Abstract
Plant morphogenesis is profoundly influenced by light (a phenomenon known as photomorphogenesis). For example, light inhibits seedling hypocotyl growth via activation of phytochromes and additional photoreceptors. Subsequently, information is transmitted through photoreceptor-linked signal transduction pathways and used (via previously unknown mechanisms) to control hypocotyl growth. Here we show that light inhibition of Arabidopsis (Arabidopsis thaliana) hypocotyl growth is in part dependent on the DELLAs (a family of nuclear growth-restraining proteins that mediate the effect of the phytohormone gibberellin [GA] on growth). We show that light inhibition of growth is reduced in DELLA-deficient mutant hypocotyls. We also show that light activation of phytochromes promotes the accumulation of DELLAs. A green fluorescent protein (GFP)-tagged DELLA (GFP-RGA) accumulates in elongating cells of light-grown, but not dark-grown, transgenic wild-type hypocotyls. Furthermore, transfer of seedlings from light to dark (or vice versa) results in rapid changes in hypocotyl GFP-RGA accumulation, changes that are paralleled by rapid alterations in the abundance in hypocotyls of transcripts encoding enzymes of GA metabolism. These observations suggest that light-dependent changes in hypocotyl GFP-RGA accumulation are a consequence of light-dependent changes in bioactive GA level. Finally, we show that GFP accumulation and quantitative modulation of hypocotyl growth is proportionate with light energy dose (the product of exposure duration and fluence rate). Hence, DELLAs inhibit hypocotyl growth during the light phase of the day-night cycle via a mechanism that is quantitatively responsive to natural light variability. We conclude that DELLAs are a major component of the adaptively significant mechanism via which light regulates plant growth during photomorphogenesis.
Plants sense specific characteristics of the light environment (including light quality, intensity, and duration of exposure) and hence can adaptively optimize growth and development in ways appropriate to prevailing environmental conditions. The potency of light as a modulator of plant developmental programs is powerfully illustrated by the extreme morphological differences that exist between plants grown in the light (and exhibiting photomorphogenesis) and plants grown in the dark (and exhibiting skotomorphogenesis; Kendrick and Kronenberg, 1994). These differences prompted the identification of Arabidopsis (Arabidopsis thaliana) mutants that are compromised in their ability to undergo normal photomorphogenesis. Many of these mutants were first identified because they display altered photomorphogenesis in an organ known as the hypocotyl. The Arabidopsis hypocotyl is an embryonic stem structure that elongates substantially during seed germination and seedling establishment. Hypocotyl elongation growth is the product of the expansion of hypocotyl cells and light inhibits hypocotyl growth by inhibiting hypocotyl cell expansion (Cowling and Harberd, 1999). Accordingly, wild-type Arabidopsis seedlings exhibit a short, relatively unexpanded hypocotyl when grown in the light, and a tall, expanded hypocotyl when grown in the dark. Mutant screens led to the identification of mutants that exhibit a tall hypocotyl in the light (for review, see Whitelam and Harberd, 1994) and initiated genetic analysis of photomorphogenesis.
These genetic studies, in combination with additional information from physiological and biochemical approaches, have brought us to our current advanced level of understanding of the mechanisms of light signaling in photomorphogenesis (for review, see Chen et al., 2004). For example, we now know that light is detected by several different families of plant photoreceptors, prominent among which are the red (R)/far-red (FR) light-detecting phytochromes and the blue light-detecting cryptochromes and phototropins (Quail, 2002; Chen et al., 2004). Downstream of photoreceptor function, signals are transduced by a network of specific (and shared) signaling pathways and then integrated at the level of growth regulation (Chen et al., 2004).
The R/FR-responsive phytochromes are perhaps the best understood plant photoreceptor system and play key roles in the regulation of growth and development throughout the plant life cycle (Quail, 2002). The Arabidopsis genome encodes five distinct phytochromes, phytochrome A to phytochrome E (phyA–phyE), among which phyA and phyB have particularly important functions in regulating seedling development in response to light (Quail, 2002). During hypocotyl growth, phyA exclusively confers responsivity to continuous FR light signals, whereas phyB primarily confers responsivity to continuous R light signals. Thus, a phyA mutant (lacking phyA function) displays an elongated hypocotyl in continuous FR light, whereas a phyB mutant displays an elongated hypocotyl in continuous white light and R light, but not in FR light (Dehesh et al., 1993; Nagatani et al., 1993; Whitelam et al., 1993). In wild-type plants, continuous FR light activates the phyA photoreceptor and thus inhibits hypocotyl growth, whereas continuous R light inhibits hypocotyl growth via activation of the phyB photoreceptor. However, the mechanisms via which hypocotyl growth is inhibited in response to photoreceptor activation remain unclear.
To further understand the mechanism of light-mediated hypocotyl growth inhibition, we focused our attention on the gibberellin (GA)-DELLA growth regulatory system. The phytohormone GA is well known to be a promoter of growth during various phases of the plant life cycle: Seed germination, vegetative growth, initiation of flowering, floral, and fruit development are all stimulated by GA (Richards et al., 2001). In addition, GA is known to control the growth of Arabidopsis hypocotyls in both light and dark (Reed et al., 1996; Cowling and Harberd, 1999). GA-deficient mutant Arabidopsis plants are dwarfed, late flowering, and infertile, whereas treatment with GA restores normal growth (Richards et al., 2001). GA promotes growth by overcoming the effects of the DELLA proteins (DELLAs; Peng et al., 1997; Silverstone et al., 1998; Dill and Sun, 2001; King et al., 2001), which are nuclear growth repressors, a subset of the GRAS family of candidate transcriptional regulators (Richards et al., 2001; Bolle, 2004). The Arabidopsis DELLAs comprise GAI, RGA, RGL1, RGL2, and RGL3 (Peng et al., 1997; Dill and Sun, 2001; Silverstone et al., 2001; Lee et al., 2002; Wen and Chang, 2002), and recent genetic analyses have revealed both distinct and overlapping functions for individual DELLAs in the regulation of plant development (Lee et al., 2002; Cheng et al., 2004; Tyler et al., 2004; Yu et al., 2004; Achard et al., 2006).
According to the relief-of-restraint model (Harberd, 2003), DELLAs restrain plant growth, whereas GA promotes growth by overcoming DELLA-mediated growth restraint (Harberd et al., 1998; Dill and Sun, 2001; King et al., 2001; Silverstone et al., 2001; Harberd, 2003). GA relieves DELLA restraint by promoting the disappearance of nuclear DELLAs (Silverstone et al., 2001; Fleck and Harberd, 2002). In Arabidopsis, GA is detected by the GA receptor AtGID1 (Nakajima et al., 2006). Binding of GA to AtGID1 promotes interaction of AtGID1 with the DELLAs (Nakajima et al., 2006), subsequent polyubiquitination of the DELLAs via the E3 ubiquitin-ligase SCFSLY1, and eventual destruction of DELLAs in the 26S proteasome (McGinnis et al., 2003; Sasaki et al., 2003; Dill et al., 2004; Fu et al., 2004). Thus, DELLAs restrain plant growth, whereas GA promotes growth by targeting the DELLAs for destruction (Harberd, 2003).
