Abstract
In mammalian cells, mRNAs with AU-rich elements (AREs) are targeted for translational silencing and rapid degradation. Here we present evidence that in human cells the proteins Tristetraprolin (TTP) and BRF-1 deliver ARE-mRNAs to processing bodies (PBs), cytoplasmic assemblies of mRNAs, and associated factors that promote translational silencing and mRNA decay. First, depletion of endogenous TTP and BRF proteins, or overexpression of dominant-negative mutant TTP proteins, impairs the localization of reporter ARE-mRNAs in PBs. Second, TTP and BRF-1 localize tethered mRNAs to PBs. Third, TTP can nucleate PB formation on untranslated mRNAs even when other mRNAs are trapped in polysomes by cycloheximide treatment. ARE-mRNA localization in PBs is mediated by the TTP N- and C-terminal domains and occurs downstream from mRNA polysome release, which in itself is not sufficient for mRNA PB localization. The accumulation of ARE-mRNAs in PBs is strongly enhanced when the mRNA decay machinery is rendered limiting by mRNA decay enzyme depletion or TTP/BRF-1 overexpression. Based on these observations, we propose that the PB functions as a reservoir that sequesters ARE-mRNAs from polysomes, thereby silencing ARE-mRNA function even when mRNA decay is delayed. This function of the PB can likely be extended to other mRNA silencing pathways, such as those mediated by microRNAs, premature termination codons, and mRNA deadenylation.
Keywords: mRNA turnover, AU-rich elements, TTP, BRF-1, processing bodies
mRNA turnover plays a key role in the regulation of gene expression. AU-rich elements (AREs) are found in the 3′ untranslated region (UTR) of many human mRNAs (ARE-mRNAs) that undergo translational silencing and rapid turnover, a number of which encode interleukins, cytokines, and proto-oncogenes (Chen and Shyu 1995; Gueydan et al. 1999; Wilusz et al. 2001; Shim and Karin 2002; Zhang et al. 2002; Mazan-Mamczarz et al. 2006). AREs have been divided into different classes (Classes I, II, and III), depending on the occurrence of the sequence “AUUUA” (Chen and Shyu 1995). The ARE sequences function as binding sites for trans-acting factors that regulate ARE-mRNA translation and stability (Chen and Shyu 1995; Gueydan et al. 1999; Piecyk et al. 2000; Wilusz et al. 2001; Shim and Karin 2002; Zhang et al. 2002; Lopez de Silanes et al. 2005; Mazan-Mamczarz et al. 2006).
One of the best-characterized ARE-binding proteins is Tristetraprolin (TTP). TTP belongs to a family of human ARE-binding proteins that contains tandem CCCH zinc finger RNA-binding domains required for interaction with the ARE (Lai et al. 2000; Blackshear 2002). The other members of the TTP family are BRF-1 (also called hsERF1/ZFP36L1/TIS11b) and BRF-2 (also called hsERF2/ZFP36L2/TIS11d) (Lai et al. 2000; Stoecklin et al. 2002). TTP has been shown to destabilize a number of Class II (containing overlapping AUUUA-repeats) ARE-containing mRNAs, including those that encode Tumor Necrosis Factor α (TNF-α), Granulocyte Macrophage-Colony-Stimulating Factor (GM-CSF), and Interleukin-3 (IL-3) proteins (Lai et al. 1999; Carballo et al. 2000; Stoecklin et al. 2000; Lai et al. 2006). The TTP paralogs BRF-1 and BRF-2 are less studied, but affect ARE-mRNA stability in a manner similar to TTP when overexpressed or depleted (Lai et al. 2000; Stoecklin et al. 2002). However, whereas TTP is primarily expressed in lymphocytes (Carballo et al. 1998), BRF-1 and BRF-2 appear to be expressed in a wider range of tissues (Ramos et al. 2004; Stumpo et al. 2004). The mechanism by which TTP and BRF proteins destabilize ARE-mRNAs is not well understood. However, TTP and BRF-1 can target a tethered non-ARE reporter mRNA for rapid decay (Lykke-Andersen and Wagner 2005). In addition, TTP and BRF-1 interact with a number of mRNA decay enzymes responsible for deadenylation, decapping, and exonucleolytic decay, as well as with the endonuclease Ago2, which is involved in RNA interference (RNAi) (Chen et al. 2001; Jing et al. 2005; Lykke-Andersen and Wagner 2005). Thus, TTP and BRF-1 appear to activate decay of ARE-mRNAs by recruiting mRNA decay enzymes. Other ARE-binding proteins that influence ARE-mediated mRNA decay have been identified and include K homology Splicing Regulatory Protein (KSRP), hnRNP D/AUF-1, and HuR (DeMaria and Brewer 1996; Fan and Steitz 1998; Peng et al. 1998; Xu et al. 2001; Gherzi et al. 2004; Chou et al. 2006). However, it is not known how the different ARE-binding proteins cooperate to regulate the turnover of ARE-mRNAs in the cell.
