Abstract
MicroRNAs (miRNAs) have important roles in diverse cellular processes, but little is known about their identity and functions during early mammalian development. Here, we show the effects of the loss of maternal inheritance of miRNAs following specific deletion of Dicer from growing oocytes. The mutant mature oocytes were almost entirely depleted of all miRNAs, and they failed to progress through the first cell division, probably because of disorganized spindle formation. By comparing single-cell cDNA microarray profiles of control and mutant oocytes, our data are compatible with the notion that a large proportion of the maternal genes are directly or indirectly under the control of miRNAs, which demonstrates that the maternal miRNAs are essential for the earliest stages of mouse embryonic development.
Keywords: Maternal microRNAs, Dicer, oocyte, zygote
MicroRNAs (miRNAs) are a large family of short noncoding RNAs (17–25 nucleotides) (He and Hannon 2004). A key function of miRNAs is to repress expression of their target genes through sequence complementation, which reduces the abundance of the target mRNAs and/or inhibits their translation (Bartel 2004; Bagga et al. 2005). MiRNA genes are first transcribed into miRNA primary transcripts by RNA polymerase II (Kim 2005). These primary transcripts are then processed into miRNA precursors by the Drosha/DGCR8 complex and transported from the nucleus to the cytoplasm. Finally, Dicer processes the miRNA precursors into mature miRNAs. From previous studies, Dicer seems to be critical for early mouse development since its loss of function is embryonic lethal at embryonic day 7.5 (E7.5) (Bernstein et al. 2003).
In this study, we have examined the role of miRNAs in the mouse oocyte. The mature oocyte contains a number of molecules that are manufactured during oocyte maturation and utilized during early stages of development before activation of the embryonic genome (Dean 2002). It is likely that miRNAs would also be present in the oocyte, but no information is yet available in the mouse. The purpose of this study was to determine if there is significant inheritance of maternal miRNAs in mammalian zygotes, and to investigate if they play a critical role in early mammalian development. We have investigated how the loss of Dicer affects synthesis of miRNA during oocyte maturation and their impact on mRNA and early development.
Results and Discussion
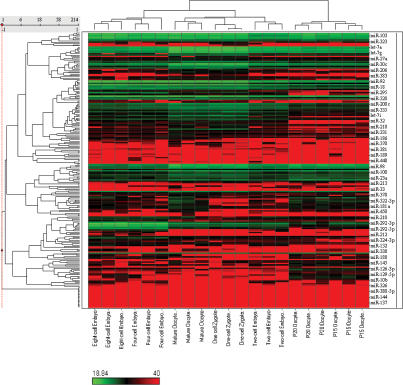
First, we decided to investigate if there is significant biogenesis of miRNAs in developing oocytes, and their inheritance in the zygote. We therefore examined expression of miRNAs in single cells during oogenesis by a real-time PCR-based miRNA expression profiling method that we recently developed (C. Chen et al. 2005; Tang et al. 2006a, b). We compared the miRNA expression profiles of growing oocytes obtained from females 15–16 d after birth (postnatal days 15–16 [P15–P16]) and at P20–P21, and of mature oocytes from adult females. This analysis reveled dynamic changes in miRNA expression during oogenesis (Fig. 1; Supplementary Table S1).
Figure 1.
Unsupervised hierarchical clustering heat map of miRNA expression profile of oocytes and early embryos. The cluster heat map was produced using expression levels (Ct value) of 214 miRNAs. Correlation coefficient was used as a similarity measure and a complete linkage method was used as a clustering method. Note that a higher Ct value means a lower expression level.
Next, we compared expression of miRNA in the mature oocyte with miRNAs in the zygote, which showed essentially the same miRNA expression pattern in both these cells (Fig. 1; Supplementary Tables S1, S2). This observation indicates that the miRNAs we detected in the zygote are probably maternally inherited from oocytes and not transcribed in the zygote itself.
