Abstract
The completion of the rice (Oryza sativa) genome draft has brought unprecedented opportunities for genomic studies of the world's most important food crop. Previous rice gene annotations have relied mainly on ab initio methods, which usually yield a high rate of false-positive predictions and give only limited information regarding alternative splicing in rice genes. Comparative approaches based on expressed sequence tags (ESTs) can compensate for the drawbacks of ab initio methods because they can simultaneously identify experimental data-supported genes and alternatively spliced transcripts. Furthermore, cross-species EST information can be used to not only offset the insufficiency of same-species ESTs but also derive evolutionary implications. In this study, we used ESTs from seven plant species, rice, wheat (Triticum aestivum), maize (Zea mays), barley (Hordeum vulgare), sorghum (Sorghum bicolor), soybean (Glycine max), and Arabidopsis (Arabidopsis thaliana), to annotate the rice genome. We developed a plant genome annotation pipeline, Plant Gene and Alternatively Spliced Variant Annotator (PGAA). Using this approach, we identified 852 genes (931 isoforms) not annotated in other widely used databases (i.e. the Institute for Genomic Research, National Center for Biotechnology Information, and Rice Annotation Project) and found 87% of them supported by both rice and nonrice EST evidence. PGAA also identified more than 44,000 alternatively spliced events, of which approximately 20% are not observed in the other three annotations. These novel annotations represent rich opportunities for rice genome research, because the functions of most of our annotated genes are currently unknown. Also, in the PGAA annotation, the isoforms with non-rice-EST-supported exons are significantly enriched in transporter activity but significantly underrepresented in transcription regulator activity. We have also identified potential lineage-specific and conserved isoforms, which are important markers in evolutionary studies. The data and the Web-based interface, RiceViewer, are available for public access at http://RiceViewer.genomics.sinica.edu.tw/.
Rice (Oryza sativa) is one of the most economically important cereal plants and a model organism for studies of crop plants. The high-quality sequencing of the entire rice genome was completed and publicly released in 2004 (International Rice Genome Sequencing Project, 2005). Although the rice genome has been extensively annotated (Bruskiewich et al., 2003; Karlowski et al., 2003; Kikuchi et al., 2003; Yuan et al., 2003; Juretic et al., 2004; Ito et al., 2005; Yuan et al., 2005; Jaiswal et al., 2006; Ohyanagi et al., 2006), the annotation results differ widely and contain a large number of predicted genes (Ito et al., 2005; Yuan et al., 2005). Although predicted genes to a great extent compensate for the limitations of expressed sequence tag (EST)-based gene annotations, many of them may be false positives. Furthermore, annotations of alternatively spliced transcripts of rice genes are underrepresented. Therefore, it is necessary to reexamine the predicted genes with use of experimental data and systematically analyze the alternatively spliced transcripts in the rice genome.
ESTs are direct evidence of gene expression. With suitable algorithms and well-curated ESTs, the inherent errors in EST information can be effectively reduced in gene/isoform annotations. Therefore, genes and alternative splicing (AS) transcripts can be simultaneously identified with high accuracy by use of experimental evidence (e.g. support from ESTs or microarray data; Zhu et al., 2003; Chuang et al., 2004; Iida et al., 2004; Ner-Gaon et al., 2004; Bonizzoni et al., 2005; Foissac and Schiex, 2005; Ner-Gaon and Fluhr, 2006). However, the number of rice ESTs is still limited, with only a small percentage (<50%) of annotated genes expressed, according to The Institute for Genomic Research (TIGR) Rice Genome Annotation (http://www.tigr.org/tdb/e2k1/osa1/riceInfo/info.shtml#Genes; Yuan et al., 2005). In animals, evolutionarily conserved ESTs can be applied with high accuracy to the prediction of genes and AS variants in EST-scarce species (Chuang et al., 2004; Kan et al., 2004; Chen et al., 2005, 2006). Because other crop plants and model organisms, such as maize (Zea mays), wheat (Triticum aestivum), and Arabidopsis (Arabidopsis thaliana) have been widely studied, inclusion of ESTs from these plant species may lead to the identification of rice genes/AS isoforms that have not previously been annotated. Furthermore, an evolutionary approach can also distinguish between conserved and lineage-specific isoforms and AS events, which are important to evolutionary studies. The Gramene Web site (http://www.gramene.org/Oryza_sativa) demonstrates that cross-species EST mapping to the rice genome may benefit evolutionary studies of the grass family, and a recent study comparing AS events between rice and Arabidopsis (Wang and Brendel, 2006) showed that important findings could be revealed with cross-species EST comparisons.
In this study, we developed a plant genome annotation pipeline, Plant Gene and Alternatively Spliced Variant Annotator (PGAA), for gene/AS prediction in the rice genome. PGAA is a comparative method that first identifies AS variants and genes by use of the same-species-EST-to-genome comparison and then curates the results with cross-species EST data conserved in the annotated genome. ESTs from seven plant species, rice, wheat, maize, barley (Hordeum vulgare), sorghum (Sorghum bicolor), soybean (Glycine max), and Arabidopsis, are used. All of the selected species have approximately 40,000 EST entries in the TIGR EST database. The PGAA annotation results are compared with those deposited in the National Center for Biotechnology Information (NCBI), Rice Annotation Project (RAP; First Rice Annotation Project Meeting; Ohyanagi et al., 2006), and TIGR (Yuan et al., 2005) databases. In this article, we describe functional analyses of PGAA-identified potentially novel genes/isoforms and discuss the evolutionary implications. To facilitate comparison of the annotation results and EST conservation, we have developed a Web-based visualization tool, RiceViewer, which is readily accessible to the public.
