Abstract
Nitric oxide and S-nitrosothiols (SNOs) are widespread signaling molecules that regulate immunity in animals and plants. Levels of SNOs in vivo are controlled by nitric oxide synthesis (which in plants is achieved by different routes) and by S-nitrosoglutathione turnover, which is mainly performed by the S-nitrosoglutathione reductase (GSNOR). GSNOR is encoded by a single-copy gene in Arabidopsis (Arabidopsis thaliana; Martínez et al., 1996; Sakamoto et al., 2002). We report here that transgenic plants with decreased amounts of GSNOR (using antisense strategy) show enhanced basal resistance against Peronospora parasitica Noco2 (oomycete), which correlates with higher levels of intracellular SNOs and constitutive activation of the pathogenesis-related gene, PR-1. Moreover, systemic acquired resistance is impaired in plants overexpressing GSNOR and enhanced in the antisense plants, and this correlates with changes in the SNO content both in local and systemic leaves. We also show that GSNOR is localized in the phloem and, thus, could regulate systemic acquired resistance signal transport through the vascular system. Our data corroborate the data from other authors that GSNOR controls SNO in vivo levels, and shows that SNO content positively influences plant basal resistance and resistance-gene-mediated resistance as well. These data highlight GSNOR as an important and widely utilized component of resistance protein signaling networks conserved in animals and plants.
Considerable evidence indicates that nitric oxide (NO) and NO-related metabolites, such as S-nitrosothiols (SNOs) play a central role in signal transduction and defense in animal and plants (Wendehenne et al., 2001). Delledonne et al. (1998) have shown that inhibitors of NO accumulation blocked the localized cell death induced in plant-pathogen interactions, called the hypersensitive response (HR), promoting disease and bacterial growth in Arabidopsis (Arabidopsis thaliana). They conclude that NO functions as a key signal in plant disease resistance. The efficient induction of HR requires an appropriate balance between reactive oxygen intermediates (ROIs) and NO production (Delledonne et al., 2001). Several defense genes, such as genes encoding Phe ammonia-lyase, glutathione S-transferase (GST), and pathogenesis-related protein 1 (PR-1), are induced by administration of NO donors or recombinant mammalian NO synthase (NOS; Delledonne et al., 1998; Durner et al., 1998).
NO can be synthesized in plants by different routes, both enzymatically and nonenzymatically (Cooney et al., 1994; Rockel et al., 2002; Modolo et al., 2005). Intracellular NO reacts with proteins and nonprotein thiols to form nitrosothiols (Stamler, 1994). Nitrosylation of proteins is achieved by reaction with sulfhydryl groups and transition metals and in many cases results in the regulation of protein activity (Stamler et al., 2001). NO reacts rapidly with glutathione (GSH), the major intracellular low-molecular-mass antioxidant, to yield S-nitrosoglutathione (GSNO). GSNO is considered to represent a functionally relevant signaling molecule that might act both as NO reservoir and NO donor (Stamler et al., 1992; Lindermayr et al., 2005) or independently of homolytic cleavage to NO (Gaston, 1999). An additional level to regulate the NO system is by breakdown of GSNO by the recently discovered GSNO reductase (GSNOR) that is conserved from bacteria to humans (Liu et al., 2001). It has been shown that the GSNOR activity controls intracellular levels of both GSNO and S-nitrosylated proteins and enhances cellular resistance to nitrosative stress in animal models (Liu et al., 2001, 2004). Innate immunity, which includes preformed barriers and induced defense responses, constitutes a broad spectrum of barriers against attempted pathogen invasion. Recent findings have revealed striking similarities in the molecular mechanisms for the activation of innate immunity response in plants, vertebrates, and insects (Nürnberger et al., 2004). Pathogen-associated molecular patterns that are characteristic of microbial organisms, such as lipopolysaccharides of gram-negative bacteria, trigger innate immune responses in animals and act as defense inducers in multiple plant species (Nürnberger et al., 2004; Zeidler et al., 2004). NO production is part of the lipopolysaccharide-induced innate immunity both in animals and plants leading to the activation of defense-related genes and synthesis of antimicrobial protein and peptides (Nathan and Shiloh, 2000; Liu et al., 2004; Zeidler et al., 2004). These findings support the idea of a common evolutionary origin of pathogen defense system in higher eukaryotes. NO bioactivity is controlled by NO synthesis (i.e. by the different routes above mentioned) and by NO degradation, which is mainly performed by the GSNOR (Liu et al., 2001, 2004). In Arabidopsis, GSNOR, previously known as GSH-dependent formaldehyde dehydrogenase (FALDH) or also class III alcohol dehydrogenase (ADH), due to its activity versus primary alcohols, is encoded by a single-copy gene (ADH2, GenBank accession no. X82647; Martínez et al., 1996). Arabidopsis transgenic plants that either overexpress or underexpress the enzyme were generated by cloning the ADH2 gene into an Agrobacterium binary vector under the control of 35S promoter, both in the sense and the antisense orientations (Achkor et al., 2003). These plants have been used in this work to study GSNOR involvement in basal disease resistance and resistance (R)-gene-mediated resistance against oomycetes (Peronospora parasitica [Pp]) and bacteria (Pseudomonas syringae pv. maculicola [Psm]). Our results highlight GSNOR as an important and widely utilized component of resistance protein signaling networks.
RESULTS
GSNOR Transgenic Lines
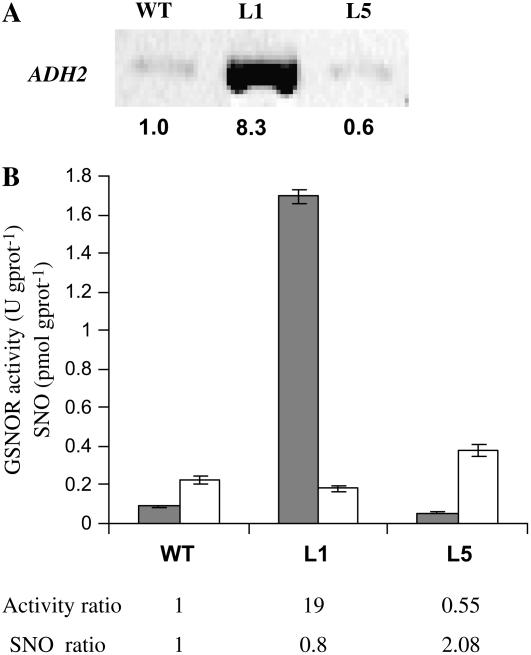
We have described elsewhere the generation of Arabidopsis transgenic lines transformed with either sense or antisense constructs of the ADH2 gene placed under the control of the 35S cauliflower mosaic virus promoter (Achkor et al., 2003). ADH2 gene codes for the GSH-dependent FALDH, also known as class III ADH, which is encoded by a single-copy gene (ADH2, GenBank accession no. X82647; Martínez et al., 1996). This enzyme has been recently shown to be the GSNOR (Liu et al., 2001; Sakamoto et al., 2002). Determination of GSNOR activity in several independent transgenic lines overexpressing the ADH2 gene revealed increments of up to 19-fold the activity in wild-type Arabidopsis, which correlated with high increments in mRNA and protein levels. At the opposite, several independent antisense lines tested showed decrements of up to 50% the GSNOR activity measured in wild-type plants. One overexpressing line (L1) and one antisense line (L5) were chosen for further experiments and deeper characterization. Figure 1, A and B show, respectively, transcript levels and GSNOR activity values for these two lines.
Figure 1.
