Abstract
Ultraviolet irradiation of fission yeast cells in G1 phase induced a delay in chromatin binding of replication initiation factors and, consistently, a transient delay in S-phase entry. The cell cycle delay was totally dependent on the Gcn2 kinase, a sensor of the nutritional status, and was accompanied by phosphorylation of the translation initiation factor eIF2α and by a general depression of translation. However, the G1-specific synthesis of factors required for DNA replication was not reduced by ultraviolet radiation. The cell cycle delay represents a novel checkpoint with a novel mechanism of action that is not activated by ionizing radiation.
Keywords: Cell cycle, checkpoint, DNA replication, translation, UV irradiation
In order to proliferate, all types of cells pass through a series of distinct stages called the cell cycle, consisting of periods for chromosome replication (S phase) and for chromosome segregation and nuclear division (mitosis), followed by cytokinesis. Before DNA duplication, in G1 phase, the decision is taken whether to enter the mitotic cell cycle or, alternatively, to enter a quiescent, nonproliferating state. This decision is of crucial importance since dysregulation of the control mechanisms at the G1/S transition may lead to mutations, chromosomal fragmentation, and genetic instability, which are events known to promote cancer development. Therefore, most types of cells contain sophisticated mechanisms to regulate the transition from G1 to S phase.
In the presence of DNA damage, the cell cycle is temporarily halted by mechanisms termed checkpoints (Hartwell and Weinert 1989). The signal from damaged DNA is transmitted via an array of proteins, mostly by phosphorylation events. The checkpoint mechanisms are similar in all eukaryotic cells, and the proteins involved are highly conserved through evolution. In the fission yeast Schizosaccharomyces pombe, the DNA damage checkpoints (intra-S and G2/M) have in common their final target, the cell cycle kinase Cdc2, which is phosphorylated and thereby inhibited upon checkpoint activation (Caspari and Carr 1999). Phosphorylation of Cdc2 is totally dependent on the involvement of the so-called checkpoint Rad proteins and the downstream kinases Chk1 and Cds1.
We have turned our interest to regulation of the G1/S transition in response to ultraviolet (UV) light. As S. pombe cells exit from mitosis, a Cdc10-dependent transcription program is initiated to produce two proteins essential for initiation of chromosome replication, namely, Cdc18 and Cdt1. These two proteins bind to the Origin Recognition Complex (ORC), which is located at future chromosomal origins of replication. The six Mcm proteins constitute the replicative helicase, and they are loaded onto the origins in a Cdc18- and Cdt1-dependent process (for reviews, see Kearsey and Labib 1998; Tye 1999), thereby forming the prereplicative complex (pre-RC). Formation of the pre-RC is a prerequisite for the initiation of chromosome replication.
We have shown earlier that fission yeast, like other eukaryotes, delays entry into S phase after exposure to UV (Nilssen et al. 2003). The exit from G1 phase is delayed by UV irradiation in a process that does not target Cdc2. Here we investigate and further characterize the mechanism of this delay and demonstrate that it fills all the requirements of a checkpoint. We show that the checkpoint delays the formation of pre-RCs by a novel mechanism involving the Gcn2 kinase, which is known to regulate translation according to the nutrient supply.
Results and Discussion
UV delays Mcm binding in G1 phase
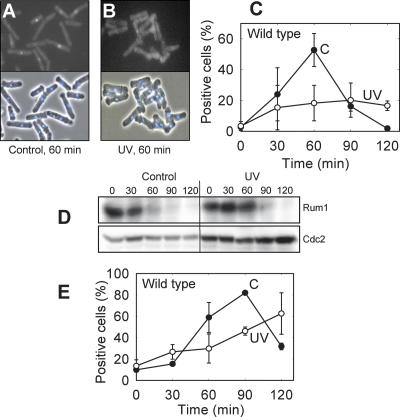
cdc10ts cells were synchronized in G1 phase, exposed to UV light (254 nm), and released into the cell cycle. The DNA histograms (Supplementary Fig. 1) show that most of the control cells had entered S phase by 60 min, whereas there is little sign of an increase in DNA content in the irradiated cells until 120 min, in agreement with our earlier data (Nilssen et al. 2003). The flow cytometry data cannot tell us whether the irradiated cells were arrested in G1 or in early S phase, so we employed molecular markers to make this distinction. The pre-RCs must be formed before any chromosome replication can take place, and pre-RC formation is therefore a convenient marker upstream of S phase. To monitor the kinetics of pre-RC formation, we used a strain expressing a GFP-tagged Mcm6 under its own promoter, allowing detection of chromatin-bound Mcm6 after extraction with detergent, as described previously (Kearsey et al. 2000). Nuclei that contained bound Mcm6-GFP after extraction could readily be distinguished from GFP-negative cells (Fig. 1A,B). We found that almost no cells, irradiated or not, had formed pre-RCs at the time of release from the temperature block. Sixty minutes after release, about half of the nonirradiated control cells were positive for Mcm6 after extraction (Fig. 1C). In contrast, very few irradiated cells contained pre-RCs at this time. Similar results were obtained by GFP-tagging Mcm2 (data not shown).
Figure 1.