ANALYSIS PROCEDURE
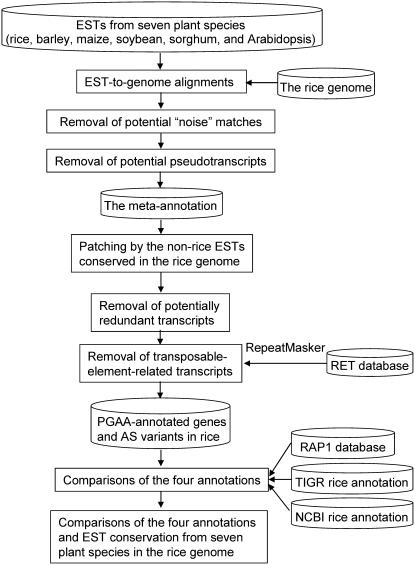
PGAA involves three consecutive steps: gene identification by use of rice ESTs (metaannotation), transcript patching by use of nonrice ESTs (the patching process), and redundancy removal. The annotation procedure is summarized in Figure 1.
Figure 1.
The analysis procedure of this study.
For metaannotation, the Complexity Reduction Algorithm for Sequence Analysis aligner (Chuang et al., 2003) is first used to search for EST fragments ≥18 bp (from the Rice Gene Index [OGI] EST and TIGR databases) that exactly match the rice genome for identification of potential gene loci. If the gap between two successive matched EST fragments is not larger than 10 bp, the gap is patched by the corresponding rice genomic sequence. Then, several criteria are used to reduce potential noise. The rice ESTs each with fewer than three rice genome-matching fragments are discarded unless the EST has at least one matching fragment longer than 100 bp or the matching fragments are also matched by a nonrice EST (Chuang et al., 2003, 2004). In addition, because ESTs may be chimeric or duplicated, two processes are used to filter out potential pseudotranscripts. First, when a rice EST has two hits in the rice genome, one hit representing a multiexon gene and the other a single-exon gene, the latter is regarded as a potential processed pseudogene and discarded. Second, following the first filter, when the remaining ESTs hit multiple times in the rice genome, only the best hit is retained for subsequent analysis. The process can remove potential pseudotranscripts that result from mapping gene duplications.
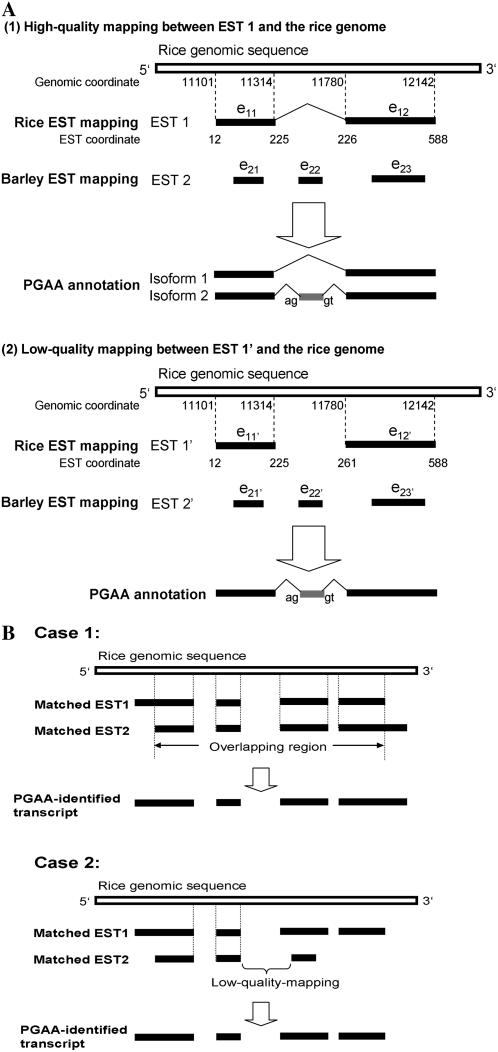
After the preliminary screening, the system patches the remaining EST matches with nonrice ESTs that are conserved in the rice genome. For example, as shown in Figure 2A (1), EST 1 (EST 2) is a rice (nonrice) EST with two (three) split fragments, e11 and e12 (e21, e22, and e23), which match the rice genomic sequence. However, e11 and e12 overlap with e21 and e23, respectively, with two possible results in our annotation. If the alignment between EST 1 and the rice genomic sequence has high quality (defined below), two isoforms will be annotated: isoform 1 with two exons (e11 and e12) and isoform 2 with three exons (e11, e22, and e12). Otherwise, only one isoform will be annotated (with three exons: e11, e22, and e12). The newly patched exon (e.g. e22) must be flanked by AG-GT/AG-GC legal splicing sites, not disrupt the reading frame, and contain no premature stop codons. Here, high-quality mapping means that e11 and e12 (which are originally contiguous on EST 1) match the rice genomic sequence in the correct order (i.e. no gap or mismatch exists between e11 and e12 on EST 1). Otherwise, the e11′-e12′ match is considered low-quality mapping because of a mismatched EST segment, which thus results in a gap in the EST-to-genome alignment between e11′ and e12′ (also see Fig. 2A [2]). Note that e22 and e22′ are not included in rice ESTs, which indicates that PGAA can identify potential missing exons or novel AS variants with use of nonrice ESTs. Also, all transcripts identified in this study are supported by evidence from expressed sequences (rice or nonrice ESTs).