A recent exciting observation is that a number of proteins involved in translational silencing and mRNA decay concentrate in discrete cytoplasmic foci called processing bodies (PBs; also called GW or Dcp bodies) (for review, see Anderson and Kedersha 2006). These proteins include the decapping enzyme Dcp2, the 5′-to-3′ exonuclease Xrn1, and factors that stimulate decapping or inhibit general translation, as well as factors involved in microRNA (miRNA)- and small interfering RNA (siRNA)-mediated mRNA silencing (Ingelfinger et al. 2002; van Dijk et al. 2002; Eystathioy et al. 2003; Andrei et al. 2005; Ding et al. 2005; Fenger-Gron et al. 2005; Ferraiuolo et al. 2005; Fillman and Lykke-Andersen 2005; Liu et al. 2005b; Pillai et al. 2005; Sen and Blau 2005). PBs are highly dynamic structures, as evidenced by the ability of proteins and mRNAs to rapidly cycle in and out of PBs (Kedersha et al. 2005). Moreover, translational inhibitors that trap mRNA in polysomes cause the rapid disappearance of PBs, whereas inhibitors that trigger polysome disassociation cause enlarged PBs (Cougot et al. 2004; Brengues et al. 2005; Teixeira et al. 2005). This suggests that PBs only form in the presence of available mRNA substrates and mRNAs exist in PBs to the exclusion of polysomes. In addition, the observation that mRNA decay intermediates can be detected in PBs suggests that mRNA decay can take place in the PB (Sheth and Parker 2003; Cougot et al. 2004). However, it remains unclear under which conditions mRNAs are sequestered in PBs and what delivers them there.
Here we present several lines of evidence that TTP and BRF-1 proteins deliver ARE-mRNAs in PBs. This is strongly enhanced when the availability of mRNA decay enzymes is limiting, suggesting that TTP and BRF proteins sequester ARE-mRNAs in PBs when mRNA decay is inefficient. Based on these observations, we propose a model for PB function in which PBs act to sequester ARE-mRNAs, as well as other PB-associated mRNAs, away from polysomes when the cellular mRNA decay machinery is limiting, thus silencing mRNA function even when mRNA degradation is slow.
Results
ARE-mRNAs are observed in PBs
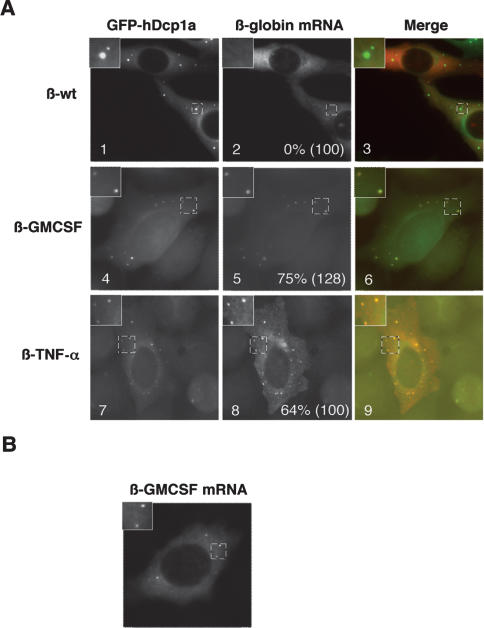
As a first step to test whether a link exists between PBs and the silencing of ARE-mRNAs, we asked whether ARE-mRNAs can be detected in PBs. β-globin reporter mRNAs that contain the AREs from the 3′ UTRs of GM-CSF (β-GMCSF) or TNF-α (β-TNF-α) mRNAs were expressed in HeLa cells and subjected to in situ hybridization. These AREs were chosen because they are both well-defined targets of TTP (Taylor et al. 1996; Carballo et al. 2000), and TTP has been observed previously in PBs at low levels (Kedersha et al. 2005). Transcription of the reporter mRNAs is controlled by a tetracycline regulatory promoter, which is activated by a tetracycline-repressible activator protein when tetracycline is absent (see Materials and Methods). hDcp1a fused to green fluorescent protein (GFP) served as a PB marker. The in situ hybridization assays in Figure 1 show that both the β-GMCSF (panel 5) and β-TNF-α (panel 8) ARE-mRNAs concentrate in PBs (observed in 75% and 64% of cells, respectively), while β-globin mRNA that contains no ARE does not (β-wt; 0% of cells observed) (panel 2). We observed that exogenously expressed GFP-hDcp1a, as well as other tested PB factors, affect PB dynamics (Fenger-Gron et al. 2005; data not shown). However, the ARE-mRNAs also localize to foci when GFP-hDcp1a is not coexpressed (Fig. 1B), excluding the possibility that ARE-mRNA localization in PBs is an artifact of hDcp1a overexpression. We conclude that ARE-mRNAs can be observed in PBs.
Figure 1.
ARE-mRNAs are observed in PBs. (A) In situ hybridization assays showing localization in HeLa cells of exogenously expressed wild-type β-globin mRNA (β-wt mRNA; panel 2), or β-globin mRNAs containing AREs from GM-CSF (β-GMCSF; panel 5) or TNF-α (β-TNF-α; panel 8) mRNAs. (Panels 1,4,7) PBs were visualized using GFP-hDcp1a. Images are merged in panels 3, 6, and 9 (GFP-hDcp1a, green; β-globin mRNAs, red). An enlarged cell section (representing the area in the dotted square) is shown in the top left corner for each image. The percentage of cells with reporter mRNAs in PBs is displayed for each experiment with the total number of cells counted indicated in parentheses. Note that the plasmid expressing β-GMCSF mRNA also expressed (unfused) GFP, thus some nuclear GFP staining is observed in panels 4 and 6, as well as in experiments using the same plasmid below. (B) In situ hybridization assay showing localization of the β-GMCSF mRNA in the absence of GFP-hDcp1a.
