Abstract
Although the morphological steps of maize (Zea mays) endosperm development are well described, very little is known concerning the coordinated accumulation of the numerous proteins involved. Here, we present a proteomic study of maize endosperm development. The accumulation pattern of 409 proteins at seven developmental stages was examined. Hierarchical clustering analysis allowed four main developmental profiles to be recognized. Comprehensive investigation of the functions associated with clusters resulted in a consistent picture of the developmental coordination of cellular processes. Early stages, devoted to cellularization, cell division, and cell wall deposition, corresponded to maximal expression of actin, tubulins, and cell organization proteins, of respiration metabolism (glycolysis and tricarboxylic acid cycle), and of protection against reactive oxygen species. An important protein turnover, which is likely associated with the switch from growth and differentiation to storage, was also suggested from the high amount of proteases. A relative increase of abundance of the glycolytic enzymes compared to tricarboxylic acid enzymes is consistent with the recent demonstration of anoxic conditions during starch accumulation in the endosperm. The specific late-stage accumulation of the pyruvate orthophosphate dikinase may suggest a critical role of this enzyme in the starch-protein balance through inorganic pyrophosphate-dependent restriction of ADP-glucose synthesis in addition to its usually reported influence on the alanine-aromatic amino acid synthesis balance.
The economic and nutritional value of maize (Zea mays) kernels is mainly due to its high starch content because it represents approximately 75% of mature seed weight. Starch is accumulated in endosperm, which, in maize as in other cereals, is persistent at seed maturity and forms a reserve used for the development of the embryo during germination. Cereal endosperm development presents five key steps (Olsen, 2001): coenocytic phase, cellularization, differentiation, reserve synthesis, and maturation. In the first place, within hours following pollination, endosperm nuclei undergo numerous divisions without any cellularization. Cell wall formation generally begins 4 d after pollination (dap) when the endosperm contains several thousand cells. Cell division still occurs, but at a lower rate until 20 dap (Larkins et al., 2001). Consistently, an important synthesis of all components involved in cell wall formation takes place during 5 to 10 dap. Next, differentiation takes place and storage products are synthesized from 12 dap to maturity. Thus, most starch present in the endosperm is synthesized within a 25-d period, from about day 12 to day 35 post pollination. During the same time, the endosperm increases in size from less than 0.1 mm to up to 10 mm. Very little is known about the beginning of cell death in the starchy endosperm, but in standard growing conditions in European Community, dry-matter accumulation levels off at about 35 dap and the dry-down process begins rapidly. Endosperm is an important tissue from the point of view of the breeder, because it is a major component of yield, and from the point of view of the physiologist and evolutionist, because it provides the ground for seedling germination and initial growth.
Whereas morphological steps of endosperm development are well described, the underlying molecular mechanisms are still largely unknown. As highlighted before, endosperm development is a complex phenomenon that must be driven by coordinate expression of numerous genes. Approaches using both spontaneous and induced mutants allow characterization of individual developmental steps. In Arabidopsis (Arabidopsis thaliana), despite the nonpersistent nature of the endosperm in seeds, screens of mutants can help to shed light on several events occurring during endosperm development (Berger, 1999). In maize, studies based on analysis of ethyl methane sulfonate mutants (Neuffer and Sheridan, 1980) or mutator transposon-induced mutants (Scanlon et al., 1994) both suggested that at least 300 genes can cause a visible endosperm phenotype. Only few of these mutants have still been molecularly characterized (Scanlon and Myers, 1998; Consonni et al., 2005). However, a higher number of genes are supposed to be expressed and play a role during endosperm development without being characterized by a visible mutant phenotype. In wheat (Triticum aestivum), gene expression in developing grain has been analyzed using microarrays. A total of 2,225 genes were identified, one-fourth of which exhibited differential expression within the 28 d post anthesis (Leader, 2005). Analysis of a cDNA library originating from mRNA extracted from wheat endosperms at 8, 10, and 12 d post anthesis indicated that 4,500 to 8,000 different genes could be expressed. Among 1,013 sequences, 601 could be assigned a function and the main categories (protein destination and storage, protein synthesis, metabolism, and cell structure) were fully consistent with the developmental stage of the tissue (Clarke et al., 2000). In maize, cDNA libraries were constructed at contrasted developmental stages (4–6 dap and 7–23 dap). Analysis of contigs and singletons suggested that at least 5,000 different genes could be expressed, excluding storage protein genes (Lai et al., 2004). This number was considered a minimal estimate following the work of Verza et al. (2005). It is interesting to note that in these studies in wheat and maize, about 35% of the sequences remained without functional identification, possibly consisting of endosperm-specific genes.
mRNAs are the primary products of gene expression, but their levels are often faintly correlated to cognate protein levels (Gygi et al., 1999; Watson et al., 2003). Moreover, a lot of posttranslational modifications can take place during protein maturation, which may result in a higher complexity at the protein level than at the mRNA level. Proteomics intends to characterize the protein complement of tissues or organs, which gives insight into the main agents of cellular biochemistry. Most proteomic studies still rely on two-dimensional (2-D) electrophoresis (2-DE) to achieve good resolution of complex protein mixtures. A major breakthrough in the methods for protein identification was the use of mass spectrometry (MS) for routine protein identification in place of Edman sequencing. It now allows extensive proteomic studies to be carried out. In plants, besides a few completely sequenced genomes, the increasing availability of large genomic and/or expressed sequence tag (EST) sequence databases, notably for grasses, increases progressively the performance of these methodologies (http://www.gramene.org).
Several proteomic analyses of seed development have been reported in the past years in dicots (Gallardo et al., 2003; Hajduch et al., 2005, 2006). In plants where endosperm is the main storage compartment, like in cereals, rice (Oryza sativa) is the model species and its now fully sequenced genome provides a favorable context for proteomic studies. Several studies were devoted to proteome analysis of whole seed or embryo and endosperm (for review, see Khan and Komatsu, 2004). The most comprehensive one is the comparison of the proteome of seed, leaf, and root using MudPit technology and 2-DE followed by tandem MS (Koller et al., 2002). Of 2,528 proteins detected over the three organs, 877 were found in the seed, 513 of which were specific. In barley (Hordeum vulgare), the salt-soluble proteome was analyzed by 2-DE during grain filling and maturation in mature and germinating seeds. About 200 protein spots were identified in successive studies, belonging to primary metabolism, cell rescue and defense, and protein folding. Specific accumulation patterns were observed during grain development and germination (Finnie et al., 2002; Ostergaard et al., 2002, 2004). A recent study in wheat focused on albumins and globulins, excluding the abundant storage proteins glutenins and gliadins; 254 proteins were identified and their accumulation profiles were compared at 10 and 36 d post anthesis. The predominant categories were different at the two stages, consistent with a shift in biochemical activity from active synthesis to maintenance, stress defense, and storage (Vensel et al., 2005).
To date, to our knowledge, there has been no proteomic study of maize endosperm development. We have established a comprehensive maize endosperm reference map, based on the proteome of 14-dap endosperms (Méchin et al., 2004); 632 proteins were identified, among which 496 were assigned a functional identification. Metabolic processes, protein destination, protein synthesis, and cell rescue, defense, cell death, and aging are the most abundant functional categories, comprising almost one-half of the identified proteins. In this article, we examine the accumulation patterns of the identified proteins in our maize endosperm reference map (Supplemental Fig. S1) at seven developmental stages, covering the five phases described on morphological grounds, to begin unraveling molecular mechanisms involved in endosperm development.
RESULTS
Accumulation of Nonstorage Proteins during Development
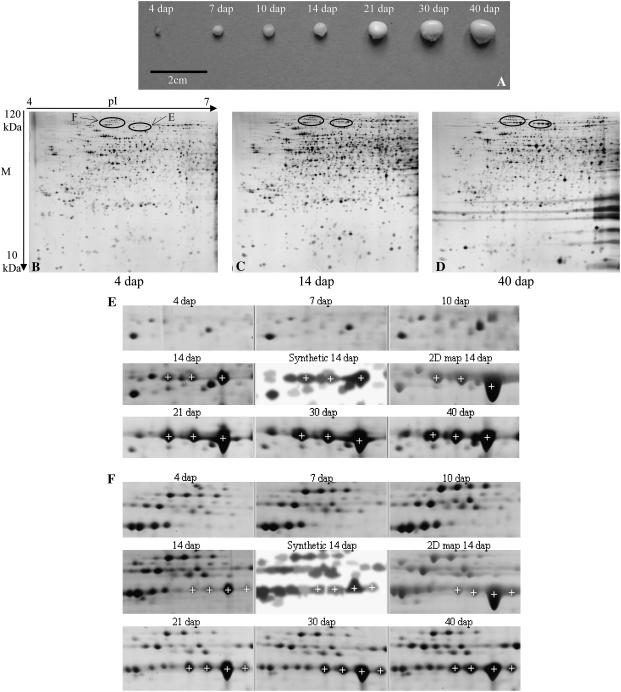
We characterized the proteome at seven developmental stages scaled on the morphoanatomical description of development. At the early stages (4, 7, and 10 dap), dissection to separate embryo and endosperm was tricky (Fig. 1A) and thus the proteome of whole kernels was analyzed. The endosperm proteome was analyzed at four later stages: 14, 21, 30, and 40 dap. Accumulated proteins ranging from 10 to 100 kD in Mr and from 4 to 7 in pI were visualized with silver staining. We chose this pH range to exclude most of the abundant storage proteins (zeins that have pIs comprised between 7 and 9) and because we intended to focus on the time course of proteins involved in the setup of the endosperm and the establishment of the enzyme equipment for starch and storage protein accumulation metabolism. Experiments comparing patterns obtained from a pH 3 to 10 range versus a pH 4 to 7 range showed that, whereas supplementary proteins could indeed be detected above pH 7, the quality of resolution, especially in the latest developmental stages because of the abundance of zeins, was better using a pH 4 to 7 range. Our 2-D map was thus based on a pI 4 to 7 2-D pattern, allowing more than 600 proteins to be processed (Méchin et al., 2004). This, in turn, dictated the pH range for developmental analysis because we wanted to work on as many identified proteins as possible.