Figure 2.
Examples of transcript annotation by PGAA. A, When a rice EST and a nonrice EST are mapped to the same rice genomic region, PGAA annotates three transcripts by high-quality rice EST matching (1). Otherwise, it patches the low-quality rice EST match with a nonrice EST segment and annotates only one transcript (2). See text for more details. B, Integration of EST matches in PGAA annotation. Case 1, The two transcripts are identical, except that the 5′ end of transcript 1 and the 3′ end of transcript 2 extend out of the overlapping part. Case 2, Transcript 2 is entirely included in transcript 1.
After the patching process, two criteria are used to reduce potentially redundant isoforms identified. First, if two identified isoforms overlap and the overlapping regions are identical, these two isoforms are assembled and replaced by the newly assembled isoform (Fig. 2B, case 1). Second, if an isoform is identified by a low-quality-mapped EST and is completely included in another isoform, it is discarded (Fig. 2B, case 2). Furthermore, to avoid potentially transposable-element-related isoforms in the PGAA annotation, we filter out repetitive elements by use of RepeatMasker (http://www.repeatmasker.org/) and Rice Transposable Element database (RTEdb; Juretic et al., 2004).
We then compare the PGAA annotated results with those from the three well-known rice annotation sources, RAP, TIGR, and NCBI (Build 2.1), and examine the four rice annotations for rice and nonrice ESTs that are conserved in the rice genome. The isoforms identified by nonrice ESTs in our annotation are singled out for analysis.
PGAA ANNOTATION RESULTS
In PGAA, the ESTs are downloaded from the TIGR gene index project (see “DATA ACCESS”). Table I illustrates the numbers of EST/tentative consensus (TC) sequences of the seven plant species analyzed in the PGAA system and the numbers (percentages) that are mapped to the rice genome. Note that TC sequences are generated by assembling ESTs into virtual transcripts, which may contain full or partial cDNA sequences (see the definition of the TIGR gene index project at http://compbio.dfci.harvard.edu/tgi/definitions.html). A large number of nonrice ESTs can be mapped to the rice genome. Particularly, as high as 32% to approximately 44% of the monocot cereal (i.e. barley, maize, sorghum, and wheat) ESTs and 48% to approximately 56% of the TCs are conserved in the rice genome. Such conserved nonrice ESTs can provide the resources for identification of potentially novel genes/AS variants in the rice genome. In addition, only ≤6% of the dicot plant (i.e. Arabidopsis and soybean) ESTs/TCs are alignable against the rice genome. This result is consistent with the phylogenetic relationships of the studied plants.
Table I.
The numbers of ESTs and TCs analyzed by PGAA and the numbers (percentages) of them mapped to the rice genome
| Total ESTsa | No. of TCs | No. of ESTs Mapped to the Rice Genome | No. of TCs Mapped to the Rice Genome | |
|---|---|---|---|---|
| Monocots | ||||
| Rice | 89,147 | 36,381 | 69,427 (78%) | 33,279 (91%) |
| Barley | 50,453 | 23,176 | 20,335 (40%) | 12,739 (55%) |
| Maize | 58,582 | 31,375 | 21,284 (36%) | 15,913 (51%) |
| Sorghum | 39,148 | 20,029 | 17,173 (44%) | 11,256 (56%) |
| Wheat | 122,282 | 44,954 | 38,629 (32%) | 21,697 (48%) |
| Dicots | ||||
| Arabidopsis | 62,010 | 28,900 | 2,225 (4%) | 1,697 (6%) |
| Soybean | 63,676 | 31,928 | 2,128 (3%) | 1,701 (5%) |
Total ESTs include TCs and other ESTs (e.g. singleton ESTs).
The PGAA system annotates a total of 34,512 genes (56,460 isoforms). The average number of isoforms per PGAA-identified gene is 1.63, and the total length of the annotated exon is 53.94 Mb (also see Table II). The average number of exons per annotated isoform is 4.1, whereas the average length of annotated exon and intron is 292 and 487 bp, respectively. For AS variant detection, 12,749 genes (36.9%) are annotated to be alternatively spliced, with a total of 34,697 isoforms that include 44,447 AS events (i.e. more than one AS event may occur to one isoform). Events include 10,131 (22.8%) exon skipping (or cassette on/off exon), 18,022 (40.5%) alternative donor/acceptor sites, and 16,294 (36.7%) intron retentions. As well, approximately 20% of the PGAA-annotated AS events are potentially novel, because they are not observed in the other three annotations (TIGR, NCBI, and RAP).
Table II.
Comparison of the TIGR, RAP, NCBI, and PGAA annotations for rice genome
| Annotations
|
||||
|---|---|---|---|---|
| TIGR | RAP | NCBI | PGAA | |
| No. of genes | 53,388 | 27,448 | 34,421 | 34,512 |
| No. of isoforms | 61,289 | 39,266 | 35,952 | 56,460 |
| Average isoforms/genes | 1.14 | 1.43 | 1.04 | 1.63 |
| No. of annotated genes (isoforms) not overlapping any of the other three annotations | 13,646 (14,272) | 2,704 (3,253) | 2,248 (2,615) | 852 (931) |
| Total length of annotated isoforms (Mb)a | 78.85 | 43.27 | 38.35 | 53.94 |
The total length of the annotated exonic sequences.