Whereas several previous studies have linked GA-mediated growth regulation with photomorphogenesis (e.g. Reed et al., 1996; Peng and Harberd, 1997; Ait-ali et al., 1999; Alabadí et al., 2004), none have systematically investigated the role of DELLAs in light-mediated growth regulation. Recent advances have led to the development of Arabidopsis mutant lines that are multiply DELLA deficient and to the use of these lines in experiments that reveal the role of DELLAs in many aspects of plant growth and development, including those that are influenced by environmental conditions (Lee et al., 2002; Cheng et al., 2004; Yu et al., 2004; Cao et al., 2005; Achard et al., 2006). Here we describe experiments that use these and other materials to determine whether DELLAs contribute to light-mediated hypocotyl growth inhibition. Essentially, we show that multiply DELLA-deficient hypocotyls have reduced capacity for light-induced hypocotyl growth inhibition and that light promotes the accumulation of the green fluorescent protein (GFP)-tagged DELLA RGA. We also show that the overlapping and distinct light-signaling pathways that originate with phyA and phyB both impact on DELLA function in the regulation of growth that characterizes photomorphogenesis. Thus, in nature, DELLAs integrate the effects on growth of the distinct signaling pathways activated following photoactivation of phyA and phyB. We also show that DELLAs regulate hypocotyl growth in response to light energy dose by integrating information that reflects irradiance (fluence rate) and duration of light exposure. Our demonstration that the abundance of transcripts encoding enzymes of GA metabolism is rapidly altered by light-dark transitions suggests that light alters bioactive GA levels and regulates DELLA activity accordingly. We conclude that DELLA restraint is a component of the mechanism via which light-signaling pathways inhibit hypocotyl elongation and propose that DELLAs contribute significantly to the regulation of growth that characterizes photomorphogenesis as a whole.
RESULTS
Light Inhibits Hypocotyl Growth via the GA-DELLA Signaling Mechanism
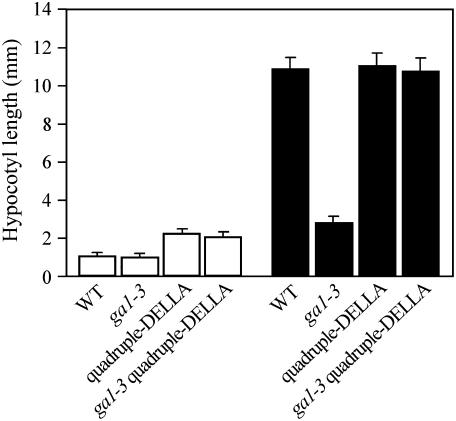
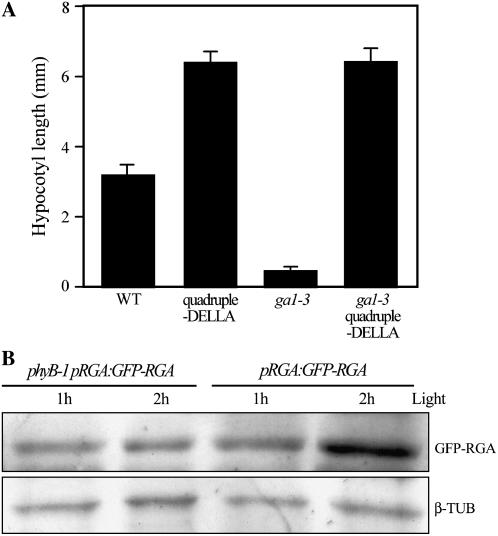
We further investigated the relationship between GA and light in the regulation of hypocotyl growth in experiments using the Arabidopsis ga1-3 mutant (a mutant that is substantially GA deficient; Sun et al., 1992; Sun and Kamiya, 1994; King et al., 2001). Whereas wild-type seedlings grown in continuous light had shorter hypocotyls than dark-grown controls, the differences between light- and dark-grown ga1-3 mutant hypocotyl lengths were considerably smaller (Fig. 1). Furthermore, ga1-3 hypocotyls grown in continuous light were of similar length to wild-type controls (Fig. 1), whereas dark-grown ga1-3 hypocotyls were markedly shorter than dark-grown wild-type hypocotyls (Fig. 1; see also Cowling and Harberd, 1999; Alabadí et al., 2004). Thus, GA plays a particularly prominent role in the regulation of hypocotyl growth in the dark.
Figure 1.
Light inhibits hypocotyl growth via a DELLA-dependent mechanism. Comparison of hypocotyl lengths of 7-d-old wild-type, ga1-3, quadruple-DELLA mutant, and ga1-3 quadruple-DELLA mutant seedlings grown in white light (white bar) or in the dark (black bar). Results are presented as means; error bars represent se.
GA promotes many aspects of the growth of plants by targeting the growth-restraining DELLAs for destruction in the cellular proteasome, thus overcoming DELLA restraint of growth (Harberd, 2003; McGinnis et al., 2003; Sasaki et al., 2003; Dill et al., 2004; Fu et al., 2004). To determine whether GA promotes hypocotyl growth by this same mechanism, we studied multiple mutant lines that are substantially lacking in DELLA function. The Arabidopsis genome encodes five DELLAs (GAI, RGA, RGL1, RGL2, and RGL3; Chen et al., 2004). The quadruple-DELLA mutant line lacks GAI, RGA, RGL1, and RGL2 (but retains RGL3), whereas the ga1-3 quadruple-DELLA line lacks the same set of DELLAs on the ga1-3 background (Cao et al., 2005; Achard et al., 2006). We found that continuous light-grown quadruple-DELLA and ga1-3 quadruple-DELLA mutant hypocotyls grew to approximately twice the length of wild type and ga1-3 controls (Fig. 1). Furthermore, substantial lack of DELLA function (as in the ga1-3 quadruple-DELLA line) completely suppressed the short-hypocotyl phenotype of dark-grown ga1-3 seedlings (Fig. 1; see also Alabadí et al., 2004). Thus, DELLA function restrains hypocotyl growth in both light and dark.
DELLA-Dependent Light-Mediated Regulation of Hypocotyl Growth Is Affected by Photoperiod Duration
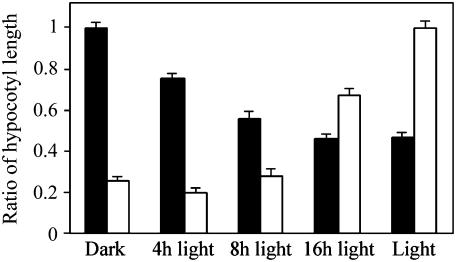
In nature, plants are exposed to the alternating periods of light and darkness that correspond to the day-night cycle. We next found that DELLAs inhibit hypocotyl growth to a degree proportionate to daylength (Fig. 2). Whereas wild-type and quadruple-DELLA mutant hypocotyl length was identical in darkness (wild-type/quadruple-DELLA hypocotyl length ratio approximating 1), the difference in hypocotyl length between these two lines increases progressively with increase in daylength (Fig. 2). Conversely, whereas the lengths of ga1-3 and wild-type hypocotyls were very different in darkness (ga1-3/wild-type hypocotyl length ratio approximating 0.2), the difference in hypocotyl length between these two lines decreases progressively as daylength increases (Fig. 2). Thus, in wild-type plants, increases in daylength cause increases in both hypocotyl growth inhibition and the relative contribution that DELLA restraint makes to that inhibition. Conversely, in GA-deficient plants, increases in daylength progressively reduce the growth-inhibitory effects of GA deficiency on hypocotyl growth, suggesting that GA becomes less critical for hypocotyl growth as the light period increases.