UV irradiation inhibits pre-RC formation and arrests the cells in G1. The cdc10ts Mcm6-GFP cells were synchronized, UV-irradiated, and released into the cell cycle. Examples of fluorescence micrographs demonstrating the identification of GFP-positive and GFP-negative cells after extraction of control (A) and irradiated (B) cells taken 60 min after release. (Top) GFP. (Bottom) DAPI and phase contrast. (C) Percentage of cells that contained chromatin-bound Mcm6-GFP (pre-RC) at different time points after release into the cell cycle. (C) Control cells; (UV) irradiated cells. Standard deviations of three independent experiments are shown. (D) Immunoblot with Rum1-specific antibody (top row) and antibody against Cdc2 as a loading control (bottom row). Time after release is given in minutes. (E) Percentage of cells that contained chromatin-bound Cdc45-YFP (pre-IC) at different times after release; otherwise as in C.
These data strongly argue that the UV-induced delay occurs in G1 and not in S phase, which is further supported by the following: The Cdc2 inhibitor Rum1, which is only expressed in G1 (Benito et al. 1998), was strongly expressed during the delay period (Fig. 1D). Moreover, the activity of the S-phase-specific checkpoint kinase Cds1 was not activated in UV-irradiated cells until long after the time when nonirradiated control cells had entered S phase (Nilssen et al. 2003). Furthermore, we measured the loading of the YFP-labeled Sna41/Cdc45 protein onto chromatin, forming the preinitiation complex (pre-IC). This step occurs after formation of the pre-RC and after the requirements for Cdk activity and for Cdc7 are fulfilled (Zou and Stillman 2000; Gregan et al. 2003), but before any DNA replication can occur. In agreement with the data for pre-RC formation, we also found that the pre-IC formation, measured as chromatin binding of Cdc45, was delayed by UV (Fig. 1E). We conclude that UV light delays the onset of DNA replication.
Our results do not demonstrate whether UV irradiation delays the loading of the Mcm proteins or stimulates their unloading. In the latter scenario, the pre-RCs may be formed with normal kinetics after UV exposure, but the Mcms are removed as they are loaded onto chromatin, for a limited period of time. It should be noted that the Mcm proteins are known to be present also at or near progressing replication forks, so the presence of chromatin-bound Mcm6 only reflects that the pre-RCs have been formed and not necessarily that Mcm6 at that particular time is part of a pre-RC.
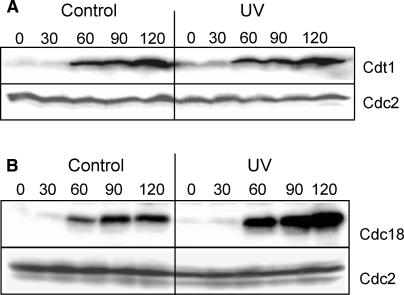
The components of the pre-RC are produced on time after UV
The delayed appearance of pre-RCs could be due to lack of one of its components. The known components are the six ORC proteins bound to chromatin together with Cdc18, Cdt1, and the Mcm proteins. The ORC and Mcm proteins are present throughout the cell cycle (Okishio et al. 1996; Lygerou and Nurse 1999; Pasion and Forsburg 1999; Kearsey et al. 2000), but both Cdc18 and Cdt1 have to be synthesized de novo before every S phase, in a Cdc10-dependent process (Kelly et al. 1993; Hofmann and Beach 1994; Nishitani et al. 2000). Immunoblotting showed that the production of Cdc18 (Fig. 2A) and Cdt1 (Fig. 2B) was not significantly delayed or reduced by UV irradiation. In fact, the level of Cdc18 was strongly increased after the UV irradiation, possibly because Cdc18 is not normally degraded until the end of S phase. We conclude that reduced expression of these critical proteins is not the reason for the observed cell cycle delay. These findings are in contrast to the situation when cells are arrested with high levels of Cdt1 (e.g., in mitosis), in which case Cdt1 is proteolyzed after UV irradiation (Ralph et al. 2006). Sporulating S. pombe cells also delay the accumulation of Cdt1 after UV irradiation (Nilssen et al. 2004). It appears that after UV irradiation, different regulatory mechanisms are used in different parts of the cell cycle.
Figure 2.
The replication initiation factors are present on time after UV irradiation. The intracellular levels of Cdt1 (A) and Cdc18 (B) were measured by immunoblotting of total cell extracts taken at different times (in minutes) after synchronization and release into the cell cycle, as described. The amount of Cdc2 serves as a loading control.
Inhibition of translation and phosphorylation of eIF2α after UV
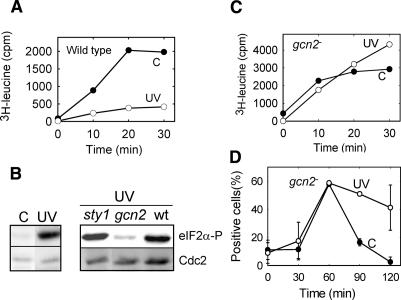
We noticed that cell growth was markedly slower after UV exposure. In vivo, incorporation of exogenously added amino acids into TCA-precipitable material was dramatically reduced by UV light (Supplementary Fig. 2), suggesting that UV irradiation down-regulated translation. To eliminate the possibility that this decrease was caused by a reduced cellular import of the tracer amino acid, we employed an established in vitro assay for translation and found an equally strong reduction in translation after UV light (Fig. 3A).
Figure 3.
