Abstract
Root hairs are long tubular outgrowths that form on the surface of specialized epidermal cells. They are required for nutrient and water uptake and interact with the soil microflora. Here we show that the Oryza sativa cellulose synthase-like D1 (OsCSLD1) gene is required for root hair development, as rice (Oryza sativa) mutants that lack OsCSLD1 function develop abnormal root hairs. In these mutants, while hair development is initiated normally, the hairs elongate less than the wild-type hairs and they have kinks and swellings along their length. Because the csld1 mutants develop the same density and number of root hairs along their seminal root as the wild-type plants, we propose that OsCSLD1 function is required for hair elongation but not initiation. Both gene trap expression pattern and in situ hybridization analyses indicate that OsCSLD1 is expressed in only root hair cells. Furthermore, OsCSLD1 is the only member of the four rice CSLD genes that shows root-specific expression. Given that the Arabidopsis (Arabidopsis thaliana) gene KOJAK/AtCSLD3 is required for root hair elongation and is expressed in the root hair, it appears that OsCSLD1 may be the functional ortholog of KOJAK/AtCSLD3 and that these two genes represent the root hair-specific members of this family of proteins. Thus, at least part of the mechanism of root hair morphogenesis in Arabidopsis is conserved in rice.
Root hairs are long tubular outgrowths from the surface of specialized epidermal cells. By greatly increasing the surface area, they are important for nutrient and water uptake and for anchorage. Each root hair is a growing extension of a single epidermal cell (Dolan et al., 1993; Schiefelbein et al., 1997). In Arabidopsis (Arabidopsis thaliana), the root hairs are formed from specific epidermal cells that are arranged in rows; these cells are located over the intercellular spaces between underlying cortical cells. In contrast, the cells located directly over cortical cells do not form hairs. The mechanisms that control this patterning in Arabidopsis have been extensively explored (Dolan et al., 1994; Lee and Schiefelbein, 1999; Dolan and Costa, 2001). The same pattern of root hair development found in Arabidopsis has also been detected in the Brassicaceae. With regard to the grasses (Poaceae), two types of root hair formation patterns have been found (for review, see Dolan and Costa, 2001). In the first, which occurs in most grass species, including the crop species rice (Oryza sativa) and maize (Zea mays), the hairs develop from any cells in the epidermis. In the second pattern, which occurs in a Pooideae subfamily that includes species such as barley (Hordeum vulgare) and wheat (Triticum aestivum), the root hairs develop from the smaller product of an asymmetric cell division.
In Arabidopsis, many of the genes involved in the initiation and elongation of root hairs have been identified. They include transcriptional regulators and cellular physiological/structural components (Wada et al., 1997; for review, see Haigler et al., 2001; Carol and Dolan, 2002; Montiel et al., 2004). Analyses of the genetic interactions between mutant genes have resulted in a conceptual framework for hair cell development (Grierson et al., 1997; Lee and Schiefelbein, 1999). The cloning of these genes has defined the many cellular activities that are required for hair growth. Particularly important are the genes that control the development of the cell wall during root hair morphogenesis. These include genes encoding a Leu-rich repeat extensin protein, a UDP-d-Glc 4 epimerase, a putative GTP-binding protein, and cellulose synthase-related proteins that play essential roles in the cell wall biosynthesis involved in root hair development (Baumberger et al., 2001; Favery et al., 2001; Wang et al., 2001; Seifert et al., 2002; Hu et al., 2003; Nguema-Ona et al., 2006). Mutation studies revealed that such root hair-specific cell wall deposition is critical for the growth of the polarized tip that is initiated by the formation of a bulge in the trichoblast.
There are 29 cellulose synthase-related genes in Arabidopsis, while rice has at least 37 putative cellulose synthase-like (CSL) genes (Hazen et al., 2002). The CSL superfamily has been classified into several subfamilies, namely, CSLA, CSLC, CSLD, CSLE, CSLF, and CSLH (http://cellwall.stanford.edu). The Arabidopsis gene KOJAK/AtCSLD3 is the first gene in the CSLD family whose loss of function results in the formation of defective root hairs. This was the only phenotype observed in the mutant plants, even though KOJAK/AtCSLD3 is expressed throughout the whole plant (Favery et al., 2001; Wang et al., 2001). This suggests that another CSLD gene may function redundantly in these tissues. A similar gene in Nicotiana alata, NaCSLD1, is abundantly expressed in the developing male gametophyte, including the pollen tube. This suggests that NaCSLD1 may be required for the formation of the cell wall in the developing pollen tubes of tobacco (Nicotiana tabacum; Doblin et al., 2001).
In this study, we used a gene trap Ds insertion mutant to define the role OsCSLD1 plays in root hair development in rice. Our observations suggest that the function of the CSLD gene is conserved between monocots and eudicots.
RESULTS
A Root Hair-Expressed Gene Trap Identifies a CSLD Family Member
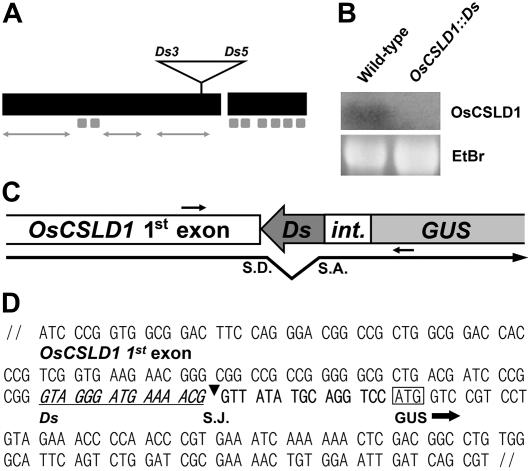
To identify genes involved in root hair development in rice, a Ds gene trap population (Japonica cv Dongjin) was screened for a root hair-specific transcription pattern (Chin et al., 1999; Kim et al., 2004). One root hair-specific gene trap was identified and resulted from an insertion of the gene trap into the OsCSLD1 gene. Sequencing of the flanking regions of the transposon insertion site indicated that the gene trap had been inserted near the end of the first exon of OsCSLD1 (Fig. 1A). Southern hybridization revealed that the mutant line contained a single copy of the Ds gene trap (data not shown). Northern hybridization with an OsCSLD1 3′ untranslated region (UTR) gene-specific probe showed that transcripts with sequences downstream of the Ds insertion site were not detected in the mutant line (Fig. 1B). To further characterize the fusion transcript from the mutant allele, the junction sequence between the first CSLD1 exon and the β-glucuronidase (GUS)-coding region was amplified by reverse transcription (RT)-PCR using mRNA from mutant roots (Fig. 1C). Figure 1D shows the sequence of the amplified DNA. The fusion transcript was produced by splicing between a donor site in the subterminal Ds end and an acceptor site in front (5′) of the GUS-coding region. The spliced junction sequence introduced an additional 10 amino acids between CSLD1 and GUS (Fig. 1, C and D). The C-terminal 314-amino acid peptide of OsCSLD1 was not present in the fusion protein. Thus, the fusion protein lacks one Asp residue, the QXXRW motif, and the six membrane-spanning domains of OsCSLD1 that are highly conserved among cellulose synthase genes.
Figure 1.