We then estimated the false-positive rate in the PGAA annotation. The emphasis here is that PGAA is an EST-based approach, which may be able to reduce false-positive predictions, thus reducing the searching scope for functional genes of rice. Of course, PGAA might yield false-positive predictions. In our early study on humans (Chuang et al., 2004), the false-positive rate (or the wrong exon rate) of such similar cross-species EST-based predictions was estimated to be 12.9%. In addition, a recent study by us on mammals (Chen et al., 2006) has shown that reverse transcription-PCR sequencing experiments validate approximately 50% to 80% of the novel AS events (or exons) identified by the cross-species EST-to-genome comparisons. In plants, however, the situation is more complicated, because plant genomes have undergone drastic changes. Therefore, the false-positive rate should be considered with caution when plants are concerned. Nevertheless, it is believed that the cross-species ESTs alignment is an invaluable resource for identification of potentially novel AS events/isoforms and that a considerable proportion of such novel predictions may be true.
ANNOTATION RESULTS AMONG TIGR, NCBI, RAP, AND PGAA
Table II shows the differences in rice genome annotation results among TIGR (Release 4), NCBI (Build 2.1), RAP (all RAP loci), and PGAA. TIGR annotates the largest number of genes and splicing isoforms, then PGAA, then NCBI, then RAP. Note that NCBI annotates only a small number of genes for the rice chromosome 11 and not chromosome 12 at all. Meanwhile, the average number of isoforms per annotated gene is the smallest in NCBI annotations but the largest in PGAA annotations. The isoform-to-gene ratio in the PGAA annotation is 14%, 43%, and 57% larger than those of the RAP, TIGR, and NCBI annotations, respectively. Because NCBI-, RAP-, and TIGR-annotated isoforms were identified with the aid of ab initio methods (e.g. FGENESH, Genscan, etc.) or full-length cDNAs, the number of AS variants identified is relatively limited. Of note, the number of TIGR-specific isoforms is high, 14,272. Because a large portion of these genes lack experimental evidence, many may be false-positive predictions (discussed in the next paragraph). However, although some ab initio predictions may be false positive, they provide an important source of potential rice genes for which expression data are still lacking. In some ab initio predictions, a considerable proportion (≥50%) of hypothetical genes were also experimentally verified (Xiao et al., 2005). Therefore, both EST-based and ab initio methods are important in gene finding.
Meanwhile, PGAA annotates genes with alignments between the rice genome and TIGR gene indices, which contain not only full-length cDNAs but also partial cDNAs and singleton ESTs (Liang et al., 2000). Although partial cDNAs and singleton ESTs are more likely than full-length cDNAs to be artificial, we have used several filters to remove potentially artificial ones in the PGAA process (discussed previously). Moreover, as shown in Figure 2A, PGAA also identifies rice isoforms with the support of nonrice ESTs. Therefore, PGAA can annotate more AS isoforms than the other three available databases.
In addition, PGAA annotates 931 isoforms that are not annotated by TIGR, RAP, or NCBI (Table II). Of these 931 isoforms, 808 (87%) are supported by both rice and nonrice EST evidence. Using the Gene Ontology (GO; Gene Ontology Consortium, 2001) and the Inter-ProScan protein domain annotations (Mulder et al., 2005; Quevillon et al., 2005), we find that about 10% of these potential novel isoforms have GO assignments or INTERPRO-annotated protein domains (Supplemental Table S1). Thus, the functions of most PGAA-specific isoforms remain unknown. We further probed the gene structures of these isoforms and discovered that 279 (652) have multiple (single) exons. Because single-exon isoforms lack the genetic property of splice junctions (i.e. AG-GT/AG-GC legal splicing sites), the accuracy of single-exon isoform prediction is generally lower than that of multiple-exon prediction. However, in the PGAA annotation, most of the PGAA-specific single-exon isoforms (576, >88%) are supported by not only rice but also nonrice ESTs. Such cross-species EST conservation strongly indicates that these PGAA-specific single-exon isoforms are likely real.
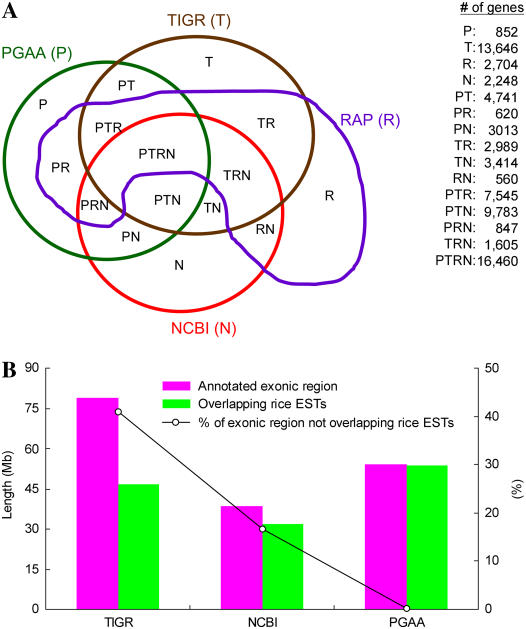
Figure 3A shows a Venn diagram to compare TIGR, NCBI, RAP, and PGAA annotation results in terms of gene number. Collectively, the four annotations annotated 71,029 genes, of which 51,763 (72.9%) are annotated by at least two methods. Note that two annotated genes/isoforms are considered the same gene in this analysis if they overlap with each other and share at least one exon. These four annotations give very different results, with only 16,460 (23%) genes in common. The TIGR-specific genes account for 20% of the collective total of annotated genes, whereas the RAP-, NCBI-, and PGAA-specific genes account for 4%, 3%, and 1%, respectively.
