Abstract
Yeast (Saccharomyces cerevisiae) Atg6/Vps30 is required for autophagy and the sorting of vacuolar hydrolases, such as carboxypeptidase Y. In higher eukaryotes, however, roles for ATG6/VPS30 homologs in vesicle sorting have remained obscure. Here, we show that AtATG6, an Arabidopsis (Arabidopsis thaliana) homolog of yeast ATG6/VPS30, restored both autophagy and vacuolar sorting of carboxypeptidase Y in a yeast atg6/vps30 mutant. In Arabidopsis cells, green fluorescent protein-AtAtg6 protein localized to punctate structures and colocalized with AtAtg8, a marker protein of the preautophagosomal structure. Disruption of AtATG6 by T-DNA insertion resulted in male sterility that was confirmed by reciprocal crossing experiments. Microscopic analyses of AtATG6 heterozygous plants (AtATG6/atatg6) crossed with the quartet mutant revealed that AtATG6-deficient pollen developed normally, but did not germinate. Because other atatg mutants are fertile, AtAtg6 likely mediates pollen germination in a manner independent of autophagy. We propose that Arabidopsis Atg6/Vps30 functions not only in autophagy, but also plays a pivotal role in pollen germination.
Autophagy is a bulk degradation mechanism by which cytoplasmic constituents are delivered to lytic compartments for degradation and recycling (Klionsky and Ohsumi, 1999; Ohsumi, 2001; Levine and Klionsky, 2004). The process involves sequestration of the cytoplasm into double-membrane vesicles called autophagosomes, which subsequently fuse with lysosomes or vacuoles. Genetic studies in yeast (Saccharomyces cerevisiae) have identified a set of ATG (autophagy) genes involved in this process (Klionsky and Ohsumi, 1999; Ohsumi, 2001). Various Atg proteins reside on the preautophagosomal structure (PAS) from which autophagosomes originate (Suzuki et al., 2001). The molecular mechanisms underlying the formation of autophagosomes, however, have not yet been fully elucidated.
Most of the yeast Atg proteins are specifically engaged in autophagy and/or the cytoplasm-to-vacuole targeting pathway, which is mechanistically similar to autophagy (Klionsky and Ohsumi, 1999; Levine and Klionsky, 2004). By contrast, ATG6 is allelic to the previously characterized VPS30 (vacuolar protein sorting), which is required for the vacuolar sorting of carboxypeptidase Y (CPY; Seaman et al., 1997; Kametaka et al., 1998). ATG6/VPS30 is conserved in higher eukaryotes and its ortholog (BECLIN 1) can restore autophagy in ATG6/VPS30-deficient yeast (Liang et al., 1999; Meléndez et al., 2003; Liu et al., 2005). Suppression of BECLIN 1 homologs blocked autophagy and disturbed various biological processes (Meléndez et al., 2003; Yu et al., 2004; Liu et al., 2005). On the other hand, the putative role of BECLIN 1 in membrane trafficking is controversial (Zeng et al., 2006).
Many Atg proteins are conserved in plants, and reverse genetic studies have demonstrated that Arabidopsis (Arabidopsis thaliana) atg mutants are hypersensitive to nutrient starvation, as well as that atg plants exhibit accelerated senescence even under favorable growth conditions (for review, see Thompson and Vierstra, 2005; Bassham et al., 2006). Here, we are interested in AtATG6, an Arabidopsis homolog of yeast ATG6/VPS30, because it is a unique autophagy gene that also participates in trafficking events in yeast. We first investigated whether AtATG6 was involved in autophagy and found that AtATG6 restored autophagy in atg6/vps30 yeast and that it localized in plant cells to a punctate structure containing Arabidopsis Atg8, a marker protein of the PAS (Suzuki et al., 2001). We also show that Arabidopsis Atg6/Vps30 partially complemented VPS function in atg6/vps30 mutant yeast. Several other Arabidopsis VPS homologs are required in pollen tubes (Hicks et al., 2004; Lobstein et al., 2004) because the polarized growth of pollen tubes requires membrane trafficking (Hepler et al., 2001; Monteiro et al., 2005). We found that disruption of AtATG6/VPS30 resulted in male sterility because the mutant pollen did not germinate. By contrast, all previously characterized atatg mutants are fertile. Our results support the idea that Atg6/Vps30 in plants has distinct functions in addition to autophagy: vesicle trafficking and pollen germination.
RESULTS
Arabidopsis Atg6 Restores Autophagy and Vacuolar Protein Sorting in atg6/vps30 Mutant Yeast
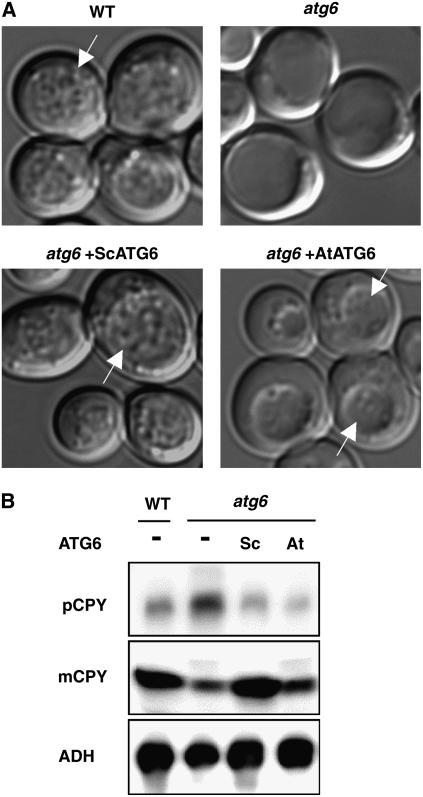
To first investigate roles of Atg6/Vps30 in higher plants, we isolated a putative Arabidopsis homolog of ATG6/VPS30 (AtATG6) encoded by a single gene (At3g61710) in the Arabidopsis genome. AtATG6-encoded protein shares 24% amino acid identity with yeast Atg6/Vps30 (Seaman et al., 1997; Kametaka et al., 1998) and 31% identity with human Beclin 1 (Liang et al., 1999). Animal homologs of ATG6/VPS30 (BECLIN 1) restored autophagy, but not CPY sorting, in atg6/vps30 mutant yeast (Liang et al., 1999; Meléndez et al., 2003). To test whether AtATG6 is a functional homolog of ATG6/VPS30, we investigated whether AtAtg6 rescues defects of yeast atg6/vps30 in autophagy and the VPS pathway. AtATG6 cDNA containing the entire open reading frame (ORF) was fused with the yeast ATG6/VPS30 promoter in a low-copy-number plasmid (pRS416) and expressed in ATG6/VPS30-deficient yeast (parental strain BY4741). Wild-type yeast cells accumulated numerous autophagic bodies within vacuoles after 5 h of nutrient starvation, whereas no such vesicles were found in the vacuoles of atg6/vps30 yeast cells (Fig. 1A). Under these conditions, expression of AtATG6 allowed atg6/vps30 cells to accumulate autophagic bodies, albeit to a lesser extent than did wild-type cells (Fig. 1A).