The accumulation of ARE-mRNAs in PBs correlates with cellular levels of TTP/BRF proteins
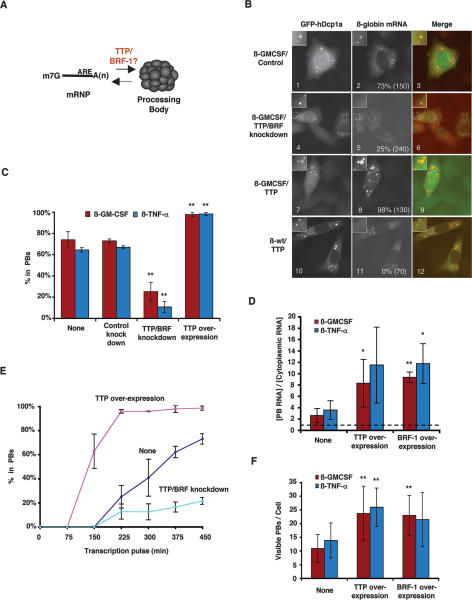
We next asked whether TTP and BRF proteins function in the delivery of the β-GMCSF and β-TNF-α ARE-mRNAs to PBs (Fig. 2A). If TTP and BRF proteins deliver substrate mRNAs to PBs, it is predicted that the accumulation of ARE-mRNA in PBs directly correlates with cellular TTP/BRF protein levels. To test this, we first depleted HeLa cells of TTP/BRF proteins (BRF-1 and BRF-2, but not TTP, could be detected in HeLa cells by Western blotting) (data not shown) by using an siRNA, which targets a sequence conserved between their mRNAs (Jing et al. 2005). Knocking down TTP/BRF proteins in this manner results in a twofold stabilization of the β-GMCSF and β-TNF-α mRNAs (Supplementary Fig. S1). As seen in Figure 2B, fewer cells concentrate β-GMCSF mRNA in PBs upon TTP/BRF protein depletion (panel 5; quantified in Fig. 2C). A similar effect is observed with the β-TNF-α reporter mRNA (Supplementary Fig. S2A, panel 5; quantified in Fig. 2C). Thus, a reduction in TTP and BRF protein levels results in reduced localization of ARE-mRNAs in PBs and slower mRNA decay.
Figure 2.
The accumulation of ARE-mRNAs in PBs correlates with cellular levels of TTP/BRF proteins. (A) Schematic displaying a possible function of TTP and BRF proteins in delivering ARE-mRNAs to PBs. (B) In situ hybridization showing localization of β-GMCSF and β-wt mRNAs in HeLa cells, in the presence of a control siRNA targeting luciferase mRNA (panel 2), an siRNA targeting TTP/BRF proteins (panel 5), or exogenously expressed TTP (panels 8,11). PBs were visualized using GFP-hDcp1a as indicated. An enlarged cell section (representing the area in the dotted square) is shown in the top left corner for each image. The percentage of cells with reporter mRNAs in PBs is displayed for each experiment with the total number of cells counted indicated in parentheses. (Panel 8) Note that, for reasons currently unknown, nuclear β-GMCSF mRNA foci are often observed in cells that overexpress TTP. (C) Graph showing the percentage of cells in which the β-GMCSF or β-TNF-α mRNAs were observed in PBs in the absence of exogenous TTP or siRNA (None), a control siRNA (Control knockdown), an siRNA targeting TTP/BRF proteins (TTP/BRF knockdown), or exogenously expressed TTP (TTP overexpression). Error bars represent standard deviation from at least three experiments. (**) P-value < 0.01. (D) Graph showing the concentration of β-GMCSF or β-TNF-α mRNA in PBs relative to the cytoplasm when no protein is overexpressed (None) or TTP (TTP overexpression) or BRF-1 (BRF-1 overexpression) protein is overexpressed. Error bars represent the standard deviation from at least three cells. (*) P-value < 0.05; (**) P-value < 0.01. (E) Graph showing the percent of cells containing the β-GMCSF mRNA substrate in PBs over time as a result of TTP overexpression, TTP/BRF depletion, or no cotransfection (None). Error bars represent standard deviation calculations from at least three experiments. (F) Graph showing the average number of visible PBs per cell when no protein is overexpressed (None), or TTP (TTP overexpression) or BRF-1 (BRF-1 overexpression) protein is overexpressed. Error bars represent standard deviation calculations obtained from averaging the PB number for 10 cells. (**) P-value < 0.01.