Figure 1.
2-D gels of nonzein proteins extracted from 4-, 14-, and 40-dap maize kernel/endosperm and cuttings of the silver-stained gels at each developmental stage focusing on the seven identified isoforms of PPDK. A, Whole kernels at the seven developmental stages. B, 4-dap kernel 2-DE gel image. C, 14-dap endosperm 2-DE gel image. D, 40-dap endosperm 2-DE gel image. Horizontal streaking due to zeins not focused in the pH range used for IEF is visible on the basic (right) side of the gels. E, Three identified CyPPDK2 isoforms (white crosses, IDs 359, 360, and 357). F, Four identified CyPPDK1 isoforms (white crosses, IDs 399, 398, 388, and 376). The synthetic image composed by Melanie 4 software from the three repeats of the 14-dap stage is shown between one silver-stained gel of the 14-dap developmental stage and the Coomassie Blue-stained gel supporting the 2-D reference map (Méchin et al., 2004). White crosses indicate the corresponding matched spots in each gel cutting.
Total protein loading was increased as development proceeded, in an effort to compensate for the increase in zein abundance in the total protein extracts. This allowed better visualization of low-abundance proteins that are supposed to take part in the biochemical processes involved in development. Actually, the compensation appeared partly successful particularly at the oldest stages, which could account for a limited decrease in the number of reproducible spots from 4 dap (1,632) to 40 dap (1,483). The distribution of standardized spot volumes appeared very similar at every stage (data not shown). Nevertheless, the global pattern changed during development, and streaking due to zeins was visible from 14 dap onward (Fig. 1, B–D).
Because we intended to follow the accumulation pattern of identified proteins, we used the 14-dap stage as the reference for matching patterns of all other stages. Among the 1,551 reproducible spots obtained at 14 dap (Fig. 1C), 94% could be matched to one or more other stages. The proportion of reproducible spots of any given stage that could be matched to the 14-dap stage was quite high, ranging from 61% at 7 dap to 88% at 21 dap. This indicates that quantitative variations, rather than large qualitative variations, probably account for the apparent global change in 2-D patterns during development.
Matching to the Endosperm Reference Map
The silver-stained 14-dap stage pattern from the developmental study was matched against the Coomassie Blue-stained 14-dap pattern supporting the reference map (Méchin et al., 2004), allowing 558 spots to be unambiguously matched to the 632 protein spots processed for the 2-D map (matches examples in Fig. 1, E and F). Among these, 54 corresponded to protein spots with multiple identifications (Méchin et al., 2004) and they were eliminated from subsequent analyses. Among the 504 remaining spots, 409 were retained for analysis of quantitative variation during development (Supplemental Table S1) on the basis of quality of detection and quantification over the seven developmental stages (see “Materials and Methods”). Among these, 211 displayed a significant (P < 0.05) linear response of their volume during development and 102 more spots displayed a significant (P < 0.05) stage effect. Thus, about 76% of the protein spots displayed a significant volume change during development. Among the 409 spots, 121 proteins had no definite function (nonidentified [NI] and not yet clear cut [NYC] function). From reinterrogation of databases, 14 of these proteins were identified (indicated by a cross in Supplemental Table S4 updated from Méchin et al. [2004]). A total of 302 identified protein spots (Supplemental Table S2) were thus available for further analysis.
Composite Expression Profiles of Functional Categories among Endosperm Development
To examine the expression tendency within functional categories (or subcategories), composite expression analysis (Hajduch et al., 2005) was performed by summing normalized spot volume for all proteins of a given functional category at each of the seven developmental stages.
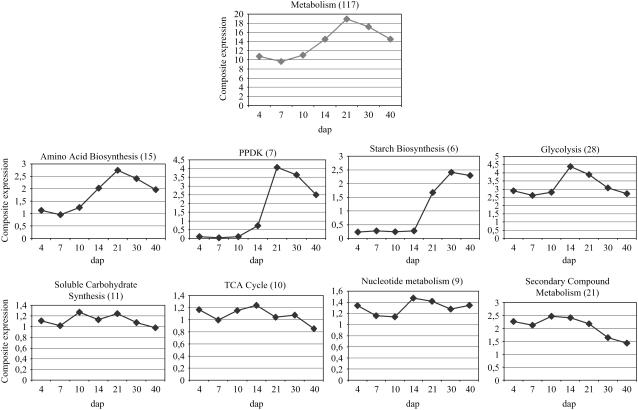
The protein destination category presented an important peak of expression at 14 dap, whereas the protein synthesis class displayed a large increase between 10 and 30 dap (data not shown). Proteins involved in metabolic processes massively increased and reached a maximum at 21 dap (Fig. 2). Between 4 and 21 dap, the relative abundance of metabolic proteins was multiplied by a factor of 2. Proteins involved in soluble carbohydrate and nucleotide metabolism did not display any particular trend in their accumulation profiles during development, whereas enzymes from secondary metabolism and, to a lesser extent, from the tricarboxylic acid (TCA) cycle presented a decrease (Fig. 2). On the other hand, proteins involved in amino acid biosynthesis and the pyruvate orthophosphate dikinase (PPDK) displayed a similar accumulation profile, with a very important peak at 21 dap (Fig. 2). Glycolysis enzymes presented their maximal level of accumulation at 14 dap. Finally, proteins involved in starch biosynthesis were at a very low level before 14 dap and reached their maximum at 30 to 40 dap (Fig. 2).
Figure 2.
Composite expression profiles of metabolism category and its 10 subcategories. The total spot number used to draw the composite profiles is indicated in parentheses.
Accumulation Profile of Identified Proteins during Development
Hierarchical Clustering Analysis
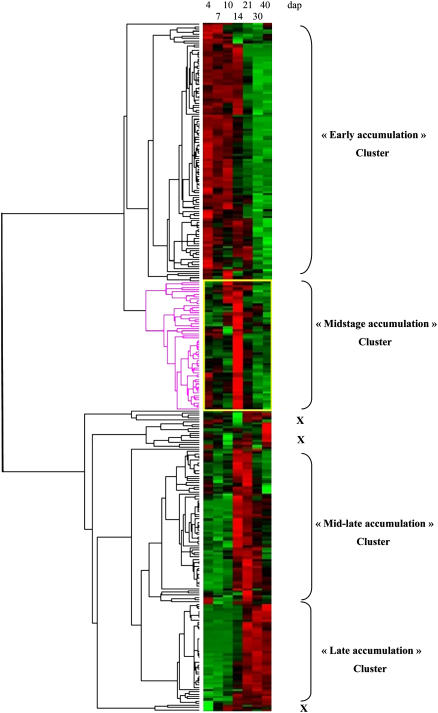
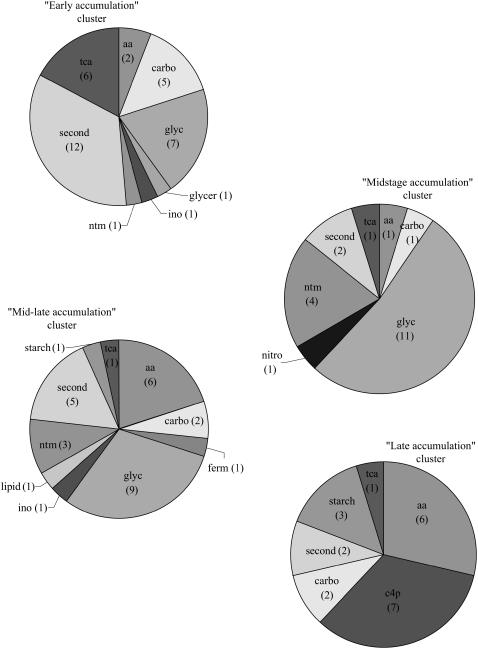
To further analyze expression patterns during development as related to protein functions, hierarchical clustering analysis was performed excluding proteins with no definite function (NI and NYC; Supplemental Table S2; Fig. 3). Four main accumulation patterns during development could be distinguished: (1) highest accumulation at the beginning of development, mostly 4 to 10 dap, but occasionally up to 14 dap (cluster early accumulation); (2) highest accumulation limited in time, at the 14-dap stage (cluster midstage accumulation); (3) highest accumulation at several intermediary stages, mostly 14 and 21 dap (cluster mid-late accumulation); and (4) highest accumulation at late developmental stages, 30 and 40 dap (cluster late accumulation). The corresponding clusters encompassed, respectively, 112, 56, 68, and 45 proteins out of the 302 included in the analysis (Supplemental Table S2); 21 proteins did not convincingly display any of the four main developmental patterns (indicated by a cross on Fig. 3): Five of them displayed low accumulations up to 30 dap and a high amount at 40 dap, 13 have erratic behavior, and three were expressed at a low level at 4 dap and almost constant afterward.
Figure 3.
Hierarchical clustering of the 302 functionally identified proteins at seven developmental stages. Two main branches can be distinguished. The first one gathers early accumulation and midstage accumulation clusters, whereas the other one associates mid-late accumulation and late accumulation clusters. A few proteins display outstanding profiles, not included in the four main clusters discussed in the text (indicated by a cross on the figure). The 302 functionally identified proteins are listed in the Supplemental Table S2, with their clustering membership.