The Gcn2 kinase is required for the UV-inducible response. (A) Incorporation of 3H-leucine into TCA-precipitable material was measured in extracts of wild-type cells prepared at different times (in minutes) after release. (B) Immunoblot of total proteins from synchronized cells probed with antibodies against phosphorylated eIF2α and Cdc2 (loading control). (Left panel) Nonirradiated and irradiated wild-type cells. (Right panel) UV-irradiated sty1 and gcn2 mutants, with wild-type cells for comparison. (C) As in A, except performed with the gcn2 strain. (D) Percentage of gcn2 mutant cells that contained chromatin-bound Mcm6-GFP (pre-RC) at different times (in minutes) after release; otherwise as in Figure 1C.
A standard pathway for regulation of translation—e.g., when cells are exposed to nutritional shifts—goes via phosphorylation of the initiation factor eIF2α (Hinnebusch 2000; Kaufman 2004). By using an antibody specific for the phosphorylated form of eIF2α, we showed that UV irradiation induced eIF2α phosphorylation (Fig. 3B, left panel). The response was rapid, since phosphorylation was at its maximum immediately after irradiation (the exposure time was ∼4 min). Control experiments with an antibody recognizing total eIF2α showed that the intracellular level was not changed after UV irradiation (data not shown). Importantly, the level of eIF2α phosphorylation after UV was as high as that found after starvation for an essential amino acid and stronger than that found after starvation for nitrogen (data not shown), emphasizing that this is a significant biological response.
UV activates the Gcn2 kinase
Several protein kinases could be responsible for the phosphorylation of eIF2α. In a database search for eIF2α-kinases in S. pombe, we found three primary candidates: Hri1 and Hri2, which are similar to the HRI kinase of mammalian cells (Zhan et al. 2002), and Gcn2, which is involved in a multitude of responses, mainly after amino acid starvation and environmental insults (for a review, see Hinnebusch 2000). Deletion of the hri genes did not reduce the phosphorylation of eIF2α or the delay of the cell cycle progression after UV irradiation (data not shown). In contrast, we found little UV-induced phosphorylation of eIF2α in the gcn2 deletion mutant (Fig. 3B, right). We conclude that the UV-induced phosphorylation of eIF2α is dependent on Gcn2.
The mitogen-activated protein (MAP) kinase pathway is activated in a variety of stress conditions in eukaryotes. However, the general stress response kinase in S. pombe, Sty1 (Degols et al. 1996), is not involved in the present response, since the level of eIF2α phosphorylation after irradiation of a sty1 mutant was similar to that found in wild-type cells (Fig. 3B, right). This is in contrast to the eIF2α phosphorylation observed after oxidative stress, which is partly Sty1 dependent (Dunand-Sauthier et al. 2005).
In accordance with the role of Gcn2 in eIF2α phosphorylation, we found that translation was not reduced after UV irradiation of a gcn2 mutant (Fig. 3C), in contrast to the findings for wild-type cells (Fig. 3A). This shows that Gcn2 regulates translation in response to UV, most likely by phosphorylating eIF2α. Furthermore, these data exclude the possibility that the cell cycle delay in wild-type cells was caused by excessive radiation damage to the translation machinery. We also found that the UV-induced delay in pre-RC formation was abolished in a gcn2 deletion mutant (Fig. 3D), demonstrating that Gcn2 activity is required for the UV-induced delay in G1 phase. These data confirm and extend our earlier observations (Nilssen et al. 2003) and establish the UV-induced delay as a bona fide checkpoint: It is totally abolished by a mutation (gcn2) and inhibited by caffeine (Nilssen et al. 2003).
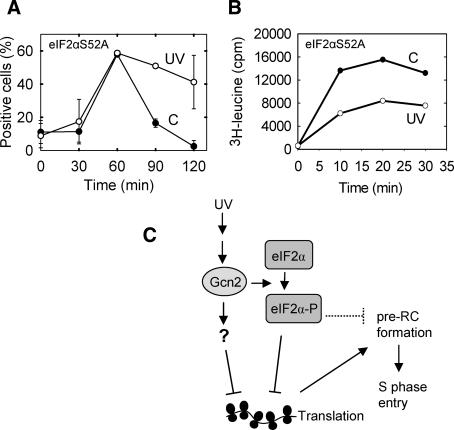
Nonphosphorylatable eIF2α
To investigate the role of eIF2α phosphorylation in checkpoint activation, we mutated the gene so that Ser 52 of the encoded protein (Ser 51 in most other species), which is the target of phosphorylation, was exchanged for the nonphosphorylatable alanine. UV irradiation of the eIF2αS52A mutant did not cause a significant delay of the pre-RC formation (Fig. 4A). Surprisingly, translation was depressed by UV light also in the mutant (Fig. 4B), although not to the same extent as in wild-type cells (Fig. 3A). Therefore, phosphorylation of Ser 52 of eIF2α is only partly required for UV-induced inhibition of translation and Gcn2 is absolutely required (Fig. 3C). It is possible that there are other sites on eIF2α that are phosphorylated by Gcn2 or that Gcn2 targets other proteins that affect translation. The lack of a UV-induced delay in the eIF2αS52A mutant suggests that phosphorylation of eIF2α per se is required for the cell cycle delay. However, it is also possible that a stronger depression of protein synthesis is required to affect pre-RC formation. Also, note that the measured level of translation is much higher in the mutant (Fig. 4B) than in wild-type cells (Fig. 3A), a phenotype expected from a cell where a negative regulator of translation has been removed.
Figure 4.