Transcript analysis of OsCSLD1∷Ds. A, Genomic structure of OsCSLD1 showing the position of the trap Ds element. The open reading frame comprised two exons that were 2,676 and 708 bp in size and a short 87-bp intron. The black boxes indicate exons, while the eight small gray boxes indicate the sequences that encode membrane-spanning domains. The arrows correspond to three variable regions. B, Northern analysis of OsCSLD1 mRNA. Total RNA was extracted from the roots of wild-type and OsCSLD1∷Ds plants. The probe was from the 3′ UTR of the OsCSLD1 gene. C, Schematic depiction of the spliced junction of the OsCSLD1∷Ds fusion transcript. Shown are part of the first exon and the GUS of the trap Ds. int., A partial intron fused to the GUS coding region. The putative splicing donor (S.D.) and acceptor sites (S.A.) are indicated by the bent arrow. The primers used to clone the cDNA are indicated by the small arrows. D, The cDNA sequence of the spliced junction of the OsCSLD1∷Ds fusion transcript. The Ds sequence is underlined and S.J. indicates the splicing junction. The ATG codon in a box is the start codon of GUS.
In rice, high frequency of Ds transposition can be achieved using tissue culture-mediated plant regeneration (Kim et al., 2004). To obtain revertant alleles, OsCSLD1∷Ds lines that are homozygous for Ac and Ds were used for tissue culture. Of the 53 regenerated plants obtained, those that lost Ds from the original site were identified by PCR using primers flanking the element and sequenced to verify the existence of empty donor sites. In the OsCSLD1∷Ds allele, the Ds element was flanked on both sides by an identical 8-bp host sequence (CCCGCGGG). A total of 18 excision sites were identified. Of these, nine new alleles restored the reading frame. Seven alleles (OsCSLD1-Rev6a) carried the same 6-bp addition (CCCGCGGCCGCGGG) and one (OsCSLD1-Rev6b) had a different 6-bp addition (CCCGCGCCCGCGGG). The other line, OsCSLD1-Rev0, had the perfect excision sequence (CCCGCGGG). Compared to the wild-type protein, the proteins generated by these eight 6-bp addition lines would contain two extra amino acids (Pro and Arg). The remaining nine excision sites had footprint sequences that failed to restore the reading frame. Seven alleles (oscsld1-7a) had the same 7-bp addition (CCCGCGGCCCGCGGG) and one (oscsld1-7b) had a different 7-bp addition (CCCGCGGGCCGCGGG). The other line, oscsld1-4, carried a 4-bp addition (CCCG). The oscsld1-7a, oscsld1-7b, and oscsld1-4 alleles would all produce the same 416-amino acid C-terminal sequences that bear no homology to the wild-type OsCSLD1 sequence.
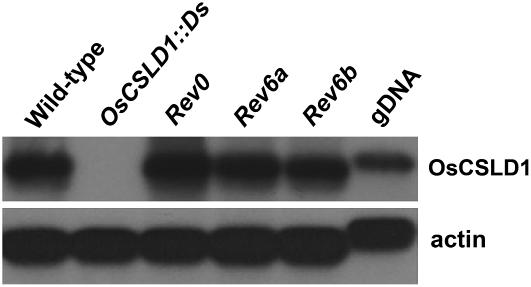
To determine if the revertants accumulate wild-type levels of OsCSLD1 mRNA, total cellular RNA was isolated from the roots of the wild type, OsCSLD1∷Ds mutant, and revertants (Rev0, Rev6a, and Rev6b). RT-PCR analysis of these RNAs using an OsCSLD1 gene-specific primer showed that wild type and revertants accumulated similar amounts of OsCSLD1 mRNA, while the mutant produced no OsCSLD1 mRNA (Fig. 2).
Figure 2.
RT-PCR analysis of OsCSLD1 mRNA expression in seedling roots of wild type, mutant, and three revertant lines (Rev0, Rev6a, and Rev6b). Total RNA was extracted from roots of 15-d-old seedlings. RT-PCR (25 cycles) was conducted with oligo(dT) primed cDNAs using OsCSLD1 gene-specific primers. The probe was the 3′ terminal genomic DNA of the OsCSLD1 gene. Actin mRNA and genomic DNA (gDNA) were used as controls.
OsCSLD1 Is Required for Root Hair Elongation
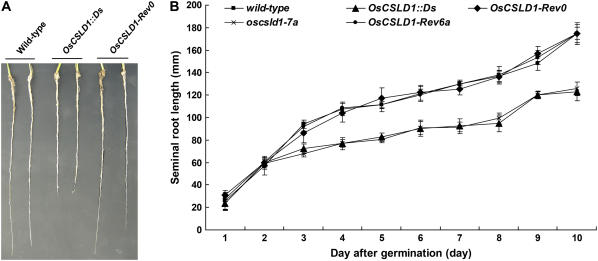
Because the OsCSLD1 gene is expressed in the root and because the gene trap insertion would be expected to result in the formation of a loss-of-function allele, we characterized the root phenotype of wild-type, OsCSLD1∷Ds, and OsCSLD1-Rev0 seedlings that were grown in soil for 10 d. At this stage, the plants produced three leaves. The seminal roots of the OsCSLD1∷Ds mutant were 30% shorter than those of the wild-type plants, while those of the revertant were the same length as the wild-type seminal roots (Table I; Fig. 3A). In contrast, the wild-type, revertant, and mutant plants had similar lengths and numbers of crown roots and lateral roots. The growth rates of the seminal roots of the wild-type, revertants, and mutant plants were compared (Fig. 3B). Overall growth rate of seminal roots of the mutant were lower than those of the wild type and revertants. Thus, OsCSLD1 predominantly affects the development of the seminal root.
Table I.
The number and length of root organs in wild-type, OsCSLD1∷Ds, OsCSLD1-Rev0 revertant, and 35S∷OsCSLD1 seedlingsa
| Genotype | Length
|
No.
|
||
|---|---|---|---|---|
| Seminal Root | Crown Rootb | Crown Root | Lateral Roots per Seminal Rootc | |
| mm | ||||
| Wild type | 175.0 ± 5.7d | 64.4 ± 13.2 | 6.4 ± 0.5 | 20.9 ± 2.0 |
| OsCSLD1∷Ds | 123.0 ± 8.3 | 62.2 ± 5.8 | 6.8 ± 0.7 | 20.2 ± 1.6 |
| OsCSLD-Rev0 | 174.6 ± 8.2 | 65.0 ± 7.0 | 6.3 ± 0.6 | 20.5 ± 0.6 |
| 35S∷OsCSLD1 | 192.0 ± 10.2 | 62.0 ± 6.8 | 6.4 ± 0.9 | 20.6 ± 1.1 |
The seeds were germinated on paper towels for 3 d in the dark and then grown in soil for an additional 10 d.
The longest crown root of each plant.
The number of lateral roots within a 10-mm distance of the lateral root closest to a root apex.
Values shown are the means ± sds from 20 plants.
Figure 3.