Functional Categories Distribution among Clusters
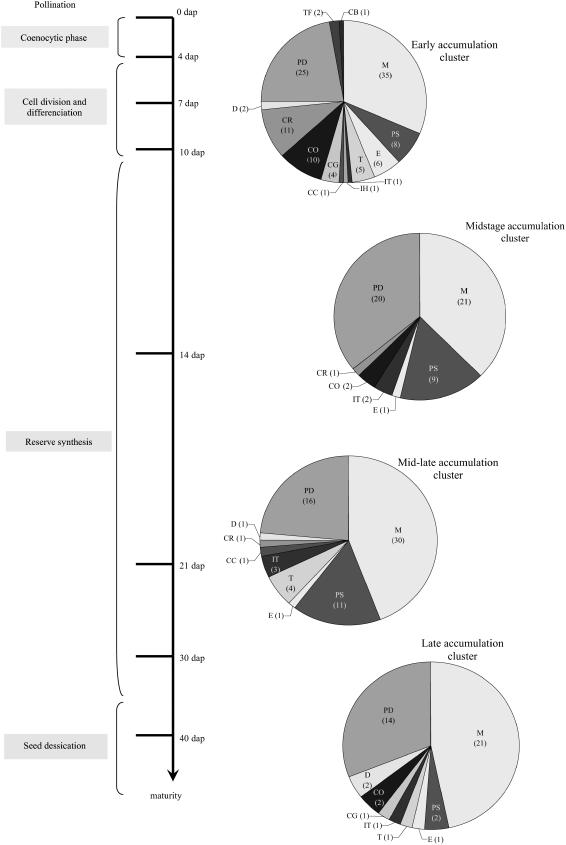
The distribution of the functional categories appeared significantly different among the four main clusters (χ2 12 degrees of freedom, 31.90, P ≤ 0.01). The early accumulation cluster encompassed the highest number of proteins and also displayed the largest functional diversity (Fig. 4). Metabolism and protein destination were the most numerous categories (53% of proteins). The midstage accumulation cluster had the lowest functional diversity (Fig. 4), with a predominance of metabolism, protein destination, and protein synthesis functional categories (91% of proteins). These categories were still important in mid-late accumulation and late accumulation clusters (84% and 82% of proteins, respectively), although protein synthesis was weakly represented in the latter (Fig. 4).
Figure 4.
Functional classification of the identified proteins involved in each of the four defined clusters. Proportions of the main functional categories in the four main clusters defined in Figure 3. The number of identified spots included in each category is indicated in parentheses. A developmental scale is indicated on the left. M, Metabolism; PS, protein synthesis; E, energy; T, transcription; IT, intracellular transport; IH, ionic homeostasis; CC, cellular communication, signal transduction; CG, cell growth, cell division, and DNA synthesis; CO, cellular organization; CR, cell rescue, defense, cell death, and aging; D, development; PD, protein destination.
Within the metabolism category itself (107 proteins), we found heterogeneity in specific function distribution among clusters (χ2 9 degrees of freedom, 22.696, P ≤ 0.01). The early accumulation cluster was characterized by a predominance of proteins involved in secondary compound metabolism (34%) and energy production (glycolysis and TCA cycle, 37%; Fig. 5). As development proceeds, maximal expression is exhibited by proteins involved in energy production (midstage accumulation cluster) then proteins involved in metabolite production, specifically amino acids, carbon skeletons, and starch (mid-late accumulation and late accumulation clusters). Proteins involved in energy production actually constituted 54% of the proteins in the midstage accumulation cluster. The TCA cycle was represented in the four clusters, with a relatively higher contribution in the early accumulation cluster, whereas the glycolytic pathway was absent from the late accumulation cluster (Fig. 5).
Figure 5.
Functional classification of the proteins involved in metabolic processes in each of the four defined clusters. Proportion of functional subcategories of metabolism category in the four clusters defined in Figure 3. The number of identified spots included in each subcategory is indicated in parentheses. aa, Amino acid biosynthesis; c4p, PPDK; starch, starch biosynthesis; glyc, glycolysis; carbo, carbohydrate synthesis; tca, TCA cycle; ntm, nucleotide metabolism; second, secondary compound metabolism; ino, inositol metabolism; ferm, fermentation; glycer, glycerol metabolism.
A remarkable feature in the second most numerous category (protein destination, 75 proteins) was the predominance of the proteins involved in degradation processes in the early accumulation cluster. This is in accordance with the disappearance of numerous functional categories from the early accumulation cluster to the midstage accumulation cluster.
Zeins that have basic pIs (above 7) were out of the range of 2-D analysis, but two low-abundance storage proteins (legumin like) were detected in the 4 to 7 pH range; as expected, they were both included in the late accumulation cluster.
Accumulation Profile of Isoforms within Functional Families
Among the 302 proteins identified, 200 grouped into 56 families defined by a unique suspected function. These functional families associated from two (26 of 56 families) up to 12 (protein disulfide isomerase [PDI]) members. These multiple forms can be posttranslational modifications of the same gene product and/or products encoded by different, but homologous, genes. For the sake of brevity, we called the multiple forms for a given function isoforms in the “Results” and “Discussion” sections. In most cases, it was not possible to sort out these hypotheses. In 19 families, however, comparison of the MS/MS peptides used for identification of the isoforms revealed homologous sequences differing by one or two amino acids (Supplemental Table S4), supporting the hypothesis that the concerned isoforms are encoded by different genes.
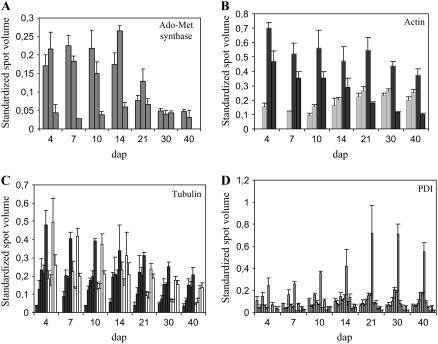
We examined possible coregulation between isoform accumulation within every family by computing Pearson correlation coefficients between spot volumes. In 16 families, no significant correlation appeared between isoforms (e.g. S-adenosyl-Met [Ado-Met] synthase; Fig. 6A). Eighty-nine of 401 correlations were significant at P < 0.01. Interestingly, these correlations were most often positive (82 of 89), indicating that when coregulation of isoforms took place, it generally resulted in an increase in protein accumulation for a given time course rather than a shift or balancing of accumulation during the considered developmental period. Indeed, these two contrasted behaviors were observed for actin and tubulins (Fig. 6, B and C). Negative coregulation between the two pairs of actin isoforms resulted in an almost constant accumulation of total amount from 4 to 40 dap, whereas the total amount of tubulins steadily decreased during development.
Figure 6.
Accumulation of isoforms of Ado-Met synthase (A), actin (B), tubulins (C), and PDI (D) during development. Mean standardized spot volumes are given for each isoform and sds are indicated as vertical bars. A, Ado-Met synthase isoforms (ID, from left to right, 568, 563, and 333). B, Actin. Light/dark gray, Isoforms displaying increased/decreased accumulation during development (ID, from left to right: 525, 505, 506, and 524). The accumulations of the two sets of isoforms are inversely correlated (−0.831; P < 0.05). C, Tubulins. Black, Five α-tubulin isoforms (ID, from left to right, 532, 551, 562, 561, and 560). White, Four β-tubulin isoforms (ID, from left to right, 543, 539, 542, and 541). D, PDI, 12 isoforms (ID, from left to right, 247, 567, 597, 201, 485, 487, 486, 484, 488, 480, 481, and 482).
Isoforms of functional families involved in protein folding (chaperones, chaperonins, and PDI) represented almost 20% of all isoforms. These families followed a similar trend of accumulation, with a maximum at about 14 to 21 dap (Fig. 6D); positive correlations involved two to six isoforms, according to the family, and negative correlations were observed between two isoforms of PDI and three isoforms of heat shock 90 family. Only for PPDK were the patterns of all isoforms in the family correlated to each other (see Fig. 7, no. 9).
Figure 7.