Protein synthesis and pre-RC formation in the eIF2αS52A mutant. The cells were synchronized and UV-irradiated as described before measurement of pre-RC formation (A) and protein synthesis (B), both as a function of time (in minutes) after release into the cell cycle. (C) A model of the molecular interactions occurring after UV irradiation to produce a depression of translation and a cell cycle delay. The question mark represents an unidentified factor. The dotted T-shape represents a hypothetical inhibitory activity.
A novel signal mechanism after UV irradiation
We have shown that the Gcn2 kinase is involved in a cell cycle checkpoint. The fact that a kinase normally dedicated to sensing the nutritional status of the cell can also regulate the cell cycle, suggests a possible mechanism of coupling between general cell growth and cell cycle progression. We have shown that UV light leads to activation of the Gcn2 kinase, which in turn mediates a cell cycle delay. Activation of Gcn2 results in phosphorylation of eIF2α, but it is not clear whether this is required for the checkpoint.
The mechanism whereby Gcn2 activation and eIF2α phosphorylation affect pre-RC formation appears not to be via an effect on translation of a known factor required for pre-RC formation, since Cdt1 and Cdc18 levels are not altered (Fig. 2) and the Mcms and ORC are stable during the mitotic cycle (see above). It has been shown that the ORC, Cdc18, and Cdt1 are sufficient for loading the Mcm proteins onto chromatin both in fission yeast (C. Houchens and T. Kelly, pers. comm.) and in budding yeast (Kawasaki et al. 2006). Therefore, all the proteins required for pre-RC formation appear to be present in the cell at the normal time also in UV-irradiated cells.
The checkpoint Rad proteins are absolutely required for the DNA damage checkpoints in S. pombe. We are currently investigating whether one or more of the checkpoint Rad proteins are involved in the present checkpoint. But in any case, the pathway presented here is clearly distinct from the previously described DNA damage checkpoints.
The translation of G1 cyclin genes has been shown to be repressed under certain stress conditions in budding yeast (Gallego et al. 1997; Polymenis and Schmidt 1997; Philpott et al. 1998), fission yeast (Grallert et al. 2000), and human cells (Hamanaka et al. 2005), thereby linking translation to regulation of progression through G1 phase. However, in the pathway presented here, the cell cycle delay occurs before any cyclin is required.
Ionizing radiation does not induce the checkpoint or eIF2α phosphorylation
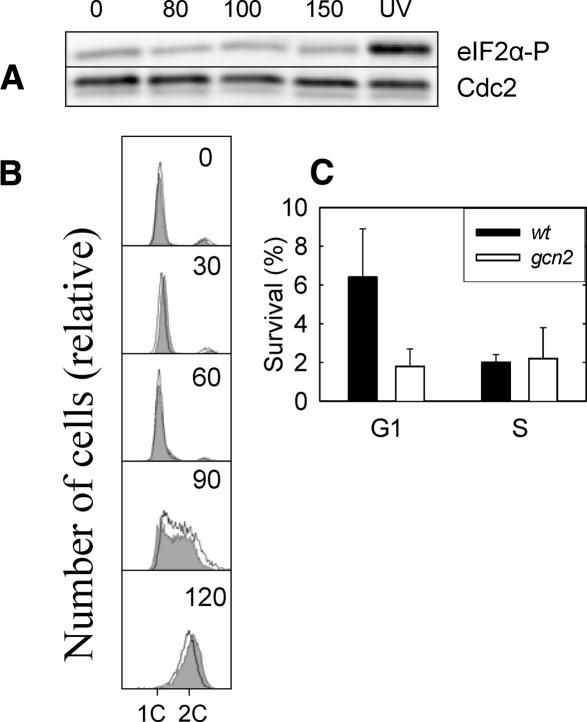
Synchronized cells exposed to relatively high doses of ionizing radiation (cell survival <5%) did not exhibit any detectable increase in the phosphorylation of eIF2α (Fig. 5A), nor did it induce any cell cycle delay, as measured by flow cytometry (Fig. 5B). Similar results were found when exposing the cells to psoralen + near-UV light. Psoralen intercalates into double-stranded DNA and forms cross-links upon exposure to near-UV light. After this DNA-specific insult, we could detect no delay in S-phase entry and no phosphorylation of eIF2α (data not shown). We conclude that the Gcn2-dependent checkpoint is not responding to DNA damage in general but appears to be at least partly specific for UV light. Therefore, our data argue that there is no general DNA damage checkpoint operating in G1 phase in S. pombe cells.
Figure 5.
Characterization of the G1 checkpoint. (A) Immunoblot of total cell extracts after different doses of ionizing radiation (Gy) was probed with antiserum against phosphorylated eIF2α (top) and Cdc2 (loading control; bottom). (B) Flow cytometry histograms of cdc10ts cells that were synchronized, exposed to 80 Gy of ionizing radiation, released into the cell cycle, and incubated for the times indicated (in minutes). Two DNA histograms are shown in each panel: one for nonirradiated control cells (filled) and one for irradiated cells (no fill). (C) Survival (with standard errors from five experiments shown) of wild-type and gcn2 mutant cells after UV irradiation in G1 or in S phase.