Figure 2.
DELLA-dependent light-mediated inhibition of hypocotyl growth is affected by photoperiod duration. Comparison of hypocotyl length ratios: wild type/quadruple-DELLA mutant (black bar) versus ga1-3/wild type (white bar) grown in different photoperiods as indicated. Results are presented as the ratio between the means of hypocotyl length; error bars represent se.
Light-Mediated Hypocotyl Growth Inhibition Is Associated with DELLA Accumulation
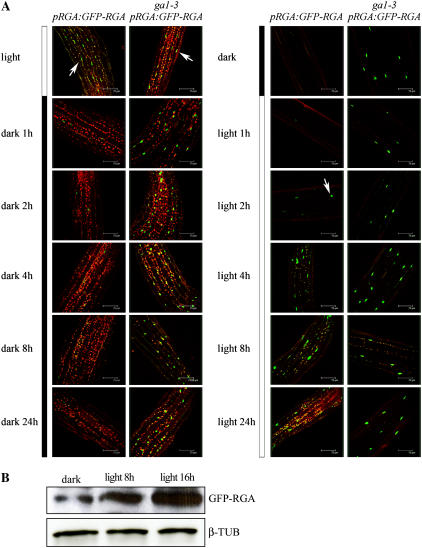
We next showed that DELLA-dependent light-induced hypocotyl growth inhibition is associated with DELLA accumulation. pRGA:GFP-RGA seedlings express a GFP-RGA fusion protein that is detectable in root cell nuclei and destroyed rapidly in response to GA treatment (Silverstone et al., 2001; Achard et al., 2003). Because light inhibits hypocotyl growth via a DELLA-dependent mechanism, we determined the effect of light on GFP-RGA in nuclei of elongating cells of the hypocotyl. We found that nuclear fluorescence attributable to GFP-RGA was readily detectable in the elongation zone of hypocotyls of light-grown pRGA:GFP-RGA seedlings, but not in hypocotyls of dark-grown controls (Fig. 3A). These observations suggest that RGA accumulates in nuclei of elongation zone cells in light-grown (but not in dark-grown) wild-type hypocotyls, thus contributing to light-mediated inhibition of hypocotyl growth. In contrast, GFP-RGA was clearly detectable in both light-grown and dark-grown ga1-3 pRGA:GFP-RGA hypocotyls (Fig. 3A). The consistency of these observations with the previous hypocotyl growth data (Fig. 1) suggests that wild-type and ga1-3 hypocotyls are of similar length when grown in continuous light because they sustain a similar level of DELLA accumulation. Conversely, dark-grown wild-type and ga1-3 hypocotyls are of different lengths largely because DELLAs accumulate in dark-grown ga1-3 hypocotyls, but not in wild-type controls.
Figure 3.
Light promotes accumulation of GFP-RGA in seedling hypocotyls. A, GFP fluorescence in nuclei of elongating cells of hypocotyls of 7-d-old pRGA:GFP-RGA or ga1-3 pRGA:GFP-RGA seedlings grown in white light or in the dark and then transferred to dark or light, respectively; times as indicated. Arrows indicate nuclear GFP-RGA fluorescence. B, Immunodetection of GFP-RGA in hypocotyls of pRGA:GFP-RGA seedlings grown in the dark for 5 d and then transferred to white light for 8 or 16 h. β-Tubulin serves as a sample-loading control.
In subsequent experiments, we determined the kinetics of change in a detectable level of nuclear GFP-RGA fluorescence following transfer of plants from light to dark (and following the reciprocal transfer; Fig. 3A). We found that GFP-RGA fluorescence in hypocotyl nuclei had largely disappeared within 1 h of transfer of pRGA:GFP-RGA seedlings from light to dark, and that GFP-RGA remained undetectable during subsequent hours of darkness (Fig. 3A). Conversely, hypocotyls of dark-grown seedlings began to exhibit detectable nuclear GFP-RGA fluorescence within 2 h of onset of light exposure, with subsequent increase in nuclear GFP-RGA level that plateaued at 4 h and beyond (Fig. 3A). Thus, the rate at which GFP-RGA is destroyed in darkness appears to be faster than the rate at which it accumulates in the light. These dynamic light-dependent changes in GFP-RGA level are not seen in ga1-3 pRGA:GFP-RGA hypocotyls (Fig. 3A), presumably because they are dependent on the modulation of GA level (Silverstone et al., 2001). In addition, we found that these effects of light on GFP-RGA accumulation were a consequence of light itself and were not circadian clock dependent (data not shown).
We confirmed that the increase in hypocotyl cell GFP-RGA fluorescence that occurred following transfer of seedlings from dark to light (Fig. 3A) was genuinely associated with an increase in GFP-RGA protein level via immunodetection of GFP-RGA (using an anti-GFP antibody; Fig. 3B). This experiment revealed a progressive increase in the level of immunologically detectable GFP-RGA that was proportionate to the duration of light exposure (Fig. 3B).
Light Regulates the Levels of Transcripts Encoding Enzymes of GA Metabolism
Reduced bioactive GA level (as in ga1-3) causes an increase in DELLA accumulation and consequent growth inhibition (e.g. Silverstone et al., 2001; Fu et al., 2004). We next investigated the possibility that light-induced accumulation of DELLAs in seedling hypocotyls might be the result of light-induced effects on GA metabolism, thus causing a reduction in the levels of bioactive GA. Indeed, several previous reports indicate that light affects GA metabolism via modulation of the levels of gene transcripts encoding GA biosynthesis enzymes (e.g. Yamaguchi et al., 1998; García-Martinez and Gil, 2001). Bioactive GA levels are increased by increasing the level of transcripts encoding GA 20-oxidase (GA20ox) and GA 3-oxidase (GA3ox), enzymes that catalyze successive steps in the synthesis of bioactive GA (Chiang et al., 1995; Phillips et al., 1995). Conversely, bioactive GA levels are reduced by increasing the level of transcripts encoding GA 2-oxidase (GA2ox), an enzyme that deactivates bioactive GA (Thomas et al., 1999).