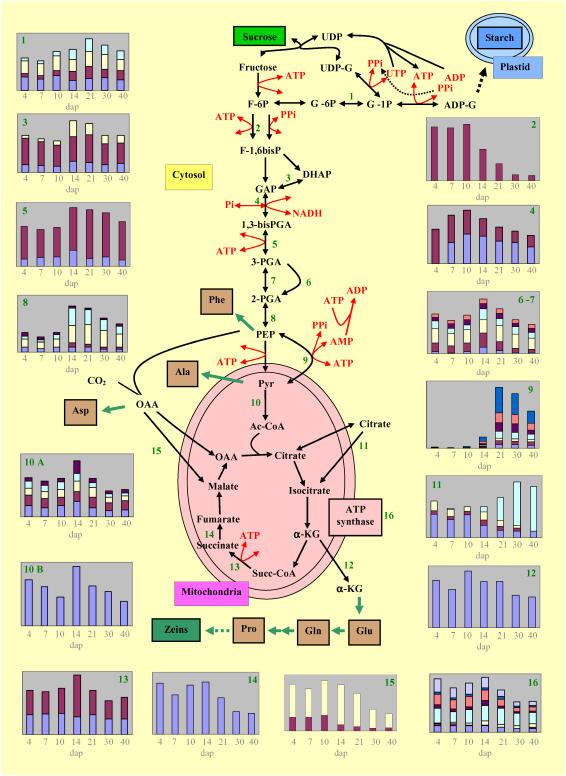
Overview of glycolysis and TCA cycle enzyme accumulation with an emphasis on the role of PPDK and pyruvate-PEP ratio in the balance between starch and protein synthesis. Schematic representation of glycolysis and TCA cycle and their connection by the intermediary complex. For every enzyme where at least one isoform was identified, a corresponding accumulation profile during development is drawn. ATP and PPi consummation or production is indicated for each step. Predicted subcellular localization was used for placing enzymes relative to the organelle (see “Materials and Methods”) of the identified enzyme isoforms. The color representation on the accumulation profile during development is given together with the corresponding ID for each isoform. 1, Phosphoglucomutase; sky blue: 402, yellow: 419, red: 418, dark blue: 421. 2, Fructokinase II; red: 172. 3, Triosephosphate isomerase; yellow: 90, red: 129, dark blue: 99. 4, Phosphoglycerate dehydrogenase; red: 199, dark blue: 211. 5, Phosphoglycerate kinase-3; red: 552, dark blue: 331. 6 and 7, Phosphoglycerate mutase; orange: 602, violet: 407, sky blue: 1,118, yellow: 601, red: 603, dark blue: 599. 8, Enolase; violet: 573, sky blue: 533, yellow: 584, red: 587, dark blue: 586. 9, PPDK; dark blue: 354, orange: 376, violet: 360, sky blue: 359, yellow: 388, red: 398, dark blue: 399. 10A, Pyruvate dehydrogenase E1 α- and β-subunit isoforms; violet: 508, sky blue: 510, yellow: 327, red: 329, dark blue: 192. 10B, Dihydrolipoamide S-acetyl transferase; dark blue: 572. 11, Aconitase hydratase; sky blue: 301, yellow: 305, red: 306, dark blue: 290. 12, NADP-specific isocitrate dehydrogenase; dark blue: 233. 13, Succinyl-CoA ligase; red: 507, dark blue: 503. 14, Succinate dehydrogenase subunit; dark blue: 366. 15, Malate dehydrogenase; yellow: 243, red: 330. 16, ATP synthase; blue: 425, dark blue: 596, orange: 592, violet: 591, sky blue: 588, yellow: 589, red: 58, dark blue: 619. UDP-G, UDP-Glc, F-6P, Fru-6P; G-6P, Glc-6P; G-1P, Glc-1P; ADP-G, ADP-Glc; F-1,6bisP, Fru-1,6BP; GAP, glyceraldehyde 3P; DHAP, dihydroxyacetone P; 1,3-bisPGA, 1,3-bisphosphoglycerate; 3PGA, 3-phosphoglycerate; 2PGA, 2-phosphoglycerate; Succ-CoA, succinyl-CoA; α-KG, α-oxoglutarate.
Most enzymes involved in carbohydrate metabolism, glycolysis, TCA cycle, and energy present several isoforms localized in different cell compartments that can be implicated in other metabolic pathways. Four software programs were used to predict the subcellular localization of the isoforms detected in our study (Supplemental Table S3) to focus on the isoforms that are likely specifically involved in glycolysis (cytosolic) and the TCA cycle (mitochondria).
Thus, seven of the 10 enzymes involved in the glycolytic pathway appeared as 23 cytosolically located isoforms. Most of them did not present sharp changes in their overall accumulation profile during development, resulting in an accumulation maximum in the middle part of development (Fig. 7). Nevertheless, this general trend hides large variation at the level of a few enzyme isoforms. Due to one of the two isoforms with a maximal accumulation at midstages, 3-phosphoglycerate kinase was present at a high level at late stages. A similar trend was observed for phosphoglocomutase. Among the five enolase isoforms, four accumulated steadily during development, whereas the fifth isoform appeared at 14 dap, which is also its maximal level of accumulation stage. Six protein spots that were identified as elements of the pyruvate dehydrogenase complex (pyruvate dehydrogenase and dihydrolipoamide S-acyltransferase) displayed an accumulation profile with a marked decrease from 14 dap onward (Fig. 7, nos. 10A and 10B).
Only one-half of the enzymes involved in the TCA cycle were identified as 10 protein spots. The accumulation profile of most isoforms showed a sustained level up to 14 dap, then a decrease, with the remarkable exception of one cytosolic isoform of aconitase that displayed late accumulation. ATP synthases had their maximal accumulation at early stages. Accumulation profiles of enzymes with their detected isoforms in parallel to the known metabolic pathways allowed us to propose a synthetic picture of cell machinery functioning during endosperm development (Fig. 7).
DISCUSSION
Changes in proteome complement as development proceeds provide clues on coregulation, crucial gene function at specific developmental phases, and relationships between phases. As deduced by the number of reproducible protein spots obtained at any stage that can be matched to the 14-dap endosperm 2-D pattern, the set of abundant gene products undergoes limited changes during development. However, the relative abundance of the same gene products appears to be different among the developmental stages. Actually, among the 632 polypeptides processed in the 14-dap endosperm 2-D map (Méchin et al., 2004), 409 were reliably quantified and matched over one or more of the seven stages, which represents the largest proteomic dataset devoted to endosperm development, as compared to other cereals. Among these proteins, 107 remained NI or had NYC function. Both NI and NYC categories probably represent a mix of all other functional categories, as can be suspected from their composite expression profile (data not shown) and scattering in a global clustering analysis (data not shown).
The choice of 14 dap as the reference stage for the identification and developmental studies probably resulted in missing proteins specifically accumulating at precocious (coenocytic phase) and late (full-storage synthesis, desiccation) stages of endosperm development, but it ensures that no embryo or pericarp-specific proteins are analyzed at the youngest stages, where no dissection was done. However, for the youngest stages, a slight contribution of embryo to protein profiles cannot be definitely excluded. Nevertheless, the temporal analysis of proteomic data by the clustering method allowed four distinct patterns of accumulation—clusters—to be recognized. Besides the metabolism, protein destination, and protein synthesis functional categories that occurred in all clusters, a few categories or subcategories appeared preferentially bound to one or the other. The early accumulation cluster grouped proteins mainly involved in cellularization, detoxification, degradation, and respiration associated with energy production. As expected, the late accumulation cluster involved the main functions related to storage product synthesis and protein folding. The midstage accumulation and mid-late accumulation clusters shared functional categories (or subcategories) such as glycolysis, protein synthesis, and destination, but differed by the number and relative abundance of the involved proteins. Each cluster corresponds to sets of similarly regulated gene products, which enables discussion on coordinated functioning of cellular processes during each major developmental phase.
Early Stages of Development
Cellularization, Cell Division, and Cell Wall Deposition
Proteins involved in cell division (transitional endoplasmic reticulum ATPases, proliferating cell nuclear antigen involved in cell proliferation), as well as five ATP synthases displayed a maximal level of accumulation during the earliest phases of endosperm development when cellularization of the syncitium (3–5 dap), cell division, and enlargement (8–12 dap) occur. Cytoskeleton proteins are expected to be largely involved in these developmental processes. They are assembled in microfilaments and microtubules, which are dynamic structures composed of polymers of actin and α- and β-tubulins, respectively (Mayer and Jürgens, 2002). Actin filaments and microtubules have two main roles, targeting organelles and vesicles, and sustaining cell growth and division (Mayer and Jürgens, 2002). In addition, actin seems to play a role in a variety of processes critical for cellular development (Meagher and Fechheimer, 2003). The almost constant accumulation of actin during development is consistent with an involvement in multiple cellular processes. In contrast, tubulins that, as components of microtubules, are more invested in cell division (Mayer et al., 1999), showed decreased accumulation. Indeed, cell division becomes limited to the subaleurone layer from 12 dap onward with a mitotic index for the whole endosperm dropping to less than 1% after 14 dap (Larkins et al., 2001). Cell wall deposition that takes place during cellularization and cell division is also cytoskeleton directed (Olsen, 2001). Accordingly, the maximal level of accumulation was observed at the earliest stages for three UDP-Glc:protein transglucosylases possibly involved in cell wall polysaccharide synthesis, as well as a Xyl isomerase.
Sustained Accumulation of Ado-Met Synthase
An important accumulation of two Ado-Met synthases was observed from 4 to 14 dap. Another isoform was expressed later, but in a relatively low amount. At 30 dap, the overall level of Ado-Met synthase dropped by about 60% of the 14-dap level and reached 14% of that level at 40 dap. Interestingly, such a sharp decrease was also observed during Medicago truncatula seed maturation (Gallardo et al., 2003). In plants, the product of Ado-Met synthase, Ado-Met, is involved in a large variety of processes: the biosynthesis of ethylene, spermine, and spermidine, the regulation of Met and other Asp-derived amino acids, and as a methyl donor in the methylation of amino acids, lipids, RNA, and DNA (Ravanel et al., 1998). Ado-Met synthase has been shown to accumulate during germination in Arabidopsis (Gallardo et al., 2002). It has been suspected to be implied in the transition from an active to a quiescent state during seed development (Gallardo et al., 2003). The highest abundance at the initial phases of maize seed growth and development and the sharp decrease observed after 14 dap is consistent with this hypothesis.
Oxidative Stress Response
At the beginning of kernel development, young pericarps are still permeable, allowing oxygen penetration in the endosperm. From 4 to 14 dap, the oxygen level is high and mitochondrial respiration is intense (Rolletschek et al., 2005). Indeed, during the stepwise, one-electron reduction of oxygen, cells continuously produce partly reduced O2 intermediates like superoxide radical, hydrogen peroxide, and hydroxyl radical, commonly known as reactive oxygen species, which can damage cell components (Scandalios, 1993), but also have important signaling roles (e.g. in cell death). To regulate concentration of reactive oxygen species, numerous antioxidant defenses are activated (Halliwell and Gutteridge, 1990; Kovtun et al., 2000). Consistently, 11 of 14 proteins grouped in the cell rescue, defense, cell death, and aging category had their maximal level of accumulation in the earliest stages. These proteins (glutathione S-transferase, superoxide dismutases, and ascorbate peroxidases) are in charge of detoxication. Consistent with the diminished oxygen level, their composite expression profile displayed a constant decrease during development.