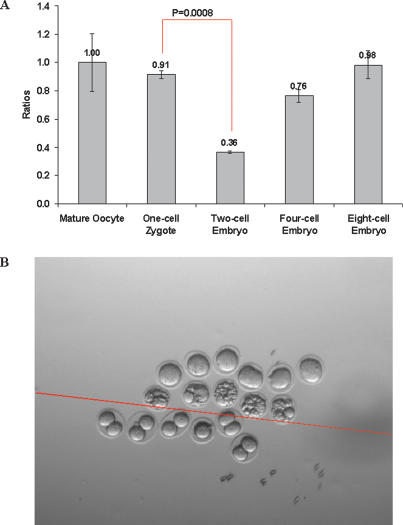
We then went on to determine if there are significant changes in miRNAs during early embryonic development. Indeed, we found that the miRNA expression profile of the zygote underwent dynamic changes during early embryonic development when examined through to the eight-cell stage (Fig. 1; Supplementary Table S2). Notably, we found that the total amount of miRNA is down-regulated by 60% between one- and two-cell-stage embryos (P < 0.001) (Fig. 2A; Supplementary Table S3), suggesting that a very significant proportion of the maternally inherited miRNAs present in the zygote is probably actively degraded during the first cell division. Some of these miRNAs were selectively lost by >95% (Supplementary Table S2). This is unexpected because there are no reports to show that miRNAs can be actively degraded in vivo under physiological conditions, although maternal mRNAs are globally degraded at this time (Hamatani et al. 2004). We confirmed our findings of the loss of maternal miRNAs by RNA in situ hybridization with LNA probes, which has been shown to be able to detect mature miRNA expression (Supplementary Fig. S2C,D; Supplementary Table S4; Kloosterman et al. 2006). Among the maternal miRNAs in the zygote, the most abundant are let-7 family miRNAs. They show dynamic regulation during oogenesis and early embryonic development (Supplementary Fig. S2A,B). The most abundant cluster is the miR-17-92 cluster. This miRNA cluster has been shown to be “oncomirs,” which is involved in cell proliferation (He et al. 2005; O’Donnell et al. 2005). Their abundance significantly increased during oogenesis and was inherited by the zygote and increased again after the two-cell embryo stage (Supplementary Fig. S3).
Figure 2.
(A) Total amount of miRNA in mature oocytes and early embryos. The error bars were the standard deviations calculated from three independent samples. (B) Morphology of mutant embryos from Dicer knockout oocytes, compared with wild-type control embryos at E1.5. Embryos above the red line are mutant, while embryos below the red line are wild-type controls.
Further investigations showed that the total miRNA in a four-cell-stage embryo was ∼2.2 times higher than the levels detected in a two-cell-stage embryo (Fig. 2A; Supplementary Table S3). This suggests that there is de novo expression of miRNAs between the two- and four-cell stages of development. Amongst the most significant miRNAs that are up-regulated in a four-cell-stage embryo are those from the miR-290 cluster, namely miR-290 to miR-295 (Houbaviy et al. 2003). Compared with the two-cell-stage embryos, these miRNAs were up-regulated by 15-fold and by 24-fold in four-cell- and eight-cell-stage embryos, respectively (Supplementary Fig. S4A,B; Supplementary Table S5). Thus, miRNAs from the miR-290 cluster are amongst the earliest to be expressed during early mouse embryonic development.
To determine if there are significant differences in miRNAs in individual blastomeres, we separated and compared the miRNA expression profiles of the two individual blastomeres from individual two-cell-stage embryos. We found that both blastomeres examined separately have essentially the same miRNA expression profile (Supplementary Fig. S5A; Supplementary Table S10). Furthermore, the total amount of miRNAs in the two blastomeres together was only ∼40% of that detected in the zygote. This shows that the miRNA profiling method is reliable and confirms that a significant proportion of the miRNAs is degraded in embryos between the one- and two-cell stages. Similarly, the miRNA expression profiles of the four individual blastomeres from a four-cell-stage embryo are essentially the same (Supplementary Fig. S5B; Supplementary Table S10). These observations indicate that the individual blastomeres at two-cell and four-cell embryo stages have similar if not identical miRNA expression profiles.
Next, we asked if these maternally inherited miRNAs are functionally important for early development. To investigate this aspect, we used mice carrying the Dicer floxed conditional allele, where exon 23 of Dicer locus is flanked by two loxP loci (Yi et al. 2006). These mice with the floxed allele of Dicer were mated with Zp3-Cre transgenic mice, which express Cre recombinase under the control of Zona pellucida glycoprotein 3 promoter (Zp3) in the growing oocyte. We thus generated [Dicer−/Flox, Zp3-Cre] animals. It is known that Zp3 expression is detected only in growing oocytes from about P5, which allowed us to delete Dicer specifically from maturing oocytes (de Vries et al. 2000). We would therefore expect loss of Dicer function in growing oocytes, and this would in turn block biogenesis of miRNAs at this stage. To establish how the loss of Dicer from the growing oocytes affects miRNAs in oocytes, we examined the miRNA profile in the mutant oocytes. We found that most, if not all, miRNAs were essentially lost from the oocytes lacking Dicer (Supplementary Fig. S6; Supplementary Table S1). We also found that the loss of the Dicer allele from oocytes rendered the females infertile. To determine the reason for infertility and when the effect manifests itself, we recovered E1.5 embryos from control and mutant oocytes following the deletion of Dicer from the growing oocytes that were fertilized by sperm from wild-type males (Fig. 2B). We found that the control females had healthy two-cell-stage embryos as expected, but all of the mutant oocytes failed to proceed through the first cell division, and about half of them were fragmented (three litters, n = 28). Thus, maternal Dicer and miRNAs in the oocyte are crucial for the earliest stages of embryonic development.