GSNOR activity and intracellular SNO levels. A, GSNOR transcript accumulation determined by semiquantitative RT-PCR. Intensity of the bands was determined by densitometry and the transcript levels were normalized in relation to those of the actin2 gene. The numbers below the bands denote relative gene expression. B, GSNOR activity (gray bars) and SNO content (white bars) in Arabidopsis 4-week-old seedlings. One unity of activity (U) corresponds to 1 μmol of coenzyme (NADH) transformed per minute. Results are the mean of three independent determinations ± sd. The numbers below the plot denote the activity and SNO ratios among the different lines. WT, Wild-type Arabidopsis accession Col-0; L1, transgenic line containing the sense construct; L5, transgenic line containing the antisense construct.
To study if GSNOR controls SNO levels in vivo, we measured SNO content in the transgenic lines. Figure 1B shows that total SNO levels in leaf extracts of 4-week-old plants were increased in L5 (antisense line; 168%) and decreased in L1 (overexpressing line; 80%), as compared to those found in wild-type Arabidopsis plants grown in the same conditions. Low Mw SNOs (<5 K) were under the limit of detection of the method. In conclusion, the expression of the transgene in the transgenic lines correlates with the expected changes in GSNOR activity and in basal SNO content, supporting the idea that GSNOR controls in vivo SNO levels. The defect in SNO accumulation detected in L1 was not due to a defect in the synthesis of NO, since we measured activation of inducible NOS (iNOS) in the local leaves of L1 after pathogen inoculation (see Supplemental Table S1).
Downexpression of GSNOR Increments Basal Resistance
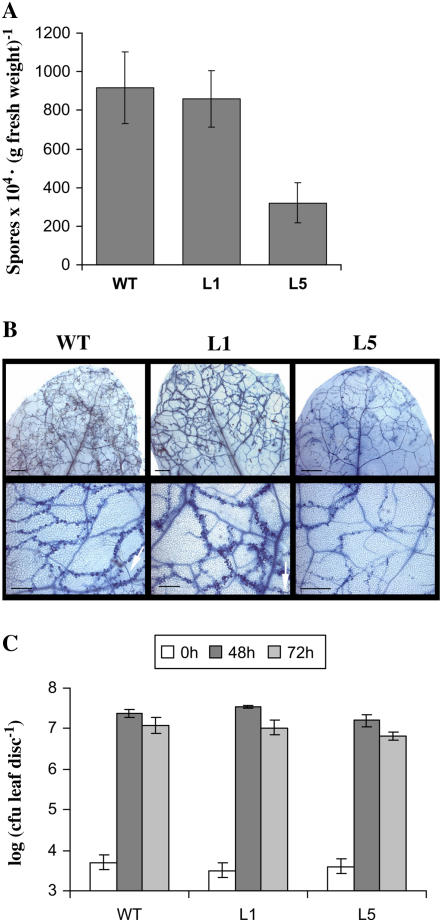
The interaction between Arabidopsis and the biotrophic oomycete Pp is a largely studied model and one of the best known systems of the race-specific pathogen resistance. Arabidopsis ecotype Columbia (Col-0) is susceptible to Pp Noco2: its mycelium is able to spread systemically through the plant tissue and to develop sexual and asexual spores within 4 to 7 d. In contrast, Arabidopsis Col-0 is resistant to isolate Cala2, due to the gene-for-gene recognition (Van der Biezen et al., 2002). We first assessed the responses between short-day-grown L1 and L5 transgenic lines to normally virulent Pp Noco2 isolate. Four-week-old plants were sprayed with an asexual spores suspension of isolate Noco2 (4 × 104 spores mL−1) and spores counting was carried out 6 d after inoculation. As can be seen in Figure 2A, number of conidia is comparable in the overexpressing (L1) and wild-type lines, whereas it is reduced by 65% in the antisense line (L5). Accordingly, we observed extensive hyphal growth as well as asexual sporulation in wild type and L1 but not in L5 (Fig. 2B, the white arrows indicate conidiophores).
Figure 2.
Pathogen growth in the different plant lines. Four-week-old plants, wild type, and same GSNOR transgenic lines as those in Figure 1 were treated by virulent pathogens Pp isolate Noco2 (A and B) or Psm (C). A, Number of asexual spores 6 d after inoculation by spray of Pp (4 × 104 spores mL−1). Values are the mean of five independent measurements ± sd. Similar results were obtained on two independent experiments. B, Development of Pp mycelium inside leaf tissues, monitored by staining with lactophenol-trypan blue, 6 d after the treatment. Bars: top section, 800 μm; bottom section, 300 μm. C, Growth of Psm in leaves at times 0, 2, and 3 d after infiltration (105 cfu mL−1). The experiment was made three times and each value is the mean of five independent measurements ± sd.
We then examined the responses to pathogenic bacteria Pseudomonas syringae, a hemibiotroph pathogen that infects through wounds and stomata and multiplies in the intercellular spaces (Glazebrook, 2005). Strains of P. syringae collectively infect a wide variety of plants including Arabidopsis. Four-week-old plants were challenged with the normally virulent Psm at low doses (105 colony forming units [cfu] mL−1) and bacterial growth was measured 2 and 3 d after infection. Figure 2C shows that no significant differences were found between the lines. From these results we conclude that the antisense line (L5) has an enhanced basal resistance to Pp Noco2 but not to Psm in the tested conditions. The difference obtained between the two pathogens tested might be the result of a difference in basal efficiency depending on the pathogen or to a differential involvement of GSNOR in each of these pathways.
GSNOR Is Required in the R-Gene-Mediated Systemic Acquired Resistance Establishment
Gene-for-gene resistance depends on the presence of single dominant genes in the pathogen (avirulence genes) that cause it to be recognized by plant hosts carrying single dominant R genes. The oomycete Pp Cala2 isolate and the bacterial Psm avrRpm1 strain are avirulent pathogens, respectively, for Arabidopsis Col-0. Plant resistance involves in both cases salicylic acid (SA)-mediated defenses and discrete necrotic lesions, but they are specified by distinct R proteins with different N-terminal domains that induce distinct early signaling networks (Parker et al., 2000; Feys et al., 2001; Rustérucci et al., 2001; Hammond-Kosack and Parker, 2003). We assessed the interaction between L1 and L5 transgenic plants and these normally avirulent pathogens.
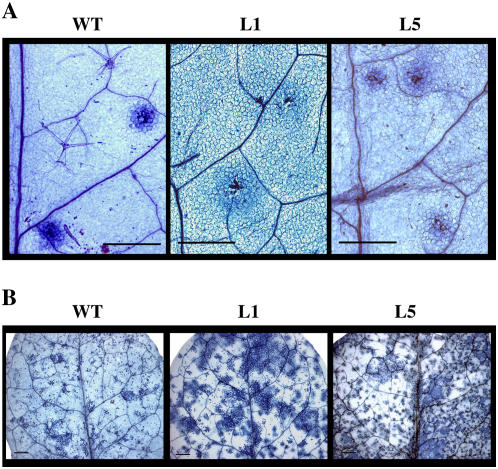
Plants were sprayed with a conidia suspension of the oomycete in the same conditions as for Noco2 isolate. No phenotypic differences between the different lines were apparent regarding the resistance to the pathogen, and all the lines exhibited localized cell death typical of the HR at the point of Pp penetration (Fig. 3A). However, a slight increment in the diameter of the microscopic necrotic spots was observed in both transgenic lines but especially in L1 (Fig. 3A) from the starting of HR establishment to up to 7 d after inoculation.
Figure 3.
HR-mediated resistance in the different Arabidopsis lines. Four-week-old plants, wild type, and same GSNOR transgenic lines as those in Figure 1 were treated by avirulent pathogens, Pp isolate Cala2 (A) or Psm avrRpm1 (B). A, 10 μL droplet of Pp Cala2 (4 × 104 spores mL−1) was placed on the leaf surface and after 3 d HR was monitored by staining with lactophenol-trypan blue. B, Plants were infiltrated with suspensions of Psm avrRpm1 (106 cfu mL−1) and after 3 d necrotic lesions were monitored by staining with lactophenol-trypan blue. Bars: 500 μm.
