmolyzed transgenic cells to distinguish the plasma membrane from the cell wall (Fig. 8, H and I). These results indicate that IGN1 protein is targeted to the plasma membrane.
Figure 8.
Confocal laser scanning micrographs of BY2 cells expressing GFP-IGN1 fusion protein. A to F, BY2 cells 20 min (A–C) and 10 h (D–F) after labeling with FM4-64. Green fluorescence from GFP-IGN1 (A and D), red fluorescence from FM4-64 (B and E), and the merged images (C and F) are shown. G, Control image from GFP only. H, GFP-IGN1 fluorescence after plasmolysis. I, Nomarsky image overlaid with H. Scale bars = 20 μm.
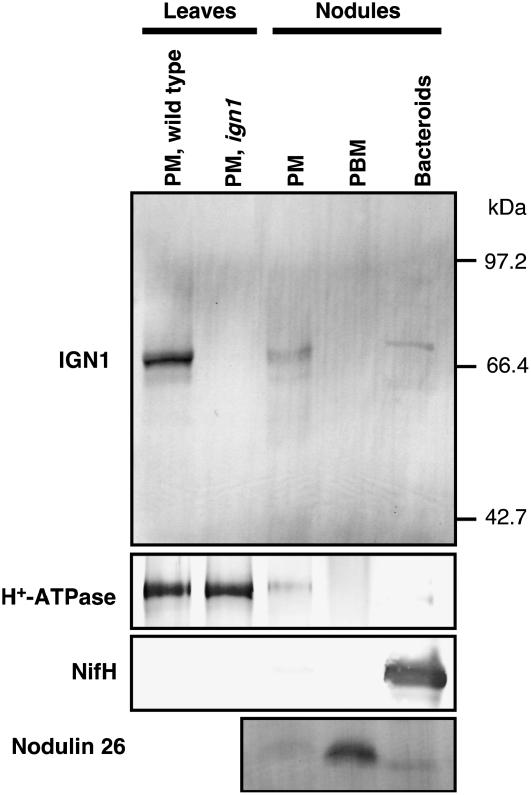
To investigate the subcellular localization of IGN1 in the tissues of Lotus plants, we performed immunological detection of IGN1 protein in the membrane fractions prepared from leaves and nodules of L. japonicus (Fig. 9). Anti-IGN1 antibody was raised against hexahistidine [(His)6]-tagged polypeptide that corresponds to the ankyrin-repeat moiety of IGN1. The IGN1 protein was clearly detected in the plasma membrane enriched fraction by means of an aqueous two-phase partitioning procedure from leaves and nodules. We further performed another fractionation of nodule cells to obtain PBM enriched fraction by means of disruption of purified symbiosomes (see “Materials and Methods”). An antibody against soybean nodulin 26 as a marker protein for PBM was used to verify the purity of the PBM fraction, indicating that the membrane fraction from purified symbiosomes was highly enriched by PBM. The IGN1 protein was not detectable in the PBM fraction and appeared to be cofractionated with H+-ATPase that serves as a plasma membrane marker. These results indicate that the IGN1 protein in leaves and nodules localizes mainly in the plasma membrane. It should be noted that the fractions from ign1 mutant plants did not show the presence of IGN1, suggesting that the mutants are virtually null for the IGN1 protein.
Figure 9.
Detection of IGN1 protein in membrane fraction from L. japonicus leaves and nodules by Anti-IGN1 antibody. Nodules were harvested from wild-type plants. Microsome fractions were isolated and further separated by means of aqueous two-phase partitioning. Upper phase after the two-phase partitioning enriches plasma membrane (PM). Purified symbiosomes were prepared from nodules by a discontinuous Percoll gradient centrifugation and fractionated into PBM enriched fraction and bacteroids. Each fraction equivalent to 3 μg protein was subjected to SDS-PAGE, blotted onto an Immobilon-P membrane, and processed with antibodies against IGN1 and H+-ATPase, nitrogenese component II (Nif H; see “Materials and Methods”).
DISCUSSION
In this article, we described characterization of a novel Fix− mutant, ign1 of L. japonicus, and molecular identification of the causal gene, IGN1, that encodes an ankyrin-repeat membrane protein. The ign1 mutant is able to form, in early stages of nodule development, apparently normal nodules containing infected cells packed with bacteroids in which nitrogen fixation actually takes place, even though at very low levels. However, the ign1 mutants cannot further develop nitrogen-fixing symbiosis because disintegration of the infected cells occurs just after the onset of nitrogen fixation (Figs. 1 and 3). Thus the ign1 nodules exhibit an obvious early or premature senescence.
A number of Fix− mutants that form ineffective nodules have been isolated from many legume species. Ineffective nodules formed on those Fix− mutants exhibit more or less the phenotype of premature senescence, in both determinate (Imaizumi-Anraku et al., 1997; Suganuma et al., 2003) and indeterminate nodules (Novak et al., 1995; Suganuma et al., 1995, 1998; Morzhina et al., 2000). One of the most well characterized Fix− mutants of L. japonicus is sen1 (Kawaguchi et al., 2002; Suganuma et al., 2003). Ineffectiveness of the sen1 nodules is presumed to be due to the defect of rhizobial differentiation to bacteroids with complete lack of nitrogenase activity and causes premature senescence, such as highly vacuolated infected cells, enlargement of symbiosomes, and disorganization of infected cells. These phenotypes of premature senescence are also observed in the sst1 mutant (Krusell et al., 2005) as well as in ineffective nodules formed by R. etli CE3 on L. japonicus (Banba et al., 2001). It has been shown that the premature senescence of nodules is not simply due to nitrogen deficiency caused by ineffective nitrogen fixation (Banba et al., 2001). Rather, it is more likely that it reflects the activation of a kind of plant defense response to nonfixing or inefficient endosymbionts that confer no benefit to the host plants (Suganuma et al., 2003). In this regard, however, it should be noted that the ign1 mutant shows disintegration of the symbiosomes followed by cytoplasmic disorganization of infected cells much earlier and more drastically than nodules formed either on the sen1, sst1 mutants, or by R. etli CE3. In addition, the nodules formed by the ΔnifH mutant of M. loti showed no ultrastructural abnormality of symbiosomes at least during early stages of nodule formation (Fig. 4F), suggesting that rapid disintegration of symbiosomes in the ign1 nodules is not simply due to the lack of nitrogen-fixing activity. Deterioration of infected cells in the ign1 nodules is not accompanied by enhanced vacuolation and occurs almost randomly in the entire infected zone, whereas it occurs in the proximal part of the sen1 nodules and those formed by R. etli CE3. Thus, the premature senescence observed in ign1 nodules is not identical, even if similar in part, with that in the ineffective nodules previously characterized.