The rationale of the G1 checkpoint
It is presumed that the main purpose of cell cycle checkpoints is to delay cell cycle progression to allow certain processes to finish before the next step in the cell cycle is initiated, thereby increasing cell survival and genetic stability. To gain some insight into the biological purpose of the present checkpoint, we irradiated wild-type and gcn2 mutant cells (lacking the checkpoint) in G1 phase and in S phase. Whereas wild-type cells are significantly less sensitive to UV irradiation in G1 than in S phase, the opposite is true for gcn2 mutant cells (Fig. 5C), indicating that cells with the G1 checkpoint intact have a specific advantage after UV irradiation in G1 phase.
It is interesting that phosphorylation of eIF2α was also observed after UV irradiation of budding yeast cells in G1 (our unpublished data) and of human cells (Deng et al. 2002; Wu et al. 2002), making it likely that this biological response is operating in these organisms and suggesting that it might be conserved through evolution.
Materials and methods
Yeast strains and cell handling
The basic growth media were as described before (Moreno et al. 1991). All our S. pombe strains are derived from the 972h− strain and are given in the Supplemental Material. Cultures growing exponentially in liquid EMM (supplied with amino acids as required) at a cell concentration of 2 × 106/mL to 4 × 106/mL (OD600 of 0.1–0.2) were used for experiments. The temperature-sensitive cdc10-M17 cells were synchronized and irradiated with UV light (254 nm) as described before (Nilssen et al. 2003). Extraction of unbound MCM (pre-RC) or Cdc45 (pre-IC) proteins was performed as described (Kearsey et al. 2000; Gregan et al. 2003). For ionizing radiation, the cells were exposed to 4 MeV electrons from a linear accelerator (Varian Clinac 2100 CD) at a dose rate of ∼80 Gy/min.
Immunoblots
Cell extracts were made either by the TCA protein extraction method (Caspari et al. 2000) or by the RIPA method. For the latter, cells were harvested by centrifugation (3000g) for 5 min at 4°C and washed with STOP buffer (150 mM NaCl, 50 mM NaF, 1 mM NaN3, 10 mM EDTA at pH 8.0). The pellet was resuspended in 20 μL of RIPA buffer (150 mM NaCl, 1% Trition X-100, 0.1% SDS, 50 mM TRIS at pH 8.0) and boiled for 5 min, followed by freezing in liquid nitrogen. Glass beads (300 μL) were added, and the cells were broken using Fast Prep (Thermo Electron Corporation) for 3 × 20 sec at a speed of 6.5. Sample buffer (400 μL) was added to each sample and boiled for 5 min before the debris was spun down and the supernatant was loaded on an SDS-PAGE gel.
The following antibodies were used: antiserum against phosphorylated eIF2-α (Biosource no. 44-7282) at a dilution of 1:2000; anti-peroxidase, PAP1, against the TAP tag (Sigma; P1291), diluted 1:1000; antiserum against Rum1 at a dilution of 1:200 (a kind gift from S. Moreno); and antibody against Cdks with the PSTAIRE motif, recognizing Cdc2 (Santa Cruz Biotechnology; sc-53), diluted 1:2000. In addition, antiserum against total eIF2α (a kind gift from S.R. Kimball) was used as a standard in the quantification of the level of phosphorylated eIF2-α. This loading control gave essentially the same results as those found with the antibody against PSTAIRE, but with somewhat lower quality, and the latter was routinely used. Detection was performed using standard ECL or ECF kits (Amersham Biosciences) and quantified with a PhosphorImager and the ImageQuant software.
Strain expressing nonphosphorylatable eIF2α protein
eIF2α is encoded by the tif211 gene (cosmid SPAC3G9.09c). Plasmid pJAS022 containing tif211 with flanking regions was used to make a strain expressing a nonphosphorylatable eIF2α protein. Site-directed mutagenesis (Stratagene, QuickChange XL) was used to exchange the Ser 52 codon (TCC) for an alanine codon (GCG, see below). Thereby a unique Esp31 restriction site was introduced, serving as a diagnostic marker for the tif211 mutation. The 1.8-kb HindIII fragment of ura4 was given blunt ends by treatment with Klenow enzyme and inserted into the EcoRV site ∼130 nucleotides downstream from tif211. A PCR product containing the mutated tif211 gene and the ura4 marker was transformed into ura4-deficient strains. The presence of the tif211 mutation in the chromosome was confirmed by PCR analysis, by Southern blot analysis, and by sequencing of relevant regions. In addition, no phosphorylation of tif211 mutant transformants was detected in immunoblots using antibody against phosphorylated eIF2α.
tif211 mutagenesis steps were as follows: pJAS022 with tif211, Ser 52: GCTCTCTGAGCTTTCCAGACGTCGTATTCG; serine to alanine: GCTCTCTGAGCTTGCCAGACGTCGTATTCG; alanine to alanine, Esp31 site: GCTCTCTGAGCTTGCGAGACGTCGTATTCG.