We then examined the responses of the same transgenic lines to the avirulent bacterial pathogen Psm avrRpm1 by infiltrating the leaves with the bacterial suspension (106 cfu mL−1). No macroscopic disease symptoms were observed 3 d after treatment that could be related to a loss of resistance in transgenic lines (results not shown). Moreover, HR was induced in both L1 and L5 transgenic lines, as well as in wild-type plants (Fig. 3B). The microscopic lesions developed by both L1 and L5 lines were more spread than in wild-type plants (Fig. 3B), as it was also observed after Pp Cala2 challenging. It has been reported that NO production is necessary for HR in concert to hydrogen peroxide (Delledonne et al., 1998). We show here that up- or down-regulation of GSNOR does not affect HR establishment in the incompatible interactions. However, the spreading of HR lesions is slightly affected but still restricted, at the opposite of lesions simulating disease 1 (lsd1) mutants that lacks up-regulation for detoxification of accumulating ROIs (Kliebenstein et al., 1999; Rustérucci et al., 2001).
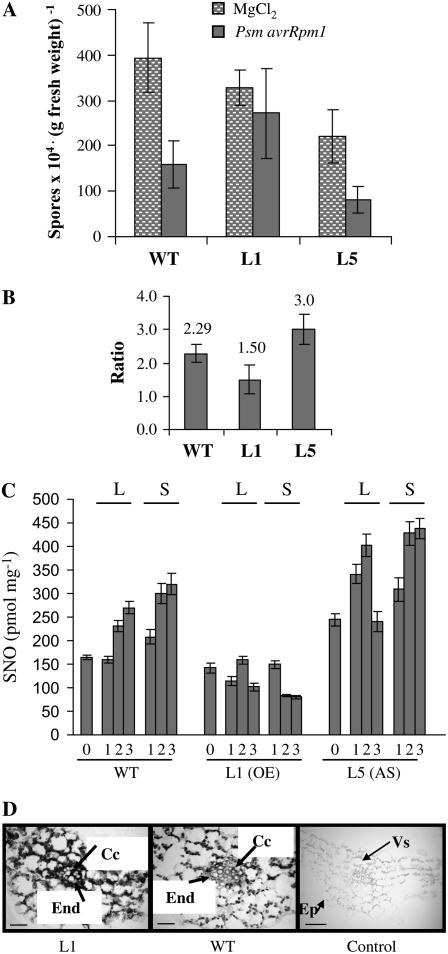
Systemic acquired resistance (SAR) is an induced defense response that is activated by avirulent pathogens and protects plants from further infections by a wide variety of pathogens (Ryals et al., 1996). SAR is accompanied by an increase of endogenous SA, both locally and systemically, and correlates tightly with the expression of genes encoding PR proteins (Durrant and Dong, 2004). To further explore the impact of the manipulation of GSNOR intracellular levels on plant defense mechanisms we tested SAR establishment in our transgenic lines. This was performed using the combination of avirulent bacteria (Psm avrRpm1) and virulent oomycete (Pp Noco2): avirulent bacteria were infiltrated in two leaves of 4-week-old plants and 48 h later these plants were sprayed with a conidia suspension of the virulent oomycete. Figure 4A shows the Pp sporulation on plants mock preinfiltrated or pretreated with the avirulent bacteria for a typical experiment. The SAR is revealed by a significant difference of sporulation between both treatments. Figure 4B shows the ratio average of two independent experiments. It is apparent that SAR establishment is enhanced in L5 (antisense line) with a significant increment of 30% compared to wild type and that it is significantly impaired (35% less) in L1 (overexpressing line).
Figure 4.
Quantification of SAR against Pp Noco2 in the different plant lines and immunolocalization of GSNOR in the phloem. SAR establishment was measured using the combination of avirulent bacteria (Psm avrRpm1) and virulent oomycete (Pp Noco2). Two rosette leaves of the different lines were infiltrated either with the avirulent bacteria (106 cfu mL−1) or mock inoculated with 10 mm MgCl2, and 48 h later were sprayed with a suspension of the virulent oomycete (4 × 104 spores mL−1). Number of conidia was counted 6 d after Pp inoculation. The average of five independent measurements ± sd is represented in A for a typical experiment. The experiment was performed twice and the mean value ± sd is represented in B as the ratio of number of spores in plants previously infiltrated with Psm avrRpm1 or with MgCl2 (bar height ratio). C, Total SNO levels in leaf extracts from the different Arabidopsis lines, either challenged or unchallenged with Psm avrRpm1 (106 cfu mL−1). Systemic and challenged leaves were collected at days 1, 2, and 3 after treatment. Values were normalized per milligram of protein. D, Immunodetection of GSNOR in the phloem. Immunolocalization was performed using specific antibodies against Arabidopsis GSNOR as described in Espunya et al. (2006). In the control panel, the antibody was preincubated with purified FALDH protein prior to the immunolocalization experiments. Bars: 25 μm. End, Endodermis; Cc, phloem companion cells; Vs, vascular system; Ep, epidermis.
Incompatible plant-pathogen interactions induce H2O2 and NO bursts, both necessary for HR establishment. The level of GSNOR activity in our transgenic lines did not interfere with HR establishment but modulated plant SNO levels in vivo. To see if the above GSNOR effect on SAR efficiency was related to SNO levels, we measured total SNO content in wild-type and transgenic Arabidopsis plants after challenging with avirulent bacteria (Psm avrRpm1). Figure 4C shows that challenging with Psm avrRpm1 induces SNO accumulation in wild-type Arabidopsis, both locally and systemically. In L5, SNO levels increase more rapidly than in wild type in the local leaves (see 1 d in Fig. 4C) and, at long times, both wild type and L5 accumulate more SNO in systemic than in local leaves (see days 2 and 3, Fig. 4C). Moreover, SNO levels in L5 are always higher than in wild type, in unchallenged and challenged plants and in systemic and local tissues. To the contrary, SNO accumulation is impaired in plants overexpressing GSNOR, both in local and systemic leaves. In L1, SNO levels were 37% and 24% those of wild type, in local and systemic leaves, respectively, at day 3 after challenging. Low Mw SNOs (<5 K) were under the limit of detection, except for L5 in systemic leaves at day 2 and 3 after challenging (data not shown), suggesting that the increase in total SNO content correlated with an increase in low Mw. In conclusion, SNO accumulated in local and systemic leaves during the incompatible interaction. The expression of the transgene correlates with SNO content before and after challenging in the transgenic lines, and has consequences for the accumulation of SNO in local and sytemic leaves during incompatible interactions. The impairment of SNO accumulation correlates with the impairment of SAR establishment in the GSNOR overexpressing line.
It has been proposed that GSNO might serve as a long distance phloem mobile for SAR establishment (Durner and Klessig, 1999). We have previously shown the localization of GSNOR in phloem companion cells and xylem parenchyma in Arabidopsis plants (Espunya et al., 2006) and as expected, L1 overexpressing line contains more GSNOR in the vascular system (Fig. 4D). We conclude that GSNOR activity is involved in SAR establishment and it is tempting to speculate that its role is linked to SAR signal transport through the vascular system. However, the use of a constitutive promoter to drive GSNOR overexpression does not allow us to determine whether perturbation of SNO levels in the systemic leaves are due to the impairment of transport or to the activity of the transgene in the distal tissues.