IGN1 encodes a novel ankyrin-repeat protein with a PEST sequence in the amino terminus and transmembrane domains at the carboxy terminus (Fig. 7). Ankyrin-repeat proteins are widely distributed in prokaryotes, eukaryotes, and some viruses (Sedgwick and Smerdon, 1999). An ankyrin motif consists of a pair of α-helices and β-hairpin, and stacking of the motifs constructs the ankyrin domain that mediates protein-protein interactions (Sedgwick and Smerdon, 1999; Mosavi et al., 2002). Ankyrin-repeat proteins are involved in diverse cellular processes and not assigned to a specialized biological function(s). In animals and yeast, ankyrin-repeat proteins appear in a wide variety of biological activities, such as cell cycle control, transcriptional regulation, cytoskeletal organization, and developmental regulation (Sedgwick and Smerdon, 1999). In higher plants, however, only a few ankyrin-repeat proteins have been characterized so far in regard to their biological functions, and thus they compose one of the largest uncharacterized gene families in the plant kingdom (Lu et al., 2003). In Arabidopsis, Becerra and coworkers found 37 ankyrin-repeat proteins containing transmembrane domains (AtANKTM; Becerra et al., 2004), which have similar domain structures with IGN1. They are the most abundant group of ankyrin-repeat proteins in Arabidopsis, but almost all of them are functionally uncharacterized. ACD6 is one among them studied previously (Lu et al., 2003). Although the exact biochemical functions of ACD6 is unknown, it is postulated to be a positive regulator of the defense responses against virulent bacteria and of salicylic acid-dependent cell death. Amino acid sequence of IGN1 does not show significant similarity to that of ACD6, but the two proteins share the identical domain structure. ACD6 is recently shown to be localized in plasma membrane (Lu et al., 2005) similarly to IGN1. NPR1/NIM1 and AKR2, both isolated from Arabidopsis, are also noteworthy because they have been shown to be involved in defense responses against pathogen infection (Cao et al., 1997; Yan et al., 2002). NPR1/NIM1 and AKR2 both contain ankyrin repeats that are preceded by a PEST sequence, though they are not membrane proteins. NPR1 is shown to be a key regulator of acquired resistance in disease resistance, while AKR2 is related to the regulation of antioxidation metabolism that is also involved in disease resistance. It is shown that both proteins exert their regulatory functions through interactions with other proteins mediated by their ankyrin domains. These previous works may provide important clues for exploring the functions of IGN1 in symbiotic interactions of legume plants with compatible Rhizobium bacteria.
We showed that IGN1 protein is targeted to the plasma membrane when transformed into tobacco BY2 cells as a fusion with GFP (Fig. 8). Furthermore, immunological analysis indicated the localization of IGN1 in plasma membrane fractions isolated from leaves and nodules of L. japonicus (Fig. 9). Localization of IGN1 in PBM that is derived from plasma membrane and serves as the boundary between endosymbionts and the host cell cytoplasm is unlikely because the IGN1 protein could not be detected in the PBM fraction prepared from purified symbiosomes. In contrast, it was clearly detected in the plasma membrane enriched fraction from root nodules. Therefore, we suppose that IGN1 functions in the plasma membrane in nodule cells. Localization in the plasma membrane has been demonstrated for ankyrins in animals, which play a role as an adaptor between cytoskeletal proteins and other proteins such as Na+/K+-ATPase (Denker and Barber, 2002). IGN1 does not seem to have such a function, because the linker function of animal ankyrins is achieved through multiple distinctive domains for protein-protein binding (Bennett, 1992). Rather, IGN1 may function as a regulator for activities of other cytoplasmic or membrane proteins. Since IGN1 contains only ankyrin repeats as functional domains, the transmembrane regions of IGN1 appear to have a role only to anchor itself to the membrane. It is noteworthy that the N-terminal part of IGN1 containing the ankyrin repeats is expected to be intracellular, because it has been demonstrated for ACD6 that shows the same domain structure as IGN1 (Lu et al., 2005). Several proteins containing only ankyrin repeats as their functional domains, such as I-κB in animals, are well studied and shown to be involved in transcriptional regulation. I-κB prevents the transcription factor NF-κB from translocating into the nucleus (Verma et al., 1995). Degradation of I-κB as a result of a series of events including infection of bacterial and viral pathogens, leads to translocation of NF-κB into nucleus and then its target genes are activated. It is notable that the degradation of I-κB requires its PEST sequence region (Brown et al., 1995; Rodriguez et al., 1995). In a similar fashion, IGN1 may function as a membrane anchorage protein to regulate subcellular localization of other proteins that are required for development and persistence of symbiosomes. Alternatively, IGN1 may interact with and regulate other membrane proteins, such as receptor kinases involved in signal transduction or membrane transporters. In any case, identification of protein(s) that interact with IGN1 is essential, and therefore, a major focus in future studies to obtain more insights regarding possible functions of IGN1.