To determine more precisely the stage at which the loss of Dicer from oocytes starts to affect oogenesis and embryonic development, we examined the expression of some of the key genes by single-cell cDNA analysis at three different stages during oogenesis (Rajkovic et al. 2004). We found that expression of a number of genes, including Oct4, Fragilis, Stella, C-mos, Bnc1, H2AX, H1foo, SCP3, Nobox, Gata4, and RFPL4, was unaffected in growing oocytes at P15–P16 (Supplementary Fig. S1). The overall morphology of the mutant oocytes was also indistinguishable from that of control oocytes at this stage. Next, we checked expression of the same genes in mature ovulated oocytes, which we obtained from [Dicer−/Flox, Zp3-Cre] females that were mated with vasectomized males. We again found that the Dicer mutant oocytes were morphologically indistinguishable from control oocytes in their appearance, maturity, and size. The overall numbers of ovulated mutant oocytes were also indistinguishable from those obtained from control females. However, we found that the Dicer mutant oocytes showed higher expression of C-mos and H2AX compared with the levels we found in control oocytes (Supplementary Fig. S1). In the E0.5 embryos, genes including H1foo and SCP3 also showed higher expression compared with the control. Therefore, mature oocytes lacking in Dicer, and consequently miRNAs, already showed an effect on mRNAs.
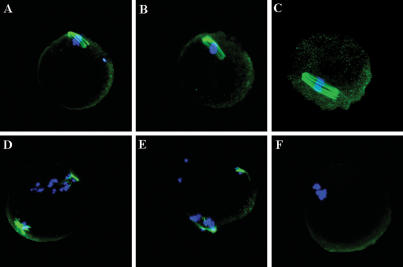
To determine if the miRNAs are involved in maturation of the oocyte, we also checked the chromosome and spindle organization (Lefebvre et al. 2002; Terret et al. 2003). We found that compared with control oocytes, the Dicer mutant oocytes showed reduced and disorganized spindles, and the chromosomes were also not aligned altogether properly (Fig. 3). This indicates that loss of Dicer and miRNAs affects the spindle organization of mature oocytes.
Figure 3.
Spindle in mutant mature oocytes (D–F) compared with wild-type mature oocytes (A–C). The spindle was stained with rat monoclonal anti-tubulin (YL1/2) antibody (green), and the chromosome was stained with DAPI (blue).
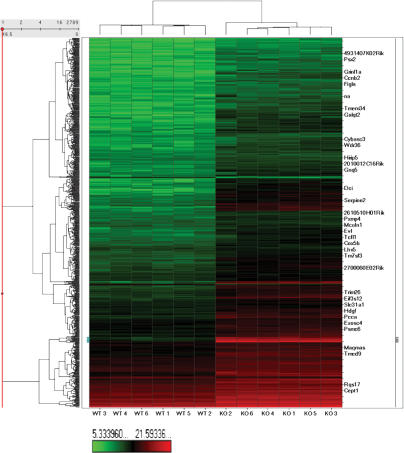
To obtain more comprehensive information on how miRNAs affect embryonic development at the whole-genome scale, we prepared single-cell cDNAs from control and Dicer mutant oocytes for microarray analysis (Kurimoto et al. 2006). We found that compared with control oocytes, Dicer knockout oocytes showed an increase in the levels of many genes that are probably important during early development. The cDNA levels of more than one-third of the genes expressed in oocytes increased (>1.5-fold) after the loss of Dicer and miRNAs (Fig. 4; Supplementary Table S7). We confirmed the microarray data by quantitative PCR on single-cell cDNAs (Supplementary Table S8). This analysis shows that miRNAs that are expressed during oogenesis profoundly shape the gene expression profile of the mature oocyte. Since we did not find significant enrichment of predicted target genes of miRNAs amongst the expression-increased genes in the Dicer knockout oocytes, our findings are in agreement with a similar study in zebrafish, where the microarray data of Dicer mutant and wild-type embryos were also compared (Giraldez et al. 2006).
Figure 4.
Unsupervised hierarchical clustering heat map of wild-type and Dicer knockout single-oocyte cDNA microarray. All genes that are differentially expressed between wild-type (left six columns) and Dicer knockout oocytes (right six columns) are shown. Clustering is based on the log2 of chemiluminescent intensities.
To further understand the molecular basis for the role of maternal miRNAs on early development, we compared our microarray data with the predicted target genes of maternal miRNAs using a bioinformatics approach. We searched the sequence complement to the “seed” region of miRNAs in the 3′ untranslated region (UTR) of all the 30,677 genes with defined 3′UTRs represented on the ABI chip. We found that the genes that have no target sites in their 3′UTRs for the 101 maternal miRNAs have 4.2-fold more chance to be coexpressed in oocytes with these miRNAs (P = 2.2E-16) (Supplementary Tables S9, S11). This indicates that miRNAs expressed in the oocyte functionally shape the gene expression profile, which is in accordance with the reports that genes expressed in a particular tissue tend to avoid being the targets of coexpressed miRNAs in the same tissue (Farh et al. 2005; Stark et al. 2005; Sood et al. 2006). By combing the analysis of the single-cell miRNA profile with single-cell cDNA microarrays, we confirmed at the resolution of single cells that some of the genes expressed in the oocyte had a strong tendency to avoid being the target sites of coexpressed miRNAs.