Figure 1.
Functional complementation by Arabidopsis ATG6/VPS30 of autophagy and vacuolar protein sorting in a yeast atg6/vps30 mutant. The Arabidopsis ATG6/VPS30 (AtATG6) gene was cloned in the low-copy-number plasmid pRS416 and expressed in atg6/vps30 yeast cells. Yeast ATG6/VPS30 (ScATG6) and the empty vector (pRS416) were used as controls. A, Cells grown to mid-log stage were collected, washed, and subsequently incubated for another 5 h in nutrient-free medium in the presence of 1 mm phenylmethylsulfonyl fluoride. Autophagic bodies accumulated within vacuoles (arrows). B, Intracellular or extracellular extracts were prepared from wild-type or atg6/vps30 cells that were transformed with the empty vector (−), yeast (Sc), or Arabidopsis (At) ATG6/VPS30. Secreted CPY (pCPY) in the extracellular extracts and vacuolar-targeted CPY (mCPY) or alcohol dehydrogenase (ADH) in the intracellular extracts were detected by immunoblotting.
We next measured the amount of intracellular and extracellular CPY protein to determine whether CPY was delivered into vacuoles or secreted. Wild-type or atg6/vps30 cells transformed with ATG6/VPS30 seemed to properly sort CPY into vacuoles, where CPY was processed to its mature form of 61 kD (Fig. 1B, mCPY). In atg6/vps30 cells, on the other hand, mis-sorted CPY was secreted (Fig. 1B, pCPY) and concomitantly lower quantities of mature CPY were detected in the intracellular fraction (Fig. 1B, mCPY). In AtATG6-transformed atg6/vps30 cells, secretion of CPY was suppressed, suggesting that AtAtg6 restores vacuolar sorting of CPY in ATG6/VPS30-disrupted yeast.
AtAtg6 Colocalizes with AtAtg8 at a Punctate Structure
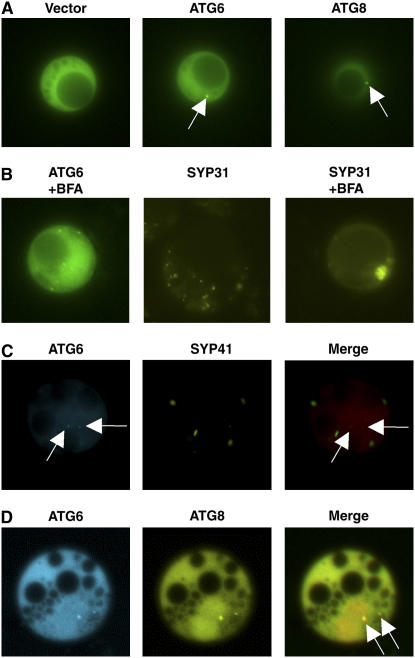
In yeast cells, green fluorescent protein (GFP)-tagged Atg6/Vps30, like other Atg proteins, partially localizes to the punctate structure called the PAS (Obara et al., 2006). To visualize AtAtg6 protein in plant cells, a GFP-AtAtg6 fusion protein was expressed in Arabidopsis protoplasts under the control of the cauliflower mosaic virus 35S promoter. As shown in Figure 2A, the fluorescent signal from GFP-AtAtg6 was distributed in the cytoplasm and to a single or a few bright dots. Similar dot signals were observed when AtAtg6 was fused to the N terminus of GFP (AtAtg6-GFP; data not shown). Such punctation was not found in cells transformed with the empty vector (35S-GFP alone; Fig. 2A; see also Supplemental Fig. S1B).
Figure 2.
Intracellular localization of AtAtg6 in Arabidopsis cells. AtAtg proteins tagged with GFP derivatives were expressed in protoplasts prepared from Arabidopsis suspension cells. Protoplasts were incubated for 24 h in Suc-free medium. A, Localization of GFP-AtAtg6 (middle) and GFP-AtAtg8 (right), a marker protein of the PAS (arrows). The vector control (pEZS-CL) is shown at left. B, Effects of BFA on the localization of GFP-AtAtg6. Cells were incubated for 2 h in the presence of 200 μm BFA. Venus-tagged SYP31 was used as a Golgi marker. C, CFP-AtAtg6 and Venus-tagged SYP41, a marker protein of TGN, were expressed together in Arabidopsis cells. Arrows indicate punctate signals of CFP-AtAtg6. D, Punctate colocalization (arrows) of CFP-AtAtg6 and YFP-AtAtg8.