Root phenotypes of wild type, OsCSLD1 mutants, and revertants. A, Gross root phenotype of soil-grown seedlings from wild-type, OsCSLD1∷Ds, and OsCSLD1-Rev0 plants. Germinated seeds were grown in soil for 10 d. B, Growth rates of seminal roots of MS-grown seedlings from wild type, the two mutants (OsCSLD1∷Ds and oscsld1-7a), and the two revertants (OsCSLD1-Rev0 and OsCSLD1-Rev6a). Sterilized seeds were germinated and grown in soil for 10 d. For measurements, germinating seeds that sprouted seminal roots to the same extent were used. oscsld1-7b and OsCSLD1-Rev6b which growth rates were similar to ones of OsCSLD1∷Ds and OsCSLD1-Rev0, respectively, were not shown. [See online article for color version of this figure.]
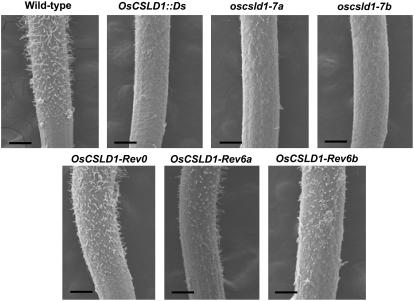
To examine the effect of the OsCSLD1 mutation on the morphology of the seminal root in more detail, seedlings were grown for 3 d on Murashige and Skoog (MS) media. Roots of the OsCSLD1∷Ds, oscsld1-7a, and oscsld1-7a mutant seedlings were compared to those of wild-type and revertant (OsCSLD1-Rev0, OsCSLD1-Rev6a, and OsCSLD1-Rev6b) seedlings using scanning electron microscopy (SEM). SEM images of the epidermis of seminal roots of these seven genotypes are shown in Figure 4. This showed that all the mutants had much shorter root hairs than either wild type or revertants. In 3-d-old roots, the root hair zone starts at 1 mm from the root apex. Root hairs at 2 and 3 mm from the apex were examined by SEM and their length and number were determined (Table II). While all lines had the same numbers of root hairs, the root hairs of all three mutants were 65% to 75% shorter than those of the wild type. In contrast, the root hairs of the OsCSLD1-Rev0 revertant were almost as long as the hairs of the wild type, while those of the OsCSLD1-Rev6a and 6b revertant were slightly shorter (73% of the wild-type length). Because the mRNA levels of OsCSLD1-Rev6a and 6b were similar to wild type (Fig. 2), incomplete reversion might be due to the modification of protein sequences in these revertants.
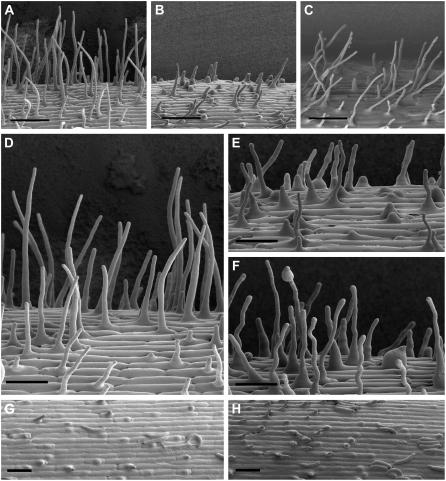
Figure 4.
SEM images of the seminal roots of wild-type, OsCSLD1∷Ds, Rev0, Rev6a, Rev6b, oscsld1-7a, and oscsld1-7 seedlings. Seedlings were grown vertically for 3 d on MS media. The seminal roots were fixed and observed by SEM. The photos show the root hair and elongation zones. Bar = 200 μm.
Table II.
The length and number of root hairs on the seminal roots of wild type, three mutants, and three revertants
| Genotype | Wild Type | OsCSLD1∷Ds | oscsld1-7a | oscsld1-7b | Rev0 | Rev6a | Rev6b |
|---|---|---|---|---|---|---|---|
| Root hair length, μmb | 117.8 ± 4.3c | 36.2 ± 0.9 | 44.0 ± 2.1 | 31.8 ± 0.6 | 110.1 ± 3.1 | 94.3 ± 7.8 | 78.0 ± 8.4 |
| Root hair no.d | 15.2 ± 1.2 | 14.6 ± 1.5 | 14.3 ± 1.8 | 13.7 ± 1.7 | 14.8 ± 1.3 | 14.5 ± 1.5 | 14.2 ± 1.9 |
Plant materials were grown on 0.5% phytogel in 0.5× MS for 3 d. The captured images were used to measure individual root hairs.
Root hairs examined were in the 2 to 3 mm from the root apex.
The values are the means ± sds from 200 root hairs (10 hairs/root).
Hairs were counted in a 200 × 200 μm2 sector that was 2 to 3 mm from the root apex. The values are the means ± sds from 20 roots.
To observe the morphology of the root hairs in more detail, the wild-type, OsCSLD1∷Ds, and OsCSLD1-Rev0 lines were examined by cryo SEM. The wild type and revertants were morphologically similar (Fig. 5, A and C). However, compared to normal root hairs (Fig. 5D), mutants exhibited bulges at the tips, swollen bases, and uneven swelling along the length of the root hair (Fig. 5, E and F), demonstrating that tip growth was defective. However, the spatial arrangement of the hair and nonhair cells in the elongation and hair-forming zones of the mutant were not altered, indicating that cell patterning is not defective in these mutants (Fig. 5, G and H).
Figure 5.
Epidermal morphology of the seminal roots from wild-type, OsCSLD1∷Ds, and OsCSLD1-Rev0 seedlings in the hair and elongation zones. A to C, The hair zone in wild type, OsCSLD1∷Ds, and OsCSLD1-Rev0, respectively. D to F, Close ups of root hairs in the wild type (D) and OsCSLD1∷Ds (E and F). G and H, The elongation zone in wild type and OsCSLD1∷Ds. Bulges are where root hairs are emerging. Seedlings were grown vertically for 3 d on MS media and the epidermal cells were observed by cryo-SEM. Bars = 100 μm (A–C) and 50 μm (D–H).
Because the seminal roots of the OsCSLD1∷Ds mutant were shorter than those of wild-type plants, we examined whether or not the OsCSLD1 mutation affects epidermal cell size or cell number in seminal roots. Cell sizes were compared between wild-type and mutant plants that were grown on MS media for 5 d. Under these growth conditions, mutant and wild-type seminal roots were 10 and 6.5 cm long, respectively. The following three zones of seminal root were measured: the elongation zone, the root hair initiation zone, and the lateral root initiation zone. In rice, epidermal cells of seminal roots exhibit distinct size differences even within the same zone. Taking such heterogeneity into account, cell sizes were estimated according to the following two parameters: (1) the surface areas occupied by a group of 20 cells; and (2) the average cell dimensions of these 20 cells that were classified into two groups, long and short. Even though long and short cells were arbitrarily classified, their size differences were easily apparent, as shown in Supplemental Figure S1. Table III shows the average surface areas of 20 epidermal cells and the average cell lengths (classified as short and long) of 10 seminal roots from the wild type and from the mutant. Cell widths were not presented because there were no differences in cell widths between wild-type and mutant roots. The average surface areas of 20 cells were 5,908.3 (wild type) and 6,031.7 μm2 (mutant) in the elongation zone; 27,840.8 (wild type) and 27,452.8 μm2 (mutant) in the root hair initiation zone; and 34,618.3 (wild type) and 33,285.5 μm2 (mutant) in the lateral root initiation zone. Even though there were distinct differences in cell sizes between these zones, there were no significant size differences between wild-type and mutant epidermis. The average lengths of long cells were 54.6 (wild type) and 52.9 μm (mutant) in the elongation zone, 122.9 (wild type) and 119.4 μm (mutant) in the root hair initiation zone, and 135.8 (wild type) and 134.0 μm (mutant) in the lateral root initiation zone. Short cells have average lengths as follows: 25.5 (wild type) and 26.1 μm (mutant) in the elongation zone, 58.7 (wild type) and 59.1 μm (mutant) in the root hair initiation zone, and 69.0 (wild type) and 69.6 μm (mutant) in the lateral root initiation zone. The differences in the average lengths of both classes fell well within the sds. As the 35% difference in seminal root length between the mutant and the wild type could not be explained by cell size, we conclude that the seminal root epidermis of the mutants had fewer cells.