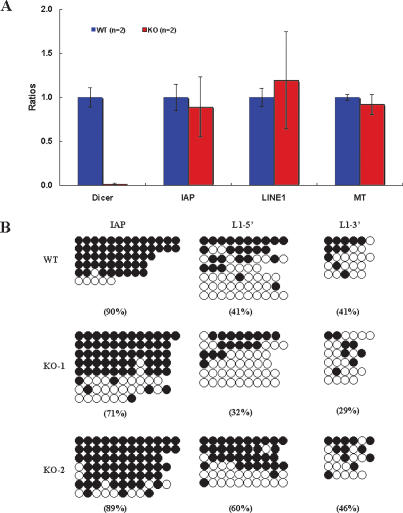
As it is known that miRNAs also apparently have an effect on the expression of repetitive elements (Fukagawa et al. 2004; Kanellopoulou et al. 2005), we examined the expression levels of IAP (intracisternal A particle element; an endogenous retrovirus), LINE1 (long interspersed nuclear element 1; a nonretrovirus retrotransposon), and MTs (mouse transcript; a nonautonomous retrotransposon), which are the most abundant repetitive elements in the mouse oocyte. However, quantitative real-time RT–PCR analysis showed no significant differences in their expression between Dicer mutant and control oocytes (Fig. 5A). We also investigated the DNA methylation status of IAP and LINE1 by the bisulphite genomic sequencing method and found no significant differences between Dicer mutant and control oocytes (Fig. 5B). Thus the phenotype of the Dicer mutant oocyte is due to the direct or indirect effects of the loss of miRNAs, not the derepression of repetitive elements.
Figure 5.
(A) Transcript abundance of repetitive elements in control and Dicer mutant oocytes measured by quantitative real-time RT–PCR. The error bars represent standard deviations calculated from two independent samples. (B) DNA methylation status of repetitive elements in control and Dicer mutant oocytes measured by bisulphite genomic sequencing. Filled circles represent methylated CpG and open circles represent nonmethylated CpG. Horizontally aligned circles represent a single DNA molecule. The overall percentage of methylated CpGs is shown below each group.
In summary, we have generated a comprehensive miRNA expression profile for growing oocytes and embryos up to the eight-cell stage. Interestingly, unlike the observations in zebrafish and Xenopus (P.Y. Chen et al. 2005; Watanabe et al. 2005), we found abundant maternally inherited miRNAs in mouse zygotes, among which let-7 family miRNAs are the most abundant. It seems that sperm-borne miRNAs do not contribute significantly to miRNAs in the zygote (Amanai et al. 2006). Furthermore, in zebrafish it was shown that maternal miRNAs are dispensable for early embryonic development, although the maternal Dicer is important because it is necessary for zygotic synthesis and expression of miR-430 (Giraldez et al. 2005, 2006). In the mouse, while there is a significant global loss of maternal miRNAs between the one- and two-cell stages of development, de novo synthesis of miRNAs commences at the two-cell stage. This includes expression of miR-290 to miR-295, which are the first embryonic miRNAs to be detected. It is noteworthy that mir-290 to mir-295 are also specifically expressed in embryonic stem cells, which may suggest their association with pluripotency (Houbaviy et al. 2003). We cannot formally rule out the possibility that the Dicer mutant phenotype is not solely due to loss of miRNAs because Dicer might directly play some unknown roles in chromatin formation. Amongst the genes that were detected at higher levels in the oocytes following the loss of Dicer, some of these effects may be due to secondary consequences of miRNA depletion. Nevertheless, this study provides evidence that the maternal inheritance of miRNAs is crucial for early mammalian development. The detailed analysis of the oocyte miRNAs and their impact on mRNAs we present here may help to elucidate their precise roles in early mouse development in the future.
Materials and methods
Embryos and knockout mice
Embryos before implantation were recovered from F1 (C57BL/6 × CBA) females mated with F1 male mice (Nagy et al. 2003). Oocytes were isolated from F1 female mice. All the “mature oocytes” we mentioned in the text are ovulated mature oocytes.
The knockout mice carrying the Dicer floxed allele was described previously (Yi et al. 2006). Basically, exon 23 of the Dicer locus was floxed by two loxP loci (referred to as DicerFlox). The DicerFlox/Flox mice were mated with Zp3-Cre transgenic mice, which express Cre recombinase under the control of the Zona pellucida glycoprotein 3 promoter (de Vries et al. 2000). Then the [DicerFlox/+, Zp3-Cre] female mice were mated with DicerFlox/Flox male mice. From this mating, we obtained [Dicer−/Flox, Zp3-Cre] mice, and following the deletion of the Floxed allele in the oocyte, we generate oocytes that are the null mutants for Dicer. The control mice we used are littermates with the genotype of [Dicer+/Flox, Zp3-Cre], Dicer+/Flox, or Dicer−/Flox.