We next attempted to determine the organelle to which AtAtg6 localized. Punctate localization of GFP-AtAtg6 was not influenced by brefeldin A (BFA; Fig. 2B). Venus-tagged SYP31, SYP41, and SYP81 served as marker proteins for the Golgi, trans-Golgi network (TGN), and endoplasmic reticulum (ER), respectively (Uemura et al., 2004), and each of these marker proteins showed intracellular distribution patterns that were clearly distinguished from that of AtAtg6 (Fig. 2, B and C; data not shown). Bright dots of cyan fluorescent protein (CFP)-AtAtg6 also did not overlap with any of these organelle markers (Fig. 2C; data not shown). In contrast, the PAS marker protein, GFP-AtAtg8, exhibited a punctate localization profile similar to that of GFP-AtAtg6 (Fig. 2A; see also Supplemental Fig. S1). We then compared the localization of CFP-AtAtg6 with that of yellow fluorescent protein (YFP)-AtAtg8. Each fusion protein again exhibited clear enrichment in the punctate structure within the cytoplasm, and the coincidence of CFP and YFP signals on the bright dots (Fig. 2D, arrows; Supplemental Fig. S2) suggested the colocalization of AtAtg6 with AtAtg8 in plant cells, similar to that shown in yeast cells (Obara et al., 2006).
Deletion of AtATG6 Causes a Male Gametophytic Mutation
To evaluate the functions of AtAtg6 in planta, we employed a reverse genetic approach. Our initial attempts to screen Arabidopsis mutants (two independent lines, SALK_051168 and SALK_109281) harboring T-DNA insertions within the AtATG6 gene identified no homozygous mutant lines. AtATG6 heterozygous plants exhibited reduced AtATG6 mRNA levels relative to wild-type plants (Supplemental Fig. S3), but appeared normal in appearance. Each AtATG6 heterozygous plant showed approximately a 1:1 segregation ratio for transmission of the T-DNA in the first selfing progeny (Table I). A Mendelian transmission ratio of 3:1 was rejected by a χ2 test (P = 7.4 × 10−25; P = 3.6 × 10−17). Embryos produced by selfing of AtATG6/atatg6 plants grew normally and no aborted seeds were found in the siliques. Therefore, failures of transmission of T-DNA to the progeny seemed to be due to defects in the gametes, not embryonic lethality. Next, we performed crossing experiments to determine the gametophytes that were affected by atg6. When AtATG6 heterozygous plants (background Columbia; the line SALK_051168 was used in the following experiments) were used as pollinators of wild-type plants (Wassilewskija [Ws] was used to make it possible to confirm crossing by PCR), no T-DNA-inserted progeny were obtained (Table I). This suggested the failure of transmission of atg6 through the male gametophyte, which was statistically supported by a χ2 test (P = 9.2 × 10−9). In contrast, pollination of AtATG6 heterozygous stigmas with wild-type pollen yielded progeny with a 1:1 segregation ratio of transmission of T-DNA (Table I). The observed ratio was nearly identical to the expected T-DNA transmission ratio of 1:1 through the female gametophyte (χ2 = 0.44; P = 0.51). Deletion of AtATG6, therefore, appeared specifically to influence male gametophytes but not to compromise the function of female reproductive structures.
Table I.
Genetic transmission analysis of atg6 mutations
Numbers of progeny containing T-DNAs (atg6-1, SALK_051168; atg6-2, SALK_109281) were counted using PCR and actual transmission efficiency (TE; the number of T-DNA-containing progeny/total number of seedlings × 100) was calculated. The χ2 value is shown for the predicted ratio of 3:1 (self-crosses) or 1:1 (reciprocal crosses).
| Genotype | Numbers
|
TE (%) | χ2 (P) | |
|---|---|---|---|---|
| T-DNA+ | T-DNA− | |||
| ATG6/atg6-1 (selfed) | 131 | 142 | 48.0 | 106 (7.4 × 10−25) |
| ATG6/atg6-2 (selfed) | 33 | 36 | 47.8 | 71 (3.6 × 10−17) |
| ♂ATG6/atg6-1 × ♀Ws | 0 | 33 | 0.0 | 33 (9.2 × 10−9) |
| ♂Ws × ♀ATG6/atg6-1 | 16 | 20 | 44.4 | 0.44 (0.51) |
Pollen Lacking AtATG6 Does Not Germinate
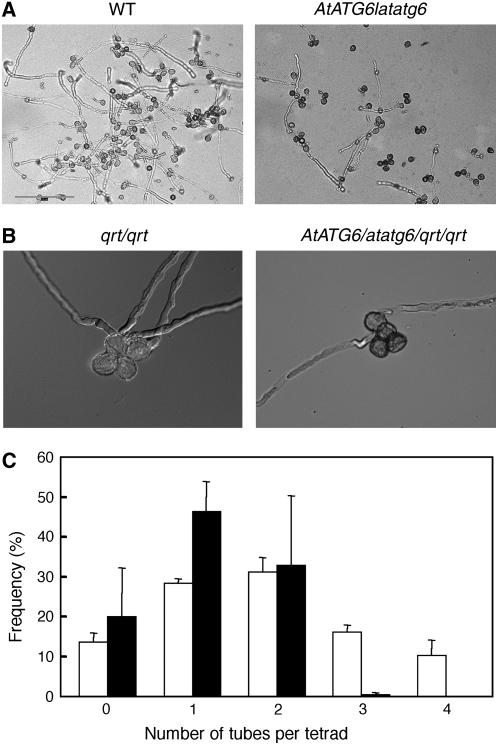
To determine the biological basis of the male gametophytic mutant phenotype of atatg6, we performed microscopic analysis of atatg6 pollen. Pollen grains from AtATG6 heterozygous flowers were indistinguishable from wild-type pollen in shape and size (data not shown). However, the efficiency of the in vitro pollen germination of AtATG6 heterozygous flowers (39.8 ± 1.4% of pollen grains germinated; n = 977) was lower than that of wild-type controls (76.7 ± 2.5%; n = 1,715; for each genotype, ses for three independent experiments are shown; Fig. 3A). Under these conditions, pollen from other atatg mutants (atatg4a4b-1 and atatg2 were tested) germinated with efficiency similar to that of wild-type controls (data not shown).