The ign1 mutant displayed no growth abnormality other than its symbiotic defect. This implies that the IGN1 gene is not crucial for ordinary growth of the plants under nonsymbiotic conditions. Nevertheless, IGN1 is expressed constitutively through all organs of L. japonicus, and is not nodule specific. It is very likely, therefore, that IGN1 makes provision for unfavorable environmental stimuli, such as pathogen attack most probably. In this regard, it will be intriguing to investigate the responses of the ign1 mutant upon infection of pathogenic bacteria and/or fungi. At present, it is too early to assume the exact functions of IGN1 in symbiotic interactions of legume plants with Rhizobium bacteria. However, drastic disorganization of symbiosomes at early stages of nodule formation of the ign1 mutant strongly suggests that IGN1 is required for differentiation and/or persistence of bacteroids and symbiosomes. On the basis of the mutant phenotype and the domain structure of the IGN1 protein, we hypothesize that IGN1 is required for preventing the host plant cells from inappropriately invoking premature senescence, or a kind of defense system, against compatible microsymbionts, thus being essential for functional symbiosis. We will focus our future studies of IGN1 to test this hypothesis.
MATERIALS AND METHODS
Plant Materials
A number of symbiotic mutants of Lotus japonicus B-129 Gifu and MG-20 Miyakojima were generated by somatic mutation through intensive culture of calli and/or suspension cells followed by regeneration of the plants (Y. Umehara, unpublished data). The ign1 mutant was isolated by the same method from a progeny of L. japonicus B-129 Gifu transformed with a nodule-specific gene according to a standard hypocotyl transformation procedure (Stiller et al., 1997). The phenotypic trait of the ign1 mutant was independent of the transgene and the mutant line was established by eliminating the transgene by segregation after backcrossing with wild-type Gifu plants.
Surface-sterilized seeds of L. japonicus B-129 Gifu and of the ign1 mutant were germinated on 0.8% agar plates in an artificially lit growth cabinet on a 16-h-light/8-h-dark cycle at 24°C for 6 d. The seedlings were transplanted onto vermiculite pots supplied with one-half-strength B & D medium with or without nitrogen (Imaizumi-Anraku et al., 1997) and grown in a growth chamber controlled at 26°C (16-h light) and 24°C (8-h night). The plants were inoculated with Mesorhizobium loti MAFF303099 (Niwa et al., 2001) 3 d after transplanting.
Acetylene Reduction Assay
An intact plant was placed in a 12 mL glass tube with a rubber cap and incubated in a water bath at 25°C. Acetylene (1 mL) was injected in the tube and the amount of ethylene formed after 15 min was determined by gas chromatography as described (Kouchi et al., 1991).
Light Microscopy
Nodules on root segments were fixed in 0.25% (w/v) glutaraldehyde and 4% paraformaldehyde in 0.1 m sodium phosphate buffer (pH 7.2) overnight. Fixed nodules were then sliced at 200 to 250 μm thickness by a razor blade and dehydrated through a graded ethanol series followed by embedding in Teknovit 7100 (Kulzer) according to the manufacturer's instructions. Thin sections (1 μm) were made using an ultramicrotome (Sorvall model MT2-B; DuPont) with a glass knife, attached on a coverslip, and stained with 0.002% (w/v) toluidine blue.
Electron Microscopy
Nodule samples were processed for electron microscopy according to the protocol described by Siddique and Bal (1992) with some modifications. In brief, fresh nodules were cut to small pieces (<1 mm thick) and fixed in 2.5% glutaraldehyde and 4.0% paraformaldehyde in 0.1 m sodium phosphate buffer (pH 7.2) for 3 h at room temperature and then overnight at 4°C. After washing extensively in the same buffer, the materials were postfixed in 2.0% osmium tetraoxide in 0.1 m sodium phosphate (pH 7.2) for 3 h at room temperature, dehydrated through graded ethanol and then acetone series, and embedded in an epoxy resin (Quetol-812, Nisshin EM Co.). Ultrathin sections were made by an ultramicrotome (UltraCut-R, Leica Microsystems) equipped with a diamond knife (Diatome). They were poststained with uranium acetate and lead citrate and viewed under a transmission electron microscope (H-7100, Hitachi).
Fine Mapping of the ign1 Locus
F2 mapping population was established by crossing the ign1 mutant that was derived from L. japonicus B-129 Gifu with the accession MG-20 Miyakojima (Kawaguchi et al., 2001). F2 plants homozygous for the ign1 mutant were screened by the greenish nodule phenotype 18 d after inoculation with M. loti Tono (Kawaguchi et al., 2002). A total of 1,501 homozygous F2 individuals were used for analysis with markers flanking IGN1.
Nucleic Acid Isolation, cDNA Cloning, and DNA/RNA Gel-Blot Analyses
Total RNA was prepared by an RNeasy plant mini kit (Qiagen) according to the manufacturer's instructions. Genomic DNA was isolated by the cetyl-trimethyl-ammonium bromide method as previously described (Li et al., 2001). IGN1 cDNA was prepared from 5′ and 3′ RACE-ready cDNA pool from RNA of 21-d-old nodules using a SMART RACE cDNA amplification kit (CLONTECH) according to the manufacturer's manual. In brief, the first and nested PCR for 5′ RACE were performed with IGN1 specific primers 5′-GGTTAATGCATTAACATTGGTGTCTGGC-3′ and 5′-CATCAGCACTTTGCCCTTTCACTGCCA-3′, respectively, in combination with universal primers supplied in the kit. RACE to amplify 3′ end was performed with an IGN1 specific primer 5′-CTGCTGTCAAAGGATTGCAGCTTGTTG-3′ and universal primer mixture in the kit. The full-length cDNA for IGN1 mRNA was obtained by conjugation of the 5′ and 3′ RACE fragments at a unique NdeI site in the overlapped region of both fragments.