Translation assays
For the in vivo translation assay, the cdc10ts cells were arrested in G1, UV-irradiated, and released into the cell cycle as described in the text. Three replicate samples of irradiated and nonirradiated cells were collected at the times indicated. To 980 μL of cell culture we added 20 μL of growth medium containing a mix of 35S-labeled methionine/cysteine (0.2 μL; GE Healthcare AGQ0080, 530 MBq/mL) and 1 mM unlabeled methionine and cysteine, and the samples were incubated for 10 min at 25°C. Thereafter, 250 μL of ice-cold 10% PCA containing 10 mM unlabeled Met and Cyt were added, and the samples were left on ice for >10 min. The precipitates were collected by filtration on Whatman GF/C glass fiber discs that were washed twice with 5 mL of cold 10% PCA, four times with 5 mL of 0.1 N HCl, and once with 3 mL of ice-cold 96% EtOH before liquid scintillation counting of the dried filters. The in vitro assay was performed as described (Abou et al. 1994), with some modifications: To the reaction buffer (125 mM NH4Cl, 3 mM/mg acetate, 0.5 mM ATP, 0.1 mM GTP, 25 mM creatine phosphate, 100 μg/mL creatine phosphokinase, 10% glycerol, 20 mM HEPES/KOH at pH 7.4), a mixture of all amino acids except leucine (Formedium) was added to an average final concentration of 40 μM. Tritiated leucine (GE Healthcare TRK170; 2.3 TBq/mmol) was mixed with the cell extracts on ice and transferred to 25°C, and aliquots were removed after different times of incubation. The reaction was stopped by addition of TCA, the precipitated material was collected on glass fiber filters and washed with ethanol, and 3H-activity was measured by liquid scintillation counting.
Cell survival assay
The cdc10 mutant cells were synchronized in G1 and UV-irradiated immediately, as described. Alternatively, the synchronized cells were incubated for 60 min at the permissive temperature before irradiation, at which time the DNA histogram showed that the vast majority of cells were in S phase. Cell survival was assayed after plating on YE agar.
Acknowledgments
We are grateful for technical assistance from I. Løbersli, M.O. Haugli, J.A. Sandvik, T. Furre, K. Monaghan, and Morigen. Thanks are also due to D. Hermand, P. Nurse, and J. Liu for strains, and to S. Moreno and S.R. Kimball for antibodies. This work was supported by grants from the Norwegian Cancer Society and from the Norwegian Research Council.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.421807
References
- Abou E.S., Good L., Melekhovets Y.F., Nazar R.N., Good L., Melekhovets Y.F., Nazar R.N., Melekhovets Y.F., Nazar R.N., Nazar R.N. Inhibition of protein synthesis by an efficiently expressed mutation in the yeast 5.8S ribosomal RNA. Nucleic Acids Res. 1994;22:686–693. doi: 10.1093/nar/22.4.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benito J., Martin-Castellanos C., Moreno S., Martin-Castellanos C., Moreno S., Moreno S. Regulation of the G1 phase of the cell cycle by periodic stabilization and degradation of the p25rum1 CDK inhibitor. EMBO J. 1998;17:482–497. doi: 10.1093/emboj/17.2.482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspari T., Carr A.M., Carr A.M. DNA structure checkpoint pathways in Schizosaccharomyces pombe. Biochimie. 1999;81:173–181. doi: 10.1016/s0300-9084(99)80050-9. [DOI] [PubMed] [Google Scholar]
- Caspari T., Dahlen M., Kanter-Smoler G., Lindsay H.D., Hofmann K., Papadimitriou K., Sunnerhagen P., Carr A.M., Dahlen M., Kanter-Smoler G., Lindsay H.D., Hofmann K., Papadimitriou K., Sunnerhagen P., Carr A.M., Kanter-Smoler G., Lindsay H.D., Hofmann K., Papadimitriou K., Sunnerhagen P., Carr A.M., Lindsay H.D., Hofmann K., Papadimitriou K., Sunnerhagen P., Carr A.M., Hofmann K., Papadimitriou K., Sunnerhagen P., Carr A.M., Papadimitriou K., Sunnerhagen P., Carr A.M., Sunnerhagen P., Carr A.M., Carr A.M. Characterization of Schizosaccharomyces pombe Hus1: A PCNA-related protein that associates with Rad1 and Rad9. Mol. Cell. Biol. 2000;20:1254–1262. doi: 10.1128/mcb.20.4.1254-1262.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degols G., Shiozaki K., Russell P., Shiozaki K., Russell P., Russell P. Activation and regulation of the Spc1 stress-activated protein kinase in Schizosaccharomyces pombe. Mol. Cell. Biol. 1996;16:2870–2877. doi: 10.1128/mcb.16.6.2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng J., Harding H.P., Raught B., Gingras A.C., Berlanga J.J., Scheuner D., Kaufman R.J., Ron D., Sonenberg N., Harding H.P., Raught B., Gingras A.C., Berlanga J.J., Scheuner D., Kaufman R.J., Ron D., Sonenberg N., Raught B., Gingras A.C., Berlanga J.J., Scheuner D., Kaufman R.J., Ron D., Sonenberg N., Gingras A.C., Berlanga J.J., Scheuner D., Kaufman R.J., Ron D., Sonenberg N., Berlanga J.J., Scheuner D., Kaufman R.J., Ron D., Sonenberg N., Scheuner D., Kaufman R.J., Ron D., Sonenberg N., Kaufman R.J., Ron D., Sonenberg N., Ron D., Sonenberg N., Sonenberg N. Activation of GCN2 in UV-irradiated cells inhibits translation. Curr. Biol. 2002;12:1279–1286. doi: 10.1016/s0960-9822(02)01037-0. [DOI] [PubMed] [Google Scholar]