The details of the microRNA expression profiling assay, RNA in situ hybridization by LNA probe, immunostaining, single-cell cDNA, real-time PCR, microarray gene expression procedures and analysis, and bisulphite genomic sequencing can be found in the Supplemental Material. The data of the single-oocyte cDNA microarray were deposited in GeneBank (http://www.ncbi.nlm.nih.gov/geo). The accession number is GSE6806.
Acknowledgments
We thank Eric Miska, Anne McLaren, Kenneth Livak, Naoki Miyoshi, Katsuhiko Hayashi, Maria Elena Torres Padilla, Jie Na, David Adams, and James Smith for their helpful discussions and generous suggestions. We also thank W.N. de Vries and B.B. Knowles for the Zp3-Cre transgenic mice. This work was supported by grants from the Wellcome Trust and BBSRC to M.A.S. M.K. is supported by the Japanese Society for the Promotion of the Science (JSPS).
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.418707
References
- Amanai M., Brahmajosyula M., Perry A.C., Brahmajosyula M., Perry A.C., Perry A.C. A restricted role for sperm-borne microRNAs in mammalian fertilization. Biol. Reprod. 2006;75:877–884. doi: 10.1095/biolreprod.106.056499. [DOI] [PubMed] [Google Scholar]
- Bagga S., Bracht J., Hunter S., Massirer K., Holtz J., Eachus R., Pasquinelli A.E., Bracht J., Hunter S., Massirer K., Holtz J., Eachus R., Pasquinelli A.E., Hunter S., Massirer K., Holtz J., Eachus R., Pasquinelli A.E., Massirer K., Holtz J., Eachus R., Pasquinelli A.E., Holtz J., Eachus R., Pasquinelli A.E., Eachus R., Pasquinelli A.E., Pasquinelli A.E. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell. 2005;122:553–563. doi: 10.1016/j.cell.2005.07.031. [DOI] [PubMed] [Google Scholar]
- Bartel D.P. MicroRNAs: Fenomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bernstein E., Kim S.Y., Carmell M.A., Murchison E.P., Alcorn H., Li M.Z., Mills A.A., Elledge S.J., Anderson K.V., Hannon G.J., Kim S.Y., Carmell M.A., Murchison E.P., Alcorn H., Li M.Z., Mills A.A., Elledge S.J., Anderson K.V., Hannon G.J., Carmell M.A., Murchison E.P., Alcorn H., Li M.Z., Mills A.A., Elledge S.J., Anderson K.V., Hannon G.J., Murchison E.P., Alcorn H., Li M.Z., Mills A.A., Elledge S.J., Anderson K.V., Hannon G.J., Alcorn H., Li M.Z., Mills A.A., Elledge S.J., Anderson K.V., Hannon G.J., Li M.Z., Mills A.A., Elledge S.J., Anderson K.V., Hannon G.J., Mills A.A., Elledge S.J., Anderson K.V., Hannon G.J., Elledge S.J., Anderson K.V., Hannon G.J., Anderson K.V., Hannon G.J., Hannon G.J. Dicer is essential for mouse development. Nat. Genet. 2003;35:215–217. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- Chen C., Ridzon D.A., Broomer A.J., Zhou Z., Lee D.H., Nguyen J.T., Barbisin M., Xu N.L., Mahuvakar V.R., Andersen M.R., Ridzon D.A., Broomer A.J., Zhou Z., Lee D.H., Nguyen J.T., Barbisin M., Xu N.L., Mahuvakar V.R., Andersen M.R., Broomer A.J., Zhou Z., Lee D.H., Nguyen J.T., Barbisin M., Xu N.L., Mahuvakar V.R., Andersen M.R., Zhou Z., Lee D.H., Nguyen J.T., Barbisin M., Xu N.L., Mahuvakar V.R., Andersen M.R., Lee D.H., Nguyen J.T., Barbisin M., Xu N.L., Mahuvakar V.R., Andersen M.R., Nguyen J.T., Barbisin M., Xu N.L., Mahuvakar V.R., Andersen M.R., Barbisin M., Xu N.L., Mahuvakar V.R., Andersen M.R., Xu N.L., Mahuvakar V.R., Andersen M.R., Mahuvakar V.R., Andersen M.R., Andersen M.R., et al. Real-time quantification of microRNAs by stem-loop RT–PCR. Nucleic Acids Res. 2005;33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P.Y., Manninga H., Slanchev K., Chien M., Russo J.J., Ju J., Sheridan R., John B., Marks D.S., Gaidatzis D., Manninga H., Slanchev K., Chien M., Russo J.J., Ju J., Sheridan R., John B., Marks D.S., Gaidatzis D., Slanchev K., Chien M., Russo J.J., Ju J., Sheridan R., John B., Marks D.S., Gaidatzis D., Chien M., Russo J.J., Ju