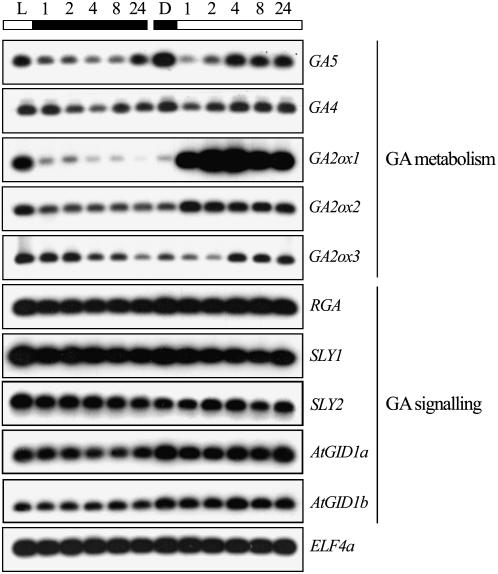
Previous experiments have shown that light (phytochrome-mediated R/FR light) regulates GA metabolism genes during Arabidopsis seed germination (Yamaguchi et al., 1998). We therefore determined the effects of light on the accumulation of selected GA metabolism gene transcripts in Arabidopsis seedling hypocotyls. We found that hypocotyls grown in the dark accumulate relatively high levels of GA20ox1 (encoded by the GA5 gene) and low levels of GA2ox transcripts (Fig. 4). Conversely, when grown in continuous light, hypocotyls contain relatively high amounts of GA2ox transcripts (particularly with respect to the GA2ox1 transcript) and accumulate relatively low levels of GA5. These observations indicate that the biosynthesis and accumulation of bioactive GA is promoted in dark-grown seedling hypocotyls (due to a combination of high GA20ox and low GA2ox activity), whereas deactivation of bioactive GA is promoted in dark-grown seedling hypocotyls (due to the combination of low GA20ox and high GA2ox activity). Thus, in dark-grown hypocotyls, the relatively high level of bioactive GA targets DELLAs for destruction (Fig. 3A) and promotes hypocotyl growth (Fig. 1). Conversely, in light-grown hypocotyls, the relatively low level of bioactive GA favors DELLA accumulation, thus promoting repression of hypocotyl growth.
Figure 4.
Light regulates the transcript level of GA metabolism genes. Levels of GA metabolism GA5 (GA20ox1), GA4 (GA3ox1), GA2ox1, GA2ox2, and GA2ox3, and GA-signaling RGA, SLY1, SLY2, AtGID1a, AtGID1b gene transcripts (determined by reverse transcription-PCR) in 7-d-old seedling hypocotyls grown in white light (L) or in the dark (D), then transferred to dark or light, respectively, for the time as indicated (1–24 h). ELF4a transcripts provide loading control.
We also examined the dynamics of GA metabolism gene transcript levels in seedlings transferred from dark to light (or vice versa). Movement of seedlings from dark to light resulted, within 1 h, in pronounced accumulation of GA2ox1 transcripts (and, to a lesser extent, GA2ox2 and GA2ox3 transcripts) and a corresponding decrease in GA5 and GA4 (which encodes for the GA3ox1 enzyme) transcript levels in seedling hypocotyls. This sudden change in gene transcript levels presumably has two consequences. First, the increase in GA2ox transcript levels results in rapid deactivation of the bioactive GA that was produced during the preceding dark period. Second, the decrease in GA5 and GA4 transcript levels reduces the production of further active GA.
Previous work has shown that increased DELLA activity (e.g. as seen in the GA-deficient ga1-3 mutant or in the constitutively DELLA-restraining gai mutant) results in increased levels of GA4 and GA5 transcripts due to perturbation of a DELLA-dependent negative feedback loop (Cowling et al., 1998; Xu et al., 1999). It is possible that the slight increase in GA4 and GA5 transcript levels observed between 2 and 4 h of light exposure (Fig. 4) is due to such a mechanism. However, the light/dark-mediated sudden changes in levels of GA metabolism gene transcripts described above are consistent with the hypothesis that light-dependent changes in bioactive GA level contribute to the DELLA-dependent effect of light on hypocotyl growth.
Light Has No Detectable Effect on the Levels of Transcripts Encoding GA-Signaling Components
In addition to the above-described changes in GA metabolism, the light-mediated GFP-RGA accumulation demonstrated in Figure 3 might also have been the consequence of light-dependent modulation of the levels of transcripts encoding components of the GA-signaling pathway. As outlined in the introduction, GA is perceived by AtGID1 receptors (Nakajima et al., 2006), thus enhancing the affinity between the DELLAs and a specific SCF E3 ubiquitin-ligase complex, involving the F-box proteins SLEEPY1 (SLY1) and SLEEPY2 (SLY2; Fu et al., 2004; Strader et al., 2004) and promoting the eventual destruction of DELLAs in the 26S proteasome. We therefore analyzed the accumulation of transcripts encoding RGA, SLY1, SLY2, AtGID1a, and AtGID1b in dark- and light-grown seedlings. As shown in Figure 4, exposure to light or dark had no detectable effect on the levels to which these transcripts accumulated. Thus, the accumulation of the GFP-RGA signal in light-treated seedlings (Fig. 3) is unlikely to be due to light-induced enhancement of GFP-RGA transcript levels or to a decrease in SLY1, SLY2, AtGID1a, or AtGID1b transcript levels.
Activated Phytochrome Inhibits Hypocotyl Growth by Promoting Accumulation of DELLAs in Response to Light
We next began to characterize those regions of the light spectrum that contribute to DELLA-dependent light-mediated inhibition of hypocotyl growth, first assaying the FR and R light regions of the spectrum that are particularly associated with the phytochrome family of photoreceptors (Quail, 2002).
As shown previously for continuous white light (Fig. 1), we found that quadruple-DELLA mutant hypocotyls were longer than wild-type hypocotyls in both FR and R (Figs. 5A and 6A). Furthermore, the short-hypocotyl phenotype exhibited by ga1-3 in R was suppressed in the ga1-3 quadruple-DELLA line (Fig. 6A). Thus, there is a DELLA-dependent component to the mechanism via which both R and FR inhibit hypocotyl growth.
Figure 5.
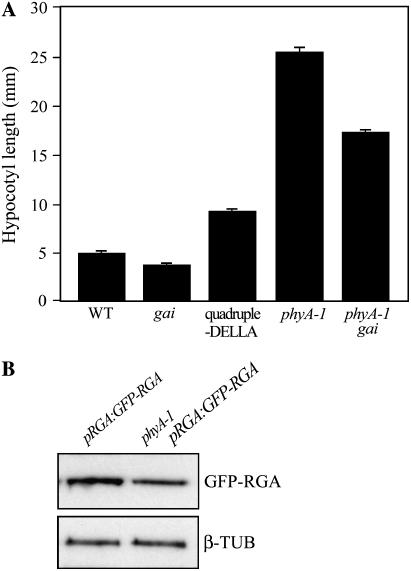
Involvement of DELLAs in PHYA-dependent FR-mediated inhibition of hypocotyl growth. A, DELLAs contribute to the activated PHYA-mediated inhibition of hypocotyl growth. Mean (±se) hypocotyl length of 7-d-old wild type; gai; quadruple-DELLA mutant; phyA-1 and phyA-1 gai seedlings grown in FR light. B, Immunodetection of GFP-RGA in 7-d-old pRGA:GFP-RGA and phyA-1 pRGA:GFP-RGA seedlings grown in continuous FR light. β-Tubulin (β-TUB) serves as a sample-loading control.
Figure 6.