Molecular Analysis
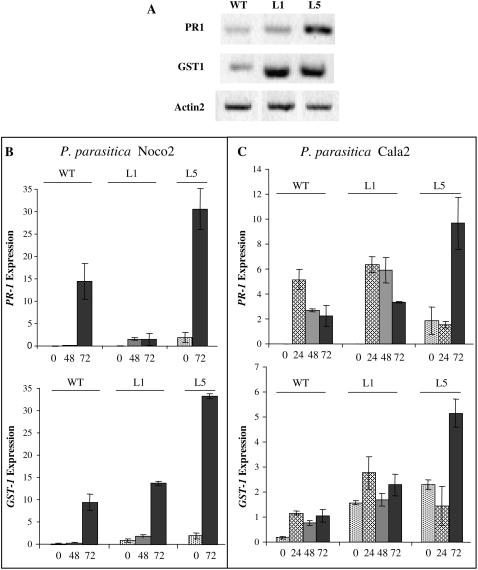
Plants respond to pathogen attack generally by activation of HR and a large number of defense effectors mechanisms, including production of both antimicrobial metabolites and proteins as well as physical reinforcement of cell walls. Many of the gene expression changes that occur in the gene-for-gene responses also occur during susceptible interactions, but with slower kinetics and reduced amplitude (Tao et al., 2003). To correlate activation of known defense genes with the phenotypic responses detected in the GSNOR transgenic plants when challenged to pathogens, we measured the transcript levels of the PR-1 and of GST-1, a protectant gene induced by ROIs during incompatible interactions (Levine et al., 1994). Both genes are also induced by NO donors (Delledonne et al., 1998; Durner et al., 1998).
As shown in Figure 5A, PR-1 is constitutively activated in the antisense plants (6-fold the transcript levels of wild-type plants), whereas the overexpressing plants exhibit similar levels as those of wild type, which were almost negligible. Moreover, GST-1 is constitutively activated in both transgenic lines (4-fold the transcript levels of wild-type plants). We also studied the kinetics of PR-1 and GST-1 induction when plants were challenged with the virulent and avirulent Pp. Both genes were induced by pathogens but the induction was faster using avirulent Pp isolate, as expected (Figs. 5, B and C). In the transgenic lines the induction pattern was nearly similar to that of wild type: a slow increase of the transcript levels from 0 to 72 h in compatible interactions, and a rapid increase within 24 h in incompatible interactions. In incompatible interactions, the level of GST-1 transcripts is maintained until 72 h, while those of PR-1 decrease with time, indicating a transient induction in the last case. What is remarkable is that 72 h after treatments both genes are further induced to very high levels in the antisense line after treatment with either one of the two pathogens tested (PR-1 and GST-1 transcripts are 4- and 5-fold, respectively, more abundant than in wild type in the incompatible interactions, and 2- and 3-fold, respectively, in the compatible interactions; Fig. 5, B and C). Furthermore, in the overexpressing line, PR-1 induction is seriously impaired at long times after the infection by the virulent pathogen but not by the avirulent one (see 72 h, Fig. 5, B and C, respectively), while GST-1 induction is similar, both in kinetics and intensity to that in wild-type plants, whatever pathogen was inoculated.
Figure 5.
Expression pattern of PR-1 and GST-1 in the different plant lines. Transcript accumulation was determined by semiquantitative RT-PCR using gene-specific primers and conditions, as described in “Materials and Methods.” A, Determination of PR-1 and GST-1 expression in the different plant lines, without any treatment. Total RNA was extracted from 4-week-old Arabidopsis plants and transcript levels for Actin2 gene, which expression is constitutive, were used to control the amount of mRNA in the reaction. B and C, Time courses of PR-1 and GST-1 expression after Pp Noco2 (B) or Pp Cala2 (C) inoculation. Four-week-old Arabidopsis lines were sprayed with the oomycete spores suspensions and samples for RNA preparations were collected at the indicated times. The values in the histograms denote the relative expression normalized in relation to that of EIF4 gene, in which expression is constitutive.
DISCUSSION
Our results show that manipulation of intracellular levels of GSNOR has important consequences for disease resistance in Arabidopsis. Decreasing cellular levels of GSNOR (using antisense strategy) enhances basal disease resistance against oomycetes (Pp), but overexpression of GSNOR using the 35S cauliflower mosaic virus promoter does not affect significantly basal plant resistance. The constitutive expression of PR-1 exhibited by the antisense plants might account for the enhanced resistance, like it happens in numerous mutants such as constitutive expressor of PR genes or lsd (Bowling et al., 1994; Dietrich et al., 1994; Clarke et al., 2001). Moreover, interaction with the virulent oomycetes results in further induction of PR-1 gene, whose transcript levels accumulate to higher amounts than in wild-type plants (Fig. 5B). To the contrary, plants overexpressing GSNOR did not show constitutive expression of PR-1, and the very limited induction observed after interaction with virulent oomycetes (much less than in wild-type plants, see Fig. 5B) did not change significantly plant susceptibility. However, basal resistance to the bacteria Psm is not affected in either one of the transgenic lines. Shah et al. (2001) showed that resistance to virulent Ps is primarily conferred by the NPR1-dependent pathway, while that to Pp is governed by the NPR1-independent pathway. The enhanced resistance to virulent oomycete but not to virulent bacteria in the GSNOR antisense line could be the result of a differential involvement of GSNOR in each of these pathways or, alternatively, to a difference in basal efficiency depending on the pathogen.
Local R-gene-mediated resistance is not significantly affected in our transgenic plants. HR and defense genes (PR-1, GST) are induced rapidly in both lines and the L1 overexpressing line expresses normal amounts of PR-1 transcript compared to wild type. Measures of SNO content before and after challenging with avirulent bacteria show that basal SNO levels are increased in the antisense plants and decreased in the overexpressing plants as compared to wild-type plants. Although plants overexpressing GSNOR (L1) exhibit no increase in SNO levels after challenging with the avirulent pathogen, they maintain a certain threshold (Fig. 1C), likely as a consequence of the activation of pathogen iNOS at the local site of infection (see Supplemental Table S1), and they are able to trigger efficiently the defense mechanisms at the local site of infection. However, a different situation occurs with SAR establishment. SAR, which requires SA and is associated to PR-1 expression, is enhanced in the antisense plants and lessened in the overexpressing plants. In the same way, Arabidopsis DIR1, a lipid transfer protein like, promotes SAR long-distance signaling and its mutation has no impact on local resistance (Maldonado et al., 2002). It is tempting to speculate that the inverse relationship between GSNOR activity and SAR establishment indicates that generation and/or transport of SAR signal is affected, since impairment of SAR cannot be explained by an impairment of the pathway producing PR-1 or HR. This hypothesis is supported by our data showing that accumulation of SNO after challenging the plants with avirulent bacteria occurs both at the infection site and in the systemic leaves of wild-type and L5 lines, reaching the highest levels in the systemic leaves of L5 at long times after infection. Moreover, in the overexpressing line (L1) SNO accumulation is more disturbed in the systemic than in the local leaves. Our data showing that increase of SNO levels in the systematic leaves of wild-type Arabidopsis plants is not associated to activation of iNOS, strongly suggests that other mechanisms beside NO synthesis control SNO levels in those tissues, i.e. transport of GSNO from the challenged leaves through the phloem, or down-regulation of GSNOR activity in the systemic leaves, or both. Other authors have proposed that GSNO might serve as a long-distance phloem mobile for SAR establishment (Durner and Klessig, 1999) and the localization of GSNOR in the vascular system makes this enzyme a promising candidate to regulate GSNO transport. However, the use of 35S promoter to drive expression of GSNOR in our transgenic plants does not allow us to determine whether perturbation of SNO levels in the systemic leaves is due to the impairment of transport or to the activity of the transgene in the distal tissues. Alternatively, we cannot discard that maintenance of high levels of PR-1 transcripts at long times in the antisense line might account for a better SAR in these plants.