Table III.
Surface dimensions of root epidermal cells in the seminal roots of wild type and the OsCSLD∷Dsa
| Root Zones Measured | Cellular Parameters | Wild-Type | OsCSLD1∷Ds | |
|---|---|---|---|---|
| Elongation zoneb | Surface area/20 cellsc | 5,908.3 ± 12.5d | 6,031.7 ± 27.7 | |
| Cell lengthe | Long cells | 54.6 ± 6.8d | 52.9 ± 6.4 | |
| Short cells | 25.5 ± 4.4 | 26.1 ± 4.3 | ||
| Root hair initiation zonef | Surface area/20 cells | 27,840.8 ± 46.3 | 27,452.8 ± 18.8 | |
| Cell length | Long cells | 122.9 ± 12.7 | 119.4 ± 16.1 | |
| Short cells | 58.7 ± 6.7 | 59.1 ± 6.8 | ||
| Lateral root initiation zoneg | Surface area/20 cells | 34,618.3 ± 19.2 | 33,285.5 ± 37.7 | |
| Cell length | Long cells | 135.8 ± 16.1 | 134.0 ± 15.1 | |
| Short cells | 69.0 ± 7.7 | 69.6 ± 8.9 |
Plant materials were grown on 0.5% phytogel in 0.5× MS for 5 d. The lengths of seminal roots of the wild type and mutant were 10 and 6.5 cm, respectively.
The zone below the root hair initiation area.
Surface areas occupied by a group of 20 cells were estimated (μm2). Captured images were used to measure cellular dimensions.
The values are the means ± sds of 10 seminal roots.
Cell width data was omitted, because cell widths were identical between wild-type and mutant roots. Length is measured in micrometers.
The zone where root hairs were initiated.
The zone where lateral roots were initiated.
OsCSLD1 Is Expressed Only in Root Hairs
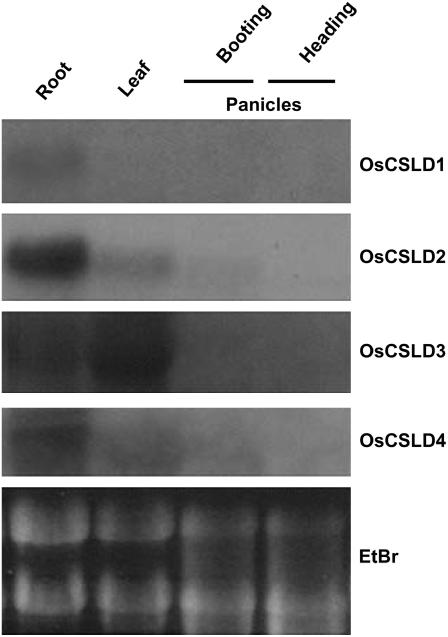
There are four genes in the rice CSLD subfamily and northern hybridization analysis was used to determine the expression of each of these genes in the roots and leaves of 15-d-old plants (Fig. 6). Their expression in the panicles of two different flowering stages (booting and heading) was also examined; booting is the stage where the young panicles are enclosed in leaves, and heading is the stage after the panicles have emerged from the leaves but before anthesis. OsCSLD1 gene transcripts were detected in the root and were not detected in the leaves and flowers. OsCSLD2 is expressed strongly in the roots, weakly in the leaves and flowers during the booting stage, and not expressed during the heading stage. OsCSLD3 and OsCSLD4 had similar expression patterns, being expressed in the roots and leaves but not in the flowers at either stage. However, OsCSLD3 was more highly expressed than OsCSLD4 in the leaves.
Figure 6.
Expression patterns of the OsCSLD subfamily. Total RNA was isolated from the roots and leaves of 15-d-old plants and the flowering tissues of mature rice plants at two different stages (booting and heading). Total RNA (30 μg) was loaded in each lane. Equal loading was confirmed by ethidium bromide staining. Northern hybridizations were performed with gene-specific probes for OsCSLD1, OsCSLD2, OsCSLD3, and OsCSLD4, as described in “Materials and Methods.”
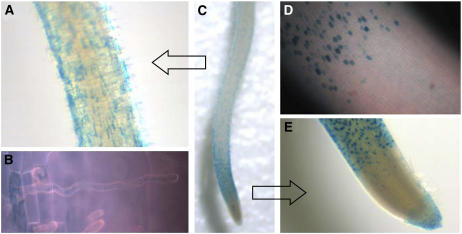
To characterize the expression pattern in more detail, we analyzed the GUS expression patterns of heterozygous OsCSLD1∷Ds/+ plants. This showed that OsCSLD1 is expressed in the epidermal cells of the root hair zone, the elongation zone, and the root cap, but not in the division zone (Fig. 7C). In the root hair zone, GUS is expressed only in the root hair cells (Fig. 7, A and B) and is never observed in hairless cells. In the elongation zone before hair outgrowth had occurred, the spatial arrangement of the GUS-positive cells is similar to the one seen in the root hair zone (Fig. 7, D and E). This implies that the epidermal cells expressing OsCSLD1 in the elongation zone are the cells that will go on to form root hairs.
Figure 7.
GUS patterns in OsCSLD1∷Ds/+ roots. A, The root hair zone under the light microscope. B, A root hair cell under high magnification (400×). The GUS stain appeared to be contained in an internal compartment. C, The GUS staining of the whole seminal root. D, GUS staining between the elongation zone and the root cap was not detected. E, Below the root hair zone, GUS expression in the elongation zone and in the root cap. The roots of 5-d-old heterozygous seedlings were stained with 0.5% 5-bromo-4-chloro-3-indoyl glucuronide.