DELLAs contribute to the activated PHYB-mediated inhibition of hypocotyl growth. A, Mean (±se) hypocotyl length of 7-d-old wild type, quadruple-DELLA, ga1-3, and ga1-3 quadruple-DELLA seedlings grown in R light. B, Immunodetection of GFP-RGA in hypocotyls of 4-d-old pRGA:GFP-RGA and phyB-1 pRGA:GFP-RGA seedlings grown in the dark and then transferred to white light for 1 or 2 h as indicated. β-Tubulin (β-TUB) serves as a sample-loading control.
The phyA photoreceptor is the predominant detector of FR during seedling hypocotyl growth. Accordingly, phyA-1 loss-of-function mutant hypocotyls are longer than wild-type hypocotyls in FR (Whitelam et al., 1993; Johnson et al., 1994; Fig. 5A). To further characterize the role of DELLAs in phyA-mediated hypocotyl growth inhibition, we studied the GA response mutant gai (Peng and Harberd, 1993). The gai mutant allele encodes a mutant gai form of GAI that is relatively resistant to the effects of GA (Peng et al., 1997; Fu et al., 2004) and thus constitutively represses growth. The length of FR-grown phyA-1 gai double-mutant hypocotyls was intermediate between that of phyA-1 and gai (Fig. 5A). Thus, the constitutive growth repression conferred by gai reduces FR-grown phyA-1 mutant hypocotyl growth, further indicating that FR-activation of phyA inhibits hypocotyl growth via a mechanism that contains a DELLA-dependent component. The fact that FR-grown gai and phyA-1 gai hypocotyls are not of identical length indicates the existence of an additional DELLA-independent mechanism via which activated phyA inhibits hypocotyl growth. In further experiments, we determined the effect of the phyA-mediated FR signal on the accumulation of GFP-RGA (Fig. 5B). GFP-RGA accumulated to higher levels in FR-treated pRGA:GFP-RGA seedlings than in FR-treated phyA-1 pRGA:GFP-RGA seedlings, thus indicating that FR modulation of GFP-RGA levels is at least partially phyA dependent.
Whereas phyA inhibits hypocotyl growth in FR, phyB plays a predominant role in inhibition of hypocotyl growth in R and white light (Reed et al., 1993; Whitelam et al., 1993). We next investigated the role of phyB in the DELLA-dependent light-mediated inhibition of seedling hypocotyl growth, using the phyB-1 mutant (which lacks phyB function; Reed et al., 1993). In these experiments, we compared the kinetics of GFP-RGA accumulation following the transfer of pRGA:GFP-RGA and phyB-1 pRGA:GFP-RGA seedlings from the dark to white light (as shown in Fig. 3). Whereas immunodetectable GFP-RGA levels did not obviously increase in phyB-1 pRGA:GFP-RGA hypocotyls in the period between 1 and 2 h following onset of light exposure, pRGA:GFP-RGA seedlings displayed a large increase in GFP-RGA level during this period (Fig. 6B). Thus, lack of phyB delays light-induced accumulation of GFP-RGA.
Taken together, the above results indicate that an activated phytochrome (both phyA and phyB) contributes to the light-promoted accumulation of DELLAs. In turn, DELLA accumulation promotes light-mediated hypocotyl growth inhibition.
Hypocotyl Growth Inhibition and GFP-RGA Accumulation Are Light Dose Dependent
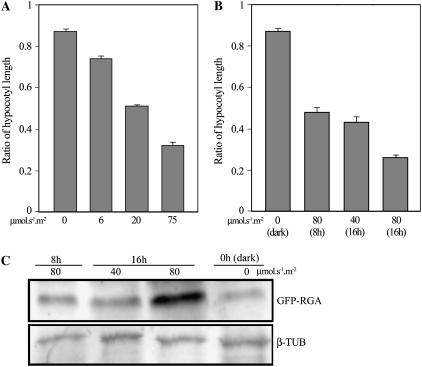
The natural light environment varies with respect to photoperiod duration and fluence rate, the total light dose that plants receive being the product of these two variables. We next investigated the effect of variation in fluence rate on light-mediated DELLA-dependent hypocotyl growth inhibition by comparing the growth of wild-type and quadruple-DELLA mutant seedling hypocotyls in different fluence rates of continuous light. At low fluence rate (6 μmol s−1 m−2), we found that wild-type hypocotyls were a little shorter than quadruple-DELLA mutant hypocotyls (wild type/quadruple-DELLA hypocotyl length ratio of approximately 0.75; Fig. 7A). As fluence rate increased, wild-type hypocotyls became progressively shorter than quadruple-DELLA mutant hypocotyls, such that at high fluence rate (75 μmol s−1 m−2) wild-type hypocotyls were considerably shorter than quadruple-DELLA hypocotyls (wild type/quadruple-DELLA hypocotyl length ratio of approximately 0.3; Fig. 7A). Thus, the DELLA-dependent component of light-mediated hypocotyl growth inhibition becomes increasingly prominent with increased fluence rate.
Figure 7.
DELLA inhibition of hypocotyl growth is proportionate to light dose. A, Comparison of wild type/quadruple-DELLA mutant hypocotyl length ratio for 7-d-old seedlings grown in dark (0 μmol s−1 m−2) or in the presence of continuous white light at fluence rates of 6, 20, or 75 μmol s−1 m−2. B, Comparison of wild type/quadruple-DELLA mutant hypocotyl length ratio for 7-d-old seedlings grown in photoperiod (light duration per 24-h cycle)/fluence rate combinations as indicated. C, Immunodetection of GFP-RGA in hypocotyls of 5-d-old pRGA:GFP-RGA seedlings grown in photoperiod (light duration per 24-h cycle)/fluence rate combinations as indicated. β-Tubulin (β-TUB) serves as a sample-loading control.
We next investigated the combined effect of variation in fluence rate and photoperiod duration on hypocotyl growth (Fig. 7B) and found that the wild type/quadruple-DELLA hypocotyl length ratio decreases proportionately with an increase in light energy dose (the product of photoperiod duration and fluence rate). For example, the wild type/quadruple-DELLA hypocotyl length ratio was approximately the same when seedlings were exposed to 80 μmol s−1 m−2 for an 8-h photoperiod or 40 μmol s−1 m−2 for a 16-h photoperiod, different combinations of fluence rate and exposure duration that result in the same energy dose (Fig. 7B). In contrast, when the photoperiod was maintained (16 h), but the fluence rate increased 2-fold (40–80 μmol s−1 m−2; Fig. 7B), the wild type/quadruple-DELLA hypocotyl length ratio decreased 2-fold (Fig. 7B). Thus, the degree to which DELLAs contribute to hypocotyl growth inhibition is proportionate to the light energy dose. In accord with this conclusion, we found that accumulation of immunodetectable GFP-RGA was also proportionate to light dose (Fig. 7C). For example, the degree of GFP-RGA accumulation remained the same when the photoperiod was increased 2-fold and fluence rate halved (compare 80 μmol s−1 m−2 per 8-h photoperiod with 40 μmol s−1 m−2 per 16-h photoperiod; Fig. 7C). Conversely, maintenance of photoperiod with increase in fluence rate (compare 40 μmol s−1 m−2 per 16-h photoperiod with 80 μmol s−1 m−2 per 16-h photoperiod; Fig. 7C) resulted in a clearly detectable increase in immunodetectable GFP-RGA. Taken together, the observations in Figure 7 indicate that DELLAs accumulate in quantitative proportion to the light energy to which seedlings are exposed and that DELLAs therefore contribute to light-mediated inhibition of hypocotyl growth in a light dose-dependent fashion.