Protein Turnover and Switch to Storage Compound Synthesis
An important number of proteins involved in proteolysis (16 of 26) had their maximal level of accumulation at the earliest stages, most of which were components of the ubiquitin-proteasome complex (13 of 16). The function of this complex is to target and degrade specific proteins. As a first step, polyubiquitinylation of substrates is achieved through the action of three enzymes: E1, a ubiquitin-activating enzyme, E2, a ubiquitin-conjugating enzyme, and E3, a ubiquitin ligase that determines the specificity of the substrate. The polyubiquitinated protein is then processed by the 26S proteasome, which consists of a core 20S protease capped at each of its ends by a regulatory 19S complex. This degradation pathway plays an important role in various aspects of plant growth and development (Vierstra, 2003; Moon et al., 2004). Four α- and five β-subunits of the 20S proteasome displayed the strongest accumulation between 4- and 14-dap, whereas only three α-isoforms accumulated later, belonging to the midstage accumulation cluster. As far as the ubiquitination enzymes are concerned, four isoforms of E1 were observed in the early accumulation cluster, whereas an E2 had its maximal level of accumulation at the mid-late accumulation phase. The number and relative abundance of proteins involved in degradation processes in the early accumulation phase suggests important protein turnover and rearrangements during or at the end of this period. This is consistent with the decreased number in functional categories observed from 14 dap onward and with a switch of the cellular machinery from cell division and tissue differentiation to storage compound synthesis. Degradation of gene products would allow a reallocation of amino acids to pathways involved in synthesis of storage products, particularly zeins. Accordingly, composite expression analysis showed that the accumulation of proteins classified in the protein synthesis category increased from 10 dap to 14 to 21 dap. Moreover, the preponderance of proteins involved in amino acid metabolism increased after 14 dap. The whole picture was consistent with the onset of storage protein synthesis between 10 and 14 dap.
Folding of nascent polypeptides into functional proteins is controlled by a number of molecular chaperones and protein-folding catalysts. Our analysis revealed 12 different isoforms of PDI, an endoplasmic reticulum-located protein that catalyzes the formation, isomerization, and reduction/oxidation of disulfide bonds (Houston et al., 2005). Twenty-seven chaperonins or chaperones were also observed, including the plant homolog of the immunoglobulin heavy-chain binding protein (BiP), which is an endoplasmic reticulum-localized member of the heat shock 70 family. BiP has been proposed to play a role in protein body assembly and retention of zeins within the endoplasmic reticulum (Boston et al., 1991; Vitale and Ceriotti, 2004). The accumulation pattern of PDI, chaperones, and chaperonins followed similar trends, with a relatively low level before 14 dap, a maximum at 14 to 21 dap, and a slight decrease up to 40 dap, in agreement with the dynamics of storage protein deposition.
Relative Changes in Glycolysis and TCA Cycle in Relation to Oxygen Supply
As compared to glycolysis for which 7 of 10 enzymatic functions were identified, we detected at most one-half of the enzymes involved in the TCA cycle. To check whether the pH 4 to 7 range used for protein separation could be responsible for such bias, we examined the function of proteins with a pI above 7 in Brassica napus grain filling (Hajduch et al., 2006). Considering the 517 listed proteins, 21% are out of the 4 to 7 range, but there are large differences according to the functional classes. As predictable, more than 50% of the storage proteins have basic pIs; thus, when excluding these proteins, the ratio falls down to 14.5%. The enzymes falling out of the 4 to 7 pH range appeared always to be supplementary isoforms of the one detected in the 4 to 7 pI range. Thus, the mitochondrial localization of the TCA cycle reactions could more satisfactorily explain that 50% of the enzymes were not detected in our analysis.
A striking observation is that glycolysis enzymes were grouped in the first three clusters, indicating relatively lower accumulation at the late stage, whereas the proportion of TCA enzymes is the highest in the early accumulation cluster and remains approximately constant afterward. The observed evolution of the global accumulation patterns of glycolysis enzymes is consistent with the absence of significant change in glycolytic intermediates from 12 to 42 dap as noted by Rolletschek et al. (2005). The pyruvate dehydrogenase complex that catalyzes the conversion of pyruvate to acetyl-CoA, a step linking the glycolytic pathway to the TCA cycle, showed decreased accumulation from 14 dap onward with a mean level twice reduced after 14 dap, probably leading to a decrease in enzymatic activity. This would suggest that, at late stages, an important part of the pyruvate synthesized by glycolysis does not join the TCA cycle.
These variations in accumulation profiles of glycolysis and TCA enzymes might be related to the heterogeneity of oxygen distribution in the developing kernel demonstrated by Rolletschek et al. (2005), keeping in mind that, because our proteome analysis was performed from whole-kernel extracts up to 10 dap and from whole endosperm from 14 dap, the profiles result from the admixture of different compartments. Most parts of maize kernels, especially endosperm, are subjected to low oxygen during development, an effect that is more drastic than in dicotyledonous seeds (Rolletschek et al., 2005). The low-oxygen condition strongly inhibits mitochondrial respiration because NADH oxidation along the electron transport chain is blocked when the final acceptor, oxygen, is missing. As a consequence, NAD dehydrogenases are rapidly inhibited, especially in the TCA cycle. NADH accumulation may be critical in endosperm because the fermentative pathway, which is the classical sink for NADH, presented no clear metabolic shift in kernel (Rolletschek et al., 2005). Accordingly, our results showed a moderate increase in alcohol dehydrogenase, a marker of fermentative glycolysis under low-oxygen stress (Sachs, 1991).
Thus, reserve synthesis takes place in the low-oxygen condition, which is likely to limit the respiration pathway to glycolysis. As highlighted by Rolletschek et al.(2005), starch synthesis is very well adapted to this condition because it does not require oxygen. As illustrated in Figure 7, the cleavage of imported Suc by Suc synthase (Susy), and the conversion of UDP-Glu into ADP-Glu, the starch synthase substrate, does not necessitate an external energy supply because the cosubstrates of Susy (UDP) and UDP-Glu pyrophosphorylase (inorganic pyrophosphate [PPi]) are regenerated through the transphosphorylation of UTP into ATP and the action of the ADP-Glu pyrophosphorylase (AGPase), respectively. The conversion of the Fru moiety into Glc-6-P requires ATP, which can be produced by the glycolysis pathway up to pyruvate formation. Actually, two pyruvates are produced from one Fru, with a net positive balance of two ATP and two NADH molecules.
Possible Role for PPDK in Starch versus Protein Balance
A main feature resulting from this proteomic analysis is the special time course of PPDK. It was nearly absent in early development, whereas its abundance is massively increased from 21 dap onward. Although this enzyme is classically involved in C4 photosynthesis, it was previously reported to be abundant in other cereal-developing kernels (Meyer et al., 1982; Aoyagi and Bassham, 1984; Gallusci et al., 1996). However, in developing rice, PPDK was mostly expressed in the syncytial/cellularization stage (Chastain et al., 2006) rather than during the second part of grain filling, as in other species. Its expression was also shown to be enhanced by low-oxygen stress in another sink organ, the rice root (Moons et al., 1998). Nonphotosynthetic PPDK catalyzes the reversible conversion of pyruvate, Pi, and ATP into phosphoenolpyruvate (PEP), AMP, and PPi. In the presence of pyruvate kinase (PK), an irreversible enzyme that converts PEP in pyruvate, PPDK, forms a so-called futile cycle. As summarized by Chastain et al. (2006), two nonexclusive roles may be assigned to this cycle (1) providing carbon skeletons for Ala-derived amino acids in the PEP-pyruvate direction; and (2) synthesizing PEP for aromatic amino acid synthesis and for PEP carboxylase reaction in the opposite direction. In the PEP-pyruvate direction, the reaction is parallel to PK reaction, but two ATPs are generated by PPDK versus one ATP by PK. In the pyruvate-PEP direction, PPDK produces one PPi and one AMP, further converted into ADP at the cost of one ATP.
Thus, the PPi-ATP balance is strongly dependent on the PPDK reaction, which may suggest a third role for PPDK. In maize kernels, expression of PPDK during the late accumulation of reserve in the endosperm could be linked to the balance of starch versus protein synthesis because it provides a means to increase PPi in the cytosol. In potato (Solanum tuberosum) tubers, this was shown to favor Susy activity and subsequently starch synthesis through the AGPase located in the amyloplast. A similar hypothesis was proposed from PPDK mutants in rice (Kang et al., 2005), but it might not be pertinent here because the enzyme is mainly expressed earlier than the filling phase (Chastain et al., 2006). The location of most of the maize endosperm AGPase activity in the cytosol totally changes the situation from potato because the AGPase reaction is fully reversible and the direction of the reaction only depends on the relative concentration of PPi and ATP. Thus, cytosolic PPi accumulation should push the reaction in the ADP-Glu degradation direction. By contrast, in the chloroplast or amyloplast, a very active pyrophosphatase directs the reaction toward ADP-Glu synthesis.
In addition to the possible restriction of starch accumulation, the onset of PPDK activity would favor PEP accumulation, which, together with pyruvate, plays a central role in amino acid metabolism. It should also be noted that, among the TCA cycle enzymes, aconitase increased twice at the end of endosperm development, mainly through the appearance of a late stage specific isoform. It can be assumed that aconitase operates at full rate to synthesize isocitrate, which could produce α-oxoglutarate because NAD(P)-specific isocitrate dehydrogenase is active. In this way, carbon skeletons would be provided to the Glu-Gln cycle and further on for Pro, an important component of zeins together with Gln (Wang and Larkins, 2001). The oxaloacetate needed for citrate synthesis could originate from the PEP carboxylase pathway, which was shown to be active in the maize kernel (Jeanneau et al., 2002). Experimental supports for the importance of the amino acid conversion in kernels have been provided by Pernollet et al. (1986). Looking for carbon fate from source leaves to kernels from photosynthetic 14CO2 labeling, they demonstrated that kernels were able to synthesize all the amino acids necessary for storage proteins from the two main incoming amino acids in phloem (i.e. Gln and Asn) and from carbon skeletons provided by carbohydrates through glycolysis. Oxygen levels may partly interact with amino acid and protein synthesis. Proteins are more concentrated in outer endosperm cells, like aleurone, where oxygen is more available. However, the zein protein deposition pattern described by Dolfini et al. (1992) extends into the anoxic region encountered very close to endosperm external layers (Rolletschek et al., 2005).