Figure 3.
Pollen germination efficiency of AtATG6/atatg6 heterozygotes. A, In vitro pollen germination of the AtATG6/atatg6 heterozygous mutant (right) and the wild-type control (left). B, In vitro pollen germination of AtATG6/atatg6/qrt/qrt (right) and AtATG6/AtATG6/qrt/qrt (left). C, Frequency (%) of wild-type (AtATG6/AtATG6/qrt/qrt; white bars) and heterozygous mutant (AtATG6/atatg6/qrt/qrt; black bars, n > 500) tetrads with zero to four pollen tubes. The values shown represent the means of three different experiments (±sd).
We crossed quartet1 (qrt1) mutant plants to AtATG6/atatg6 plants to further address the role of AtAtg6 in pollen germination. The qrt mutation yielded four microspores derived from a single pollen mother cell that stay attached to one another for the duration of pollen development, which makes it easy to perform tetrad analyses (Preuss et al., 1994; McCormick, 2004). We expected that if AtAtg6 were essential for pollen germination, at most two pollen grains in each AtATG6/atatg6/qrt/qrt tetrad would germinate. This prediction was correct; only one or two pollen grains germinated from each AtATG6/atatg6/qrt/qrt tetrad, whereas up to four pollen grains germinated from each wild-type (AtATG6/AtATG6/qrt/qrt) tetrad (Fig. 3, B and C). Apparently, there were no severe defects in the elongation of germinated pollen tubes of AtATG6/atatg6/qrt/qrt tetrads (Fig. 3B; data not shown), possibly because only the pollen-carrying wild-type AtATG6 could germinate. These results strongly suggested that atatg6 mutant pollen fails to germinate.
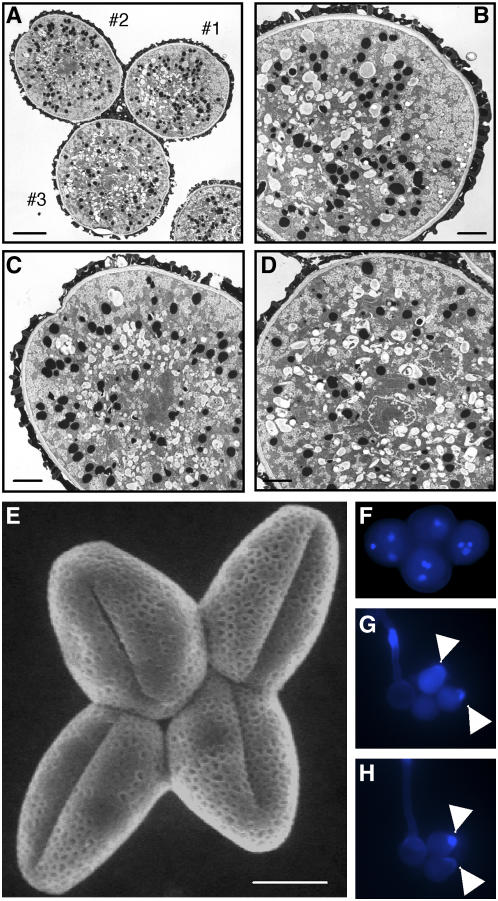
Despite the defects in pollen germination, transmission electron microscopy and environmental scanning electron microscopy (ESEM) showed that all four pollen grains in an AtATG6/atatg6/qrt/qrt tetrad were normal in appearance and indistinguishable from one another (Fig. 4). Each mutant pollen grain contains three normal nuclei: one vegetative and two sperm nuclei (Fig. 4F). We also stained pollen with aniline blue to investigate whether disorganization of cell wall composition (e.g. callose) was associated with failures of pollen germination (Johnson and McCormick, 2001). Each pollen grain of an AtATG6/atatg6/qrt/qrt tetrad, even if not germinated, was able to perform local outgrowth of the pollen wall or polarized deposition of callose in a manner similar to that of AtATG6/AtATG6/qrt/qrt pollen (Fig. 4, G and H). It was therefore likely that the polarity of the pollen germination site was determined normally in atatg6 pollen. Taken together, these results suggested that AtAtg6 is not essential for male meiosis or pollen development prior to germination.
Figure 4.
Morphology of AtATG6/atatg6/qrt/qrt pollen grains. A, Transmission electron micrograph of an AtATG6/atatg6/qrt/qrt tetrad. The pollen grains (1, 2, and 3) are enlarged in B, C, and D, respectively. The fourth pollen grain is out of focus. Bars = 5 μm in A; 2 μm in B to D. E, ESEM image of the AtATG6/atatg6/qrt/qrt tetrad. Bar = 10 μm. F, DAPI staining of the AtATG6/atatg6/qrt/qrt tetrad. G and H, Aniline blue staining of tetrads from AtATG6/AtATG6/qrt/qrt (G) and AtATG6/atatg6/qrt/qrt (H) plants. Note that the fourth pollen grain in H is out of focus. In both cases, more than three pollen grains in each tetrad either germinated or exhibited polarized accumulation of callose (arrowheads).
Expression of ATG6 in Arabidopsis
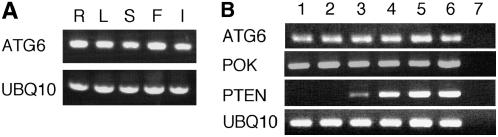
We previously showed that the AtATG4 and AtATG8 genes were ubiquitously expressed in whole plants (Yoshimoto et al., 2004). Similarly, reverse transcription (RT)-PCR revealed that AtATG6 mRNA was expressed in all organs tested (Fig. 5A). Expression of AtATG6 mRNA was not up-regulated during floral development (Fig. 5B). Likewise, POK/AtVPS52, another Arabidopsis VPS homolog implicated in pollen tube elongation (Lobstein et al., 2004), was expressed in different tissues and continuously expressed throughout pollen development (Lobstein et al., 2004; Fig. 5B). These results suggest the occurrence of autophagy and vesicle trafficking in various tissues.