DISCUSSION
It has recently become apparent that DELLAs integrate endogenous and environmental cues in the regulation of plant growth (Achard et al., 2003, 2006; Fu and Harberd, 2003; Alvey and Harberd, 2005). DELLAs are nuclear growth repressors and GA relieves DELLA restraint by promoting the destruction of DELLAs in the proteasome (Silverstone et al., 2001; Dill et al., 2004; Fu et al., 2004). Thus, DELLAs integrate a growth response output to the diverse signaling pathway inputs that alter GA levels.
Light plays a major role in the environmental regulation of growth and development, controlling growth at almost every stage of the life cycle from seed germination to floral induction. The effects of light are particularly dramatic during seedling establishment when light inhibits hypocotyl growth. Previous studies have shown GAs promote skotomorphogenetic growth of Arabidopsis and pea (Pisum sativum) seedlings grown in darkness (Cowling and Harberd, 1999; Alabadí et al., 2004). Furthermore, a rapid decrease in the levels of bioactive GA1 has been observed upon transfer of etiolated pea seedlings into light (Ait-ali et al., 1999; Gil and García-Martinez, 2000; O'Neill et al., 2000). Here we have shown that Arabidopsis GA metabolism gene transcript levels change rapidly in response to changes in the light environment. Whereas GA4 and GA5 transcripts (encoding GA3ox1 and GA20ox1 enzymes, respectively) are down-regulated within 1 h of exposure of seedling hypocotyls to light, transcripts encoding the GA2ox1 enzyme are up-regulated within the same period of time. Conversely, GA4 and GA5 transcripts are up-regulated and transcripts encoding GA2ox1 are down-regulated when seedling hypocotyls are moved from light to dark. Accordingly, bioactive GA levels are expected to be relatively low in light-grown and relatively high in dark-grown seedling hypocotyls. In consequence, DELLAs accumulate to higher levels in light-grown than in dark-grown hypocotyls, causing light-grown hypocotyls to be shorter than dark-grown hypocotyls. We therefore conclude that DELLAs account for a major component of the growth regulation that characterizes photomorphogenesis. This conclusion is confirmed by the observation that light-grown quadruple-DELLA mutant hypocotyls are longer than light-grown wild-type hypocotyls (Fig. 1). However, because light-grown quadruple-DELLA mutant hypocotyls are not as long as dark-grown wild-type hypocotyls (Fig. 1) and FR-grown gai hypocotyls are shorter than FR-grown phyA-1 gai hypocotyls, it is likely that additional (DELLA-independent) growth regulatory components also mediate photomorphogenetic growth regulation.
We have shown that quadruple-DELLA mutant hypocotyls are longer than wild-type hypocotyls when grown in continuous FR or continuous R light (where activated phyA or phyB are, respectively, the most predominantly acting phytochromes). We therefore conclude that there is a DELLA-dependent component of photomorphogenesis and that this component is a downstream consequence of the light activation of photoreceptors and their associated signal transduction cascades. In essence, our results indicate that activated phytochromes cause a decrease in hypocotyl levels of bioactive GAs, consequent accumulation of DELLAs, and resultant inhibition of hypocotyl growth. It is possible (but currently untested) that additional photoreceptors (e.g. the cryptochrome and phototropin photoreceptors and their respective signal transduction cascades; Chen et al., 2004) also regulate photomorphogenesis via effects on DELLA function.
We observed rapid changes in GFP-RGA level as a consequence of the change from a dark to a light environment (or vice versa), suggesting that plants use the DELLA restraint mechanism to rapidly adapt their growth rate in response to the prevailing light environment. In the specific case of Arabidopsis hypocotyls grown in day/night cycles, onset of light inhibits growth by promoting DELLA accumulation; onset of dark promotes growth by reducing DELLA accumulation. We also found that DELLAs enable plants to integrate information about the light environment (duration of exposure, fluence rate), allowing responses appropriate to the total energy of light to which they are exposed. Thus, DELLA integration likely conditions appropriate response to natural variation in the light environment (e.g. shade, cloud cover, etc.).
Recent years have seen impressive progress in the genetic and molecular definition of the mechanisms of plant photoperception and subsequent signaling events (Chen et al., 2004). However, until now, it has not been clear how these initial signaling steps connect during photomorphogenesis to the basic growth machinery of the plant. Our results indicate that light regulates growth in part by modulating the level of accumulation of nuclear DELLAs (by altering GA levels). Thus, growth regulatory DELLAs are a key component of the mechanism via which light signals are transduced into changes in growth.
MATERIALS AND METHODS
Plant Material
All experiments used the Landsberg erecta laboratory strain of Arabidopsis (Arabidopsis thaliana) as genetic background. The ga1-3, gai, quadruple-DELLA (gai-t6 rga-t2 rgl1-1 rgl2-1), ga1-3 quadruple-DELLA, pRGA:GFP-RGA, phyA-1, and phyB-1 lines were as described previously (Sun et al., 1992; Reed et al., 1993; Whitelam et al., 1993; Chen et al., 2004; Fu et al., 2004; Cao et al., 2005; Achard et al., 2006). The phyA-1 gai, phyA-1 pRGA:GFP-RGA, phyB-1 pRGA:GFP-RGA, and ga1-3 pRGA:GFP-RGA lines were isolated from F3 progeny of the appropriate crosses.
Hypocotyl Growth Experiments
All seeds were surface sterilized and placed on germination medium (GM) plates (Achard et al., 2003) at 4°C for 4 d to synchronize germination. For experiments involving ga1-3, all lines were GA treated (10 μm GA3) for 3 d at 4°C, then washed three times with distilled water before sterilization. Subsequently, plates were placed in vertical orientation in controlled environment chambers (22°C) for 2 d in darkness before transfer to white or R light at 75 μmol s−1 m−2 (or as indicated). After 5 d, plates were scanned and hypocotyl length was measured using Image J software (public domain; http://rsb.info.nih.gov/ij) or SigmaScan Pro 5 software (Systat Software). FR growth experiments were performed as previously described (Desnos et al., 2001). For each set of experiments, at least 30 seedlings were measured.
Detection of GFP Fluorescence in Seedling Hypocotyls
Fluorescence due to GFP-RGA in the nuclei of cells of the elongation zone of seedling hypocotyls (at a point roughly one-fourth the length of hypocotyls below the cotyledons) was determined as follows. pRGA:GFP-RGA-containing lines were grown on GM plates for 2 d in darkness and subsequently either maintained in the dark or transferred to white light for 5 d. Seedlings were then exposed to dark or light for the time as indicated before observation of hypocotyls by confocal microscopy (Achard et al., 2003).