Thus, PPDK may play a central role in storage product composition, both through the PEP-pyruvate balance and the production of PPi. Maize cytosolic PPDK is encoded by two genes, one of which (CyPPDK1) is activated by the Opaque-2 transcription factor (Maddaloni et al., 1996). Actually, the products of both genes are most probably identified in our study because the seven identified spots were located in two groups of different pIs and slightly different Mr. The presence in each group of isoforms slightly differing in pI strongly suggests posttranslational modification by phosphorylation. Indeed, examination of the sequences of both gene products revealed several potential phosphorylation sites. Comparative analysis of 2-D gels of wild-type and o2 mutant endosperms supports the hypothesis that the more acidic group of spots is encoded by the CyPPDK1 gene, which is down-regulated by the mutation (Damerval and Le Guilloux, 1998). Opaque-2 is a basic Leu zipper transcriptional factor (Hartings et al., 1989; Schmidt et al., 1990) that is specifically expressed during endosperm development (Gallusci et al., 1994) and controls the transcription of several zein genes, namely, the 22-kD α-zeins and the 14-kD β-zeins (Schmidt et al., 1992; Cord Neto et al., 1995), and a variety of nonzein genes, including CyPPDK1 (Habben et al., 1993, 1995; Damerval and Le Guilloux, 1998). Thus, Opaque-2 would play a key role in storage compound accumulation during endosperm development by (1) activating PPDK synthesis, which in turn would control the pyruvate-PEP cycling and PPi production, thus restraining starch synthesis in the hypoxic endosperm region; and (2) activating zein synthesis in oxygen-richer regions, which would result in an increased demand in amino acids, especially pyruvate and PEP-derived amino acids. The time course of PPDK expression is also consistent with the continuation of protein synthesis when starch synthesis has slowed down. Zein accumulation begins around 12 dap and, whereas the rate declines from 21 dap, the accumulation does not stop before 70 dap. In addition, proteins involved in seed maturation like late-embryogenesis abundant proteins and dehydrins are mainly synthesized after 40 dap. The importance of this late protein synthesis activity is further supported by the sustained abundance of the chaperonins and folding proteins at late stages.
The data on Opaque-2 are in favor of the role of PPDK on storage protein composition, which could be explained, in a first step, through the regulation of the relative accumulation of Ala-Phe balance. However, the suggested additional role on the starch-protein balance could reinforce the effect. This second effect could be tested by several means: comparison of PPDK accumulation in high- or low-protein lines or in recombinant inbred lines (quantitative trait loci approach), analysis of PPDK mutator insertion mutants, and search for associations between natural genetic variability of PPDK genes and seed starch and protein content in a large set of inbred lines. Actually, search for colocalization between PPDK loci and quantitative trait loci for seed protein and starch give positive results in the maize database (www.maizegdb.org).
To conclude, examination of endosperm proteomic data along development in light of the physiological and biochemical function of the proteins leads to a very consistent interpretation of the observed changes and enables introduction of new testable hypotheses concerning the critical role of some processes. The importance of anoxia in understanding the observed shift of the biochemical pathways appears essential and a pivotal role of PPDK is put forward.
MATERIALS AND METHODS
Sampling
The Institut National de la Recherche Agronomique maize (Zea mays) inbred line F2 is becoming a reference genotype for many maize studies in Europe. F2 plants were grown in the field at Clermont-Ferrand (France, 63) in 2002. Plants were manually selfed in August. Three ears were harvested at each of the following developmental stages: 4, 7, 10, 14, 21, 30, and 40 dap to cover the main phases of endosperm development. At every stage, fertilized kernels from the middle part of each ear were sampled (Fig. 1A). Starting from 14 dap (as soon as kernels were developed sufficiently to allow easy dissection), kernels were dissected to remove the embryo and pericarp. At every stage, three samples were prepared by mixing an equal number of kernels from the three cobs. Samples were frozen in liquid nitrogen and stored at −80°C until protein extraction.
Total Protein Extraction, Solubilization, and Quantification
Total proteins of each of the 21 samples (seven developmental stages and three repetitions per stage) described above were extracted according to Damerval et al. (1986). Briefly, whole kernels or endosperms were ground in a mortar with a pestle in liquid nitrogen. The powder was resuspended in acetone with 0.07% (v/v) β-mercaptoethanol and 10% (w/v) TCA. Proteins were allowed to precipitate for 1 h at −20°C. Then the pellet was washed overnight with acetone containing 0.07% (v/v) β-mercaptoethanol. The supernatant was discarded and the pellet dried under vacuum.
Protein resolubilization was performed according to Méchin et al. (2003) using 50 μL/mg of R2D2 buffer (urea 5 m, thiourea 2 m, CHAPS 2%, SB3-10 2%, dithiothreitol 20 mm, Tris(2-carboxyethyl)phosphine hydrochloride 5 mm, ampholytes 0.75%). After resolubilization, samples were centrifuged and the supernatant was transferred to an Eppendorf tube prior to protein quantification.
Total protein content of each of the 21 samples was evaluated using the 2-D Quant kit (Amersham Biosciences).
2-DE, Gel Staining, and Image Analysis
Isoelectric focusing (IEF) was performed using 24-cm immobilized pH gradient (IPG) strips (Amersham Biosciences). Solubilized proteins were applied on an IPG strip for in-gel rehydration. Focusing was achieved using a Protean IEF cell (Bio-Rad). Active rehydration was performed at 22°C during 12 h at 50 V; then the focusing itself was achieved. For improved sample entry, the voltage was increased step by step from 50 to 10,000 V (0.5 h at 200 V, 0.5 h at 500 V, 1 h at 1,000 V, then 10,000 V for a total of 84,000 Vh).
After IEF, strips were equilibrated to improve protein transfer to the 2-D gel. The equilibrated strips were sealed at the top of the 1-mm-thick 2-D gel (24 × 24 cm) with the help of 1% low-melting agarose in SDS electrophoresis buffer (Tris 25 mm, Gly 0.2 m, and SDS 0.1%). Continuous gels (11% T, 2.67% C gels with piperazine diacrylyl as cross-linking agent) were used. Separation was carried out at 20 V for 1 h and subsequently at a maximum of 30 mA/gel, 120 V overnight, until bromphenol blue front had reached the end of the gel.
Following SDS-PAGE, gels were stained with silver nitrate, according to the procedure described in Méchin et al. (2003). Scanning was carried out at 300 dpi with a 16-bit grayscale pixel depth using an image scanner (Amersham Biosciences), and gel images were analyzed with Melanie 4 software (Swiss Institute of Bioinformatics). Automatically detected spots were manually checked and some modifications were done when necessary by modifying area selection, adding, or deleting spots. Following detection, the normalized volume (individual spot volume divided by the total volume of all detected spots in the considered gel) of every spot was computed for further analyses.
Selecting Nonzein Proteins
Our objective in this work was to analyze the developmental patterns of proteins involved in cell functioning and not to look at the accumulation of storage proteins. Zeins, the maize main storage proteins, progressively accumulate during endosperm development and thus gradually appear on the basic (pI > 7) side of the gels in a Mr range of 27 to 10 kD (Consoli and Damerval, 2001). To deposit the same quantity of nonzein proteins, we performed at each stage preliminary IEF, applying an equal amount of solubilized total proteins on an IPG strip with a linear pH gradient from 4 to 7, where zeins were not focused. We quantified the total spot volume on each 2-D gel by image analysis. This allowed us to determine the amount of nonzein protein at every stage and thus to ensure an equivalent loading of gels in nonzein proteins at all stages. Analytical IEF was then performed using 24-cm IPG strips with a linear pH gradient from 4 to 7 and equivalent nonzein protein loading of about 30 μg for every stage. This protein loading allowed saturation with silver staining to be limited to few very big spots, mainly at the latest developmental stages.
Protein Identification by Liquid Chromatography-MS/MS
A total of 632 protein spots were manually excised from the 14-dap endosperm Coomassie Blue-stained gel (Méchin et al., 2004). In-gel digestion was performed with a Progest system (Genomic Solution) according to a standard trypsin protocol. Briefly, after a washing step, gel particles were digested during 5 h with 125 ng of modified trypsin (Promega). The resulting peptides were extracted with 30 μL of 5% trifluoroacetic acid (TFA), 10% acetonitrile (ACN), then 30 μL of 0.2% TFA, 83% ACN. After drying in a vacuum centrifuge, peptide extracts were resuspended in 20 μL of 0.1% TFA, 3% ACN.
HPLC was performed with Ultimate liquid chromatography system combined with Famos autosample and Switchos II microcolumn switching for preconcentration (LC Packings). The sample was loaded on the column (PEPMAP C18, 5 μm, 75 μm i.d., 15 cm; LC Packings) using a preconcentration step on a micro precolumn cartridge (300 μm i.d., 5 mm). Five microliters of sample were loaded on a precolumn at 5 μL/min. After 3 min, the precolumn was connected with the separating column and the gradient was started at 200 nL/min. Buffers were 0.1% HCOOH, 3% ACN (A), and 0.1% HCOOH, 95% ACN (B). A linear gradient from 5% to 30% B for 25 min was applied. Including the regeneration step, one run was 60 min in length. The LCQ deca xp+ (Thermofinnigan) was used with a nano electrospray interface. Ionization (1.2–1.4 kV ionization potential) was performed with liquid junction and noncoated capillary probe (New Objective). Peptide ions were analyzed by the nth-dependent method as follows: (1) full MS scan (mass-to-charge ratio 500–1,500); (2) ZoomScan (scan of the two major ions with higher resolution); and (3) MS/MS of these two ions.