Total RNAs (6 μg) from various organs were subjected to denaturing 1% (w/v) agarose gel electrophoresis. Genomic DNA (3 μg) was digested by restriction enzymes to completion and subjected to 0.7% (w/v) agarose gel electrophoresis. They were transferred onto nylon membranes and hybridized with 32P-labeled IGN1 cDNA probe. A cDNA fragment (416 nucleotides in length) for a portion of IGN1 mRNA was amplified from genomic DNA of wild-type Gifu plants by PCR with a primer set of 5′-GGATCCTCGAGGTGCATACAGAAGCCA-3′ (forward) and 5′-ATCGATGGTACCAAGGGATGAACTGAG-3′ (reverse) and used to generate the probe with a random prime DNA labeling kit (Takara).
Plant Transformation
An IGN1 genomic clone was isolated from a genomic library of wild-type Gifu plants prepared in EMBL3 lambda vector (Stratagene) by screening with 32P-labeled IGN1 cDNA as a probe. For the complementation test, an NheI-XbaI fragment (8.6 kb) that contains the entire IGN1 gene was excised, subcloned into a binary vector pCAMBIA1300 (Cambia), and transformed into the ign1 mutant by an Agrobacterium tumefaciens-mediated hypocotyl transformation procedure (Stiller et al., 1997). Plants from the T1 progeny were analyzed for the presence of the transgene in their genome by detecting the hygromycin-resistant gene by PCR using a primer set of 5′-CTGAAATCACCAGTCTCTCTCTAC-3′ (forward) and 5′-CCTATAGGAACCCTAATTCCCTTA-3′ (reverse) and 12 transformants were analyzed for the nodule phenotypes.
DNA Sequencing and Computer Analysis
DNA sequencing was done by the dye-termination method using an automated DNA sequencer (Gene Analyzer 3700, Applied Biosystems). Sequences were analyzed by BLAST (http://www.ncbi.nlm.nih.gov/BLAST/) and FASTA (http://www.ddbj.nig.ac.jp/search/fasta-j.html). For domain analysis, Pfam (http://www.sanger.ac.uk/Software/Pfam/) was applied. Secondary structures, transmembrane regions, and subcellular targeting peptide were predicted by PredictProtein (http://www.embl-heidelberg.de/predictprotein/predictprotein.html) and TMHMM v2.0 (http://www.cbs.dtu.dk/services/TMHMM-2.0/), respectively. PEST sequences were defined by PEST-FIND (http://www.at.embnet.org/embnet/tools/bio/PESTfind/).
Expression of IGN1 Fused to GFP in BY2 Cells
To generate the GFP-IGN1 fusion construct, a coding region of IGN1 cDNA was amplified with a primer set of 5′-TGTACATGGCTTCCAACTTCAACCAA-3′ (forward) and 5′-GCGGCCGCTCAAAGCGCGTAAATTGGAT-3′ (reverse) and the amplified fragment was fused to sGFP (S65T) cDNA in frame at the BsrGI site (Niwa et al., 1999). This fusion construct was placed between the cauliflower mosaic virus 35S promoter and the nopaline synthetase terminator and inserted into pCAMBIA1300. The resultant plasmid was transformed into A. tumefaciens LBA4404 by direct transfer with a freeze-thaw procedure.
Tobacco (Nicotiana tabacum) BY2 cells were grown in a BY2 medium at 28°C in the dark with continuous shaking (Nagata et al., 1992). Cells subcultured for 3 d were cocultured with Agrobacterium for 3 d and then the transformed BY2 cells were grown on solid medium containing 30 μg/mL hygromycin and 400 μg/mL carbenicillin. The selected transgenic cells were further subcultured for 3 to 4 d and observed by confocal laser microscopy. The plasma membrane was visualized by incubating the cells in the medium containing 20 μm FM4-64 (Molecular Probes) for 5 min followed by washing once with fresh medium and observation within 20 min. For staining vacuoles, cells were incubated with 20 μm FM4-64 for 30 min, washed once, and further incubated for 10 h before microscopic observation. Plasmolysis was induced by incubating the cells in 0.5 m mannitol for 10 min. The fluorescence of GFP and FM4-64 in the cells was analyzed using a Bio-Rad Micro Radiance 2000 confocal laser microscope (Bio-Rad) excited at 488 nm and 514 nm, respectively, with an argon laser.
Preparation of Plasma Membrane Enriched Fraction
Leaves (7 g fresh weight) or nodules (5 g fresh weight) were harvested from L. japonicus and homogenized at 4°C in a mortar with a pestle in 50 mm MOPS-KOH buffer (pH7.5) containing 250 mm Suc, 5 mm EDTA, 5 mm EGTA, 2.5 mm potassium disulfite, 1.5% (w/v) polyvinylpyrrolidone K-30, and 1 mm phenylmethanesulfonyl fluoride. The homogenate was filtered through two layers of Miracloth, centrifuged at 10,000g for 10 min, and the supernatant was further centrifuged at 126,000g for 40 min. The precipitate was washed with 10 mm potassium phosphate and 250 mm Suc (pH 7.8; PS buffer) and resuspended in 1 mL of PS buffer to obtain the microsome fraction, which was further processed by aqueous two-phase partitioning to enrich for plasma membranes (Uemura and Yoshida, 1983). In brief, the microsome suspension (1 mL) was mixed with the polymer suspension to make an aqueous two-phase system containing 5.6% (w/w) dextran T-500, 5.6% (w/w) polyethylene glycol 3350, 30 mm NaCl, 250 mm Suc, and 10 mm potassium phosphate (pH 7.8). Separation of the two phases was achieved by centrifugation at 3,000g for 10 min. The upper phase was subjected to a second partitioning. The upper phase in the second partitioning was diluted with 5 mm Tris-2-morpholinoethanesulfonic acid and 250 mm Suc (pH 7.0; TS buffer). The diluted fraction was centrifuged at 126,000g for 60 min and the precipitate was suspended in TS buffer.