Figure 5.
Expression of AtATG6 in Arabidopsis. RT-PCR analyses of AtATG6 mRNA levels in various organs (A) and during floral development (B). A, Each organ was collected from plants grown hydroponically for 4 to 5 weeks. R, Roots; L, leaves; S, stems; F, fruits; I, inflorescences. B, Flowers at different stages of development (Gupta et al., 2002) and leaves were collected from plants grown in soil for 5 to 6 weeks. 1, Leaves; 2, small green buds; 3, large green buds; 4, white buds; 5, opening flowers; 6, opened flowers; 7, leaf mRNA (prior to RT). AtATG6 and POK/VPS52 cDNAs were amplified with 25 cycles of PCR. AtPTEN1, a pollen-specific cDNA (Gupta et al., 2002), was amplified with 27 cycles of PCR. UBQ10 cDNA was amplified with 20 cycles.
DISCUSSION
We have identified AtATG6 as a functional homolog of yeast ATG6/VPS30 and characterized its function in plants. First, a yeast complementation assay demonstrated that expression of AtATG6 partially restored autophagy in atg6/vps30 yeast. Liu et al. (2005) recently showed that Arabidopsis BECLIN 1 (AtATG6) rescued autophagy in atg6/vps30 yeast cells, in agreement with our results. However, it has thus far been unclear as to whether plant or mammal BECLIN 1 homologs are effective in trafficking events, such as CPY sorting. In this article, we show that expression of AtATG6 rescued atg6/vps30 yeast from the mis-sorting of CPY protein. Although it has not been determined in plant cells how AtAtg6 regulates vesicular trafficking, we found that AtATG6-deleted plants displayed a male gametophytic mutant phenotype, which was similarly exemplified by another atvps mutant (see below). In contrast, none of the previously characterized atatg mutants has exhibited male gametophytic defects. Taken together, our results raise the possibility that Arabidopsis Atg6/Vps30 may be involved in other biological pathways (e.g. vesicle trafficking) in addition to autophagy.
In Arabidopsis cells, GFP-AtAtg6 proteins were predominantly distributed throughout the cytoplasm in accordance with biochemical studies identifying cytosolic Atg6/Vps30 in yeast cells (Seaman et al., 1997; Kihara et al., 2001a). In addition, recent microscopic observations revealed that Atg6/Vps30 is partially targeted to the punctate structure upon which the PAS marker protein Atg8 resides (Obara et al., 2006). As expected, CFP-AtAtg6 colocalized with YFP-AtAtg8 at the dot-like structures, which were reminiscent of the PAS. By contrast, in whole plants, GFP-AtAtg8 localizes to many dot structures in each cell, some of which may represent autophagosomes (Yoshimoto et al., 2004). The appearance of autophagosomes might depend on cell type: An isolated cell, such as a yeast cell, contains only a few autophagosomes and Atg8 predominantly localizes at a single dot (PAS) rather than to autophagosomes (Suzuki et al., 2001), mimicking the punctate localization of AtAtg proteins in Arabidopsis suspension cells.
In addition to the PAS, yeast Atg6/Vps30 also resides at the endosome and on the vacuolar membrane (Obara et al., 2006), and mammalian Beclin 1 localizes to the TGN (Kihara et al., 2001b). Although the localization of AtAtg6 to endomembranes was not detected under our experimental conditions, we cannot exclude the possibility that plant Atg6/Vps30/Beclin 1 may function in part through the endomembrane system to control vesicle trafficking.
Precise molecular mechanisms by which Atg6/Vps30/Beclin 1 proteins regulate autophagosome formation have not yet been described in any organisms. In yeast, Atg6/Vps30 associates with Vps34, a phosphatidylinositol 3-kinase (PI3-kinase; Kihara et al., 2001a). Recent studies have suggested that targeting of Atg6/Vps30 and Vps34 to the PAS is critical for subsequent autophagosome formation (Obara et al., 2006). Vps34 may be recruited to the PAS by Atg14, a connector protein that binds Vps34 and Atg6/Vps30, and then phosphatidlylinositol-3 phosphate (PI3-P) produced by PI3-kinase at the PAS could subsequently recruit other proteins involved in autophagosome formation (Obara et al., 2006). Atg6/Vps30 seems indispensable for Atg14 function because Atg14 is unstable in atg6/vps30 yeast cells (Kihara et al., 2001a). Although no ATG14 homologs have been found in plants or animals, it is tempting to speculate that AtAtg6 may interact with an unidentified PI3-kinase regulatory protein that would direct the specificity of PI3-kinase activity toward autophagosome formation, possibly by recruiting the PI3-kinase complex to the putative PAS in Arabidopsis cells.
A number of Arabidopsis atg mutants have been isolated thus far, each of which has exhibited retarded growth under nutrient starvation and accelerated senescence even under nutrient-rich conditions (Thompson and Vierstra, 2005; Bassham et al., 2006). Similarly, virus-induced gene silencing of tobacco (Nicotiana tabacum) autophagy genes, including a BECLIN 1/ATG6 homolog (NbATG6), enhanced yellowing of leaves under environmental stress, such as pathogen attack or dark-induced senescence (Liu et al., 2005). In this particular gene-silencing system, no difference was apparent between the phenotypes of NbATG6- and other NbATG-silenced plants. In contrast, we assumed that the atg6/vps30 mutant would reveal more severe phenotypes than other atg mutants because yeast Atg6/Vps30 mediates various biological pathways, including autophagy. Accordingly, of the atatg plants examined thus far, only atatg6 mutants exhibited pollen tube germination abnormalities. This phenotype was not ascribed to autophagy because none of the atatg mutants (except atatg6) has exhibited male gametophytic defects (Thompson and Vierstra, 2005; Bassham et al., 2006; this study). As described above, this finding supports our hypothesis that AtAtg6/Vps30 may have an autophagy-independent role responsible for pollen germination.