It is striking that our results are in contradiction with those reported by Feechan et al. (2005). The approach used to modify the intracellular levels of GSNOR is different in each case (expression of transgenes in this work and T-DNA insertion mutants in the other case), and so are plant growth conditions. Some authors have reported that 3-week-old plants grown either on short or long day exhibit analogous responses against compatible and incompatible interactions (Greenberg et al., 2000). However, age-related resistance is established in the transition from vegetative to reproductive development (Kus et al., 2002; Rustérucci et al., 2005) that involves signaling pathways independent of those in compatible and incompatible interactions. The age of plants used in Feechan et al. (2005) is not reported, and defense response is extremely dependent on plant age. On the other hand, the measured SNO levels in Feechan et al. (2005) mutants and those of this work are roughly equivalent, both before and after challenging the plants with avirulent pathogens, and in both cases inversely correlated to GSNOR activity. Highly significant is the finding that Feechan et al. (2005) detect a substantial reduction of SA content in atgsnor1-3 mutant (reduced GSNOR activity), whereas the measured contents of free SA and SA glucoside in our transgenic lines were not significantly different from those in wild-type plants (results not shown). Several lines of evidence point to a tight interrelationship between NO and SA in plant defense and in the wounding/jasmonic acid signaling pathway. NO donors produce SA accumulation (Durner et al., 1998) and many NO-regulated enzymes, including aconitase or catalase, are likewise regulated by SA (Durner et al., 1997; Clark et al., 2000). In tobacco (Nicotiana tabacum) cells, NO is required for the full action of SA but the activity of NO is fully dependent on SA in the SAR signaling pathway (Song and Goodman, 2001). Thus, it has been postulated that SA acts both upstream and downstream of NO (Durner and Klessig, 1999). SA signaling in plant defense implies a complex network where multiple stimuli can activate SA synthesis/signaling and positive and negative feedback loops allow for the tighter regulation of SA accumulation (Shah, 2003). Moreover, the precise role of, and the interrelationship between, the key partners of plant defense, H2O2, SA, and NO, is far from being completely understood, likely due to the complexity of the response. It is still controversial whether NO and H2O2 bursts are coincidental and act in concert to initiate cell death (Delledonne et al., 2001) or, to the contrary, NO synthesis is initiated by activation of apoplastic NOS by a lipid-derived signal after membrane breakdown due to the initial programmed cell death (PCD) of the first cells (Zhang et al., 2003; Shapiro, 2005). Lipid-based molecules are likely to play important signaling functions in plant defense (Maldonado et al., 2002; Zeidler et al., 2004). In the model postulated by Zhang et al. (2003), NO accumulates initially in the apoplastic space and acts as a cell-to-cell signal that spreads PCD in the HR, but it is not the initiator of PCD. Moreover, NO inhibits the activity of ascorbate peroxidase and catalase, which are H2O2 scavenging enzymes, potentiating the HR (Shapiro, 2005). A picture is emerging where SA could potentiate the effects of triggering factors (such as calcium fluxes, NO production, caspase-like protease activity, and others) of PCD acting in agonist-dependent gain control (Zhang et al., 2004). The increased sensitivity to pathogens of atgsnor1-3 plants shown in Feechan et al. (2005) might be related to their impairment in SA accumulation, providing an explanation for the differences between their results and the results presented in this article. The advantage of our approach is that SA accumulation is not compromised in the transgenic plants, which allows us to draw conclusions strictly based on the changes of SNO levels as a consequence of changes of GSNOR expression. The other point of concern is the pattern of expression of GSNOR in adult Arabidopsis plants. GSNOR has a constitutive pattern of expression and the enzyme is ubiquitously present in plant cells (Martínez et al., 1996; Dolferus et al., 1997; Espunya et al., 2006). Our transgenic plants show the same qualitative pattern of GSNOR expression, both spatially and temporary (Martínez et al., 1996; Díaz et al., 2004; Espunya et al., 2006; this work). It is important to know if T-DNA insertions in the promoter of GSNOR-encoding gene, as in atgsnor1-1 and atgsnor1-2 lines used in Feechan et al. (2005), change substantially gene expression specificity. Nevertheless, it has been reported that NO-induced gene expression can also be independent of SA (Durner et al., 1998; Grün et al., 2006).
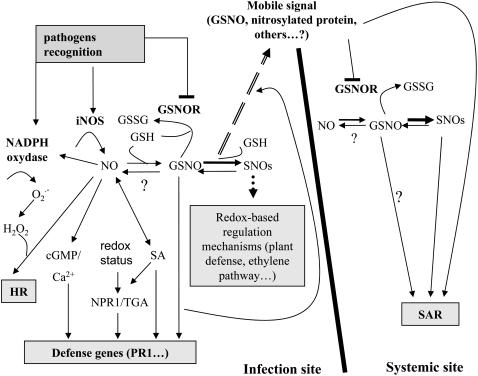
We propose here a model that integrates our data regarding GSNOR involvement in plant defense and data previously published by other authors that supports the idea of NO/GSNO as positive regulators of plant resistance, HR, and defense gene activation (Fig. 6). Based on the data presented in this article and from previous work (Díaz et al., 2003) we propose that down-regulation of GSNOR, with its concomitant increments of intracellular SNO levels, enhances plant immunity. We also show that high levels of SNO in the systemic leaves, which correlate with SAR establishment, are independent of iNOS activation during incompatible interactions and, thus, are likely to be regulated by the activity of GSNOR. This model is in agreement with evolutionary conserved mechanisms in pathogen defense system in plants and animals, which extends to the role of GSNOR in this pathway.
Figure 6.
Hypothetical model to explain the role of GSNOR in incompatible interactions and SAR. Pathogen recognition initiates H2O2 and NO bursts by activation of NADPH and iNOS, respectively. NO, H2O2, and SA act synergistically in triggering HR and other defense responses (Delledonne et al., 2001; Glazebrook, 2005). Different branches, which are SA dependent or independent, and mediated or not by NPR1, lead to amplification of the initial signal and effective activation of downstream defense responses (Durner et al., 1998; Wendehenne et al., 2001; Nimchuk et al., 2003; Glazebrook, 2005). Key regulators of NO and GSNO levels, which are mutually influenced, are the enzymes involved in NO synthesis and in GSNO catabolism, respectively (Liu et al., 2001; Wendehenne et al., 2001). Whereas it is well established that GSNO acts as an NO donor, the role of GSNO as a NO sink is controversial and it is still unknown if GSNO acts as a NO donor in these processes, or if it is a bioactive molecule in itself. GSNOR activity is the main regulator of intracellular GSNO and SNO levels as well (Liu et al., 2001; this work), with an emerging pivotal role in regulating important processes such as plant resistance or the ethylene pathway (Lindermayr et al., 2006). We propose that down-regulation of GSNOR, with its concomitant increments of intracellular SNO levels enhances plant immunity. Maintaining appropriate levels of GSNO is also important for SAR. There is no induction of iNOS in the systemic leaves after challenging the plants with avirulent bacteria, but SNO levels rise in those tissues and this is concomitant with SAR establishment. GSNOR might have a key role in the regulation of SNO levels in the systemic leaves, either because GSNO is the phloem-mobile signal for SAR activation and/or because the mobile signal down-regulates GSNOR in the systemic leaves, slowing down GSNO breakdown. In favor of the first hypothesis, there is evidence that GSNOR localizes into the phloem, and could regulate signal transport. However, both hypotheses are not exclusive; for instance, down-regulation of GSNOR gene after wounding, both at local and systemic leaves, has been reported (Díaz et al., 2003).