Preparation of PBM Enriched Fraction
Symbiosomes were prepared from 4-week-old nodules of L. japonicus by the method developed for soybean (Glycine max) nodules (Udvardi et al., 1988) with some modifications. In brief, nodules from approximately 1,600 L. japonicus plants were homogenized very gently in a mortar with a pestle in 100 mm MOPS-KOH buffer (pH 7.2) containing 375 mm mannitol, 5 mm MgSO4, 10 mm EGTA, 3 mm 4-aminobenzamide, 10 mm dithiothreitol, 1% (w/v) polyvinylpyrrolidone-40, 4% (w/v) Dextran T40, and 1% bovine serum albumin. The homogenate was filtered through two layers of Miracloth, centrifuged at 160g for 10 min to remove unbroken tissues and debris, and then the supernatant was further centrifuged at 670g for 15 min. The pellet was suspended in 20 mm MOPS-1,3-bis[tris(hydroxymethyl)-methylamino]propane buffer (pH 7.2) containing 550 mm mannitol, 2 mm MgSO4, and 10 mm KCl (washing buffer), placed on a stepwise gradient of 45%, 60%, and 80% (v/v) Percoll, and centrifuged at 4,200g for 20 min. The pellet at the interface of 60% and 80% (v/v) Percoll was collected in washing buffer and applied onto 80% (v/v) Percoll cushion. After centrifugation at 670g for 10 min, the pellet on the Percoll cushion was suspended in washing buffer and centrifuged again at 400g for 5 min. The final pellet contained symbiosomes of high purity. The purified symbiosomes were suspended in washing buffer containing a protease inhibitor cocktail for plant cell and tissue extracts (Sigma) diluted in 10 volumes of 20 mm Tris-Cl, 1 mm MgSO4, 2 mm EGTA (pH 7.2), and disrupted by a voltex mixer. After removing bacteroids by centrifugation at 8,000g for 10 min, the PBM fraction was obtained by centrifugation at 126,000g for 90 min.
Immunoblotting
Recombinant IGN1 protein was prepared by a PET system (Novagen). A cDNA fragment corresponding to the N terminus of IGN1 protein including seven ankyrin repeats was PCR amplified from the IGN1 full-length cDNA with a primer pair of 5′-CCATGGCTTCCAACTTCAACCAACC-3′ (forward) and 5′-CTCGAGCTCTTTGGAAATATTATGA-3′ (reverse). The amplified fragment was digested by NcoI and XhoI, subcloned into expression vector pET21d, and was introduced into Escherichia coli BL21(DE3). An overnight preculture was transferred into Luria-Bertani medium at a 1:10 ratio and incubated at 37°C for 90 min with vigorous shaking. Production of recombinant protein was induced by addition of 1 mm isopropyl d-thiogalactopyranoside and accumulation of expressed protein was achieved by further incubation for 2 h. The cells were harvested by centrifugation, washed with 100 mm Tris-Cl (pH 8.0), and resuspended with the same buffer containing 300 mm NaCl, 1 mm phenylmethanesulfonyl fluoride, and proteinase inhibitor cocktail for His tag (Sigma). The cells were disrupted by sonication and centrifuged at 10,000g for 30 min to remove debris. The supernatant was applied on a nickel-nitrilotriacetic acid agarose (Qiagen) column. The column was washed and developed essentially according to the manufacturer's instructions. The eluate with 100 mm Tris-Cl, 300 mm NaCl, and 400 mm imidazole was diluted in 10 volumes of 100 mm Tris-Cl (pH 8.0) and passed through Q-Sepharose (Amersham) column. The unbound fraction was concentrated by Centricon YM-10 (Millipore) and applied to a Sephacryl S-200 (Amersham) column. The column was developed with 50 mm sodium phosphate and 200 mm NaCl (pH 7.0). The sample after gel filtration was subjected to SDS-PAGE and purified protein with apparent size of 45 kD was detected. This protein was confirmed to be the truncated IGN1 protein by mass spectroscopy.
Antiserum against the purified truncated IGN1 protein was raised in rabbit. For affinity purification of the antibody, the antigen was coupled to an NHS-activated Hi-Trap column (Amersham) according to the manufacturer's instructions. The anti-IGN1 antiserum was applied to the antigen column. The column was washed with 50 mm sodium phosphate (pH 7.0) containing 150 mm NaCl and then the bound antibody was eluted with 100 mm Gly-Cl buffer (pH 2.7) followed by immediate neutralization with 1.5 m Tris.
For immunodetection of IGN1 protein, membrane fractions were dissolved in SDS-PAGE sample buffer (62.5 mm Tris-Cl [pH 6.8], 2% [w/v] SDS, 50 mm dithiothreitol, and 8 m urea) and subjected to SDS-PAGE with a 10% (w/v) polyacrylamide gel (Kadenbach et al., 1983). After electrophoresis, the proteins were blotted onto Immobilon-P filter (Millipore) and reacted with purified anti-IGN1 antibody. The membranes were further treated with alkaline phosphatase-conjugated goat anti-rabbit immunoglobulin antibody. The phosphatase activity was detected by mixture of 4-nitroblue tetrazolium and 5-bromo-4-chloro-3-indolyl-phosphate. An antibody against Zea mays P-type H+-ATPase, nitrogenase from pea (Pisum sativum) bacteroids, and soybean nodulin 26 was used to confirm the enrichment of plasma membrane, bacteroids, and PBM, respectively, in each fraction.
Accession Numbers
The DNA data bank of Japan accession numbers for IGN1 genomic and cDNA sequences are AB251641 and AB251640, respectively.