The compelling question, then, is how AtAtg6 mediates pollen germination. Extensive studies have identified several key components for pollen tube growth; one of these is vesicular trafficking (Hepler et al., 2001; Monteiro et al., 2005). The tip of the pollen tube contains numerous vesicles that could provide precursor materials for the rapidly expanding cell wall of the growing cells. Arabidopsis VPS homologs, including VCL1 (VPS16) and POK (VPS52), seem to play roles in germinating pollen tubes (Hicks et al., 2004; Lobstein et al., 2004). Similarly, it is possible that AtATG6/VPS30 may be involved in the trafficking machinery that is crucial for pollen tube elongation.
Another possible explanation is that AtAtg6-mediated PI3-kinase signaling might be involved in pollen germination because PI3-P is known to act as a second messenger and modulate a variety of cellular activities in plants (Meijer and Munnik, 2003). Previous studies described roles of another phosphoinositide, phosphatidylinositol 4,5-bisphosphate, during cytoskeletal organization and membrane trafficking, which may regulate cell elongation of root hairs and of pollen tubes (Kost et al., 1999; Vincent et al., 2005). To our knowledge, however, the involvement of PI3-P in pollen tube germination has not been reported. AtATG6-disrupted pollen, which completely lacks AtATG6 but is still viable, would serve as a useful system to further investigate the molecular mechanisms underlying PI3-kinase-regulated polarized cell growth.
Finally, despite its inhibitory effects on pollen germination, the atatg6 mutation affected neither the functions of female gametophytes nor the viability of pollen, suggesting that AtAtg6 is not essential for growth of germ cells. Nevertheless, this does not necessarily exclude the possibility that disruption of AtATG6 drastically affects various developmental processes in plants. For instance, if atatg6 homozygous plants were available, their early embryonic development would be severely damaged, as shown in AtVPS34-suppressed transgenic Arabidopsis (Welters et al., 1994). In animals, suppression of BECLIN 1 affected dauer development (Meléndez et al., 2003) and cell death (Yu et al., 2004). Suppression of AtATG6 by antisense or RNAi techniques would unveil another unanticipated physiological significance of ATG6/VPS30/BECLIN1 genes, in addition to AtATG6-mediated pollen germination.
MATERIALS AND METHODS
Complementation of Yeast Mutants
An ORF of Arabidopsis (Arabidopsis thaliana) ATG6 (At3g61710; The Arabidopsis Information Resource [TAIR]; http://www.arabidopsis.org) was fused with the 5′ flanking sequence (−816 to −1 relative to the initiation codon) of yeast (Saccharomyces cerevisiae) ATG6/VPS30 in the pRS416 vector. A genomic fragment of yeast ATG6/VPS30 (−952 to +2,325; Kametaka et al., 1998) was inserted into pRS416 as a control. Wild-type yeast (strain BY4741) and an atg6/vps30 deletion mutant (Y2132; Giaever et al., 2002) were then transformed with these plasmids.
For autophagy complementation experiments, cells were grown at 30°C to the mid-log stage (OD600 of 1.0–1.5) in synthetic medium (0.67% [w/v] yeast nitrogen base without amino acids and ammonium) supplemented with 2% (w/v) Glc, 0.5% (w/v) ammonium sulfate, 0.5% (w/v) casamino acid, and 0.002% (w/v) Trp. Cells were then collected, washed, and incubated for another 5 h in the synthetic medium in the presence of 1 mm phenylmethylsulfonyl fluoride. Accumulation of autophagic bodies was observed using a differential interference contrast microscope (IX81; Olympus).
For VPS complementation, cells under 5 h of nitrogen starvation were collected by centrifugation and used to prepare intracellular extracts, whereas the supernatant was collected as the extracellular fraction. Preparation of cell extracts and immunoblot analyses were performed as described previously (Hamasaki et al., 2005).
Plant Materials
Wild-type Arabidopsis (strain Columbia) plants were grown at 22°C with 16-h-light/8-h-dark photoperiods. Two distinct T-DNA insertion lines of AtATG6 (SALK_051168 and SALK_109281) were generated by the SALK Institute, and AtATG6 heterozygous plants were identified by PCR. The primer 5′-TGGTTCACGTAGTGGGCCATCG-3′ was designed for the T-DNA sequence and others were used for AtATG6 (see below). AtATG6 heterozygous plants (SALK_051168) were crossed with qrt mutant plants (qrt1-2; Arabidopsis Biological Resource Center [ABRC] CS8846).
Arabidopsis suspension-cultured cells were maintained as described (Ueda et al., 2001).
Subcellular Localization of AtAtg6 Protein Fused with GFP Derivatives
The ORF of AtATG6 or AtATG8a (Yoshimoto et al., 2004) was fused to the C terminus of enhanced GFP in the pEZS-CL plasmid (Carnegie Institution of Washington). The ORFs of AtATG6 and AtATG8i were C-terminally fused to enhanced CFP or enhanced YFP (p2CGW7 and p2YGW7, Karimi et al., 2005) using Gateway technology (Invitrogen). Venus-tagged SYP31, SYP41, and SYP81 were used as the Golgi, TGN, and ER marker proteins, respectively (Uemura et al., 2004). Transient expression of fluorescence proteins in Arabidopsis suspension cells was performed as described in Ueda et al. (2001). BFA treatment was performed as described previously (Uemura et al., 2004). Cells were observed using an epifluorescence microscope (Olympus IX81).