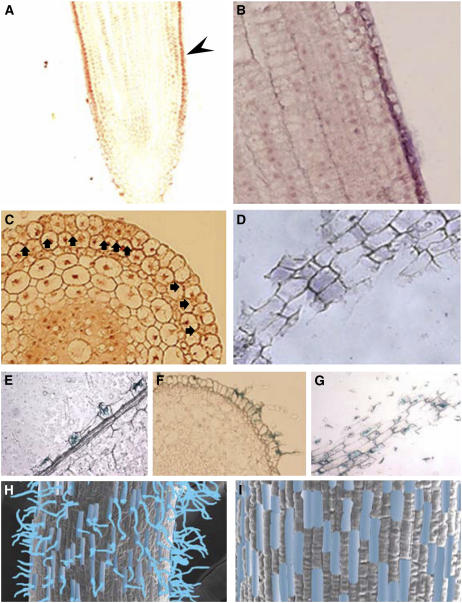
To verify the expression patterns of OsCSLD1 that were revealed by the gene trap line, the mRNA distribution in wild-type roots was examined by in situ hybridization analysis. Thus, longitudinal, transverse, and tangential sections of wild-type roots were hybridized with OsCSLD1-specific antisense probes. The in situ hybridization patterns on median longitudinal sections are shown in Figure 8, A and B. Strong signals were detected along the epidermal layer in the root hair zone and the elongation zone but signals were no longer detectable in the division zone near the root apex (Fig. 8A). Expression in the epidermis was patchy, with only some cells expressing the transcript (Fig. 8B). The transverse and tangential in situ-hybridized sections are shown in Figure 8, C and D, respectively. The tangential sections were made through the single cell layer of the epidermis. Hybridization signals were detected in only some of the cells along the epidermal layer. A similar expression pattern was also observed in the sections of GUS-stained roots that were heterozygous for OsCSLD1∷Ds, as only some cells in the longitudinal, transverse, and tangential sections showed GUS staining (Fig. 8, E–G, respectively). Figure 8, H and I compare the distribution of the GUS-positive cells in the root hair zone and the elongation zone. The positive cells are indicated by blue shading. OsCSLD1 was expressed in similar patterns in not only the root hair zone but also in the elongation zone. This indicates that OsCSLD1 is expressed only in trichoblasts before initiation of root hair growth and during hair elongation.
Figure 8.
In situ localization of OsCSLD1 mRNA and GUS staining of 5-d-old roots from wild-type and OsCSLD1∷Ds/+ seedlings, respectively. A, In situ hybridization with a medial longitudinal section. Sections of 5-d-old wild-type roots were probed with digoxigenin-labeled antisense RNA and viewed under a light microscope. The arrowhead indicates the signal in the epidermal cell layer of the medial longitudinal section. B, Higher magnification of this part of the epidermal layer. C, In situ hybridization with transverse sections. Signal-positive cells are marked with arrowheads. D, In situ hybridization with tangential sections. Sections through the epidermal cell layer were used for hybridization. E, GUS staining of a longitudinal section. GUS-stained roots of 5-d-old of OsCSLD1∷Ds/+ seedlings were embedded in paraffin and sections were viewed under a light microscope. The section shows GUS staining in root hair cells of the epidermis. F and G, GUS staining of transverse and tangential sections, respectively. GUS-positive cells were clearly distinguishable from GUS-negative cells along the epidermal cell layer. H and I, The GUS-positive epidermal cells in the root hair zone and the elongation zone, respectively. GUS-positive cells are indicated by blue shading. GUS-stained roots were fixed and examined by SEM.
Overexpression of OsCSLD1 Results in the Development of Longer Root Hairs
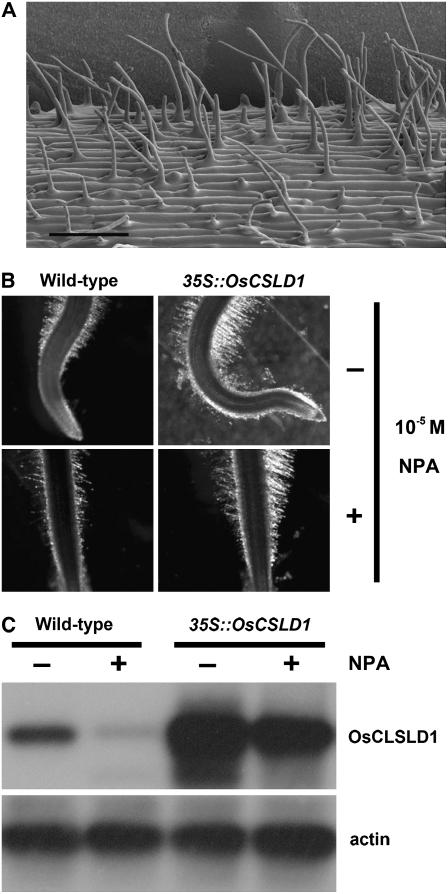
Because loss of OsCSLD1 function results in the development of short root hairs, we hypothesized that overexpression of this gene would result in the formation of longer hairs. To generate OsCSLD1-overexpressing lines, we first isolated the full-length cDNA of OsCSLD1 by using RT-PCR employing gene-specific primers. The longest OsCSLD1 cDNA of 4,023 bp was fused with green fluorescent protein (GFP) at its 3′ end and was used to generate overexpression transgenics in the same Japonica cultivar (Dongjin) from which the Ds mutant was derived. Eight independent 35S∷OsCSLD1 lines were confirmed to be overexpressing lines by Southern-blot probing with DNA from the hygromycin phosphotransferase gene and by examining GFP expression (Supplemental Fig. S2). The independent transformants contained one or two copies of the T-DNA insert (data not shown). The independent transgenic lines showed similar phenotypes. In the 10-d-old seedlings, the seminal roots of the overexpressing lines were about 10% longer than those of the wild type (Table I). However, the wild-type and overexpressing lines had similar numbers of crown roots and lateral roots and the length of these roots was indistinguishable between genotypes (Table I). SEM analysis shows that the root hairs of the overexpressing lines were morphologically normal and had a normal distribution along the root (that patterning was normal; Fig. 9A). However, the root hairs on the seminal roots of the overexpressing lines were almost twice (183%) as long as the root hairs from the normal plants (Table IV). Extended elongation of root hairs and seminal root in overexpressing plants further supports the functional role of OsCSLD1 in root morphogenesis.
Figure 9.
Seminal root morphology and expression of the OsCSLD1-overexpressing line grown in the presence or absence of NPA. A, The epidermal cells of the hair zone observed by cryo-SEM. An OsCSLD1-overexpressing line was grown vertically on MS media. B, Seminal roots grown without or with NPA. Germinated seeds were grown for 5 d in MS media without or with 10−5 m NPA. C, RT-PCR analysis of OsCSLD1 mRNA. The mRNAs of the roots shown in B were subjected to 25 cycles of RT-PCR using OSCSLD1 gene-specific primers.
Table IV.
The length of the root hairs (μm) on seminal roots of wild-type and 35S∷OsCSLD1 seedlings grown in the presence or absence of NPAa
| Wild Type | 35S∷CSLD1 | |
|---|---|---|
| No treatment | 178.5 ± 13.3b | 328.1 ± 18.2 |
| 10−5m NPA | 93.7 ± 9.1 | 286.6 ± 10.2 |
Plant materials were grown on 0.5% phytogel in 0.5× MS for 5 d. Seedlings were exposed to 10−5 m NPA. SEM-captured images were used to measure individual root hairs.
The lengths of root hairs 2 to 3 mm from the root apex were measured. The values are the means ± sds from 200 root hairs (10 hairs/root).
To verify that overexpression of OsCSLD1 results in the development of longer root hairs than wild type, we took advantage of the fact that root hairs fail to elongate when exposed to 1-N-naphthylphthalamic acid (NPA), an auxin efflux inhibitor. Thus, we measured the lengths of the root hairs of wild-type and OsCSLD1 overexpressing seedlings that had been grown in the presence of 10−5 m NPA. NPA treatment suppressed the growth of seminal roots to the same extent in both wild-type and overexpression lines. However, this treatment dramatically decreased the wild-type root hair length (Fig. 9B), even though root hairs of the NPA-treated wild type maintained a normal morphology (data not shown). In contrast, the effect was much less pronounced in 35S∷OsCSLD1 plants (Fig. 9B; Table IV). While the root hairs of the treated wild-type plants (93.7 μm ± 9.1) were 53% shorter than those of the untreated wild-type plants (178.5 μm ± 13.3), the root hairs of NPA-treated overexpressing plants (286.6 μm ± 10.2) were 88% of the length of those of the untreated plants (328.1 μm ±18.2).