A consistent phenotype of mutants with constitutive expression of PR-1 is the presence of spontaneous necrotic lesions (Dietrich et al., 1994; Greenberg et al., 1994; Shah et al., 2001). However, there are some exceptions (Bowling et al., 1994; Li et al., 2001) suggesting that constitutive activation of PR-1 is independent of cell death. The antisense plants reported in this work do not show spontaneous lesions but they consistently exhibit shorter root length (Espunya et al., 2006). This phenotype could not be due to the fitness costs associated with the constitutive activation of SAR (Heil and Baldwin, 2002), because it is present both in the overexpressing and the antisense lines, but could be the result of an alteration in GSH homeostasis as we have previously postulated (Espunya et al., 2006). The constitutive activation of GST-1 shown in this work supports our previous results of a disturbance of redox balance in the transgenic lines, since GST-1 is a marker of ROS. This might also be related to the microscopic spreading of HR lesions observed in our mutants. Due to the required balance of H2O2 and NO production in HR establishment (see “Discussion” above), disturbing NO/GSNO levels by means of GSNOR might influence cell death propagation and affect the size of the lesions. Moreover, proteins involved in maintaining redox homeostasis, such as catalase or γ-glutamylcysteinyl synthetase are regulated by nitrosylation (Han et al., 1995; Foster and Stamler, 2004). The two pathogens tested in this work use different receptors to trigger HR. In one case, Pp, the R-gene product belongs to the TIR-NB-LRR group of proteins, and in the other case, P. syringae, the R genes code for CC-NB-LRR proteins. Our results suggest that GSNOR activity affects HR development in both pathways, acting downstream where the two pathways converge or by an independent pathway.
In summary, our results highlight the importance of the enzyme GSNOR in plant basal resistance and R-gene-mediated resistance. The constitutive activation of plant resistance, as observed for the L5 antisense line, confers an important advantage regarding the approach for developing disease-resistant plants exploiting naturally occurring defense mechanisms. Our results are in concordance with those reported in eukaryotes, vertebrates, and invertebrates, which establish NO accumulation as important for full display of innate immunity (Liu et al., 2004; Zeidler et al., 2004). GSNOR is present in all organisms and much conserved during evolution. Its proposed role in controlling intracellular SNO levels that affect nitrosylation of thiols in proteins and other important cellular molecules, places it at a key point to control cellular homeostasis. We also show that SAR is affected by manipulation of GSNOR content in the cell. GSNOR, which localizes in the phloem, could regulate SAR signal transport through the vascular system.
MATERIALS AND METHODS
Plant Growth
Arabidopsis (Arabidopsis thaliana) ecotype Col-0 and the transgenic lines were grown in Intercept 5GR soil (Scotts Co.) at 22°C, 55% humidity, under 9-h photoperiod light (140 μE m−2 s−1).
Reverse Transcription-PCR Analysis
Total RNA was extracted from frozen leaves with Trizol (Invitrogen). Four micrograms of total RNAs were subjected to first-strand cDNA synthesis in a 20 μL reaction, using SuperScript II RT reverse transcriptase (Invitrogen). Two microliters of the cDNA diluted 1:10 were used as template in the reverse transcription (RT)-PCR amplifications (Biotools DNA polymerase). The gene-specific primers and annealing temperatures used were 5′gctactggtgttgggattatgatgaatgac3′ and 5′tctccttgttcatgtacttttcaacaagcc3′ for amplification of FALDH gene at 50°C; 5′ctaggatccaaaatggccgatggtgagg3′ and 5′gaaactcaccaccacgaaccag3′ for actin2 gene at 60°C; 5′ctctcgcaatcttcgctcttctcttt3′ and 5′tcatagatctggtccttgaaac3′ for eIF4A gene at 42°C; 5′ctttgtagctcttgtaggtgctcttgttc3′ and 5′tcctgcatatgatgctccttattgaaatactgat3′ for PR-1 gene at 50°C; and 5′aaagcttgtttgggagcaagtcttaaagc3′ and 5′aacactcggcagcagaaaaacagagtaaac3′ for GST-1 gene at 55°C. The linear range of PCR product synthesis was established for each primer pair and the number of cycles was accordingly chosen to reflect the midpoint of this range. PCR products were visualized in ethidium bromide-stained agarose gels. The intensity of the bands was determined by densitometry and the expression of each gene was normalized in relation to that of the actin2 gene or the eIF4A gene.
Determination of GSNOR and NOS Activities
GSNO was purchased from Sigma (Sigma Aldrich). GSNOR activity was determined at 25°C in 0.1 m sodium phosphate, pH 7.5, by monitoring the consumption of NADH and GSNO (ɛ340 = 7.06 mm−1 cm−1) in a Cary 400 Bio spectrophotometer. One unit of activity corresponds to 1 μmol of coenzyme transformed per minute. NOS activity was determined using the NOS detection system (Sigma Aldrich).
Determination of SNO Content
Total SNO levels were determined by Saville's method (Saville, 1958). Proteins were extracted in 100 mm Tris HCl, pH 6.8. The extracts were incubated for 5 min with an equivalent volume of solution A (1% sulfanilamide dissolved in 0.5 m HCl) in the presence or absence of solution B (solution A plus 0.2% HgCl2), allowing the development of the diazonium salt. The formation of the azo dye product was obtained by reacting the two samples for an additional 5 min with an equal volume of solution C [0.02% of N-(1-naphthyl) ethylenediamine dihydrochloride dissolved in 0.5 m HCl], and the absorbance was subsequently read at 550 nm with a spectrophotometer (Ultrospec 1100 pro, AmershamPharmacia Biotech). S-NOHCy was quantified as the difference of absorbance between solution B and A (B − A), comparing the values with a standard curve made from a solution of GSNO (Sigma-Aldrich). Low Mr SNOs were determined in the fraction passing through a 5 K cut of ultrafiltration membrane. The results were normalized against whole cell-lysate protein content, measured by the Bradford method.
Pathogen Growth and Inoculation Procedures
Pp isolates Noco2 and Cala2 were maintained on the genetically susceptible Arabidopsis accession Col-0 and Landsberg erecta, respectively. Pp conidia were suspended in water (4 × 104 spores mL−1) and sprayed onto 4-week-old plants. Inoculated plants were kept under a sealed propagator as described in Rustérucci et al. (2001) for up to 7 d. Growth of Pp was monitored by determining the number of conidia per gram fresh weight of leaves. For each measurement, five seedlings (rosettes) were harvested, vortexed with 2 mL water, and the spores counted at least twice in a hemocytometer. Alternatively, 10 μL droplet of Pp conidia (4 × 104 spores mL−1) was placed on the leaf surface, and plants were incubated for up to 7 d under the same conditions. Psm strain ES4326 carrying the plasmid pLAFR3 ± avrRPM1 was grown at 28°C in King's B medium containing the appropriate antibiotics (rifampicin 50 μg mL−1, tetracyclin 15 μg mL−1). Overnight log phase cultures were diluted to 105 (virulent) or 106 (avirulent) cfu mL−1 in 10 mm MgCl2, pressure infiltrated into leaves and bacterial growth in plants determined 2 d after infiltration, as in Maldonado et al. (2002).
SAR Assay
SAR was monitored by comparing growth of virulent oomycete Pp Noco2 in plants previously inoculated either with the avirulent bacteria Psm avrRpm1 (106 cfu mL−1) or with 10 mm MgCl2 (mock inoculation). Two rosette leaves of the different lines were infiltrated and 48 h later rosettes were sprayed with a spores suspension of the virulent Pp (4 × 104 spores mL−1). Plant conditioning and spore counting were similar to the one described above.
Histochemical Analysis of Plant Cell Death and Pp Development
Plant cell necrosis induced by pathogen inoculation, as well as the development of Pp mycelium inside leaf tissues was monitored by staining with lactophenol trypan blue and destaining in saturated chloral hydrate as described by Koch and Slusarenko (1990). Material was mounted on a slide in 60% glycerol and examined using a light microscope (Axiophot; Zeiss).