Microscopic Observation of Pollen
Pollen grains were germinated in the dark at 22°C on an agar plate containing 0.6% agar, 13.5% Suc, 30 μg mL−1 myoinositol, 0.4 mm boric acid, 10 mm CaCl2, and 1 mm KCl.
Sections of mature pollen were observed using a JEM-2000EX transmission electron microscope (JEOL) as described by Yoshimoto et al. (2004).
Pollen was observed in the chamber of an ESEM (PHILIPS XL30; FEI Electron Optics) at 2°C under 650 to 660 Pa of pressure with an acceleration voltage of 15 kV. Images were processed using Adobe Photoshop 6.0 (Adobe Systems).
Staining of pollen with 4′,6-diamidino-2-phenylindole (DAPI) or aniline blue was performed as described previously (Regan and Moffatt, 1990; Shimizu and Okada, 2000).
RT-PCR
Arabidopsis plants were grown hydroponically for 4 to 5 weeks (Hanaoka et al., 2002) and each organ was harvested. Flowers at different developmental stages were collected as described by Gupta et al. (2002). RNA was extracted from each sample using TRIzol reagent (GIBCO-BRL) and treated with DNase I. cDNA was synthesized using a ProSTAR first-strand RT-PCR kit (Stratagene). The primers used were AtATG6, 5′-GCTGAAGTAAACCATCAACTG-3′ and 5′-GTTACACATCGTATGGAGGAG-3′; UBIQUITIN10 (UBQ10, At4g05320), 5′-GCAGATCTTTGTTAAGACTC-3′ and 5′-CCAAAGTGATAGTTTTCCCAG-3′; POK (At1g71270), 5′-GGTGTTAAGTGCACATTTTCGTG-3′ and 5′-TCACCAAAGAAATCATCACAAAA-3′; and AtPTEN1 (At5g39400), 5′-ATGGGTCTCAAGCTCTCACG-3′ and 5′-ACCGTAAACAAGGTACGCCGA-3′.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers AY039613 (AtATG6) and BT024554 (AtATG8i).
Supplemental Data
The following materials are available in the online version of this manuscript.
Supplemental Figure S1. Intracellular localization of AtAtg8 in Arabidopsis cells.
Supplemental Figure S2. Colocalization of AtAtg6 and AtAtg8 proteins in Arabidopsis cells.
Supplemental Figure S3. Reduced AtATG6 mRNA levels in AtATG6 heterozygous plants.
Supplementary Material
Acknowledgments
We thank Dr. Hirokazu Tsukaya at the NIBB for helpful comments; Dr. Christiane Genetello (VIB-Ghent University) for the Gateway plasmids; Drs. Masaaki Umeda and Takashi Ueda (The University of Tokyo) for Arabidopsis suspension cells; Dr. Masa H. Sato (Kyoto Prefectural University) for Venus-SYP plasmids; and Ms. Masami Miwa, Rie Ichikawa, and Chieko Nanba for technical assistance. ESEM was performed at the NIBB Center for Analytical Instruments. Mutant seeds were obtained from ABRC.
This work was supported by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Yoshinori Ohsumi (yohsumi@nibb).
The online version of this article contains Web-only data.
References
- Bassham DC, Laporte M, Marty F, Moriyasu Y, Ohsumi Y, Olsen LJ, Yoshimoto K (2006) Autophagy in development and stress responses of plants. Autophagy 2 2–11 [DOI] [PubMed] [Google Scholar]
- Giaever G, Chu AM, Ni L, Connelly C, Riles L, Véronneau S, Dow S, Lucau-Danila A, Anderson K, André B, et al (2002) Functional profiling of the Saccharomyces cerevisiae genome. Nature 418 387–391 [DOI] [PubMed] [Google Scholar]
- Gupta R, Ting JT, Sokolov LN, Johnson SA, Luan S (2002) A tumor suppressor homolog, AtPTEN1, is essential for pollen development in Arabidopsis. Plant Cell 14 2495–2507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamasaki M, Noda T, Baba M, Ohsumi Y (2005) Starvation triggers the delivery of the endoplasmic reticulum to the vacuole via autophagy in yeast. Traffic 6 56–65 [DOI] [PubMed] [Google Scholar]
- Hanaoka H, Noda T, Shirano Y, Kato T, Hayashi H, Shibata D, Tabata S, Ohsumi Y (2002) Leaf senescence and starvation-induced chlorosis are accelerated by the disruption of an Arabidopsis autophagy gene. Plant Physiol 129 1181–1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepler PK, Vidali L, Cheung AY (2001) Polarized cell growth in higher plants. Annu Rev Cell Dev Biol 17 159–187 [DOI] [PubMed] [Google Scholar]
- Hicks GR, Rojo E, Hong S, Carter DG, Raikhel NV (2004) Geminating pollen has tubular vacuoles, displays highly dynamic vacuole biogenesis, and requires VACUOLESS1 for proper function. Plant Physiol 134 1227–1239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson SA, McCormick S (2001) Pollen germinates precociously in the anthers of raring-to-go, an Arabidopsis gametophytic mutant. Plant Physiol 126 685–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kametaka S, Okano T, Ohsumi M, Ohsumi Y (1998) Apg14p and Apg6/Vps30p form a protein complex essential for autophagy in the yeast, Saccharomyces cerevisiae. J Biol Chem 273 22284–22291 [DOI] [PubMed] [Google Scholar]
- Karimi M, De Meyer B, Hilson P (2005) Modular cloning in plant cells. Trends Plant Sci 10 103–105 [DOI] [PubMed] [Google Scholar]
- Kihara A, Kabeya Y, Ohsumi Y, Yoshimori T (2001. b) Beclin-phosphatidylinositol 3-kinase complex functions at the trans-Golgi network. EMBO Rep 2 330–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kihara A, Noda T, Ishihara N, Ohsumi Y (2001. a) Two distinct Vps34 phosphatidylinositol 3-kinase complexes function in autophagy and carboxypeptidase Y sorting in Saccharomyces cerevisiae. J Cell Biol 152 519–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ, Ohsumi Y (1999) Vacuolar import of proteins and organelles from the cytoplasm. Annu Rev Cell Dev Biol 15 1–32 [DOI] [PubMed] [Google Scholar]