SEQUEST software (Thermofinnigan) was used to interpret MS/MS. Identification was performed with SEQUEST using protein sequences databases downloaded from the National Center for Biotechnology Information (NCBI; http://www.ncbi.nlm.nih.gov) and maize EST databases from Plant GDB (http://www.plantgdb.org). Peptides identified by SEQUEST were filtered according to their charge state, cross-correlation score (Xcorr > 1.7 for n + 1 and > 2.2 for n + 2), normalized difference in correlation score (ΔCn ≥ 0.2), and the tryptic nature of each peptide.
Among 409 protein spots retained for quantitative variation analysis during development, 121 were NI or considered as NYC function in our previous analysis (Méchin et al., 2004). A reinterrogation was done with SEQUEST using updated protein sequences downloaded from the NCBI and maize EST databases from Plant GDB. Peptides identified were filtered as described above. Fourteen protein spots were thus identified and assigned to functional categories following Schoof et al. (2002; see Supplemental Table S4).
Subcellular Localization
Subcellular localization of proteins presented in Figure 7 were predicted using four different programs: TargetP (Emanuelsson et al., 2000), iPSORT (Bannai et al., 2002), Predotar (Small et al., 2004), and WoLF PSORT (Horton et al., 2006). Information about subcellular localization was incorporated in Supplemental Table S3 if at least three programs predicted the same subcellular destination.
Experimental Design and Matching with the Reference Map
Each developmental stage was represented by three well-defined 2-D gels corresponding to the three independent protein extracts. For every stage, the image of the best-resolved gel was defined as a reference and the spots were automatically matched against this image; the quality of the matches was visually checked; a synthetic image was then built, which included all the spots that were present in at least two of the three repeats. The synthetic image obtained from the 14-dap stage was then used as a pivotal reference for matching against (1) the 14-dap endosperm Coomassie Blue-stained gel image used for the 2-D map (Méchin et al., 2004; Supplemental Fig. S1) and (2) the synthetic images of the six other stages.
Assessment of Quality of Quantification during Development
A total of 504 spots could be unambiguously matched between the 14-dap stage synthetic image and the 2-D map image. For every spot, a coefficient of variation was calculated at every stage. Three classes of quantification quality were defined: A (RSD ≤ 0.2), B (RSD between 0.2 and 0.45), and C (RSD > 0.45). Spots with three or more Cs were discarded (36 spots), and spots with 1 or 2 Cs (82) were visually checked over the whole experiment for quality of detection and matching. At the end of checking, 409 protein spots with good quantification properties at every stage were retained for statistical analyses (Supplemental Table S1).
Statistical Analyses
SAS package (Procedure GLM) was used to examine properties of individual spot volumes during development. For every spot, the mean normalized volume was then computed at each stage. Missing data at intermediary stages were estimated as linear interpolation between the adjacent stages.
Hierarchical clustering analysis was performed on standardized spot volumes using Gene Cluster software (M. Eisen; http://rana.stanford.edu/software) using centered correlation and the average linkage procedure. The resulting tree was visualized using the associated TreeView software, which gives the opportunity to visualize and export lists of individuals (in our case, protein spots) belonging to specific clusters. Homogeneity of clusters for distribution of functional categories was tested by χ2, grouping categories where theoretical numbers were below 5.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. 2-D reference map.
Supplemental Table S1. Individual expression data for 409 retained protein spots.
Supplemental Table S2. The 302 identified protein spots with their allocation to functional categories and cluster membership.
Supplemental Table S3. Proteins presented on Figure 7 with their subcellular localization prediction.
Supplemental Table S4. Summary of all proteins processed in the reference 2-D map, with allocation to functional categories.
Supplementary Material
Acknowledgments
We are grateful to Dr. Michael Hodges and Pr. Graham Noctor for their helpful and critical feedback on a draft of this manuscript. We also thank two anonymous reviewers for constructive comments on an earlier version of the manuscript.
This work was supported by the European Community in the context of the Zeastar European program (QLRT–2000–00020) and by a grant within the same program (to V.M.).
The authors responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) are: Valérie Méchin (mechin@grignon.inra.fr) and Catherine Damerval (damerval@moulon.inra.fr).
The online version of this article contains Web-only data.
References
- Aoyagi K, Bassham JA (1984) Pyruvate orthophosphate dikinase of C3 seeds and leaves as compared to the enzyme from maize. Plant Physiol 75 387–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannai H, Tamada Y, Maruyama O, Nakai K, Miyano S (2002) Extensive feature detection of N-terminal protein sorting signals. Bioinformatics 18 298–305 [DOI] [PubMed] [Google Scholar]
- Berger F (1999) Endosperm development. Curr Opin Plant Biol 2 28–32 [DOI] [PubMed] [Google Scholar]
- Boston RS, Fontes EBP, Shank BB, Wrobel RL (1991) Increased expression of the maize immunoglobulin binding protein homolog b-70 in three zein regulatory mutants. Plant Cell 3 497–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chastain CJ, Heck JW, Colquhoum TA, Voge DG, Gu XY (2006) Posttranslational regulation of pyruvate orthophosphate dikinase in developing rice (Oryza sativa) seeds. Planta 224 924–934 [DOI] [PubMed] [Google Scholar]
- Clarke BC, Hobbs M, Skylas D, Appels R (2000) Genes active in developing wheat endosperm. Funct Integr Genomics 1 44–55 [DOI] [PubMed] [Google Scholar]
- Consoli L, Damerval C (2001) Quantification of individual zein isoforms resolved by two-dimensional electrophoresis: genetic variability in 45 maize inbred lines. Electrophoresis 22 2983–2989 [DOI] [PubMed] [Google Scholar]
- Consonni G, Gavazzi G, Dolfini S (2005) Genetic analysis as a tool to investigate the molecular mechanisms underlying seed development in maize. Ann Bot (Lond) 96 353–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cord Neto G, Yunes JA, daSilva MJ, Vettore AL, Arruda P, Leite A (1995) The involvement of Opaque 2 on beta-prolamin gene regulation in maize and Coix suggests a more general role for this transcriptional activator. Plant Mol Biol 27 1015–1029 [DOI] [PubMed] [Google Scholar]
- Damerval C, de Vienne D, Zivy M, Thiellement H (1986) Technical improvements in two-dimensional electrophoresis increase the level of genetic variation detected in wheat seedling proteins. Electrophoresis 7 52–54 [Google Scholar]
- Damerval C, Le Guilloux M (1998) Characterization of novel target proteins of the o2 mutation expressed during maize endosperm development. Mol Gen Genet 257 354–361 [DOI] [PubMed] [Google Scholar]
- Dolfini SF, Landoni M, Tonelli C, Bernard L, Viotti A (1992) Spatial regulation in the expression of structural and regulatory storage-protein genes in Zea mays endosperm. Dev Genet 13 264–276 [Google Scholar]
- Emanuelsson O, Nielsen H, Brunak S, von Heijne G (2000) Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol 300 1005–1016 [DOI] [PubMed] [Google Scholar]
- Finnie C, Melchior S, Roepstorff P, Svensson B (2002) Proteome analysis of grain filling and seed maturation in barley. Plant Physiol 129 1308–1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallardo K, Job C, Groot SP, Puype M, Demol H, Vandekerckhove J, Job D (2002) Importance of methionine biosynthesis for Arabidopsis seed germination and seedling growth. Physiol Plant 116 238–247 [DOI] [PubMed] [Google Scholar]
- Gallardo K, Le Signor C, Vandekerckhove J, Thompson RD, Burstin J (2003) Proteomics of Medicago truncatula seed development establishes the time frame of diverse metabolic processes related to reserve accumulation. Plant Physiol 133 664–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallusci P, Salamini F, Thompson RD (1994) Differences in cell type-specific expression of the gene Opaque 2 in maize and transgenic tobacco. Mol Gen Genet 244 391–400 [DOI] [PubMed] [Google Scholar]
- Gallusci P, Varotto S, Matsuoko M, Maddaloni M, Thompson RD (1996) Regulation of cytosolic pyruvate, orthophosphate dikinase expression in developing maize endosperm. Plant Mol Biol 31 45–55 [DOI] [PubMed] [Google Scholar]
- Gygi SP, Rochon Y, Franza BR, Aebersold R (1999) Correlation between protein and mRNA abundance in yeast. Mol Cell Biol 19 1720–1730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habben JE, Kirleis AW, Larkins BA (1993) The origin of lysine-containing proteins in opaque-2 maize endosperm. Plant Mol Biol 23 825–838 [DOI] [PubMed] [Google Scholar]