Immunolocalization
Immunolocalization of GSNOR in leaf sections of Arabidopsis was performed as described in Espunya et al. (2006).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table S1. NOS activity.
Supplementary Material
Acknowledgments
We sincerely thank Dr. C. Lamb (John Innes Centre) because part of this work was carried out at his laboratory.
This work was supported by grants from the Dirección General de Enseñanza Superior (grant nos. BMC 2003–00393 and BFU2004–00383) and the Comissionat per a Universitats i Recerca (grant no. 2001SGR00198). M.D. was awarded the Agencia Española de Cooperación Iberoamericana and was also supported by a European Molecular Biology Organization short-term fellowship. M.C. was supported by the United Kingdom Biotechnology and Biological Sciences Research Council (grant no. BB/C51565511).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: M. Carmen Martínez (carmen.martinez@uab.es).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Achkor H, Díaz M, Fernández MR, Biosca JA, Parés X, Martínez MC (2003) Enhanced formaldehyde detoxification by overexpression of glutathione-dependent formaldehyde dehydrogenase from Arabidopsis. Plant Physiol 132 2248–2255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowling SA, Guo A, Cao H, Gordon AS, Klessig DF, Dong X (1994) A mutation in Arabidopsis that leads to constitutive expression of systemic acquired resistance. Plant Cell 6 1845–1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark D, Durner J, Navarre DA, Klessig DF (2000) Nitric oxide inhibition of tobacco catalase and ascorbate peroxidase. Mol Plant Microbe Interact 13 1380–1384 [DOI] [PubMed] [Google Scholar]
- Clarke JD, Aarts N, Feys BJ, Dong X, Parker JE (2001) Constitutive disease resistance requires EDS1 in the Arabidopsis mutants cpr1 and cpr6 and is partially EDS1-dependent in cpr5. Plant J 26 409–420 [DOI] [PubMed] [Google Scholar]
- Cooney RV, Harwood PJ, Custer LJ, Franke AA (1994) Light-mediated conversion of nitrogen dioxide to nitric oxide by carotenoids. Environ Health Perspect 102 460–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delledonne M, Xia Y, Dixon RA, Lamb C (1998) Nitric oxide functions as a signal in plant disease resistance. Nature 394 585–588 [DOI] [PubMed] [Google Scholar]
- Delledonne M, Zeier J, Marocco A, Lamb C (2001) Signal interactions between nitric oxide and reactive oxygen intermediates in the plant hypersensitive disease resistance response. Proc Natl Acad Sci USA 6 13454–13459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz M, Achkor H, Titarenko E, Martínez MC (2003) The gene encoding glutathione-dependent formaldehyde dehydrogenase/GSNO reductase is responsive to wounding, jasmonic acid and salicylic acid. FEBS Lett 543 136–139 [DOI] [PubMed] [Google Scholar]
- Díaz M, Fernández MR, Martínez MC (2004) Histochemical assay to detect class III ADH activity in situ in Arabidopsis seedlings. Biotech Histochem 79 91–94 [DOI] [PubMed] [Google Scholar]
- Dietrich RA, Delaney TP, Uknes SJ, Ward ER, Ryals JA, Dangl JL (1994) Arabidopsis mutants simulating disease resistance response. Cell 77 565–577 [DOI] [PubMed] [Google Scholar]
- Dolferus R, Osterman JC, Peacock WJ, Dennis ES (1997) Cloning of the Arabidopsis and rice formaldehyde dehydrogenas genes: implications of the origin of plant ADH enzymes. Genetics 146 1131–1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durner J, Klessig DF (1999) Nitric oxide as a signal in plants. Curr Opin Plant Biol 2 369–374 [DOI] [PubMed] [Google Scholar]
- Durner J, Shah J, Klessig DF (1997) Salicylic acid and disease resistance in plants. Trends Plant Sci 2 266–274 [Google Scholar]
- Durner J, Wendehenne D, Klessig DF (1998) Defense gene-induction in tobacco by nitric oxide, cyclic GMP, and cyclic ADP-ribose. Proc Natl Acad Sci USA 95 10328–10333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durrant WE, Dong X (2004) Systemic acquired resistance. Annu Rev Phytopathol 42 185–209 [DOI] [PubMed] [Google Scholar]
- Espunya MC, Díaz M, Moreno-Romero J, Martínez MC (2006) Modification of intracellular levels of glutathione-dependent formaldehyde dehydrogenase alters glutathione homeostasis and root development. Plant Cell Environ 29 1002–1011 [DOI] [PubMed] [Google Scholar]
- Feechan A, Kwon E, Yun BW, Wang Y, Pallas JA, Loake GJ (2005) A central role for S-nitrosothiols in plant disease resistance. Proc Natl Acad Sci USA 102 8054–8059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feys BJ, Moisan LJ, Newman MA, Parker JE (2001) Direct interaction between the Arabidopsis disease resistance signaling proteins, EDS1 and PAD4. EMBO J 20 5400–5411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster MW, Stamler JS (2004) New insights into protein S-nitrosylation: mitochondria as a model system. J Biol Chem 279 25891–25897 [DOI] [PubMed] [Google Scholar]
- Gaston B (1999) Nitric oxide and thiol groups. Biochim Biophys Acta 1411 323–333 [DOI] [PubMed] [Google Scholar]
- Glazebrook J (2005) Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu Rev Phytopathol 43 205–227 [DOI] [PubMed] [Google Scholar]
- Greenberg JT, Guo A, Klessig DF, Ausubel FM (1994) Programmed cell death in plants: a pathogen-triggered response activated co-ordinately with multiple defense functions. Cell 77 551–563 [DOI] [PubMed] [Google Scholar]
- Greenberg JT, Silverman FP, Liang H (2000) Uncoupling salicylic acid-dependent cell death and defense-related responses from disease resistance in the Arabidopsis mutant acd5. Genetics 156 341–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grün S, Lindermayr C, Sell S, Durner J (2006) Nitric oxide and gene regulation in plants. J Exp Bot 57 507–516 [DOI] [PubMed] [Google Scholar]
- Hammond-Kosack KE, Parker JE (2003) Deciphering plant-pathogen communication: fresh perspectives for molecular resistance breeding. Curr Opin Biotechnol 14 177–193 [DOI] [PubMed] [Google Scholar]
- Han J, Stamler JS, Li H-L, Griffith O (1995). Inhibition of γ-glutamylcysteine synthetase by S-nitrosylation. In JS Stamler, S Gross, S Moncada, A Higgs, eds, Biology of Nitric Oxide (IV). Portland Press, London, p 114
- Heil M, Baldwin IT (2002) Fitness costs of induced resistance: emerging experimental support for a slippery concept. Trends Plant Sci 7 61–67 [DOI] [PubMed] [Google Scholar]
- Kliebenstein DJ, Dietrich RA, Martin AC, Last RL, Dangl JL (1999) LSD1 regulates salicylic acid induction of copper zinc superoxide dismutase in Arabidopsis thaliana. Mol Plant Microbe Interact 12 1022–1026 [DOI] [PubMed] [Google Scholar]
- Koch E, Slusarenko A (1990) Arabidopsis is susceptible to infection by a downy mildew fungus. Plant Cell 2 437–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kus JV, Zaton K, Sarkar R, Cameron RK (2002) Age-related resistance in Arabidopsis is a developmentally regulated defense response to Pseudomonas syringae. Plant Cell 14 479–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine A, Tenhaken R, Dixon RA, Lamb C (1994) H2O2 from the oxidative burst orchestrates the plant hypersensitive response. Cell 79 583–593 [DOI] [PubMed] [Google Scholar]