Figure 3.
A, A Venn diagram of the number of genes annotated by PGAA (P), TIGR (T), NCBI (N), and RAP (R) annotations. For example, PT means that the genes are annotated by both PGAA and TIGR but not by NCBI or RAP. B, Comparison of rice EST coverage by the PGAA, TIGR, and NCBI annotations.
Because some TIGR-, NCBI-, and PGAA-annotated genes are not fully supported by rice ESTs, we compared these three annotations in terms of proportion of rice EST coverage. Figure 3B shows that 32.29 Mb (approximately 41%) and 6.37 Mb (approximately 17%) of TIGR- and NCBI-annotated exonic sequences, respectively, do not overlap with any rice ESTs. Despite the limitations in rice EST information, the high proportion of non-rice-EST-overlapping isoforms in the TIGR and NCBI databases is questionable. Figure 3B also shows that only a small fraction (approximately 0.1%, or 58 kb) of PGAA-identified isoforms do not overlap with any rice ESTs (i.e. are supported only by nonrice ESTs). Note that the RAP gene loci are identified on the basis of either full-length rice cDNA matches or ab initio prediction plus rice EST coverage (see the RAP annotation document at http://rapdownload.lab.nig.ac.jp/index.html). Therefore, all of the RAP-identified loci are rice-EST supported and are not illustrated in the figure.
CROSS-SPECIES EST CONSERVATION IN THE RICE GENOME
Table III indicates the same-species and cross-species EST conservation in rice genes annotated in TIGR, NCBI, RAP, and PGAA. In terms of length, only 67%, 60%, and 6% of the annotated isoforms overlap with either rice or nonrice ESTs, rice ESTs only, and both rice and nonrice ESTs, respectively. The absence of overlap with nonrice ESTs most likely results from inadequate EST information. However, some of the isoforms might be rice specific. Therefore, the isoforms may have been conserved across species or have different splicing forms in the orthologous genes in different species. Further analyses are required to determine which of the above two explanations is true. However, about 1% of the isoforms overlap with only nonrice ESTs, and 33% overlap with no ESTs at all. The authenticity of these isoforms needs further validation. Note that, by the definitions of RAP and PGAA, all RAP- and PGAA-annotated isoforms are EST supported. Therefore, the annotated isoforms not supported by ESTs must come from either the TIGR or the NCBI annotations. Meanwhile, some of the nonrice ESTs conserved in the rice genome do not overlap with any annotated isoforms or the rice ESTs. These ESTs might represent some gain or loss events in the crop plants after domestication. Because crop plants are known to have undergone whole-genome duplications and large-scale gene loss events (Paterson et al., 2004), lineage- or species-specific genes/isoforms may have contributed to the phenotype divergences.
Table III.
EST conservation in annotated transcripts
| Size | |
|---|---|
| Mb | |
| Collective annotated transcripts of four annotationsa | 108.00 |
| Overlapping rice or nonrice ESTs | 72.52 (67%) |
| Overlapping both rice and nonrice ESTs | 6.79 (6%) |
| Overlapping rice ESTs only | 64.61 (60%) |
| Overlapping nonrice ESTs only | 1.12 (1%) |
| Overlapping no ESTs | 35.48 (33%) |
| Total nonrice ESTs conserved in the rice genome but overlapping none of the annotated isoforms or rice ESTs | 2.81 |
| From barley | 1.41 |
| From wheat | 0.80 |
| From maize | 0.56 |
| From soybean | 0.11 |
| From sorghum | 0.45 |
| From Arabidopsis | 0.13 |
The total length of genomic sequences covered by all annotated isoforms.
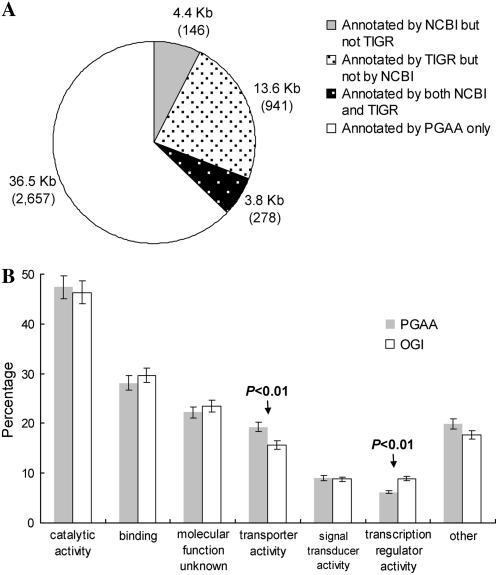
Figure 4A illustrates the lengths and numbers of PGAA-annotated rice isoforms that contain exonic regions supported by nonrice but not rice ESTs. The total length of those non-rice-EST-supported exonic regions is approximately 58 kb, of which 36.5 kb is annotated by PGAA only, 4.4 kb is also annotated by NCBI but not by TIGR, 13.6 kb by TIGR but not by NCBI, and 3.8 kb by both TIGR and NCBI. Notably, the 36.5 kb accounts for 2,657 isoforms, which contain at least one PGAA-unique exonic region. Because at least one of the exons (partial or complete) in each annotated gene locus is supported by nonrice EST(s), these isoforms are likely evolutionarily conserved. Moreover, such identified exons may also represent novel AS events. Also note that RAP annotations are not considered in Figure 4, because all of the RAP-annotated loci are supported by rice ESTs.