It has been observed previously that overexpression of TTP and BRF-1 results in enhancement of ARE-mediated mRNA decay (Lai et al. 2000; Stoecklin et al. 2002; Lykke-Andersen and Wagner 2005). We therefore tested the effect of TTP and BRF-1 overexpression on the localization of the reporter mRNAs. The in situ hybridization assay in Figure 2B (panel 8) shows that overexpression of TTP causes the β-GMCSF mRNA to accumulate in PBs at a higher concentration (approximately fourfold, quantified in Fig. 2D) and in an increased number of cells (quantified in Fig. 2C). Similar results were observed with overexpressed BRF-1 (Supplementary Fig. S2B; quantified in Fig. 2D) and when TTP was coexpressed with the β-TNF-α mRNA (Supplementary Fig. S2A, panel 8; quantified in Fig. 2C,D). Importantly, these effects are specific to ARE-mRNAs because overexpression of TTP does not lead to the accumulation in PBs of β-globin mRNA with no ARE (Fig. 2B, panel 11). The time-course experiment in Figure 2E, in which the accumulation of β-GMCSF mRNA in PBs was followed over time after its transcriptional induction, shows that TTP overexpression stimulates rapid accumulation of β-GMCSF mRNA in PBs, whereas TTP/BRF depletion has the opposite effect. Interestingly, the number of visible PBs per cell also increased ∼2.5-fold upon TTP or BRF-1 overexpression (Fig. 2F). While overexpression of TTP has been observed previously to induce the formation of stress granules (Kedersha et al. 2005), the ARE-mRNA foci observed here correspond to PBs and not stress granules as they colocalize with GFP-hDcp1a, but not with the stress granule marker TIA-1 (Supplementary Fig. S2C).
We conclude that a strong correlation exists between cellular TTP/BRF protein levels and the rate and extent of localization of β-GMCSF and β-TNF-α mRNAs in PBs. This suggests that TTP and BRF proteins function to directly or indirectly mediate the localization of target ARE-mRNAs in PBs. In addition, the extent of ARE-mRNA PB localization directly correlates with the rate of ARE-mRNA decay.
TTP and BRF-1 can target a tethered mRNA to PBs
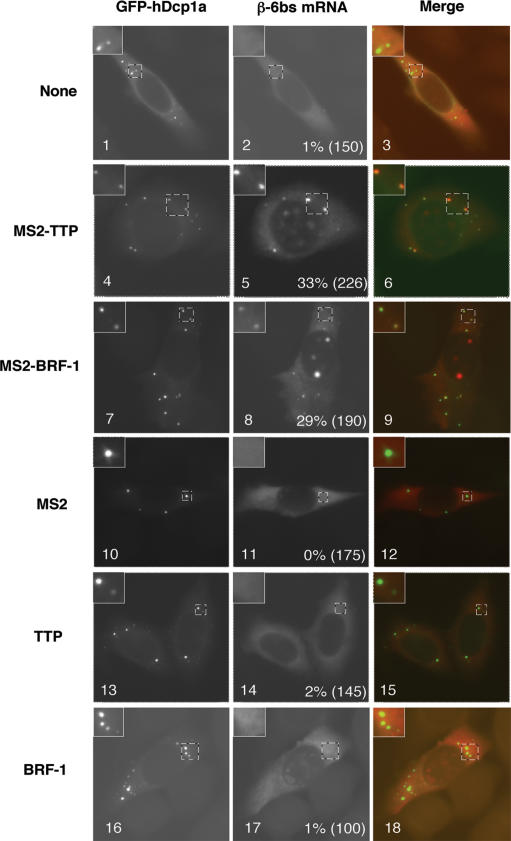
To further test the hypothesis that mRNAs that associate with TTP and BRF-1 are targeted to PBs, we used previously characterized tethering assays (Lykke-Andersen et al. 2000) to ask whether association with TTP and BRF-1 is sufficient to bring an mRNA to the PB in the absence of an ARE. A β-globin reporter mRNA that contains six MS2 coat protein-binding sites in the 3′ UTR (β-6bs) was coexpressed with TTP or BRF-1 fused to the MS2 coat protein (MS2-TTP and MS2-BRF-1). The localization of the β-6bs mRNA was subsequently monitored by in situ hybridization. As seen in Figure 3, in the presence of MS2-TTP or MS2-BRF-1, the β-6bs mRNA localizes in PBs in 29%–33% of cells (panels 5,8). In contrast, the β-6bs mRNA does not significantly localize to PBs when expressed alone (Fig. 3, panel 2) or when coexpressed with unfused MS2 (Fig. 3, panel 11), TTP (Fig. 3, panel 14), or BRF-1 proteins (Fig. 3, panel 17). The enhancement of β-6bs localization in PBs correlates with a sevenfold to 7.5-fold increase in the rate of decay of the mRNA in the presence of MS2-TTP or MS2-BRF-1 (Lykke-Andersen and Wagner 2005). These results show that TTP and BRF-1 are capable of targeting a tethered non-ARE-mRNA to PBs. Interestingly, however, AREs are more efficient at targeting mRNAs to PBs than are MS2-TTP and MS2-BRF-1 (cf. panels 5,8 in Figs. 3 and 1). This suggests that tethered TTP and BRF proteins may not completely recapitulate ARE function (see below).
Figure 3.
TTP and BRF-1 can target a tethered mRNA to PBs. In situ hybridization assays showing localization of a β-globin mRNA with six MS2 coat protein-binding sites in the 3′ UTR (β-6bs) in HeLa cells in the presence of exogenous MS2-TTP (panel 5), MS2-BRF-1 (panel 8), MS2 (panel 11), TTP (panel 14), or BRF-1 (panel 17) proteins, or no exogenous protein (panel 2). PBs were visualized by GFP-hDcp1a as indicated. An enlarged cell section (representing the area in the dotted square) is shown in the top left corner for each image. The percentage of cells with reporter mRNAs in PBs is displayed for each experiment with the total number of cells counted indicated in parentheses.