- Dunand-Sauthier I., Walker C.A., Narasimhan J., Pearce A.K., Wek R.C., Humphrey T.C., Walker C.A., Narasimhan J., Pearce A.K., Wek R.C., Humphrey T.C., Narasimhan J., Pearce A.K., Wek R.C., Humphrey T.C., Pearce A.K., Wek R.C., Humphrey T.C., Wek R.C., Humphrey T.C., Humphrey T.C. Stress-activated protein kinase pathway functions to support protein synthesis and translational adaptation in response to environmental stress in fission yeast. Eukaryot. Cell. 2005;4:1785–1793. doi: 10.1128/EC.4.11.1785-1793.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego C., Gari E., Colomina N., Herrero E., Aldea M., Gari E., Colomina N., Herrero E., Aldea M., Colomina N., Herrero E., Aldea M., Herrero E., Aldea M., Aldea M. The Cln3 cyclin is down-regulated by translational repression and degradation during the G1 arrest caused by nitrogen deprivation in budding yeast. EMBO J. 1997;16:7196–7206. doi: 10.1093/emboj/16.23.7196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grallert B., Kearsey S.E., Lenhard M., Carlson C.R., Nurse P., Boye E., Labib K., Kearsey S.E., Lenhard M., Carlson C.R., Nurse P., Boye E., Labib K., Lenhard M., Carlson C.R., Nurse P., Boye E., Labib K., Carlson C.R., Nurse P., Boye E., Labib K., Nurse P., Boye E., Labib K., Boye E., Labib K., Labib K. A fission yeast general translation factor reveals links between protein synthesis and cell cycle controls. J. Cell Sci. 2000;113:1447–1458. doi: 10.1242/jcs.113.8.1447. [DOI] [PubMed] [Google Scholar]
- Gregan J., Lindner K., Brimage L., Franklin R., Namdar M., Hart E.A., Aves S.J., Kearsey S.E., Lindner K., Brimage L., Franklin R., Namdar M., Hart E.A., Aves S.J., Kearsey S.E., Brimage L., Franklin R., Namdar M., Hart E.A., Aves S.J., Kearsey S.E., Franklin R., Namdar M., Hart E.A., Aves S.J., Kearsey S.E., Namdar M., Hart E.A., Aves S.J., Kearsey S.E., Hart E.A., Aves S.J., Kearsey S.E., Aves S.J., Kearsey S.E., Kearsey S.E. Fission yeast Cdc23/Mcm10 functions after pre-replicative complex formation to promote Cdc45 chromatin binding. Mol. Biol. Cell. 2003;14:3876–3887. doi: 10.1091/mbc.E03-02-0090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamanaka R.B., Bennett B.S., Cullinan S.B., Diehl J.A., Bennett B.S., Cullinan S.B., Diehl J.A., Cullinan S.B., Diehl J.A., Diehl J.A. PERK and GCN2 contribute to eIF2α phosphorylation and cell cycle arrest after activation of the unfolded protein response pathway. Mol. Biol. Cell. 2005;16:5493–5501. doi: 10.1091/mbc.E05-03-0268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell L.H., Weinert T.A., Weinert T.A. Checkpoints: Controls that ensure the order of cell cycle events. Science. 1989;246:629–634. doi: 10.1126/science.2683079. [DOI] [PubMed] [Google Scholar]
- Hinnebusch A.G. Mechanism and regulation of initiator methionyl-tRNA binding to ribosomes. In: Sonenberg N, et al., editors. Translational control of gene expression. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2000. pp. 185–243. [Google Scholar]
- Hofmann J.F., Beach D., Beach D. cdt1 is an essential target of the Cdc10/Sct1 transcription factor: Requirement for DNA replication and inhibition of mitosis. EMBO J. 1994;13:425–434. doi: 10.1002/j.1460-2075.1994.tb06277.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman R.J. Regulation of mRNA translation by protein folding in the endoplasmic reticulum. Trends Biochem. Sci. 2004;29:152–158. doi: 10.1016/j.tibs.2004.01.004. [DOI] [PubMed] [Google Scholar]
- Kawasaki Y., Kim H.D., Kojima A., Seki T., Sugino A., Kim H.D., Kojima A., Seki T., Sugino A., Kojima A., Seki T., Sugino A., Seki T., Sugino A., Sugino A. Reconstitution of Saccharomyces cerevisiae prereplicative complex assembly in vitro. Genes Cells. 2006;11:745–756. doi: 10.1111/j.1365-2443.2006.00975.x. [DOI] [PubMed] [Google Scholar]
- Kearsey S.E., Labib K., Labib K. MCM proteins: Evolution, properties, and role in DNA replication. Biochim. Biophys. Acta. 1998;1398:113–136. doi: 10.1016/s0167-4781(98)00033-5. [DOI] [PubMed] [Google Scholar]
- Kearsey S.E., Montgomery S., Labib K., Lindner K., Montgomery S., Labib K., Lindner K., Labib K., Lindner K., Lindner K. Chromatin binding of the fission yeast replication factor mcm4 occurs during anaphase and requires ORC and cdc18. EMBO J. 2000;19:1681–1690. doi: 10.1093/emboj/19.7.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly T.J., Martin G.S., Forsburg S.L., Stephen R.J., Russo A., Nurse P., Martin G.S., Forsburg S.L., Stephen R.J., Russo A., Nurse P., Forsburg S.L., Stephen R.J., Russo A., Nurse P., Stephen R.J., Russo A., Nurse P., Russo A., Nurse P., Nurse P. The fission yeast cdc18+ gene product couples S phase to START and mitosis. Cell. 1993;74:371–382. doi: 10.1016/0092-8674(93)90427-r. [DOI] [PubMed] [Google Scholar]