J., Sheridan R., John B., Marks D.S., Gaidatzis D., Russo J.J., Ju J., Sheridan R., John B., Marks D.S., Gaidatzis D., Ju J., Sheridan R., John B., Marks D.S., Gaidatzis D., Sheridan R., John B., Marks D.S., Gaidatzis D., John B., Marks D.S., Gaidatzis D., Marks D.S., Gaidatzis D., Gaidatzis D., et al. The developmental miRNA profiles of zebrafish as determined by small RNA cloning. Genes & Dev. 2005;19:1288–1293. doi: 10.1101/gad.1310605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries W.N., Binns L.T., Fancher K.S., Dean J., Moore R., Kemler R., Knowles B.B., Binns L.T., Fancher K.S., Dean J., Moore R., Kemler R., Knowles B.B., Fancher K.S., Dean J., Moore R., Kemler R., Knowles B.B., Dean J., Moore R., Kemler R., Knowles B.B., Moore R., Kemler R., Knowles B.B., Kemler R., Knowles B.B., Knowles B.B. Expression of Cre recombinase in mouse oocytes: A means to study maternal effect genes. Genesis. 2000;26:110–112. [PubMed] [Google Scholar]
- Dean J. Oocyte-specific genes regulate follicle formation, fertility and early mouse development. J. Reprod. Immunol. 2002;53:171–180. doi: 10.1016/s0165-0378(01)00100-0. [DOI] [PubMed] [Google Scholar]
- Farh K.K., Grimson A., Jan C., Lewis B.P., Johnston W.K., Lim L.P., Burge C.B., Bartel D.P., Grimson A., Jan C., Lewis B.P., Johnston W.K., Lim L.P., Burge C.B., Bartel D.P., Jan C., Lewis B.P., Johnston W.K., Lim L.P., Burge C.B., Bartel D.P., Lewis B.P., Johnston W.K., Lim L.P., Burge C.B., Bartel D.P., Johnston W.K., Lim L.P., Burge C.B., Bartel D.P., Lim L.P., Burge C.B., Bartel D.P., Burge C.B., Bartel D.P., Bartel D.P. The widespread impact of mammalian microRNAs on mRNA repression and evolution. Science. 2005;310:1817–1821. doi: 10.1126/science.1121158. [DOI] [PubMed] [Google Scholar]
- Fukagawa T., Nogami M., Yoshikawa M., Ikeno M., Okazaki T., Takami Y., Nakayama T., Oshimura M., Nogami M., Yoshikawa M., Ikeno M., Okazaki T., Takami Y., Nakayama T., Oshimura M., Yoshikawa M., Ikeno M., Okazaki T., Takami Y., Nakayama T., Oshimura M., Ikeno M., Okazaki T., Takami Y., Nakayama T., Oshimura M., Okazaki T., Takami Y., Nakayama T., Oshimura M., Takami Y., Nakayama T., Oshimura M., Nakayama T., Oshimura M., Oshimura M. Dicer is essential for formation of the heterochromatin structure in vertebrate cells. Nat. Cell Biol. 2004;6:784–791. doi: 10.1038/ncb1155. [DOI] [PubMed] [Google Scholar]
- Giraldez A.J., Cinalli R.M., Glasner M.E., Enright A.J., Thomson J.M., Baskerville S., Hammond S.M., Bartel D.P., Schier A.F., Cinalli R.M., Glasner M.E., Enright A.J., Thomson J.M., Baskerville S., Hammond S.M., Bartel D.P., Schier A.F., Glasner M.E., Enright A.J., Thomson J.M., Baskerville S., Hammond S.M., Bartel D.P., Schier A.F., Enright A.J., Thomson J.M., Baskerville S., Hammond S.M., Bartel D.P., Schier A.F., Thomson J.M., Baskerville S., Hammond S.M., Bartel D.P., Schier A.F., Baskerville S., Hammond S.M., Bartel D.P., Schier A.F., Hammond S.M., Bartel D.P., Schier A.F., Bartel D.P., Schier A.F., Schier A.F. MicroRNAs regulate brain morphogenesis in zebrafish. Science. 2005;308:833–838. doi: 10.1126/science.1109020. [DOI] [PubMed] [Google Scholar]
- Giraldez A.J., Mishima Y., Rihel J., Grocock R.J., Van Dongen S., Inoue K., Enright A.J., Schier A.F., Mishima Y., Rihel J., Grocock R.J., Van Dongen S., Inoue K., Enright A.J., Schier A.F., Rihel J., Grocock R.J., Van Dongen S., Inoue K., Enright A.J., Schier A.F., Grocock R.J., Van Dongen S., Inoue K., Enright A.J., Schier A.F., Van Dongen S., Inoue K., Enright A.J., Schier A.F., Inoue K., Enright A.J., Schier A.F., Enright A.J., Schier A.F., Schier A.F. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science. 2006;312:75–79. doi: 10.1126/science.1122689. [DOI] [PubMed] [Google Scholar]