Figure 4.
A, The lengths and numbers of PGAA-annotated rice isoforms supported by nonrice ESTs. B, Distributions of the 2,657 isoforms (Fig. 4A) that contain the PGAA-specific exonic regions supported by nonrice ESTs and ESTs of the TIGR OGI in molecular function subcategories of GO. The error bars indicate 95% confidence interval. Statistical significance evaluated by two-tailed Fisher's exact test.
PGAA differs from other EST-based annotation tools in that it uses cross-species EST information to compensate for the insufficiency and correct the errors of the use of same-species ESTs. These rescued genes/isoforms may have important functions in terms of evolutionary conservation. We performed a GO-based functional analysis (Gene Ontology Consortium, 2001) for the PGAA-specific annotations (i.e. 2,657 isoforms in Fig. 4A) and found only 1,195 isoforms (45%) with GO assignments. Compared with the GO annotations of OGI (downloaded from the TIGR database; Fig. 4B), these 1,195 isoforms were significantly enriched in transporter activity (P < 0.01 by two-tailed Fisher's exact test) but significantly underrepresented in transcription regulator activity (P < 0.01) of the molecular function subcategories. Either a considerable number of transporter-protein AS isoforms have not been discovered or interspecies AS divergence is particularly significant in this functional group. The latter scenario suggests that the newly identified isoforms supported by nonrice ESTs have contributed to the divergence between rice and other nonrice species at the RNA level. However, the interspecies conservation of AS isoforms in transcription regulator activity indicates that this basic cellular activity has undergone only minor RNA-level changes during the evolutionary diversification of the grass family. In contrast, no significant differences were observed between the newly identified isoforms and the OGI isoforms in the other two main GO categories, biological process and cellular component (Supplemental Fig. S1).
DESCRIPTION OF RICEVIEWER WEB INTERFACE
All data generated in this study are accessed through a Web-based interactive interface, RiceViewer (http://RiceViewer.genomics.sinica.edu.tw/). The RiceViewer presents the structure and AS variants of rice genes on the basis of four annotation sources: NCBI, TIGR, RAP, and PGAA. For each annotated gene, EST matches from the seven studied species are also provided.
The interface supports three types of queries. It can accept rice mRNA or protein accession numbers, rice genomic coordinates, and nucleotide sequences for BLAST searching against the rice genome (Fig. 5A). In the first type of query, the coordinates of the queried gene are shown in the query results, whereas in the second type, the information of all annotated genes located within the specified region is displayed in small-to-large genomic coordinate order (Fig. 5B). By clicking on one of the gene regions, the gene structures and AS variants of the selected gene region are displayed according to TIGR, NCBI, RAP, and PGAA annotations (Fig. 5C). Note that if an annotation identifies no transcripts within the selected region, then the corresponding slot will be empty. Moreover, in the PGAA annotation, the accession identifier is highlighted if it also belongs to a clone of the Knowledge-based Oryza Molecular biological Encyclopedia (KOME; Fig. 5C). By clicking on the PGAA ID, the related descriptions of the isoforms (including KOME clone ID, nucleotide sequence, and GO annotation) are shown in a pop-up window (Fig. 5D). Users can click on the visit KOME button to link to the corresponding KOME report. Also note that the colors of exons in Figure 5C indicate the direction and level of EST coverage for the exons presented. Light gray represents exons that do not have EST evidence from rice or the six other species, likely predicted exons with no current EST evidence. Green and red indicate exons that are encoded in direct and complement strands, respectively. Each color is further divided into four shade levels according to EST coverage levels: dark green for exons with EST evidence from both rice and at least one of the six other species, light green for those with only rice EST evidence, and medium green for those with only EST evidence from at least one of the six other species but not from rice. The color-coding scheme thus enables users to distinguish exons with different directions and EST coverage levels at a glance. We also provide a comparison of the four annotations (see Fig. 5E). The interface also shows the sizes and proportions of ESTs from the seven species that overlap with the four annotations in the user-specified gene region. Moreover, the lengths of ESTs not overlapping with any one of the four annotations are also illustrated (Fig. 5E). These ESTs are likely noise and hence are filtered out in the PGAA annotation process.
Figure 5.
The RiceViewer interface. A, Types of queries. Users can query by gene accession numbers, genomic regions, or nucleotide sequences. B, Gene regions found by specifying a genomic region. C, TIGR, NCBI, RAP, and PGAA annotations of the selected gene. D, The related descriptions of the PGAA-annotated isoforms. By clicking on the visit KOME button, users can link to the KOME report. E, Comparison of the four annotations and the proportion of ESTs overlapping the annotated isoforms in the four annotations. See text for more details. F, Cross-species EST conservation in the selected gene. The interface automatically displays all the exons of the selected isoform if the user clicks on the Automatic Display button. Users can record in (G) the coordinates of the gene(s) that they have selected to track their analyses.