Acknowledgments
We express our sincere thanks to Drs. Masayuki Ishikawa, Yuka Hagiwara, Tetsuo Meshi, and Atsushi Tamai of the National Institute of Agrobiological Sciences for their suggestions and help for expression analysis of GFP-IGN1 fusion protein in BY2 cells. We are also grateful to Dr. Yoshiyuki Tanaka and Ms. Atsuko Nakamura of the National Institute of Agrobiological Sciences for their technical suggestions for aqueous two-phase partitioning. Dr. Hitoshi Sakakibara of Riken Institute and Prof. Dan Roberts of the University of Tennessee are thanked for providing the antibodies against Zea mays H+-ATPase and soybean nodulin 26, respectively. Prof. Robert W. Ridge of International Christian University is thanked for his critical reading of the manuscript.
This work was supported by the Special Coordination Funds for Promoting Science and Technology from the Ministry of Education, Culture, Sports, Science and Technology, Japan.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Hiroshi Kouchi (kouchih@nias.affrc.go.jp).
Open Access articles can be viewed online without a subscription.
References
- Ané JM, Kiss GB, Riely BK, Penmetsa RV, Oldroyd GED, Ayax C, Levy J, Debelle F, Baek JM, Kalo P, et al (2004) Medicago truncatula DMI1 required for bacterial and fungal symbioses in legumes. Science 303 1364–1367 [DOI] [PubMed] [Google Scholar]
- Asamizu E, Kato T, Sato S, Nakamura Y, Kaneko T, Tabata S (2003) Structural analysis of a Lotus japonicus genome. IV. Sequence features and mapping of seventy-three TAC clones which cover the 7.5 mb regions of the genome. DNA Res 10 115–122 [DOI] [PubMed] [Google Scholar]
- Asamizu E, Nakamura Y, Sato S, Tabata S (2005) Comparison of the transcript profiles from the root and the nodulating root of the model legume Lotus japonicus by serial analysis of gene expression. Mol Plant Microbe Interact 18 487–498 [DOI] [PubMed] [Google Scholar]
- Banba M, Siddique AB, Kouchi H, Izui K, Hata S (2001) Lotus japonicus forms early senescent root nodules with Rhizobium etli. Mol Plant Microbe Interact 14 173–180 [DOI] [PubMed] [Google Scholar]
- Becerra C, Jahrmann T, Puigdomenech P, Vicient CM (2004) Ankyrin repeat-containing proteins in Arabidopsis: characterization of a novel and abundant group of genes coding ankyrin-transmembrane proteins. Gene 340 111–121 [DOI] [PubMed] [Google Scholar]
- Bennett V (1992) Ankyrins: adaptors between diverse plasma membrane proteins and the cytoplasm. J Biol Chem 267 8703–8706 [PubMed] [Google Scholar]
- Bolte S, Talbot C, Boutte Y, Catrice O, Read ND, Satiat-Jeunemaitre B (2004) FM-dyes as experimental probes for dissecting vesicle trafficking in living plant cells. J Microsc 214 159–173 [DOI] [PubMed] [Google Scholar]
- Brown K, Gerstberger S, Carlson L, Franzoso G, Siebenlist U (1995) Control of I-κBα proteolysis by site-specific, signal-induced phosphorylation. Science 267 1485–1488 [DOI] [PubMed] [Google Scholar]
- Cao H, Glazebrook J, Clarke JD, Volko S, Dong X (1997) The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell 88 57–63 [DOI] [PubMed] [Google Scholar]
- Colebatch G, Desbrosses G, Ott T, Krusell L, Montanari O, Kloska S, Kopka J, Udvardi MK (2004) Global changes in transcription orchestrate metabolic differentiation during symbiotic nitrogen fixation in Lotus japonicus. Plant J 39 487–512 [DOI] [PubMed] [Google Scholar]
- Colebatch G, Kloska S, Trevaskis B, Freund S, Altmann T, Udvardi MK (2002) Novel aspects of symbiotic nitrogen fixation uncovered by transcript profiling with cDNA arrays. Mol Plant Microbe Interact 15 411–420 [DOI] [PubMed] [Google Scholar]
- Cook DR (1999) Medicago truncatula-a model in the making! Curr Opin Plant Biol 2 301–304 [DOI] [PubMed] [Google Scholar]
- D'Haeze W, Holsters M (2002) Nod factor structures, responses, and perception during initiation of nodule development. Glycobiology 12 79R–105R [DOI] [PubMed] [Google Scholar]
- Denker SP, Barber DL (2002) Ion transport proteins anchor and regulate the cytoskeleton. Curr Opin Cell Biol 14 214–220 [DOI] [PubMed] [Google Scholar]
- Endre G, Kereszt A, Kevei Z, Mihacea S, Kalo P, Kiss GB (2002) A receptor kinase gene regulating symbiotic nodule development. Nature 417 962–966 [DOI] [PubMed] [Google Scholar]
- Fedorova M, van de Mortel J, Matsumoto PA, Cho J, Town CD, VandenBosch KA, Gantt JS, Vance CP (2002) Genome-wide identification of nodule-specific transcripts in the model legume Medicago truncatula. Plant Physiol 130 519–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain MS, Umehara Y, Kouchi H (2006) A novel Fix− symbiotic mutant of Lotus japonicus, Ljsym105, shows impaired development and premature deterioration of nodule infected cells and symbiosomes. Mol Plant Microbe Interact 19 780–788 [DOI] [PubMed] [Google Scholar]
- Imaizumi-Anraku H, Kawaguchi M, Koiwa H, Akao S, Syono K (1997) Two ineffective-nodulating mutants of Lotus japonicus: different phenotypes caused by the blockage of endocytotic bacterial release and nodule maturation. Plant Cell Physiol 38 871–881 [Google Scholar]