- Kost B, Lemichez E, Spielhofer P, Hong Y, Tolias K, Carpenter C, Chua NH (1999) Rac homologues and compartmentalized phosphatidylinositol 4,5-bisphosphate act in a common pathway to regulate polar pollen tube growth. J Cell Biol 145 317–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B, Klionsky DJ (2004) Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev Cell 6 463–477 [DOI] [PubMed] [Google Scholar]
- Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, Levine B (1999) Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature 402 672–676 [DOI] [PubMed] [Google Scholar]
- Liu Y, Schiff M, Czymmek K, Tallóczy Z, Levine B, Dinesh-Kumar SP (2005) Autophagy regulates programmed cell death during the plant innate immune response. Cell 121 567–577 [DOI] [PubMed] [Google Scholar]
- Lobstein E, Guyon A, Ferault M, Twell D, Pelletier G, Bonhomme S (2004) The putative Arabidopsis homolog of yeast Vps52p is required for pollen tube elongation, localizes to Golgi, and might be involved in vesicle trafficking. Plant Physiol 135 1480–1490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick S (2004) Control of male gametophyte development. Plant Cell (Suppl) 16 S142–S153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijer HJG, Munnik T (2003) Phospholipid-based signaling in plants. Annu Rev Plant Biol 54 265–306 [DOI] [PubMed] [Google Scholar]
- Meléndez A, Tallóczy Z, Seaman M, Eskelinen EL, Hall DH, Levine B (2003) Autophagy genes are essential for dauer development and life-span extension in C. elegans. Science 301 1387–1391 [DOI] [PubMed] [Google Scholar]
- Monteiro D, Coelho PC, Rodrigues C, Camacho L, Quader H, Malhó R (2005) Modulation of endocytosis in pollen tube growth by phosphoinositides and phospholipids. Protoplasma 226 31–38 [DOI] [PubMed] [Google Scholar]
- Obara K, Sekito T, Ohsumi Y (2006) Assortment of phosphatidylinositol 3-kinase complexes—Atg14p directs association of complex I to the preautophagosomal structure in Saccharomyces cerevisiae. Mol Biol Cell 17 1527–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohsumi Y (2001) Molecular dissection of autophagy: two ubiquitin-like systems. Nat Rev Mol Cell Biol 2 211–216 [DOI] [PubMed] [Google Scholar]
- Preuss D, Rhee SY, Davis RW (1994) Tetrad analysis possible in Arabidopsis with mutation of the QUARTET (QRT) genes. Science 264 1458–1460 [DOI] [PubMed] [Google Scholar]
- Regan SM, Moffatt BA (1990) Cytochemical analysis of pollen development in wild-type Arabidopsis and a male-sterile mutant. Plant Cell 2 877–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaman MNJ, Marcusson EG, Cereghino JL, Emr SD (1997) Endosome to Golgi retrieval of the vacuolar protein sorting receptor, Vps10p, requires the function of the VPS29, VPS30, and VPS35 gene products. J Cell Biol 137 79–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu KK, Okada K (2000) Attractive and repulsive interactions between female and male gametophytes in Arabidopsis pollen tube guidance. Development 127 4511–4518 [DOI] [PubMed] [Google Scholar]
- Suzuki K, Kirisako T, Kamada Y, Mizushima N, Noda T, Ohsumi Y (2001) The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J 20 5971–5981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson AR, Vierstra RD (2005) Autophagic recycling: lessons from yeast help define the process in plants. Curr Opin Plant Biol 8 165–173 [DOI] [PubMed] [Google Scholar]
- Ueda T, Yamaguchi M, Uchimiya H, Nakano A (2001) Ara6, a plant-unique novel type Rab GTPase, functions in the endocytic pathway of Arabidopsis thaliana. EMBO J 20 4730–4741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura T, Ueda T, Ohniwa RL, Nakano A, Takeyasu K, Sato MH (2004) Systematic analysis of SNARE molecules in Arabidopsis: dissection of the post-Golgi network in plant cells. Cell Struct Funct 29 49–65 [DOI] [PubMed] [Google Scholar]
- Vincent P, Chua M, Nogue F, Fairbrother A, Mekeel H, Xu Y, Allen N, Bibikova TN, Gilroy S, Bankaitis VA (2005) A Sec14p-nodulin domain phosphatidylinositol transfer protein polarizes membrane growth of Arabidopsis thaliana root hairs. J Cell Biol 168 801–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welters P, Takegawa K, Emr SD, Chrispeels MJ (1994) AtVPS34, a phosphatidylinositol 3-kinase of Arabidopsis thaliana, is an essential protein with homology to a calcium-dependent lipid binding domain. Proc Natl Acad Sci USA 91 11398–11402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto K, Hanaoka H, Sato S, Kato T, Tabata S, Noda T, Ohsumi Y (2004) Processing of ATG8s, ubiquitin-like proteins, and their deconjugation by ATG4s are essential for plant autophagy. Plant Cell 16 2967–2983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L, Alva A, Su H, Dutt P, Freundt E, Welsh S, Baehrecke EH, Lenardo MJ (2004) Regulation of an ATG7-beclin 1 program of autophagic cell death by caspase-8. Science 304 1500–1502 [DOI] [PubMed] [Google Scholar]
- Zeng X, Overmeyer JH, Maltese WA (2006) Functional specificity of the mammalian Beclin-Vps34 PI 3-kinase complex in macroautophagy versus endocytosis and lysosomal enzyme trafficking. J Cell Sci 119 259–270 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.