Transcript Analysis
Total RNA was obtained from hypocotyls of 7-d-old seedlings grown on GM in either continuous white light (at 75 μmol s−1 m−2) or darkness. Seedlings were then transferred to dark or white light for the time as indicated. Total RNA was extracted using TRIzol reagent (Gibco-BRL). Generation of complementary DNA, PCR amplification (18 cycles), and blot analyses were as described previously (Achard et al., 2003). The primer pairs used for PCR amplification were as previously described (Achard et al., 2003) or as follows: GA5, 5′-cgcaagcagtttcaccggac-3′ and 5′-gagctgcttgcaccgggcgg-3′; GA4, 5′-catcacctcaactactgcga-3′ and 5′-tcacctgtaccgaagctggt-3′; GA2ox1, 5′-ccgaggaacacacttagcaag-3′ and 5′-ggcttcaacaattcgaaagg-3′; GA2ox2, 5′-gagtgactcgtgcctgagac-3′ and 5′-ccttgtatgagagtagtcat-3′; GA2ox3, 5′-tggtagaggaagagctaaag-3′ and 5′-ctaagcttggtgactatagg-3′; SLY1, 5′-gcgcagtactaccgactctg-3′ and 5′-cgagaagatgagtttcactaag-3′; SLY2, 5′-gtcgtcggagaaacgtgtag-3′ and 5′-cgttgtctgaaccggaaccg-3′; AtGID1a (At3g05120), 5′-gtttggtgggaatgagagaacg-3′ and 5′-ctaaacgcctcactgttcttcc-3′; AtGID1b (At3g63010), 5′-catccagcatgtaatccctttgg-3′ and 5′-cacttgtggaaactgtacacaacc-3′.
Immunoblot Analyses
pRGA:GFP-RGA seedlings were grown on GM in the presence of white light (fluence rate and photoperiod as indicated) for 4 d. phyB-1 pRGA:GFP-RGA seeds were germinated on GM plates in the dark for 4 d, then exposed to light of 75 μmol s−1 m−2 for the time indicated. The hypocotyls were then harvested and frozen in liquid nitrogen. phyA-1 pRGA:GFP-RGA (and control) seeds were germinated on GM plates for 2 d in light then transferred to FR for 5 d (Desnos et al., 2001). Whole seedlings were frozen in liquid nitrogen. Total proteins were extracted with buffer (250 mm Tris-Hcl, pH 6.8; 4% SDS; 20% glycerol; 0.2 m dithiothreitol), with subsequent western-blot analysis as previously described (Achard et al., 2003; Fu et al., 2004).
Acknowledgments
We thank Tai-ping Sun for the pRGA:GFP-RGA line, Garry Whitelam for phyA-1, Yuki Yasumura for AtGID1a and AtGID1b primers, and the Nottingham Arabidopsis Stock Centre for phyB-1.
This work was supported by funding from the European Union (RTN1–2000–00090 INTEGA), the Biotechnology and Biological Sciences Research Council (Core Strategic grant to the John Innes Centre and response modes grant nos. 208/P18610 and 208/P19972), and the National Natural Science Foundation of China (grant nos. 30525003 and 30521001).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Nicholas P. Harberd (nicholas.harberd@bbsrc.ac.uk).
References
- Achard P, Cheng H, De Grauwe L, Decat J, Schoutteten H, Moritz T, Van Der Straeten D, Peng J, Harberd NP (2006) Integration of plant responses to environmentally activated phytohormonal signals. Science 331 91–94 [DOI] [PubMed] [Google Scholar]
- Achard P, Vriezen WH, Van Der Straeten D, Harberd NP (2003) Ethylene regulates Arabidopsis development via the modulation of DELLA protein growth repressor function. Plant Cell 15 2816–2825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ait-ali T, Frances S, Weller JL, Reid JB, Kendrick RE, Kamiya Y (1999) Regulation of gibberellin 20-oxidase and gibberellin 3β-hydroxylase transcript accumulation during de-etiolation of pea seedlings. Plant Physiol 121 783–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alabadí D, Gil J, Blazquez MA, Garcia-Martinez J (2004) Gibberellins repress photomorphogenesis in darkness. Plant Physiol 134 1050–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvey L, Harberd NP (2005) DELLA proteins: integrators of multiple plant growth regulatory inputs? Physiol Plant 123 153–160 [Google Scholar]
- Bolle C (2004) The role of GRAS proteins in plant signal transduction and development. Planta 218 683–692 [DOI] [PubMed] [Google Scholar]
- Cao D, Hussain A, Cheng H, Peng J (2005) Loss of function of four DELLA genes leads to light- and gibberellin-independent seed germination in Arabidopsis. Planta 223 105–113 [DOI] [PubMed] [Google Scholar]
- Chen M, Chory J, Fankhauser C (2004) Light signal transduction in higher plants. Annu Rev Genet 38 87–117 [DOI] [PubMed] [Google Scholar]
- Cheng H, Qin L, Lee S, Fu X, Richards DE, Cao D, Luo D, Harberd NP, Peng J (2004) Gibberellin regulates Arabidopsis floral development via suppression of DELLA protein function. Development 131 1055–1064 [DOI] [PubMed] [Google Scholar]
- Chiang HH, Hwang I, Goodman HM (1995) Isolation of the Arabidopsis GA4 locus. Plant Cell 7 195–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowling R, Harberd NP (1999) Gibberellins control Arabidopsis hypocotyl growth via regulation of cellular elongation. J Exp Bot 50 1351–1357 [Google Scholar]
- Cowling RJ, Kamiya Y, Seto H, Harberd NP (1998) Gibberellin dose-response regulation of GA4 gene transcript levels in Arabidopsis. Plant Physiol 117 1195–1205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehesh K, Franci C, Parks BM, Seeley KA, Short TW, Tepperman JM, Quail PH (1993) Arabidopsis HY8 locus encodes phytochrome A. Plant Cell 5 1081–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desnos T, Puente P, Whitelam GC, Harberd NP (2001) FHY1: a phytochrome A-specific signal transducer. Genes Dev 15 2980–2990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dill A, Sun T-p (2001) Synergistic derepression of gibberellin signaling by removing RGA and GAI function in Arabidopsis thaliana. Genetics 159 777–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dill A, Thomas SG, Hu J, Steber CM, Sun T-p (2004) The Arabidopsis F-box protein SLEEPY1 targets gibberellin signaling repressors for gibberellin-induced degradation. Plant Cell 16 1392–1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleck B, Harberd NP (2002) Evidence that the Arabidopsis nuclear gibberellin signalling protein GAI is not destabilised by gibberellin. Plant J 32 935–949 [DOI] [PubMed] [Google Scholar]
- Fu X, Harberd NP (2003) Auxin promotes Arabidopsis root growth by modulating gibberellin response. Nature 421 740–743 [DOI] [PubMed] [Google Scholar]
- Fu X, Richards DE, Fleck B, Xie D, Burton N, Harberd NP (2004) The Arabidopsis mutant sleepy1gar2-1 protein promotes plant growth by increasing the affinity of the SCFSLY1 E3 ubiquitin ligase for DELLA protein substrates. Plant Cell 16 1406–1418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Martinez JL, Gil J (2001) Light regulation of gibberellin biosynthesis and mode of action. J Plant Growth Regul 20 354–368 [DOI] [PubMed] [Google Scholar]
- Gil J, García-Martinez JL (2000) Light-regulation of gibberellin A1 content and expression of genes coding for GA 20-oxidase and GA 3β-hydroxylase in etiolated pea seedlings. Physiol Plant 180 223–229 [Google Scholar]
- Harberd NP (2003) Relieving DELLA restraint. Science 299 1853–1854 [DOI] [PubMed] [Google Scholar]