- Imaizumi-Anraku H, Takeda N, Charpentier M, Perry J, Miwa H, Umehara Y, Kouchi H, Murakami Y, Mulder L, Vickers K, et al (2005) Plastid proteins crucial for symbiotic fungal and bacterial entry into plant roots. Nature 433 527–530 [DOI] [PubMed] [Google Scholar]
- Kadenbach B, Jarausch J, Hartmann R, Merle P (1983) Separation of mammalian cytochrome c oxidase into 13 polypeptides by a sodium dodecyl sulfate-gel electrophoretic procedure. Anal Biochem 129 517–521 [DOI] [PubMed] [Google Scholar]
- Kawaguchi M, Imaizumi-Anraku H, Koiwa H, Niwa S, Ikuta A, Syono K, Akao S (2002) Root, root hair, and symbiotic mutants of the model legume Lotus japonicus. Mol Plant Microbe Interact 15 17–26 [DOI] [PubMed] [Google Scholar]
- Kawaguchi M, Motomura T, Imaizumi-Anraku H, Akao S, Kawasaki S (2001) Providing the basis for genomics in Lotus japonicus: the accessions Miyakojima and Gifu are appropriate crossing partners for genetic analyses. Mol Genet Genomics 266 157–166 [DOI] [PubMed] [Google Scholar]
- Kouchi H, Fukai K, Kihara A (1991) Metabolism of glutamate and aspartate in bacteroids isolated from soybean root nodules. J Gen Microbiol 137 2901–2910 [Google Scholar]
- Kouchi H, Shimomura K, Hata S, Hirota A, Wu GJ, Kumagai H, Tajima S, Suganuma N, Suzuki A, Aoki T, et al (2004) Large-scale analysis of gene expression profiles during early stages of root nodule formation in a model legume, Lotus japonicus. DNA Res 11 263–274 [DOI] [PubMed] [Google Scholar]
- Krusell L, Krause K, Ott T, Desbrosses G, Kramer U, Sato S, Nakamura Y, Tabata S, James EK, Sandal N, et al (2005) The sulfate transporter SST1 is crucial for symbiotic nitrogen fixation in Lotus japonicus root nodules. Plant Cell 17 1625–1636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai H, Kouchi H (2003) Gene silencing by expression of hairpin RNA in Lotus japonicus roots and root nodules. Mol Plant Microbe Interact 16 663–668 [DOI] [PubMed] [Google Scholar]
- Kuppusamy KT, Endre G, Prabhu R, Penmetsa RV, Veereshlingam H, Cook DR, Dickstein R, Vandenbosch KA (2004) LIN, a Medicago truncatula gene required for nodule differentiation and persistence of rhizobial infections. Plant Physiol 136 3682–3691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy J, Bres C, Geurts R, Chalhoub B, Kulikova O, Duc G, Journet EP, Ane JM, Lauber E, Bisseling T, et al (2004) A putative Ca2+ and calmodulin-dependent protein kinase required for bacterial and fungal symbioses. Science 303 1361–1364 [DOI] [PubMed] [Google Scholar]
- Li H, Luo J, Hemphill JK, Wang JT, Gould JH (2001) A rapid and high yielding DNA miniprep for cotton (Gossypium spp.). Plant Mol Biol Rep 19 183a–183e [Google Scholar]
- Limpens E, Franken C, Smit P, Willemse J, Bisseling T, Geurts R (2003) LysM domain receptor kinases regulating rhizobial Nod factor-induced infection. Science 302 630–633 [DOI] [PubMed] [Google Scholar]
- Liu YG, Shirano Y, Fukaki H, Yanai Y, Tasaka M, Tabata S, Shibata D (1999) Complementation of plant mutants with large genomic DNA fragments by a transformation-competent artificial chromosome vector accelerates positional cloning. Proc Natl Acad Sci USA 96 6535–6540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H, Liu Y, Greenberg JT (2005) Structure-function analysis of the plasma membrane-localized Arabidopsis defense component ACD6. Plant J 44 798–809 [DOI] [PubMed] [Google Scholar]
- Lu H, Rate DN, Song JT, Greenberg JT (2003) ACD6, a novel ankyrin protein, is a regulator and an effector of salicylic acid signaling in the Arabidopsis defense response. Plant Cell 15 2408–2420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen EB, Madsen LH, Radutoiu S, Olbryt M, Rakwalska M, Szczyglowski K, Sato S, Kaneko T, Tabata S, Sandal N, et al (2003) A receptor kinase gene of the LysM type is involved in legume perception of rhizobial signals. Nature 425 637–640 [DOI] [PubMed] [Google Scholar]
- Morzhina EV, Tsyganov VE, Borisov AY, Lebsky VK, Tikhonovich IA (2000) Four developmental stages identified by genetic dissection of pea (Pisum sativum L.) root nodule morphogenesis. Plant Sci 155 75–83 [DOI] [PubMed] [Google Scholar]
- Mosavi LK, Minor DL, Peng ZY (2002) Consensus-derived structural determinants of the ankyrin repeat motif. Proc Natl Acad Sci USA 99 16029–16034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata T, Nemoto Y, Hasezawa S (1992) Tobacco BY-2 cell-line as the HeLa-cell in the cell biology of higher plants. Int Rev Cytol 132 1–30 [Google Scholar]
- Niwa S, Kawaguchi M, Imazumi-Anraku H, Chechetka SA, Ishizaka M, Ikuta A, Kouchi H (2001) Responses of a model legume Lotus japonicus to lipochitin oligosaccharide nodulation factors purified from Mesorhizobium loti JRL501. Mol Plant Microbe Interact 14 848–856 [DOI] [PubMed] [Google Scholar]