In addition, the coordinates of each exon can be shown by pointing the cursor to the exon of interest. By double clicking on the exon of interest, the nucleotide sequence and supporting ESTs for the selected exon are displayed in a pop-up window (Fig. 5F). Users can click on the accession number shown in Figure 5C to link to the NCBI Entrez Gene database for more information about the gene. The genomic coordinates of the gene regions for which users have browsed the corresponding EST conservation are shown on the right side in the Collection column in Figure 5G for users to track their analysis.
In the third type of query, the submitted nucleotide sequences are BLASTN aligned against the rice genome, and the alignment outputs are shown in a pop-up window. The interface simultaneously retrieves the coordinates of the three best hits from each of the three top-score rice chromosomes (if applicable) in the BLAST output file and displays genes located within these coordinates, as described above. Again, all the genes identified by the four annotations within the specified region are shown unless no annotated gene is available. Given such a condition, the gene display region (Fig. 5B) will be blank.
DATA ACCESS
The RAP, TIGR, and NCBI annotations of the rice genome were downloaded from http://rapdownload.lab.nig.ac.jp (RAP1, based on the International Rice Genome Sequencing Project genome sequence Build 3), http://rice.tigr.org/tdb/e2k1/osa1/data_download.shtml (release 3.0), and http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=genomeprj&cmd=Retrieve&dopt=Overview&list_uids=122 (Build 2.1), respectively. RTEdb (Juretic et al., 2004), created by combining the TIGR Oryza Repeat Database (Ouyang and Buell, 2004) with other published and unpublished RTE sequences, was kindly provided by Dr. Nikoleta Juretic. The original rice genomic sequences were also downloaded from NCBI (Build 2.1). The original EST databases are publicly available at the TIGR database (i.e. the gene index project; http://compbio.dfci.harvard.edu/tgi/). The EST databases of Arabidopsis, barley, maize, rice, sorghum, soybean, and wheat used in this study were AGI Release 12.1 (67 Mb), Hordeum vulgare Gene Index Release 9 (39 Mb), Zea mays Gene Index Release 15 (16 Mb), OGI Release 16 (99 Mb), Sorghum bicolor Gene Index Release 8 (30 Mb), Glycine max Gene Index Release 12 (41 Mb), and Triticum aestivum Gene Index Release 10 (86 Mb), respectively.
CONCLUDING REMARKS
In this study, we designed a cross-species EST-based pipeline for rice genome annotations and provided a Web-based interface for comparative studies. We found remarkable differences in results from current annotations and identified a large number of potentially novel genes and AS isoforms in rice with our system. Many of the isoforms are supported by nonrice ESTs, so they are interesting targets for future functional and evolutionary studies. As the numbers of crop plant ESTs increase, our system can help with detailed investigation of the AS isoforms of rice and AS evolution in the grass family.
Sequence data from this article can be found in the GenBank/EMBL/DDBJ data libraries under accession number AAAA00000000 or AACV00000000.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Distributions of the 2,657 transcripts (see Fig. 4A) that contain the PGAA-specific exonic regions supported by nonrice ESTs and ESTs of the TIGR OGI in the Biological Process and Cellular Component subcategories of GO. The error bars indicate 95% confidence interval.
Supplemental Table S1. Top 10 GO assignments of the 2,657 isoforms in Figure 4A.
Supplementary Material
Acknowledgments
We especially thank Dr. Nikoleta Juretic for providing RTEdb. In addition, we extend our deep gratitude to all the administrators of the cited sequencing centers and scientists who made their sequence data and annotation results available to the public. Without their efforts, this study would not be possible. We also gratefully acknowledge the critical and valuable criticism of the two anonymous reviewers.
This work was supported by the Genomics Research Center, Academia Sinica, Taiwan, by the National Health Research Institutes, Taiwan (contract no. NHRI–EX95–9408PC to T.-J.C., National Health Research Institutes intramural funding to F.-C.C.), and by the Research Center for Biodiversity, Academia Sinica (to S.-M.C.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Trees-Juen Chuang (trees@gate.sinica.edu.tw).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Bonizzoni P, Rizzi R, Pesole G (2005) ASPIC: a novel method to predict the exon-intron structure of a gene that is optimally compatible to a set of transcript sequences. BMC Bioinformatics 6 244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruskiewich RM, Cosico AB, Eusebio W, Portugal AM, Ramos LM, Reyes MT, Sallan MA, Ulat VJ, Wang X, McNally KL, et al (2003) Linking genotype to phenotype: the International Rice Information System (IRIS). Bioinformatics (Suppl 1) 19 i63–65 [DOI] [PubMed] [Google Scholar]
- Chen FC, Chen CJ, Ho JY, Chuang TJ (2006) Identification and evolutionary analysis of novel exons and alternative splicing events using cross-species EST-to-genome comparisons in human, mouse and rat. BMC Bioinformatics 7 136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen FC, Wang SS, Chen CJ, Li WH, Chuang TJ (2005) Alternatively and constitutively spliced exons are subject to different evolutionary forces. Mol Biol Evol 23 675–682 [DOI] [PubMed] [Google Scholar]