- Lygerou Z., Nurse P., Nurse P. The fission yeast origin recognition complex is constitutively associated with chromatin and is differentially modified through the cell cycle. J. Cell Sci. 1999;112:3703–3712. doi: 10.1242/jcs.112.21.3703. [DOI] [PubMed] [Google Scholar]
- Moreno S., Klar A., Nurse P., Klar A., Nurse P., Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- Nilssen E.A., Synnes M., Kleckner N., Grallert B., Boye E., Synnes M., Kleckner N., Grallert B., Boye E., Kleckner N., Grallert B., Boye E., Grallert B., Boye E., Boye E. Intra-G1 arrest in response to UV irradiation in fission yeast. Proc. Natl. Acad. Sci. 2003;100:10758–10763. doi: 10.1073/pnas.1833769100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilssen E.A., Synnes M., Tvegard T., Vebo H., Boye E., Grallert B., Synnes M., Tvegard T., Vebo H., Boye E., Grallert B., Tvegard T., Vebo H., Boye E., Grallert B., Vebo H., Boye E., Grallert B., Boye E., Grallert B., Grallert B. Germinating fission yeast spores delay in G1 in response to UV irradiation. BMC Cell Biol. 2004;5:40. doi: 10.1186/1471-2121-5-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishitani H., Lygerou Z., Nishimoto T., Nurse P., Lygerou Z., Nishimoto T., Nurse P., Nishimoto T., Nurse P., Nurse P. The Cdt1 protein is required to license DNA for replication in fission yeast. Nature. 2000;404:625–628. doi: 10.1038/35007110. [DOI] [PubMed] [Google Scholar]
- Okishio N., Adachi Y., Yanagida M., Adachi Y., Yanagida M., Yanagida M. Fission yeast Nda1 and Nda4, MCM homologs required for DNA replication, are constitutive nuclear proteins. J. Cell Sci. 1996;109:319–326. doi: 10.1242/jcs.109.2.319. [DOI] [PubMed] [Google Scholar]
- Pasion S.G., Forsburg S.L., Forsburg S.L. Nuclear localization of Schizosaccharomyces pombe Mcm2/Cdc19p requires MCM complex assembly. Mol. Biol. Cell. 1999;10:4043–4057. doi: 10.1091/mbc.10.12.4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philpott C.C., Rashford J., Yamaguchi-Iwai Y., Rouault T.A., Dancis A., Klausner R.D., Rashford J., Yamaguchi-Iwai Y., Rouault T.A., Dancis A., Klausner R.D., Yamaguchi-Iwai Y., Rouault T.A., Dancis A., Klausner R.D., Rouault T.A., Dancis A., Klausner R.D., Dancis A., Klausner R.D., Klausner R.D. Cell-cycle arrest and inhibition of G1 cyclin translation by iron in AFT1-1(up) yeast. EMBO J. 1998;17:5026–5036. doi: 10.1093/emboj/17.17.5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polymenis M., Schmidt E.V., Schmidt E.V. Coupling of cell division to cell growth by translational control of the G1 cyclin CLN3 in yeast. Genes & Dev. 1997;11:2522–2531. doi: 10.1101/gad.11.19.2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralph E., Boye E., Kearsey S.E., Boye E., Kearsey S.E., Kearsey S.E. DNA damage induces Cdt1 proteolysis in fission yeast through a pathway dependent on Cdt2 and Ddb1. EMBO Rep. 2006;7:1134–1139. doi: 10.1038/sj.embor.7400827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tye B.K. MCM proteins in DNA replication. Annu. Rev. Biochem. 1999;68:649–686. doi: 10.1146/annurev.biochem.68.1.649. [DOI] [PubMed] [Google Scholar]
- Wu S., Hu Y., Wang J.L., Chatterjee M., Shi Y., Kaufman R.J., Hu Y., Wang J.L., Chatterjee M., Shi Y., Kaufman R.J., Wang J.L., Chatterjee M., Shi Y., Kaufman R.J., Chatterjee M., Shi Y., Kaufman R.J., Shi Y., Kaufman R.J., Kaufman R.J. Ultraviolet light inhibits translation through activation of the unfolded protein response kinase PERK in the lumen of the endoplasmic reticulum. J. Biol. Chem. 2002;277:18077–18083. doi: 10.1074/jbc.M110164200. [DOI] [PubMed] [Google Scholar]
- Zhan K., Vattem K.M., Bauer B.N., Dever T.E., Chen J.J., Wek R.C., Vattem K.M., Bauer B.N., Dever T.E., Chen J.J., Wek R.C., Bauer B.N., Dever T.E., Chen J.J., Wek R.C., Dever T.E., Chen J.J., Wek R.C., Chen J.J., Wek R.C., Wek R.C. Phosphorylation of eukaryotic initiation factor 2 by heme-regulated inhibitor kinase-related protein kinases in Schizosaccharomyces pombe is important for fesistance to environmental stresses. Mol. Cell. Biol. 2002;22:7134–7146. doi: 10.1128/MCB.22.20.7134-7146.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L., Stillman B., Stillman B. Assembly of a complex containing Cdc45p, replication protein A, and Mcm2p at replication origins controlled by S-phase cyclin-dependent kinases and Cdc7p–Dbf4p kinase. Mol. Cell. Biol. 2000;20:3086–3096. doi: 10.1128/mcb.20.9.3086-3096.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]