The N- and C-terminal domains of TTP are PB localization domains
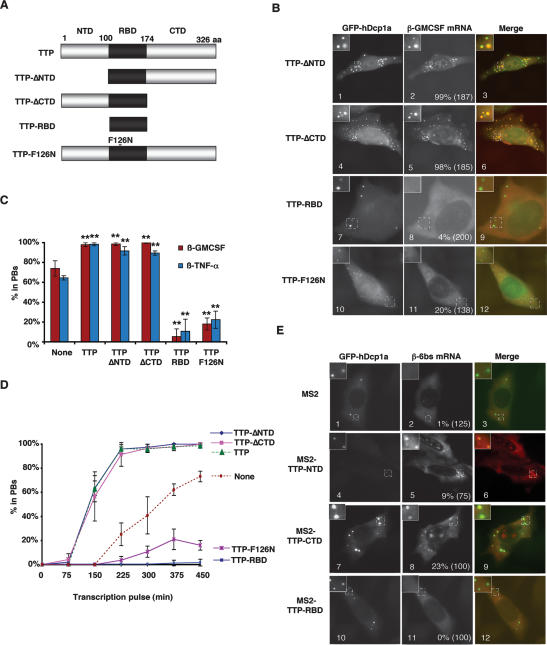
If TTP can target ARE-mRNAs to PBs, it is expected that one or more domains of TTP are required to mediate this process. We observed earlier that the N-terminal domain (NTD) and C-terminal domain (CTD) of TTP and BRF-1 can each activate decay of an associated mRNA (Lykke-Andersen and Wagner 2005), whereas the RNA-binding domain (RBD) inhibits ARE-mediated mRNA decay in a dominant-negative manner when overexpressed (Lai et al. 2000; Lykke-Andersen and Wagner 2005). We therefore tested whether removal of the NTD or CTD of TTP reduces the ability of TTP to enhance the localization of β-GMCSF mRNA in PBs. The β-GMCSF mRNA was coexpressed with either a TTP-ΔNTD (TTP100–326) or a TTP-ΔCTD (TTP1–174) mutant protein (Fig. 4A), and PB localization was visualized by in situ hybridization. The results in Figure 4B show that neither the NTD (panel 2) nor the CTD (panel 5) of TTP is required to enhance the accumulation of the β-GMCSF mRNA in PBs (quantified in Fig. 4C). This is corroborated by the time-course assay in Figure 4D. In contrast, β-GMCSF localization in PBs was strongly inhibited upon expression of the TTP RBD (TTP100–174), which lacks both the NTD and CTD (Fig. 4B, panel 8, quantified in C,D). In addition, exogenous expression of a mutant TTP protein that is incapable of binding the ARE (TTP-F126N) (Fig. 4A; Lai et al. 2002; Lykke-Andersen and Wagner 2005) also inhibits accumulation of β-GMCSF mRNA in PBs (Fig. 4B panel 11, quantified in C,D). Similar results were observed with the β-TNF-α ARE-mRNA substrate (Fig. 4C; Supplementary Fig. S2A). These results suggest that the NTD and CTD of TTP function as redundant PB localization domains. Moreover, the ability of the overexpressed RNA-binding-deficient TTP-F126N protein to inhibit PB localization of β-GMCSF and β-TNF-α mRNAs in a dominant-negative manner suggests that titratable trans-acting factors that interact with TTP are implicated in the process.
Figure 4.
The NTD and CTD of TTP are PB localization domains. (A) Schematic showing TTP and mutants thereof. The NTD and CTD are indicated in gray, while the RBD is indicated in black. TTP-F126N contains a phenylalanine-to-asparagine mutation at position 126. (B) In situ hybridization showing localization of β-GMCSF mRNA in HeLa cells in the presence of exogenously expressed TTP proteins lacking the NTD (TTP-ΔNTD; panel 2), the CTD (TTP-ΔCTD; panel 5), or both the NTD and CTD (TTP-RBD; panel 8), or in the presence of the TTP RNA-binding mutant protein (TTP-F126N; panel 11). PBs were visualized using GFP-hDcp1a. An enlarged cell section (representing the area in the dotted square) is shown in the top left corner for each image. The percentage of cells with reporter mRNAs in PBs is displayed for each experiment with the total number of cells counted indicated in parentheses. (C) Graph showing the percentage of cells in which β-GMCSF or β-TNF-α mRNAs were observed in PBs in the absence of exogenous TTP protein (None) or TTP, TTP-ΔNTD, TTP-ΔCTD, TTP-RBD, or TTP-F126N proteins. Error bars represent standard deviation from at least three separate experiments. (**) P-value < 0.01. (D) Graph showing the percent of cells that contain the β-GMCSF mRNA substrate in PBs over time as a result of the expression of wild-type and mutant TTP proteins. Error bars represent standard deviation calculations from at least three experiments. (E) In situ hybridization showing localization of β-6bs mRNA in HeLa cells in the presence of MS2 (panel 2), MS2-TTP-NTD (panel 5), MS2-TTP-CTD (panel 8), or MS2-TTP-RBD (panel 11). PBs were visualized using GFP-hDcp1a. An enlarged cell section (representing the area in the dotted square) is shown in the top left corner for each image. The percentage of cells with reporter mRNAs in PBs is displayed for each experiment with the total number of cells counted indicated in parentheses.