- Harberd NP, King KE, Carol P, Cowling RJ, Peng J, Richards DE (1998) Gibberellin: inhibitor of an inhibitor of…? Bioessays 20 1001–1008 [DOI] [PubMed] [Google Scholar]
- Johnson E, Bradley M, Harberd NP, Whitelam GC (1994) Photoresponses of light-grown phyA mutants of Arabidopsis: phytochrome A is required for the perception of daylength extensions. Plant Physiol 105 141–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendrick RE, Kronenberg GHM (1994) Photomorphogenesis in Plants. Kluwer Academic Publishers, Dordrecht, The Netherlands
- King K, Moritz T, Harberd NP (2001) Gibberellins are not required for normal stem growth in Arabidopsis thaliana in the absence of GAI and RGA. Genetics 159 767–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Cheng H, King KE, Wang W, Husssain A, Lo J, Harberd NP, Peng J (2002) Gibberellin regulates Arabidopsis seed germination via RGL2, a GAI/RGA-like gene whose expression is up-regulated following imbibition. Genes Dev 16 646–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinnis KM, Thomas SG, Soule JD, Strader LC, Zale JM, Sun T-p, Steber CM (2003) The Arabidopsis SLEEPY1 gene encodes a putative F-box subunit of an SCF E3 ubiquitin ligase. Plant Cell 15 1120–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagatani A, Reed JW, Chory J (1993) Isolation and initial characterization of Arabidopsis mutants that are deficient in phytochrome A. Plant Physiol 102 269–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima M, Shimada A, Takashi Y, Kim YC, Park SH, Ueguchi-Tanaka M, Suzuki H, Katoh E, Luchi S, Kobayashi M, et al (2006) Identification and characterization of Arabidopsis gibberellin receptors. Plant J 46 880–889 [DOI] [PubMed] [Google Scholar]
- O'Neill DP, Ross JJ, Reid JB (2000) Changes in gibberellin A1 levels and response during de-etiolation of pea seedlings. Plant Physiol 124 805–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J, Carol P, Richards DE, King KE, Cowling RJ, Murphy GP, Harberd NP (1997) The Arabidopsis GAI gene defines a signaling pathway that negatively regulates gibberellin responses. Genes Dev 11 3194–3205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J, Harberd NP (1993) Derivative alleles of the Arabidopsis gibberellin-insensitive (gai) mutation confer a wild-type phenotype. Plant Cell 5 351–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J, Harberd NP (1997) Gibberellin deficiency and response mutations suppress the stem elongation phenotype of phytochrome deficient mutants of Arabidopsis. Plant Physiol 113 1051–1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips AL, Ward DA, Uknes S, Appleford NE, Lange T, Huttly AK, Gaskin P, Graebe JE, Hedden P (1995) Isolation and expression of three gibberellin 20-oxidase cDNA clones from Arabidopsis. Plant Physiol 108 1049–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quail PH (2002) Phytochrome photosensory signalling networks. Nat Rev Mol Cell Biol 3 85–93 [DOI] [PubMed] [Google Scholar]
- Reed JW, Foster KR, Morgan PW, Chory J (1996) Phytochrome B affects responsiveness to gibberellins in Arabidopsis. Plant Physiol 112 337–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed JW, Nagpal P, Poole DS, Furuya M, Chory J (1993) Mutations in the gene for the red/far-red light receptor phytochrome B alter cell elongation and physiological responses throughout Arabidopsis development. Plant Cell 5 147–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards DE, King KE, Ait-ali T, Harberd NP (2001) How gibberellin regulates plant growth and development: a molecular genetic analysis of gibberellin signaling. Annu Rev Plant Physiol Plant Mol Biol 52 67–88 [DOI] [PubMed] [Google Scholar]
- Sasaki A, Itoh H, Gomi K, Ueguchi-Tanaka M, Ishiyama K, Kobayashi M, Jeong DH, An G, Kitano H, Ashikari M, et al (2003) Accumulation of phosphorylated repressor for gibberellin signaling in an F-box mutant. Science 299 1896–1898 [DOI] [PubMed] [Google Scholar]
- Silverstone AL, Ciampaglio CN, Sun T-p (1998) The Arabidopsis RGA gene encodes a transcriptional regulator repressing the gibberellin signal transduction pathway. Plant Cell 10 155–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstone AL, Jung HS, Dill A, Kawaide H, Kamiya Y, Sun T-p (2001) Repressing a repressor: gibberellin-induced rapid reduction of the RGA protein in Arabidopsis. Plant Cell 13 1555–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strader LC, Ritchie S, Soule JD, McGinnis KM, Steber CM (2004) Recessive-interfering mutations in the gibberellin signaling gene SLEEPY1 are rescued by overexpression of its homologue, SNEEZY. Proc Natl Acad Sci USA 101 12771–12776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun T-p, Goodman HM, Ausubel FM (1992) Cloning the Arabidopsis GA1 locus by genomic subtraction. Plant Cell 4 119–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun T-p, Kamiya Y (1994) The Arabidopsis GA1 locus encodes the cyclase ent-kaurene synthetase A of gibberellin biosynthesis. Plant Cell 6 1509–1518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas SG, Phillips AL, Hedden P (1999) Molecular cloning and functional expression of gibberellin 2-oxidases, multifunctional enzymes involved in gibberellin deactivation. Proc Natl Acad Sci USA 96 4638–4703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler L, Thomas SG, Hu J, Dill A, Alonso JM, Ecker JR, Sun T-p (2004) DELLA proteins and gibberellin-regulated seed germination and floral development in Arabidopsis. Plant Physiol 135 1008–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen C-K, Chang C (2002) Arabidopsis RGL1 encodes a negative regulator of gibberellin responses. Plant Cell 14 87–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitelam GC, Harberd NP (1994) Action and function of phytochrome family members revealed through the study of mutant and transgenic plants. Plant Cell Environ 17 615–625 [Google Scholar]
- Whitelam GC, Johnson E, Peng J, Carol P, Anderson ML, Cowl JS, Harberd NP (1993) Phytochrome A null mutants of Arabidopsis display a wild-type phenotype in white light. Plant Cell 5 757–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu YL, Li L, Gage DA, Zeevaart JA (1999) Feedback regulation of GA5 expression and metabolic engineering of gibberellin levels in Arabidopsis. Plant Cell 11 927–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S, Smith MW, Brown RGS, Kamiya Y, Sun T-p (1998) Phytochrome regulation and differential expression of gibberellin 3β-hydroxylase genes in germinating Arabidopsis seeds. Plant Cell 10 2115–2126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Ito T, Zhao Y, Peng J, Kumar P, Meyerowitz EM (2004) Floral homeotic genes are targets of gibberellin signaling in flower development. Proc Natl Acad Sci USA 101 7827–7832 [DOI] [PMC free article] [PubMed] [Google Scholar]