- Hamatani T., Carter M.G., Sharov A.A., Ko M.S., Carter M.G., Sharov A.A., Ko M.S., Sharov A.A., Ko M.S., Ko M.S. Dynamics of global gene expression changes during mouse preimplantation development. Dev. Cell. 2004;6:117–131. doi: 10.1016/s1534-5807(03)00373-3. [DOI] [PubMed] [Google Scholar]
- He L., Hannon G.J., Hannon G.J. MicroRNAs: Small RNAs with a big role in gene regulation. Nat. Rev. Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- He L., Thomson J.M., Hemann M.T., Hernando-Monge E., Mu D., Goodson S., Powers S., Cordon-Cardo C., Lowe S.W., Hannon G.J., Thomson J.M., Hemann M.T., Hernando-Monge E., Mu D., Goodson S., Powers S., Cordon-Cardo C., Lowe S.W., Hannon G.J., Hemann M.T., Hernando-Monge E., Mu D., Goodson S., Powers S., Cordon-Cardo C., Lowe S.W., Hannon G.J., Hernando-Monge E., Mu D., Goodson S., Powers S., Cordon-Cardo C., Lowe S.W., Hannon G.J., Mu D., Goodson S., Powers S., Cordon-Cardo C., Lowe S.W., Hannon G.J., Goodson S., Powers S., Cordon-Cardo C., Lowe S.W., Hannon G.J., Powers S., Cordon-Cardo C., Lowe S.W., Hannon G.J., Cordon-Cardo C., Lowe S.W., Hannon G.J., Lowe S.W., Hannon G.J., Hannon G.J., et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houbaviy H.B., Murray M.F., Sharp P.A., Murray M.F., Sharp P.A., Sharp P.A. Embryonic stem cell-specific microRNAs. Dev. Cell. 2003;5:351–358. doi: 10.1016/s1534-5807(03)00227-2. [DOI] [PubMed] [Google Scholar]
- Kanellopoulou C., Muljo S.A., Kung A.L., Ganesan S., Drapkin R., Jenuwein T., Livingston D.M., Rajewsky K., Muljo S.A., Kung A.L., Ganesan S., Drapkin R., Jenuwein T., Livingston D.M., Rajewsky K., Kung A.L., Ganesan S., Drapkin R., Jenuwein T., Livingston D.M., Rajewsky K., Ganesan S., Drapkin R., Jenuwein T., Livingston D.M., Rajewsky K., Drapkin R., Jenuwein T., Livingston D.M., Rajewsky K., Jenuwein T., Livingston D.M., Rajewsky K., Livingston D.M., Rajewsky K., Rajewsky K. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes & Dev. 2005;19:489–501. doi: 10.1101/gad.1248505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim V.N. MicroRNA biogenesis: Coordinated cropping and dicing. Nat. Rev. Mol. Cell Biol. 2005;6:376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- Kloosterman W.P., Wienholds E., de Bruijn E., Kauppinen S., Plasterk R.H., Wienholds E., de Bruijn E., Kauppinen S., Plasterk R.H., de Bruijn E., Kauppinen S., Plasterk R.H., Kauppinen S., Plasterk R.H., Plasterk R.H. In situ detection of miRNAs in animal embryos using LNA-modified oligonucleotide probes. Nat. Methods. 2006;3:27–29. doi: 10.1038/nmeth843. [DOI] [PubMed] [Google Scholar]
- Kurimoto K., Yabuta Y., Ohinata Y., Ono Y., Uno K.D., Yamada R.G., Ueda H.R., Saitou M., Yabuta Y., Ohinata Y., Ono Y., Uno K.D., Yamada R.G., Ueda H.R., Saitou M., Ohinata Y., Ono Y., Uno K.D., Yamada R.G., Ueda H.R., Saitou M., Ono Y., Uno K.D., Yamada R.G., Ueda H.R., Saitou M., Uno K.D., Yamada R.G., Ueda H.R., Saitou M., Yamada R.G., Ueda H.R., Saitou M., Ueda H.R., Saitou M., Saitou M. An improved single-cell cDNA amplification method for efficient high-density oligonucleotide microarray analysis. Nucleic Acids Res. 2006;34:e42. doi: 10.1093/nar/gkl050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre C., Terret M.E., Djiane A., Rassinier P., Maro B., Verlhac M.H., Terret M.E., Djiane A., Rassinier P., Maro B., Verlhac M.H., Djiane A., Rassinier P., Maro B., Verlhac M.H., Rassinier P., Maro B., Verlhac M.H., Maro B., Verlhac M.H., Verlhac M.H. Meiotic spindle stability depends on MAPK-interacting and spindle-stabilizing protein (MISS), a new MAPK substrate. J. Cell Biol. 2002;157:603–613. doi: 10.1083/jcb.200202052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy A., Gertsenstein M., Vintersten K., Behringer R., Gertsenstein M., Vintersten K., Behringer R., Vintersten K., Behringer R., Behringer R. Manipulating the mouse embryo. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2003. pp. 194–200. [Google Scholar]