If the NTD and CTD of TTP are in fact PB localization domains, it is expected that these protein fragments can target a tethered mRNA to PBs, similarly to TTP. We therefore tested whether the NTD (TTP1–100), CTD (TTP176–326), or RBD (TTP100–174) of TTP (Fig. 4A) are sufficient to recruit the tethered β-6bs mRNA to PBs. The in situ hybridization assays in Figure 4E demonstrate that, when fused to the MS2 coat protein, both the NTD and CTD of TTP are sufficient to target the β-6bs mRNA to PBs (panels 5,8), while the MS2 coat protein (panel 2) and the tethered RBD of TTP are not (panel 11). However, it is important to note that the NTD of TTP was much less efficient than the CTD and the full-length protein. This correlates with a previously observed weaker activation of mRNA decay by tethered NTD (Lykke-Andersen and Wagner 2005).
TTP and BRF-1 deliver nontranslating mRNAs to PBs
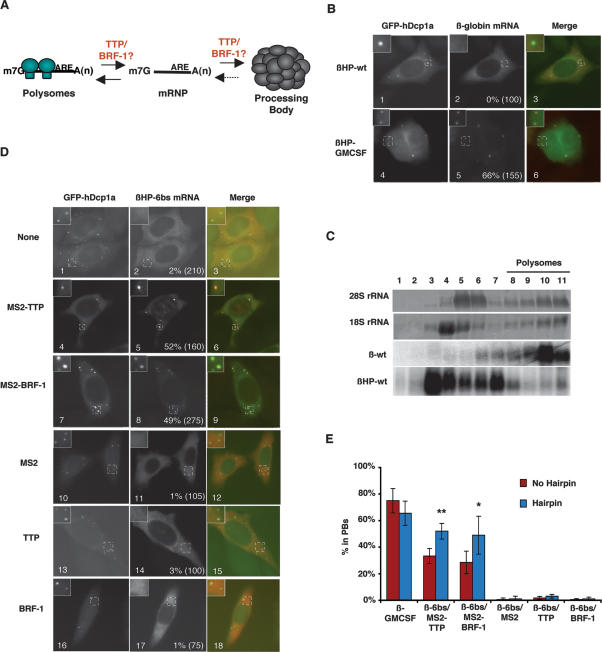
We next wished to gain insight into the mechanism by which TTP and BRF-1 shift the pool of ARE-mRNAs from polysomes to PBs. Previous studies have shown that mRNAs exist in PBs to the exclusion of polysomes (Brengues et al. 2005; Coller and Parker 2005; Teixeira et al. 2005). We considered the possibilities that TTP and BRF-1 could facilitate the localization of ARE-mRNAs to PBs either directly, or indirectly by stimulating the release of ARE-mRNAs from polysomes (Fig. 5A). It was observed previously that mRNAs that are released from polysomes in yeast through cell starvation (Brengues et al. 2005) or in mammalian cells through miRNA-mediated translational silencing accumulate in PBs (Liu et al. 2005b; Pillai et al. 2005). It was therefore possible that TTP/BRF proteins mediate the localization of ARE-mRNAs in PBs solely through inhibition of polysome formation. If this was the case, it would be predicted that inhibition of mRNA polysome association stimulates PB localization even in the absence of TTP/BRF association. To test this, we asked whether insertion of a hairpin that inhibits translation initiation (Kozak 1989) into the 5′ UTR of our β-globin reporter mRNAs results in PB localization. However, as seen in Figure 5B, insertion of the 5′ UTR hairpin does not stimulate PB localization of wild-type β-globin mRNA (βHP-wt) (panel 2) and does not enhance the localization in PBs of the β-GMCSF reporter mRNA (βHP-GMCSF) (cf. panel 5 in Figs. 5B and 1). Importantly, the hairpin efficiently inhibits polysome formation, as evidenced by sucrose gradient polysome fractionation assays (Fig. 5C). We conclude that inhibition of polysome formation in itself is insufficient for PB localization of mRNA in human HeLa cells.
Figure 5.
TTP and BRF-1 deliver nontranslating mRNAs to PBs. (A) Schematic showing possible steps at which TTP/BRF proteins may act to facilitate the localization of ARE-mRNAs in PBs. (B) In situ hybridization assays showing localization in HeLa cells of wild-type β-globin (βHP-wt; panel 2) or ARE-containing β-globin mRNA (βHP-GMCSF; panel 5) that each contain a stable hairpin in their 5′ UTR. PBs were visualized using GFP-hDcp1a. An enlarged cell section (representing the area in the dotted square) is shown in the top left corner for each image. The percentage of cells with reporter mRNAs in PBs is displayed for each experiment with the total number of cells counted indicated in parentheses. (C) Polysome fractionation analysis showing exclusion of hairpin-containing mRNAs from polysomes in HeLa tet-off cells. Polysomes were differentially sedimented by sucrose gradient and fractionated into 11 fractions. Ribosomal rRNA and mRNA content was analyzed by methylene blue staining and Northern blot analysis, respectively. (D) In situ hybridization assay showing localization in HeLa cells of a β-globin mRNA with six MS2 coat protein-binding sites in the 3′ UTR and a stable hairpin in the 5′ UTR (βHP-6bs) in the presence of exogenous MS2-TTP (panel 5), MS2-BRF-1 (panel 8), MS2 (panel 11), TTP (panel 14), or BRF-1 (panel 17) proteins, or no exogenous protein (panel 2). PBs were visualized using GFP-hDcp1a. (E) Graph showing the percentage of cells in which the β-GMCSF and β-6bs mRNAs localize to PBs in the absence (red bars) or presence (blue bars) of a stable hairpin in the 5′ UTR. Error bars represent standard deviation calculations from at least three experiments. (*) P-value < 0.05; (**) P-value < 0.01.