We then used RT-PCR with gene-specific primers to measure the relative expression of OsCSLD1 in root RNAs from NPA-treated or untreated wild-type and transgenic lines (Fig. 9C). In wild-type seedlings, NPA treatment severely suppressed the accumulation of OsCSLD1 mRNA. In contrast, both treated and untreated overexpressing seedlings maintained the high level of OsCSLD1 mRNA even though there was slight difference in the mRNA level. Therefore, the maintenance of root hair length in the presence of NPA is likely to be due to the constitutive expression of OsCSLD1mRNA in transgenic lines. However, because there is a morphological difference between oscsld1 root hairs and NPA-treated ones, further study will be required to address the relationship between the OsCSLD1-mediated root hair development and auxin action. In summary, these observations together indicate that overexpression of OsCSLD1 can extend the length of root hairs in rice.
DISCUSSION
We show here that OsCSLD1 is specifically expressed in trichoblasts before the onset of root hair growth and during hair elongation, where its activity is required for the elongation of root hairs during rice development. Analysis of mutant and overexpression lines suggests that specification of root hair fate coincides with OsCSLD1 expression but is independent of OsCSLD1 activity. OsCSLD1 expression in the elongation zone had a similar pattern to that in the root hair zone. This observation suggests that the specification of trichoblasts is initiated in the elongation zone, where cells elongate in the direction of the root axis but where root hair growth has not yet initiated. However, OsCSLD1 is not required for the specification of cell identity in the epidermis but is instead involved in the later process of hair morphogenesis. The strongest evidence for this was obtained from the transgenic lines that overexpressed OsCSLD1 under the control of the 35S promoter. While these transgenic plants produced longer root hairs than the wild type, the number and distribution pattern of the root hair cells were similar to wild type. Thus, high ectopic expression of OsCSLD1 does not induce atrichoblasts to develop into root hairs. Similarly, loss of OsCSLD1 function was not accompanied by a defect in the pattern of cell differentiation in the epidermis. We speculate that, as in Arabidopsis, OsCSLD1 may be one of the effector genes that are probably targets of the transcription factor cascade that specifies cell identity in the root epidermis (Montiel et al., 2004).
OsCSLD1 shows 77% amino acid identity and 85% similarity to KOJAK/AtCSLD3 (Supplemental Fig. S3). Like OsCSLD1∷Ds, both Arabidopsis csld3-1 and kojak-1 (kjk-1) mutants presumably produce truncated proteins lacking the C-terminal six membrane-spanning domains. Genetic lesions in the two genes led to almost identical phenotypic defects in the root hairs. It has been proposed that the KOJAK/AtCSLD3 mutation causes the failure of building polymers to be delivered to the primary cell wall in the growing root hair tip, although the biochemical defect in the cell wall that results from this has not yet been elucidated. It is possible that the membrane-spanning domains that are missing in both mutants may form a central channel through which the nascent glucan chain could be secreted (Doblin et al., 2002). Notably, in Arabidopsis, the mutant root hairs were observed to leak cytoplasm from their tips. However, such cytoplasmic leaks were never observed in the root tips of the OsCSLD1 mutant, despite the similarities in the morphology of both species. This discrepancy may relate to subtle differences in the enzymatic activity of the CSLD proteins of the two species. Alternatively, because cell wall compositions exhibit species-specific characteristics (Séné et al., 1994), it is possible that this discrepancy is simply due to differences in cell wall strength.
Two distinct differences between OsCSLD1 and KOJAK/AtCSLD3 were observed in this study. One is the growth rate of the seminal roots. In kojak/csld3 mutants, no noticeable retardation in the growth of primary roots was reported. The OsCSLD1 mutant exhibits slow growth of seminal roots. Conversely, OsCSLD1-overexpressing seedlings developed both longer root hairs and seminal roots. Because there was a correlation between root hair length and seminal root growth and root hairs are an important organ for nutrient uptake (Lauter et al., 1996; Ivashikina et al., 2001), it is reasonable to argue that the slow root growth might be a consequence of reduced efficiency in the uptake of nutrients by root hairs or their flux through the epidermal layer into the underlying layers of OsCSDL1 mutants. Because there was no noticeable difference in cell size between the wild type and mutants, the apparent shortening of the seminal roots must be due to the reduced numbers of cells in the mutants, which could result from lower cellular activity, especially in the cell division rate. Together these data indicate that OsCSLD1 is required for the growth of cells in the rice root. The other difference between OsCSLD1 and KOJAK/AtCSLD3 plants was their tissue-specific expression patterns; OsCSLD1 was expressed only in the root, whereas KOJAK/AtCSLD3 was expressed in both roots and shoots. Of the four OsCSLD subfamily members that have been identified in rice, only OsCSLD1 was specifically expressed in roots. While OsCSLD2 and 4 were strongly expressed in roots as well, they were also expressed in other tissues. These expression patterns more closely resemble that of KOJAK/AtCSLD3 than the OsCSLD1 expression pattern. Additional studies are necessary to determine whether OsCSLD1 is the only one of the four subfamily genes to affect root hair growth.
This study showed that overexpression of OsCSLD1∷GFP led to the development of longer root hairs. This suggests that OsCSLD1 is a rate-limiting factor in root hair growth. However, it is possible that root hairs could be extended even further once other genetic factors affecting root hair growth are appropriately manipulated. This is suggested by the fact that in Arabidopsis, while COW1 and KOJAK/AtCSLD3 operate in the same developmental pathway, RHD1, 3, and 4, and TIP1 participate in an independent pathway (Grierson et al., 1997; Parker et al., 2000).
In rice, the root hairs develop in random patterns along the epidermis. This is in contrast to the alternating patterns seen in members of the Pooideae subfamily of grasses (e.g. in barley and wheat) and the striped pattern observed in Arabidopsis. We have shown here that OsCSLD1 in rice is specific to trichoblasts and is involved in the elongation of root hairs. Because the Arabidopsis gene KOJAK/AtCSLD3 is known to be required for root hair elongation and is expressed in the root hair, OsCSLD1 may be the functional ortholog of KOJAK/AtCSLD3. Thus, at least part of the mechanism of root hair morphogenesis is conserved between rice and Arabidopsis. Because OsCSLD1∷Ds can serve as a trichoblast and root hair marker for expression and phenotype, this gene trap mutant will help us to understand how other genetic factors identified in Arabidopsis play the functional roles in rice. In conclusion, OsCSLD1 may be a platform for not only understanding root hair development in rice but also for launching comparative studies between different species.