- Habben JE, Moro GL, Hunter BG, Hamaker BR, Larkins BA (1995) Elongation factor 1α concentration is highly correlated with lysine content of maize endosperm. Proc Natl Acad Sci USA 92 8640–8644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajduch M, Casteel JE, Hurrelmeyer KE, Song Z, Agrawal GK, Thelen JJ (2006) Proteomic analysis of seed feeling in Brassica napus: developmental characterization of metabolic isozymes using high-resolution two-dimensional gel electrophoresis. Plant Physiol 141 32–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajduch M, Ganapathy A, Stein JW, Thelen JJ (2005) A systematic proteomic study of seed filling in soybean: establishment of high-resolution two-dimensional reference maps, expression profiles, and an interactive proteome database. Plant Physiol 137 1397–1419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halliwell B, Gutteridge JM (1990) The antioxidants of human extracellular fluids. Arch Biochem Biophys 280 1–8 [DOI] [PubMed] [Google Scholar]
- Hartings H, Maddaloni M, Lazzaroni N, Di Fonzo N, Motto M, Salamini F, Thompson R (1989) The O2 gene which regulates zein deposition in maize endosperm encodes a protein with structural homologies to transcriptional activators. EMBO J 8 2795–2801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton P, Park KJ, Obayashi T, Nakai K (2006) Protein subcellular localization prediction with WoLF PSORT. Proceedings of the Fourth Annual Asia Pacific Bioinformatics Conference APBC06, Taipei, Taiwan, pp 39–48
- Houston NL, Fan C, Xiang Q-Y, Schulze J-M, Jung R, Boston RS (2005) Phylogenetic analyses identify 10 classes of the protein disulfide isomerase family in plants, including single-domain protein disulfide isomerase-related proteins. Plant Physiol 137 762–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeanneau M, Vidal J, Gousset-Dupont A, Lebouteiller B, Hodges M, Gerentes D, Perez P (2002) Manipulating PEPC levels in plants. J Exp Bot 53 1837–1845 [DOI] [PubMed] [Google Scholar]
- Kang HG, Park S, Matsuoka M, An G (2005) White-core endosperm floury endosperm-4 in rice is generated by knockout mutations in the C-type pyruvate orthophosphate dikinase gene (OsPPDKB). Plant J 42 901–911 [DOI] [PubMed] [Google Scholar]
- Khan MM, Komatsu S (2004) Rice proteomics: recent developments and analysis of nuclear proteins. Phytochemistry 65 1671–1681 [DOI] [PubMed] [Google Scholar]
- Koller A, Washburn MP, Lange BM, Andon NL, Deciu C, Haynes PA, Hays L, Schieltz D, Ulaszek R, Wei J, et al (2002) Proteomic survey of metabolic pathways in rice. Proc Natl Acad Sci USA 99 11969–11974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovtun Y, Chiu WL, Tena G, Sheen J (2000) Functional analysis of oxidative stress-activated mitogen-activated protein kinase cascade in plants. Proc Natl Acad Sci USA 97 2940–2945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai J, Dey N, Kim CS, Bharti AK, Rudd S, Mayer KF, Larkins BA, Becraft P, Messing J (2004) Characterization of the maize endosperm transcriptome and its comparison to the rice genome. Genome Res 14 1932–1937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkins BA, Dilkes BP, Dante RA, Coelho CM, Woo YM, Liu Y (2001) Investigating the hows and whys of DNA endoreduplication. J Exp Bot 52 183–192 [PubMed] [Google Scholar]
- Leader DJ (2005) Transcriptional analysis and functional genomics in wheat. J Cereal Sci 41 149–163 [Google Scholar]
- Maddaloni M, Donini G, Balconi C, Rizzi E, Gallusci P, Forlani F, Lohmer S, Thompson R, Salamini F, Motto M (1996) The transcriptional activator Opaque-2 controls the expression of a cytosolic form of pyruvate orthophosphate dikinase-1 in maize endosperms. Mol Gen Genet 250 647–654 [DOI] [PubMed] [Google Scholar]
- Mayer U, Herzog U, Berger F, Inze D, Jürgens G (1999) Mutations in the PILZ group genes disrupt the microtubule cytoskeleton and uncouple cell cycle progression from cell division in Arabidopsis embryo and endosperm. Eur J Cell Biol 78 100–108 [DOI] [PubMed] [Google Scholar]
- Mayer U, Jürgens G (2002) Microtubule skeleton: a track record. Curr Opin Plant Biol 5 494–501 [DOI] [PubMed] [Google Scholar]
- Meagher RB, Fechheimer M (2003) The Arabidopsis cytoskeletal genome. In CR Somerville, EM Meyerowitz, eds, The Arabidopsis Book. American Society of Plant Biologists, Rockville, MD, pp 1–26 [DOI] [PMC free article] [PubMed]
- Méchin V, Balliau T, Chateau-Joubert S, Davanture M, Langella O, Negroni L, Prioul JL, Thevenot C, Zivy M, Damerval C (2004) A two-dimensional proteome map of maize endosperm. Phytochemistry 65 1609–1618 [DOI] [PubMed] [Google Scholar]
- Méchin V, Consoli L, Le Guilloux M, Damerval C (2003) An efficient solubilization buffer for plant proteins focused in immobilized pH gradients. Proteomics 3 1299–1302 [DOI] [PubMed] [Google Scholar]
- Meyer AO, Kelly GJ, Latzko E (1982) Pyruvate orthophosphate dikinase from the immature grains of cereal grasses. Plant Physiol 69 7–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon J, Parry G, Estelle M (2004) The ubiquitin-proteasome pathway and plant development. Plant Cell 16 3181–3195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moons A, Valcke R, Van Montagu M (1998) Low-oxygen stress and water deficit induce cytosolic pyruvate orthophosphate dikinase (PPDK) expression in roots of rice, a C3 plant. Plant J 15 89–98 [DOI] [PubMed] [Google Scholar]
- Neuffer MG, Sheridan WF (1980) Defective kernel mutants of maize. I. Genetic and lethality studies. Genetics 95 929–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen OA (2001) Endosperm development: cellularization and cell fate specification. Annu Rev Plant Physiol Plant Mol Biol 52 233–267 [DOI] [PubMed] [Google Scholar]
- Ostergaard O, Finnie C, Laugesen S, Roepstorff P, Svennson B (2004) Proteome analysis of barley seeds: identification of major proteins from two-dimensional gels (pI 4-7). Proteomics 4 2437–2447 [DOI] [PubMed] [Google Scholar]
- Ostergaard O, Melchior S, Roepstorff P, Svensson B (2002) Initial proteome analysis of mature barley seeds and malt. Proteomics 2 733–739 [DOI] [PubMed] [Google Scholar]
- Pernollet JC, Huet JC, Moutot F, Morotgaudry JF (1986) Relationships between photosynthesis and protein-synthesis in maize. 2. Interconversions of the photoassimilated carbon in the ear leaf and in the intermediary organs to synthesize the seed storage proteins and starch. Plant Physiol 80 216–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravanel S, Gakiere B, Job D, Douce R (1998) The specific features of methionine biosynthesis and metabolism in plants. Proc Natl Acad Sci USA 95 7805–7812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolletschek H, Koch K, Wobus U, Borisjuk L (2005) Positional cues for the starch/lipid balance in maize kernels and resource partitioning to the embryo. Plant J 42 69–83 [DOI] [PubMed] [Google Scholar]
- Sachs MM (1991) Molecular responses to anoxic stress in maize. In MB Jackson, DD Davies, H Lambers, eds, Plant Life under Oxygen Deprivation. Ecology, Physiology and Biochemistry. SPB Academic Publishing, The Hague, The Netherlands, pp 129–139
- Scandalios JG (1993) Oxygen stress and superoxide sismutases. Plant Physiol 101 7–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scanlon MJ, Myers AM (1998) Phenotypic analysis and molecular cloning of discolored-1 (dsc1), a maize gene required for early kernel development. Plant Mol Biol 37 483–493 [DOI] [PubMed] [Google Scholar]
- Scanlon MJ, Stinard PS, James MG, Myers AM, Robertson DS (1994) Genetic analysis of 63 mutations affecting maize kernel development isolated from mutator stocks. Genetics 136 281–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt RJ, Burr FA, Aukerman MJ, Burr B (1990) Maize regulatory gene opaque-2 encodes a protein with a “leucine-zipper” motif that binds to zein DNA. Proc Natl Acad Sci USA 87 46–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt RJ, Ketudat M, Aukerman MJ, Hoschek G (1992) Opaque-2 is a transcriptional activator that recognizes a specific target site in 22-kD zein genes. Plant Cell 4 689–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoof H, Zaccaria P, Gundlach H, Lemcke K, Rudd S, Kolesov G, Arnold R, Mewes HW, Mayer KF (2002) MIPS Arabidopsis thaliana database (MAtDB): an integrated biological knowledge resource based on the first complete plant genome. Nucleic Acids Res 30 91–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small I, Peeters N, Legeai F, Lurin C (2004) Predotar: a tool for rapidly screening proteomes for N-terminal targeting sequences. Proteomics 4 1581–1590 [DOI] [PubMed] [Google Scholar]
- Vensel WH, Tanaka CK, Cai N, Wong JH, Buchanan BB, Hurkman WJ (2005) Developmental changes in the metabolic protein profiles of wheat endosperm. Proteomics 5 1594–1611 [DOI] [PubMed] [Google Scholar]
- Verza NC, Neto TRES, Nogueira GC, Fisch FT, de Rosa PH, Jr VE, Rebello MM, Vettore AL, da Silva FR, Arruda P (2005) Endosperm-preferred expression of maize genes as revealed by transcriptome-wide analysis of expressed sequence tags. Plant Mol Biol 59 363–374 [DOI] [PubMed] [Google Scholar]
- Vierstra RD (2003) The ubiquitin/26S proteasome pathway, the complex last chapter in the life of many proteins. Trends Plant Sci 8 135–142 [DOI] [PubMed] [Google Scholar]
- Vitale A, Ceriotti A (2004) Protein quality control mechanisms and protein storage in the endoplasmic reticulum: a conflict of interests? Plant Physiol 136 3420–3426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Larkins BA (2001) Genetic analysis of amino acid accumulation in opaque-2 maize endosperm. Plant Physiol 125 1766–1777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson BS, Asirvatham VS, Wang L, Sumner LW (2003) Mapping the proteome of barrel medic (Medicago truncatula). Plant Physiol 131 1104–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.