We next asked whether TTP and BRF-1 mediate the localization of nontranslating mRNAs to PBs. If this is the case, it is expected that the translationally repressed βHP-6bs mRNA, which contains six MS2 coat protein-binding sites in the 3′ UTR and a 5′ UTR hairpin, will localize to PBs only when coexpressed with MS2-TTP or MS2-BRF-1. As seen in Figure 5D, both MS2-TTP and MS2-BRF-1 stimulate the localization of βHP-6bs mRNA in PBs (cf. panels 5,8 and 2). In contrast, unfused MS2 (Fig. 5D, panel 11), TTP (Fig. 5D, panel 14), or BRF-1 (Fig. 5D, panel 17) proteins do not stimulate PB localization of the βHP-6bs mRNA. We conclude that TTP/BRF proteins (Fig. 5D) and AREs (Fig. 5B) stimulate delivery of mRNAs to PBs downstream from the release from polysomes (Fig. 5A, right arrow). However, it cannot be excluded that TTP/BRF proteins and AREs also play a role in polysome release (Fig. 5A, left arrow). In fact, AREs have been observed previously to inhibit translation (Zhang et al. 2002). Interestingly, quantifications revealed that the translationally repressed βHP-6bs mRNA was more efficiently recruited to PBs by MS2-TTP and MS2-BRF-1 than was the corresponding translated β-6bs mRNA (Fig. 5E). A similar enhancement was observed with the βHP-6bs mRNA as compared with the β-6bs mRNA tethered to the NTD or the CTD of TTP (data not shown). In contrast, localization of the β-GMCSF mRNA in PBs was not enhanced by translational repression (Fig. 5E). This suggests that polysome release is a limiting step in localization of an mRNA to PBs by tethered TTP and BRF-1, but not by an ARE. Thus, either tethered TTP and BRF-1 may not fully recapitulate endogenous TTP/BRF function, or ARE-binding proteins other than TTP and BRF-1 stimulate ARE-mRNA polysome release.
The accumulation of ARE-mRNAs in PBs is strongly enhanced upon mRNA decay enzyme depletion
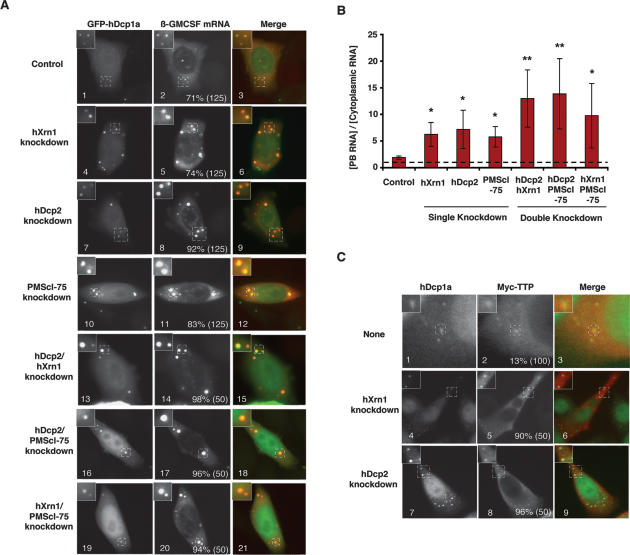
The observations described above provide evidence that TTP and BRF proteins sequester ARE-mRNAs in PBs away from the translational machinery. To ask whether a correlation exists between the accumulation of ARE-mRNAs in PBs and the efficiency of mRNA decay, we tested the effect of depleting mRNA decay enzymes. HeLa cells were transfected with plasmids expressing the β-GMCSF mRNA along with short hairpins targeting the mRNAs for the 5′-to-3′ exonuclease hXrn1 or the decapping enzyme hDcp2, both catalytic components of the PB. As seen in Figure 6A, a dramatically enhanced accumulation of β-GMCSF mRNA in PBs is observed in hXrn1- and hDcp2-depleted cells (panels 5,8), whereas no enhanced accumulation was observed with a control hairpin targeting luciferase mRNA (panel 2). In contrast to the β-GMCSF mRNA, the β-wt mRNA, which lacks an ARE, accumulates in PBs in <10% of cells following mRNA decay factor depletion (data not shown). The concentration of the β-GMCSF mRNA in PBs was quantified relative to its concentration in the cytoplasm and revealed a three- to fourfold increase upon hXrn1 or hDcp2 knockdown (Fig. 6B). This rules out the possibility that the enhanced localization of ARE-mRNAs in PBs following hXrn1 and hDcp2 knockdown is simply a result of elevated ARE-mRNA levels (1.5-fold to twofold higher levels of β-GMCSF mRNA were observed) (data not shown). In addition, we observed that localization of exogenously expressed TTP (Fig. 6C) and BRF-1 (data not shown) to PBs is also strongly enhanced following hXrn1 or hDcp2 depletion (Fig. 6C, cf. panels 5,8 and 2). We conclude that TTP/BRF proteins and ARE-mRNAs accumulate in PBs following hDcp2 and hXrn1 depletion.