- Li X, Clarke JD, Zhang Y, Dong X (2001) Activation of an EDS1-mediated R-gene pathway in the snc1 mutant leads to constitutive, NPR1-independent pathogen resistance. Mol Plant Microbe Interact 14 1131–1139 [DOI] [PubMed] [Google Scholar]
- Liu L, Hausladen A, Zeng M, Que L, Heitman J, Stamler JS (2001) A metabolic enzyme for S-nitrosothiol conserved from bacteria to humans. Nature 410 490–494 [DOI] [PubMed] [Google Scholar]
- Liu L, Yan Y, Zeng M, Zhang J, Hanes MA, Ahearn G, McMahon TJ, Dickfeld T, Marshall HE, Que LG, et al (2004) Essential roles of S-nitrosothiols in vascular homeostasis and endotoxic shock. Cell 116 617–628 [DOI] [PubMed] [Google Scholar]
- Lindermayr C, Saalbach G, Bahnweg G, Durner J (2006) Differential inhibition of Arabidopsis methionine adenosyltransferases by protein S-nitrosylation. J Biol Chem 281 4285–4291 [DOI] [PubMed] [Google Scholar]
- Lindermayr C, Saalbach G, Durner J (2005) Proteomic identification of S-nitrosylated proteins in Arabidopsis. Plant Physiol 137 921–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maldonado AM, Doerner P, Dixon RA, Lamb CJ, Cameron RK (2002) A putative lipid transfer protein involved in systemic resistance signalling in Arabidopsis. Nature 419 399–403 [DOI] [PubMed] [Google Scholar]
- Martínez MC, Achkor H, Persson B, Fernández MR, Shafqat J, Farrés J, Jörnvall H, Parés X (1996) Arabidopsis formaldehyde dehydrogenase: molecular properties of plant class III alcohol dehydrogenase provide further insight into the origins, structure and function of plant class P and liver class I alcohol dehydrogenase. Eur J Biochem 241 849–885 [DOI] [PubMed] [Google Scholar]
- Modolo LV, Augusto O, Almeida IMG, Magalhaes JR, Salgado I (2005) Nitrite as the major source of nitric oxide production by Arabidopsis thaliana in response to Pseudomonas syringae. FEBS Lett 579 3814–3820 [DOI] [PubMed] [Google Scholar]
- Nathan C, Shiloh MU (2000) Reactive oxygen and nitrogen intermediates in the relationship between mammalian hosts and microbial pathogens. Proc Natl Acad Sci USA 97 8841–8848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimchuk Z, Eulgem T, Holt BF III, Dangl JL (2003) Recognition and response in the plant immune system. Annu Rev Genet 37 579–609 [DOI] [PubMed] [Google Scholar]
- Nürnberger Z, Brunner F, Kemmerling B, Piater L (2004) Innate immunity in plants and animals: striking similarities and obvious differences. Immunol Rev 198 249–266 [DOI] [PubMed] [Google Scholar]
- Parker JE, Feys BJ, Van der Biezen EA, Noël L, Aarts N, Austin MJ, Botella MA, Frost LN, Daniels MJ, Jones JD (2000) Unravelling R gene-mediated disease resistance pathways in Arabidopsis. Mol Plant Pathol 1 17–24 [DOI] [PubMed] [Google Scholar]
- Rockel P, Strube F, Rockel A, Wildt J, Kaiser WM (2002) Regulation of nitric oxide (NO) production by plant nitrate reductase in vivo and in vitro. J Exp Bot 53 103–110 [PubMed] [Google Scholar]
- Rustérucci C, Aviv DH, Holt BF, Dangl JL, Parker JE (2001) The disease resistance signaling components EDS1 and PAD4 are essential regulators of the cell death pathway controlled by LSD1 in Arabidopsis. Plant Cell 13 2211–2224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rustérucci C, Zhao Z, Haines K, Mellersh D, Neumann M, Cameron RK (2005) Age-related resistance to Pseudomonas syringae pv. tomato is associated with the transition to flowering in Arabidopsis and is effective against Peronospora parasitica. Physiol Mol Plant Pathol 66 222–231 [Google Scholar]
- Ryals JA, Neuenschwander UH, Willits MG, Molina A, Steiner HY, Hunt MD (1996) Systemic acquired resistance. Plant Cell 8 1809–1819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto A, Ueda M, Morikawa H (2002) Arabidopsis glutathione-dependent formaldehyde dehydrogenase is an S-nitrosoglutathione reductase. FEBS Lett 515 20–24 [DOI] [PubMed] [Google Scholar]
- Saville B (1958) A scheme for the colorimetric determination of microgram amounts of thiols. Analyst 83 670–672 [Google Scholar]
- Shah J (2003) The salicylic acid loop in plant defense. Curr Opin Plant Biol 6 365–371 [DOI] [PubMed] [Google Scholar]
- Shah J, Kachroo P, Nandi A, Klessig DF (2001) A recessive mutation in the Arabidopsis SSI2 gene confers SA- and NPR1-independent expression of PR genes and resistance against bacterial and oomycete pathogens. Plant J 25 563–574 [DOI] [PubMed] [Google Scholar]
- Shapiro AD (2005) Nitric oxide signaling in plants. Vitam Horm 72 339–398 [DOI] [PubMed] [Google Scholar]
- Song F, Goodman RM (2001) Activity of nitric oxide is dependent on, but is partially required for function of, salicylic acid in the signaling pathway in tobacco systemic acquired resistance. Mol Plant Microbe Interact 14 1458–1462 [DOI] [PubMed] [Google Scholar]
- Stamler JS (1994) Redox signaling; nitrosylation and related target interactions of nitric oxide. Cell 78 931–936 [DOI] [PubMed] [Google Scholar]
- Stamler JS, Jaraki O, Osborne J, Simon DI, Keaney K, Vita J, Singel D, Valeri CR, Loscalzo J (1992) Nitric oxide circulates in mammalian plasma primarily as an S-nitroso adduct of serum albumin. Proc Natl Acad Sci USA 89 7674–7677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamler JS, Lamas S, Fang FC (2001) Nitrosylation: the prototypic redox-based signaling mechanism. Cell 116 617–628 [DOI] [PubMed] [Google Scholar]
- Tao Y, Xie Z, Chen W, Glazebrook J, Chang HS, Han B, Zhu T, Zou G, Katagiri F (2003) Quantitative nature of Arabidopsis responses during compatible and incompatible interactions with the bacterial pathogen Pseudomonas syringae. Plant Cell 15 317–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Biezen EA, Freddie CT, Kahn K, Parker JE, Jones JDG (2002) Arabidopsis RPP4 is a member of the RPP5 multigene family of TIR-NB-LRR genes and confers downy mildew resistance through multiple signalling components. Plant J 29 439–451 [DOI] [PubMed] [Google Scholar]
- Wendehenne D, Pugin A, Klessig DF, Durner J (2001) Nitric oxide: comparative synthesis and signaling in animal and plant cells. Trends Plant Sci 6 177–183 [DOI] [PubMed] [Google Scholar]
- Zeidler D, Zähringer U, Gerber I, Dubery I, Hartung T, Bors W, Hutzler P, Durner J (2004) Innate immunity in Arabidopsis thaliana: lipopolysaccharides activate nitric oxide synthase (NOS) and induce defense genes. Proc Natl Acad Sci USA 101 15811–15816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Czymmek KJ, Shapiro AD (2003) Nitric oxide does not trigger early programmed cell death events but may contribute to cell-to-cell signaling governing progression of the Arabidopsis hypersensitive response. Mol Plant Microbe Interact 16 962–972 [DOI] [PubMed] [Google Scholar]
- Zhang C, Gutsche AT, Shapiro AD (2004) Feedback control of the Arabidopsis hypersensitive response. Mol Plant Microbe Interact 17 357–365 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.