- Niwa Y, Hirano T, Yoshimoto K, Shimizu M, Kobayashi H (1999) Non-invasive quantitative detection and applications of non-toxic, S65T-type green fluorescent protein in living plants. Plant J 18 455–463 [DOI] [PubMed] [Google Scholar]
- Novak K, Pesina K, Nebesarova J, Skrdleta V, Lisa L, Nasinec V (1995) Symbiotic tissue degradation pattern in the ineffective nodules of three nodulation mutants of pea (Pisum sativum L.). Ann Bot (Lond) 76 303–313 [Google Scholar]
- Radutoiu S, Madsen LH, Madsen EB, Felle HH, Umehara Y, Gronlund M, Sato S, Nakamura Y, Tabata S, Sandal N, et al (2003) Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature 425 585–592 [DOI] [PubMed] [Google Scholar]
- Rechsteiner M, Rogers SW (1996) PEST sequences and regulation by proteolysis. Trends Biochem Sci 21 267–271 [PubMed] [Google Scholar]
- Rodriguez MS, Michalopoulos I, Arenzana-Seisdedos F, Hay RT (1995) Inducible degradation of I-kappaB-alpha in vitro and in vivo requires the acidic C-terminal domain of the protein. Mol Cell Biol 15 2413–2419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandal N, Petersen TR, Murray J, Umehara Y, Karas B, Yano K, Kumagai H, Yoshikawa M, Saito K, Hayashi M, et al (2006) Genetics of symbiosis in Lotus japonicus: recombinant inbred lines, comparative genetic maps and map position of 35 symbiotic loci. Mol Plant Microbe Interact 19 80–91 [DOI] [PubMed] [Google Scholar]
- Schauser L, Handberg K, Sandal N, Stiller J, Thykjaer T, Pajuelo E, Nielsen A, Stougaard J (1998) Symbiotic mutants deficient in nodule establishment identified after T-DNA transformation of Lotus japonicus. Mol Gen Genet 259 414–423 [DOI] [PubMed] [Google Scholar]
- Sedgwick SG, Smerdon SJ (1999) The ankyrin repeat: a diversity of interactions on a common structural framework. Trends Biochem Sci 24 311–316 [DOI] [PubMed] [Google Scholar]
- Siddique ABM, Bal AM (1992) Morphological and biochemical changes in peanut nodules during photosynthate stress. Can J Microbiol 38 526–533 [Google Scholar]
- Spaink HP, Lugtenberg BJ (1994) Role of rhizobial lipo-chitin oligosaccharide signal molecules in root nodule organogenesis. Plant Mol Biol 26 1413–1422 [DOI] [PubMed] [Google Scholar]
- Stiller J, Martirani L, Tuppale S, Chian R, Chiurazzi M, Gresshoff P (1997) High frequency transformation and regeneration of transgenic plants in the model legume Lotus japonicus. J Exp Bot 48 1357–1365 [Google Scholar]
- Stracke S, Kistner C, Yoshida S, Mulder L, Sato S, Kaneko T, Tabata S, Sandal N, Stougaard J, Szczyglowski K, et al (2002) A plant receptor-like kinase required for both bacterial and fungal symbiosis. Nature 417 959–962 [DOI] [PubMed] [Google Scholar]
- Suganuma N, Nakamura Y, Yamamoto M, Ohta T, Koiwa H, Akao S, Kawaguchi M (2003) The Lotus japonicus Sen1 gene controls rhizobial differentiation into nitrogen-fixing bacteroids in nodules. Mol Genet Genomics 269 312–320 [DOI] [PubMed] [Google Scholar]
- Suganuma N, Sonoda N, Nakane C, Hayashi K, Hayashi T, Tamaoki M, Kouchi H (1998) Bacteroids isolated from ineffective nodules of Pisum sativum mutant E135 (sym13) lack nitrogenase activity but contain the two protein components of nitrogenase. Plant Cell Physiol 39 1093–1098 [Google Scholar]
- Suganuma N, Tamaoki M, Kouchi H (1995) Expression of nodulin genes in plant-determined ineffective nodules of pea. Plant Mol Biol 28 1027–1038 [DOI] [PubMed] [Google Scholar]
- Szczyglowski K, Shaw RS, Wopereis J, Copeland S, Hamburger D, Kasiborski B, Dazzo FB, de Bruijin FJ (1998) Nodule organogenesis and symbiotic mutants of the model legume Lotus japonicus. Mol Plant Microbe Interact 11 684–697 [Google Scholar]
- Tansengco ML, Hayashi M, Kawaguchi M, Imaizumi-Anraku H, Murooka Y (2003) crinkle, a novel symbiotic mutant that affects the infection thread growth and alters the root hair, trichome, and seed development in Lotus japonicus. Plant Physiol 131 1054–1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirichine L, Imaizumi-Anraku H, Yoshida S, Murakami Y, Madsen LH, Miwa H, Nakagawa T, Sandal N, Albrektsen AS, Kawaguchi M, et al (2006) Deregulation of a Ca2+/calmodulin dependent kinase leads to spontaneous nodule development. Nature 441 1153–1156 [DOI] [PubMed] [Google Scholar]
- Udvardi MK, Price GD, Gresshoff PM, Day DA (1988) A dicarboxylate transporter on the peribacteroid membrane of soybean nodules. FEBS Lett 231 36–40 [Google Scholar]
- Uemura M, Yoshida S (1983) Isolation and identification of plasma membrane from light-grown winter rye seedlings (Secale cereale L. cv. Puma). Plant Physiol 73 586–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma IM, Stevenson JK, Schwarz EM, Van Antwerp D, Miyamoto S (1995) Rel/NF-κB/I-κB family: intimate tales of association and dissociation. Genes Dev 9 2723–2735 [DOI] [PubMed] [Google Scholar]
- Yan J, Wang J, Zhang H (2002) An ankyrin repeat-containing protein plays a role in both disease resistance and antioxydation metabolism. Plant J 29 193–202 [DOI] [PubMed] [Google Scholar]