- Chuang TJ, Chen FC, Chou MY (2004) A comparative method for identification of gene structures and alternatively spliced variants. Bioinformatics 20 3064–3079 [DOI] [PubMed] [Google Scholar]
- Chuang TJ, Lin WC, Lee HC, Wang CW, Hsiao KL, Wang ZH, Shieh D, Lin SC, Ch'ang LY (2003) A complexity reduction algorithm for analysis and annotation of large genomic sequences. Genome Res 13 313–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foissac S, Schiex T (2005) Integrating alternative splicing detection into gene prediction. BMC Bioinformatics 6 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gene Ontology Consortium (2001) Creating the gene ontology resource: design and implementation. Genome Res 11 1425–1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iida K, Seki M, Sakurai T, Satou M, Akiyama K, Toyoda T, Konagaya A, Shinozaki K (2004) Genome-wide analysis of alternative pre-mRNA splicing in Arabidopsis thaliana based on full-length cDNA sequences. Nucleic Acids Res 32 5096–5103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Rice Genome Sequencing Project (2005) The map-based sequence of the rice genome. Nature 436 793–800 [DOI] [PubMed] [Google Scholar]
- Ito Y, Arikawa K, Antonio BA, Ohta I, Naito S, Mukai Y, Shimano A, Masukawa M, Shibata M, Yamamoto M, et al (2005) Rice Annotation Database (RAD): a contig-oriented database for map-based rice genomics. Nucleic Acids Res 33 D651–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaiswal P, Ni J, Yap I, Ware D, Spooner W, Youens-Clark K, Ren L, Liang C, Zhao W, Ratnapu K, et al (2006) Gramene: a bird's eye view of cereal genomes. Nucleic Acids Res 34 D717–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juretic N, Bureau TE, Bruskiewich RM (2004) Transposable element annotation of the rice genome. Bioinformatics 20 155–160 [DOI] [PubMed] [Google Scholar]
- Kan Z, Castle J, Johnson JM, Tsinoremas NF (2004) Detection of novel splice forms in human and mouse using cross-species approach. Pac Symp Biocomput 9 42–53 [DOI] [PubMed] [Google Scholar]
- Karlowski WM, Schoof H, Janakiraman V, Stuempflen V, Mayer KF (2003) MOsDB: an integrated information resource for rice genomics. Nucleic Acids Res 31 190–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi S, Satoh K, Nagata T, Kawagashira N, Doi K, Kishimoto N, Yazaki J, Ishikawa M, Yamada H, Ooka H, et al (2003) Collection, mapping, and annotation of over 28,000 cDNA clones from japonica rice. Science 301 376–379 [DOI] [PubMed] [Google Scholar]
- Liang F, Holt I, Pertea G, Karamycheva S, Salzberg SL, Quackenbush J (2000) An optimized protocol for analysis of EST sequences. Nucleic Acids Res 28 3657–3665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulder NJ, Apweiler R, Attwood TK, Bairoch A, Bateman A, Binns D, Bradley P, Bork P, Bucher P, Cerutti L, et al (2005) InterPro, progress and status in 2005. Nucleic Acids Res 33 D201–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ner-Gaon H, Fluhr R (2006) Whole-genome microarray in Arabidopsis facilitates global analysis of retained introns. DNA Res 13 111–121 [DOI] [PubMed] [Google Scholar]
- Ner-Gaon H, Halachmi R, Savaldi-Goldstein S, Rubin E, Ophir R, Fluhr R (2004) Intron retention is a major phenomenon in alternative splicing in Arabidopsis. Plant J 39 877–885 [DOI] [PubMed] [Google Scholar]
- Ohyanagi H, Tanaka T, Sakai H, Shigemoto Y, Yamaguchi K, Habara T, Fujii Y, Antonio BA, Nagamura Y, Imanishi T, et al (2006) The Rice Annotation Project Database (RAP-DB): hub for Oryza sativa ssp. japonica genome information. Nucleic Acids Res 34 D741–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang S, Buell CR (2004) The TIGR Plant Repeat Databases: a collective resource for the identification of repetitive sequences in plants. Nucleic Acids Res 32 D360–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson AH, Bowers JE, Chapman BA (2004) Ancient polyploidization predating divergence of the cereals, and its consequences for comparative genomics. Proc Natl Acad Sci USA 101 9903–9908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quevillon E, Silventoinen V, Pillai S, Harte N, Mulder N, Apweiler R, Lopez R (2005) InterProScan: protein domains identifier. Nucleic Acids Res 33 W116–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang BB, Brendel V (2006) Genomewide comparative analysis of alternative splicing in plants. Proc Natl Acad Sci USA 103 7175–7180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao YL, Smith SR, Ishmael N, Redman JC, Kumar N, Monaghan EL, Ayele M, Haas BJ, Wu HC, Town CD (2005) Analysis of the cDNAs of hypothetical genes on Arabidopsis chromosome 2 reveals numerous transcript variants. Plant Physiol 139 1323–1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Q, Ouyang S, Liu J, Suh B, Cheung F, Sultana R, Lee D, Quackenbush J, Buell CR (2003) The TIGR rice genome annotation resource: annotating the rice genome and creating resources for plant biologists. Nucleic Acids Res 31 229–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Q, Ouyang S, Wang A, Zhu W, Maiti R, Lin H, Hamilton J, Haas B, Sultana R, Cheung F, et al (2005) The institute for genomic research osa1 rice genome annotation database. Plant Physiol 138 18–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W, Schlueter SD, Brendel V (2003) Refined annotation of the Arabidopsis genome by complete expressed sequence tag mapping. Plant Physiol 132 469–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.