MATERIALS AND METHODS
Plant Growth Conditions
Wild-type, OsCSLD1∷Ds, and 35S∷OsCSLD1 lines were from the Japonica cultivar Dongjin. For germination on culture media, seeds were surface sterilized with 10% NaClO and 0.1% Triton X-100 and gently shaken at 37°C for 30 min. After thorough washing in sterile water, seeds were sown embryo side up in bottles containing 0.5 × MS medium with 0.5% Phytagel (Sigma). The sample bottles were incubated in a growth chamber at 30°C in 16 h light (250 μE s−1 m−2) and 22°C in 8 h dark. To suppress root hair growth, seeds were germinated on 0.5% Phytagel in 0.5× MS (without Suc) containing 10−5 m NPA. For soil growth, seeds were surface sterilized and germinated in the dark for 3 d before transfer to soil in the growth chamber under the above conditions.
Isolation of RNA and Northern Analysis
For RNA preparation, samples were harvested and immediately ground in liquid nitrogen. Total cellular RNAs were extracted using easy-BLUE reagents (iNtRON Biotechnology) according to the manufacturer's instructions. Formaldehyde (1.3%) gels were prepared in MOPS/EDTA buffer (0.5 m MOPS, pH 7.0, 0.01 m Na2EDTA, pH 7.5). Vacuum-dried 30-μg RNA samples were dissolved and heat denatured in formaldehyde/formamide buffer. After electrophoresis, gels were washed with water and 10× SSC and blotted with Hybond N+ (Amersham Pharmacia Biotech) prewetted with 10× SSC. Hybridization was performed at 65°C with Church buffer (1% bovine serum albumin, 200 μm EDTA, 0.5 m sodium phosphate, 7% SDS) containing a 32P-labeled probe. Probe DNAs specific for OsCSLD1, OsCSLD2, OsCSLD3, and OsCSLD4 were obtained from genomic DNA by PCR using the following gene-specific primers: for OsCSLD1, forward 5′-TGGAGTCTTCTTCAGCTTCTG-3′ and reverse 5′-CAGGTTCCAAGTTCCGAGC-3′; for OsCSLD2, forward 5′-GACTTCTCGATGCTTCCAC-3′ and reverse 5′-CAGATAGGTCAGGAAGGTC-3′; for OsCSLD3, forward 5′-AGATCTCCTTCACGCTGAC-3′ and reverse 5′-GTGACACACCTATCGACTG-3′; for OsCSLD4, forward 5′-GCTGTTCCTCACCAAGAAG-3′ and reverse 5′-GGGAGAAGAAGATCTCGAC-3′.
RT-PCR Analysis
Total mRNAs were isolated from roots with easy-BLUE reagent (iNtRON Biotechnology). RNA samples were treated with DNAse at 37°C for 1 h and 65°C for 20 min. For RT-PCR, the first-strand cDNA was synthesized with Moloney murine leukemia virus reverse transcriptase (NEB) in 20 μL of reaction mixture containing 1 μg total RNA and an oligo(dT)25 primer, according to the manufacturer's instructions, and 2 μL of the reaction mixture was used for PCR amplification. To detect OsCSLD1 cDNA, gene-specific primers (forward 5′-TCGCCGCCGAACAAGATC-3′ and reverse 5′-CGGACCACTTGATCTCCAG-3′) were used. The PCR reaction was performed at 94°C for 5 min, followed by 25 cycles at 94°C for 30 s, 58°C for 30 s, and 72°C for 1 min. PCR products were fractionated on agarose gels, stained with ethidium bromide, blotted to Hybond N+ (Amersham Pharmacia Biotech), and hybridized with a 32P-labeled OsCSLD1 probe.
In Situ Hybridization
A standard tissue embedding method was used to obtain sections of root samples. Root samples were fixed in 0.25% glutaraldehyde, 4% paraformaldehyde, and 100 mm sodium phosphate, pH 7.5, for 1 d at 4°C (Kouchi and Hata, 1993). Samples were vacuum infiltrated in the fixative until the tissue sank. For paraffin embedding, tissues were dehydrated through a graded ethanol series, followed by a tert-butanol series. Samples were embedded in paraffin for 3 d in a 58°C oven. For in situ RNA hybridization, OsCSLD1-specific DNA was prepared from the coding and 3′ UTRs of the OsCSLD1 cDNA by PCR using three sets of primers (set 1, forward 5′-GCATCGAGATCTCCTTCAC-3′ and reverse 5′-ATCATGAGGGACGTCCACT-3′; set 2, forward 5′-CATCGTCTACGTCTGGTCTG-3′ and reverse 5′-TGTGAGTTGGCTTGTGCAG-3′; set 3, forward 5′-CATATGCTAGCTGGTCGATC-3′ and reverse 5′-TGGGGTGAAGTGCAGTTTG-3′). PCR fragments were cloned into pGEM. Three OsCSLD1-specific antisense probes were 101, 89, and 98 bp in length. Antisense RNA probes were generated using the digoxigenin RNA labeling kit (Roche Applied Science). Sections of embedded tissues (8 μm thick) were mounted on slides pretreated with poly lysine. Mixtures of three RNA probes were added to final concentrations of 0.4 to 4.0 μg mL−1 and the slides were incubated for 12 to 16 h at 50°C. Hybridization signals were visualized with an anti-digoxigenin-alkaline-phosphatase-coupled antibody (Roche Applied Science).
Isolation of Full-Length cDNA and Agrobacterium Transformation
The 4.0-kb full-length OsCSLD1 cDNA were isolated by PCR amplification with the primers OsCSLD1-5′SpeI (ACTAGTATGGCGTCGAAGGGCATCCTCAAG) and OsCSLD1-3′SpeI (ACTAGTCCAGGGGAAAGAGAAGGATCCTCC). The PCR product was ligated into the pGEM-T vector (Promega) and sequenced. A 3.4-kb fragment was excised from the vector by SpeI digestion and ligated into the corresponding site of pCAMBIA1302. The full-length OsCSLD1 cDNA was fused with GFP at its 3′ end and expressed under the cauliflower mosaic virus 35S promoter and nopaline synthase 3′ terminator. Rice (Oryza sativa) calli were transformed with Agrobacteria LEA4404 carrying pCAMBIA (35S∷OsCSLD1). Rice calli were transformed with T-DNA carrying the hygromycin phosphotransferase gene, by a previously described method (Hiei et al., 1994), with slight modifications.
SEM
Seedling roots on 0.5× MS media were immediately fixed in solution (2.5% glutaraldehyde and 0.1 m sodium cacodylate, pH 7.4) for 12 h and dehydrated in a graded ethanol series. Dehydrated materials were critical-point dried in liquid CO2 and mounted on metallic stubs. Mounted samples were shadowed with gold before viewing under the SEM (Philips XL30 S FEG). Cryo-SEM was done as described previously (Favery et al., 2001). Briefly, plants grown on phytagel medium were placed on moist nitrocellulose paper mounted on a stub and immersed in liquid nitrogen slush. Roots were transferred to a cold stage. After removal of water by sublimation, roots were sputter coated with gold and observed using a JEOL SEM.
Histochemical Analysis
Histochemical staining for GUS activity was performed by incubation in a 5-bromo-4-chloro-3-indoyl glucuronide solution (1 mg mL−1). The solution consisted of 50 mm sodium phosphate buffer, pH 7.0, 10 mm EDTA, 0.1% Triton, 2 mm potassium ferrocyanide, and 200 μg mL−1 chloramphenicol. Samples were incubated at 37°C for 2 d in the dark and then dehydrated in a 30% to 70% graded ethanol series. To obtain tissue sections, GUS-stained samples were dehydrated and embedded in paraffin as described above. For toluidine blue staining, roots grown on 0.5× MS media were stained with 0.1% aqueous toluidine blue for 2 min and washed with distilled water. Roots on slides were inspected under the light microscope.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Epidermal cells of seminal roots of wild type and the OsCSLD1∷Ds mutant.