- O’Donnell K.A., Wentzel E.A., Zeller K.I., Dang C.V., Mendell J.T., Wentzel E.A., Zeller K.I., Dang C.V., Mendell J.T., Zeller K.I., Dang C.V., Mendell J.T., Dang C.V., Mendell J.T., Mendell J.T. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- Rajkovic A., Pangas S.A., Ballow D., Suzumori N., Matzuk M.M., Pangas S.A., Ballow D., Suzumori N., Matzuk M.M., Ballow D., Suzumori N., Matzuk M.M., Suzumori N., Matzuk M.M., Matzuk M.M. NOBOX deficiency disrupts early folliculogenesis and oocyte-specific gene expression. Science. 2004;305:1157–1159. doi: 10.1126/science.1099755. [DOI] [PubMed] [Google Scholar]
- Sood P., Krek A., Zavolan M., Macino G., Rajewsky N., Krek A., Zavolan M., Macino G., Rajewsky N., Zavolan M., Macino G., Rajewsky N., Macino G., Rajewsky N., Rajewsky N. Cell-type-specific signatures of microRNAs on target mRNA expression. Proc. Natl. Acad. Sci. 2006;103:2746–2751. doi: 10.1073/pnas.0511045103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark A., Brennecke J., Bushati N., Russell R.B., Cohen S.M., Brennecke J., Bushati N., Russell R.B., Cohen S.M., Bushati N., Russell R.B., Cohen S.M., Russell R.B., Cohen S.M., Cohen S.M. Animal microRNAs confer robustness to gene expression and have a significant impact on 3′UTR evolution. Cell. 2005;123:1133–1146. doi: 10.1016/j.cell.2005.11.023. [DOI] [PubMed] [Google Scholar]
- Tang F., Hajkova P., Barton S.C., Lao K., Surani M.A., Hajkova P., Barton S.C., Lao K., Surani M.A., Barton S.C., Lao K., Surani M.A., Lao K., Surani M.A., Surani M.A. MicroRNA expression profiling of single whole embryonic stem cells. Nucleic Acids Res. 2006a;34:e9. doi: 10.1093/nar/gnj009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang F., Hajkova P., Barton S.C., O’Carroll D., Lee C., Lao K., Surani M.A., Hajkova P., Barton S.C., O’Carroll D., Lee C., Lao K., Surani M.A., Barton S.C., O’Carroll D., Lee C., Lao K., Surani M.A., O’Carroll D., Lee C., Lao K., Surani M.A., Lee C., Lao K., Surani M.A., Lao K., Surani M.A., Surani M.A. 220-plex microRNA expression profile of a single cell. Nat. Protoc. 2006b;1:1154–1159. doi: 10.1038/nprot.2006.161. [DOI] [PubMed] [Google Scholar]
- Terret M.E., Lefebvre C., Djiane A., Rassinier P., Moreau J., Maro B., Verlhac M.H., Lefebvre C., Djiane A., Rassinier P., Moreau J., Maro B., Verlhac M.H., Djiane A., Rassinier P., Moreau J., Maro B., Verlhac M.H., Rassinier P., Moreau J., Maro B., Verlhac M.H., Moreau J., Maro B., Verlhac M.H., Maro B., Verlhac M.H., Verlhac M.H. DOC1R: A MAP kinase substrate that control microtubule organization of metaphase II mouse oocytes. Development. 2003;130:5169–5177. doi: 10.1242/dev.00731. [DOI] [PubMed] [Google Scholar]
- Watanabe T., Takeda A., Mise K., Okuno T., Suzuki T., Minami N., Imai H., Takeda A., Mise K., Okuno T., Suzuki T., Minami N., Imai H., Mise K., Okuno T., Suzuki T., Minami N., Imai H., Okuno T., Suzuki T., Minami N., Imai H., Suzuki T., Minami N., Imai H., Minami N., Imai H., Imai H. Stage-specific expression of microRNAs during Xenopus development. FEBS Lett. 2005;579:318–324. doi: 10.1016/j.febslet.2004.11.067. [DOI] [PubMed] [Google Scholar]
- Yi R., O’Carroll D., Pasolli H.A., Zhang Z., Dietrich F.S., Tarakhovsky A., Fuchs E., O’Carroll D., Pasolli H.A., Zhang Z., Dietrich F.S., Tarakhovsky A., Fuchs E., Pasolli H.A., Zhang Z., Dietrich F.S., Tarakhovsky A., Fuchs E., Zhang Z., Dietrich F.S., Tarakhovsky A., Fuchs E., Dietrich F.S., Tarakhovsky A., Fuchs E., Tarakhovsky A., Fuchs E., Fuchs E. Morphogenesis in skin is governed by discrete sets of differentially expressed microRNAs. Nat. Genet. 2006;38:356–362. doi: 10.1038/ng1744. [DOI] [PubMed] [Google Scholar]