Supplemental Figure S2. Expression of OsCSLD1∷GFP in root hairs of transgenic rice plants.
Supplemental Figure S3. A phylogenic tree and protein sequence alignment.
Supplementary Material
This work was supported by the Crop Functional Genomics Center of the 21st Century Frontier Research Program, BioGreen 21 Program (Rural Development Administration; grant no. CG151), by the Korea Research Foundation (grant no. KRF–2003–015–C00636), by the Korea Science and Engineering Foundation/Ministry of Science and Technology to the Environmental Biotechnology National Core Research Center (grant no. R15–2003–012–01001–0), and by the Brain Korea21 project (fellowships to B.I.J., S.J.P., and H.L.P.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Chang-deok Han (cdhan@gsnu.ac.kr).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
References
- Baumberger N, Ringli C, Keller B (2001) The chimeric leucine-rich repeat/extensin cell wall protein LRX1 is required for root hair morphogenesis in Arabidopsis thaliana. Genes Dev 15 1128–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carol RJ, Dolan L (2002) Building a hair: tip growth in Arabidopsis thaliana root hairs. Philos Trans R Soc Lond B Biol Sci 357 815–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin HG, Choe MS, Lee SH, Park SH, Park SH, Koo JC, Kim NY, Lee JJ, Oh BG, Yi GH, et al (1999) Molecular analysis of rice plants harboring an Ac/Ds transposable element mediated gene trapping system. Plant J 19 615–623 [DOI] [PubMed] [Google Scholar]
- Doblin M, Melis LD, Newbigin E, Bacic A, Read SM (2001) Pollen tubes of Nicotiana alata express two genes from different β-glucan synthase families. Plant Physiol 125 2040–2052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doblin MS, Kurek I, Jacob-Wilk D, Delmer DP (2002) Cellulose biosynthesis in plants: from genes to rosettes. Plant Cell Physiol 43 1407–1420 [DOI] [PubMed] [Google Scholar]
- Dolan L, Costa S (2001) Evolution and genetics of root hair stripes in the root epidermis. J Exp Bot 52 413–417 [DOI] [PubMed] [Google Scholar]
- Dolan L, Duckett CM, Grierson C, Linstead P, Schneider K, Lawson E, Dean C, Poethig S, Roberts K (1994) Clonal relationships and cell patterning in the root epidermis of Arabidopsis. Development 120 2465–2474 [Google Scholar]
- Dolan L, Janmaat K, Willemsen V, Linstead P, Poethig S, Roberts K, Scheres B (1993) Cellular organisation of Arabidopsis thaliana root. Development 119 71–84 [DOI] [PubMed] [Google Scholar]
- Favery B, Ryan E, Foreman J, Linstead P, Boudonck K, Steer M, Shaw P, Dolan L (2001) KOJAK encodes a cellulose synthase-like protein required for root haircell morphogenesis in Arabidopsis. Genes Dev 15 79–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grierson CS, Roberts K, Feldmann KA, Dolan L (1997) The COW1 locus of Arabidopsis acts after RHD2, and in parallel with RHD3 and TIP1 to determine the shape, rate of elongation, and number of root hairs produced from each site of hair formation. Plant Physiol 115 981–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haigler CH, Datcheva MI, Hogan PS, Salnikov VV, Hwang S, Martin K, Delmer DP (2001) Carbon partitioning to cellulose synthesis. Plant Mol Biol 47 29–51 [PubMed] [Google Scholar]
- Hazen SP, Scott-Craig JS, Walton JD (2002) Cellulose synthase-like genes of rice. Plant Physiol 128 336–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiei Y, Ohta S, Komari T, Kumashiro T (1994) Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J 6 271–282 [DOI] [PubMed] [Google Scholar]
- Hu Y, Zhong R, Morrison WH III, Ye ZH (2003) The Arabidopsis RHD3 gene is required for cell wall biosynthesis and actin organization. Planta 217 912–921 [DOI] [PubMed] [Google Scholar]
- Ivashikina N, Becker D, Ache P, Meyerho O, Felle HH, Hedrich R (2001) K+ channel profile and electrical properties of Arabidopsis root hairs. FEBS Lett 508 463–469 [DOI] [PubMed] [Google Scholar]
- Kim CM, Piao HL, Park SJ, Chon NS, Je BI, Sun B, Park SH, Park JY, Lee EJ, Kim MJ, et al (2004) Rapid, large-scale generation of Ds transposant lines and analysis of Ds insertion sites in rice. Plant J 39 252–263 [DOI] [PubMed] [Google Scholar]
- Kouchi H, Hata S (1993) Isolation and characterization of novel nodulin cDNAs representing genes expressed at early stages of soybean nodule development. Mol Gen Genet 238 106–119 [DOI] [PubMed] [Google Scholar]
- Lauter F-R, Ninnemann O, Bucher M, Riesmeier JW, Frommer WB (1996) Preferential expression of an ammonium transporter and of two putative nitrate transporters in root hairs of tomato. Proc Natl Acad Sci USA 93 8139–8144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MM, Schiefelbein J (1999) WEREWOLF, a MYB-related protein in Arabidopsis, is a position-dependent regulator of epidermal cell patterning. Cell 99 473–483 [DOI] [PubMed] [Google Scholar]
- Montiel G, Gantet P, Allemand CJ, Breton C (2004) Transcription factor networks: pathways to the knowledge of root development. Plant Physiol 136 3478–3485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguema-Ona E, Andeme-Onzighi C, Aboughe-Angone S, Bardor M, Ishii T, Lerouge P, Driouich A (2006) The reb1-1 mutation of Arabidopsis: effect on the structure and localization of galactose-containing cell wall polysaccharides. Plant Physiol 140 1406–1417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker JS, Cavell AC, Dolan L, Roberts K, Grierson CS (2000) Genetic interactions during root hair morphogenesis in Arabidopsis. Plant Cell 12 1961–1974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiefelbein JW, Masucci JD, Wang H (1997) Building a root: the control of patterning and morphogenesis during root development. Plant Cell 9 1089–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert GJ, Barber C, Wells B, Dolan L, Roberts K (2002) Galactose biosynthesis in Arabidopsis: genetic evidence for substrate channeling from UDP-d-galactose into cell wall polymers. Curr Biol 12 1840–1845 [DOI] [PubMed] [Google Scholar]
- Séné CFB, McCann MC, Wilson RH, Crinter R (1994) Fourier-transform raman and Fourier-transform infrared spectroscopy: an investigation of five higher plant cell walls and their components. Plant Physiol 106 1623–1631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada T, Tachibana T, Shimura Y, Okada K (1997) Epidermal cell differentiation in Arabidopsis determined by a Myb Homolog, CPC. Science 277 1113–1116 [DOI] [PubMed] [Google Scholar]
- Wang X, Cnops G, Vanderhaeghen R, Block SD, Montagu MV, Van Lijsebettens M (2001) AtCSLD3, a cellulose synthase-like gene important for root hair growth in Arabidopsis. Plant Physiol 126 575–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.