Figure 6.
ARE-mRNAs accumulate in PBs when mRNA decay enzymes are depleted. (A) In situ hybridization assays showing accumulation of the β-GMCSF mRNA in PBs in HeLa cells expressing siRNAs targeting luciferase (panel 2), hXrn1 (panel 5), hDcp2 (panel 8), PMScl-75 (panel 11), hXrn1 and hDcp2 (panel 14), hDcp2 and PMScl-75 (panel 17), or hXrn1 and PMScl-75 (panel 20). PBs were visualized using GFP-hDcp1a. An enlarged cell section (representing the area in the dotted square) is shown in the top left corner for each image. The percentage of cells with reporter mRNAs in PBs is displayed for each experiment with the total number of cells counted indicated in parentheses. (B) Graph showing the average concentration of β-globin mRNA in PBs relative to the cytoplasm. Error bars represent standard deviation from measurements of at least three cells. (*) P-value < 0.05; (**) P-value < 0.01. (C) Indirect immunofluorescence assays showing localization of endogenous hDcp1a (panels 1,4,7) and exogenous Myc-tagged TTP (panels 2,5,8) in untreated cells, or cells treated with siRNAs against hXrn1 or hDcp2, as indicated. The percentage of cells in which Myc-tagged TTP protein was observed colocalizing with hDcp1a is shown with the total number of cells counted indicated in parentheses.
The exosome has been implicated previously in ARE-mRNA decay (Chen et al. 2001; Mukherjee et al. 2002), and knockdown of the exosome component PMScl-75 was observed to impair ARE-mRNA decay (Stoecklin et al. 2006). In contrast to hXrn1 and hDcp2, it is not clear whether the exosome associates with PBs, although exosome subunits have been observed in cytoplasmic foci in Drosophila cells (Graham et al. 2006). As seen in Figure 6A (panel 11), knockdown of PMScl-75 causes enhanced accumulation of the β-GMCSF reporter in PBs similar to what was observed following hDcp2 and hXrn1 depletion (quantified in Fig. 6B). Double knockdowns of mRNA decay enzymes revealed even stronger accumulation of the β-GMCSF mRNA in PBs (Fig. 6A, panels 14,17,20, quantified in B). We conclude that the β-GMCSF mRNA strongly accumulates in PBs upon depletion of mRNA decay enzymes. This suggests that ARE-mRNAs are sequestered in PBs when mRNA decay enzymes become limiting for the ARE-mRNA decay pathway. The importance of deadenylation for the localization of ARE-mRNAs in PBs is currently unclear, because overexpression of catalytically inactive deadenylases (hPan2 D1083A, hCcr4b D525A, hCaf1b D40A/E42A) showed no significant effect on the accumulation of β-GMCSF mRNA in PBs (data not shown), perhaps due to the redundance of deadenylases in the human cell (Yamashita et al. 2005; Wagner et al. 2006).
TTP nucleates PB formation on substrate mRNAs
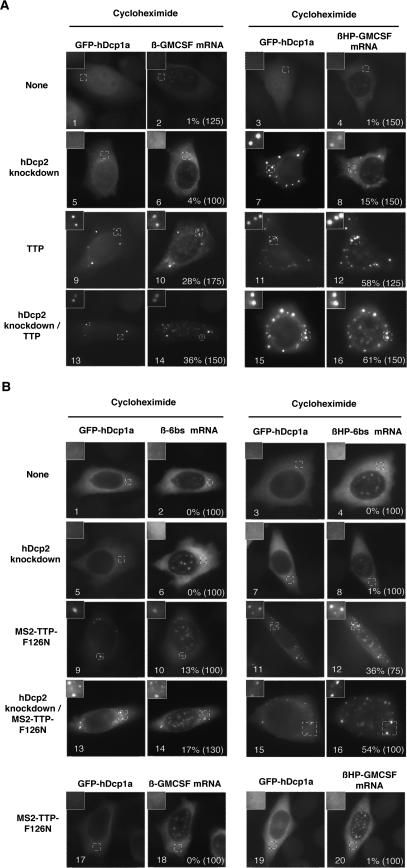
Based on the observations in Figure 6, we hypothesized that mRNAs only accumulate in visible PBs when they are targeted for mRNA decay but their degradation is slow or absent. If this is the case, it is predicted that PBs can be re-created under conditions in which PBs are normally absent (such as during cycloheximide treatment) if the mRNA decay machinery is rendered limiting. To test this, we asked whether PBs containing ARE-mRNAs can be generated in the presence of cycloheximide when hDcp2 is knocked down or TTP is overexpressed. As seen in Figure 7A, the β-GMCSF ARE-mRNA does not localize in PBs in the presence of cycloheximide (panel 2). Similarly, the translationally silenced βHP-GMCSF ARE-mRNA was not observed in PBs upon cycloheximide treatment (Fig. 7A, panel 4), even though this mRNA is not trapped in polysomes under these conditions (Fig. 5C; data not shown). However, upon depletion of the decapping enzyme hDcp2, 15% of cells form PBs containing βHP-GMCSF mRNA in the presence of cycloheximide (Fig. 7A, panel 8). In contrast, little PB formation is observed with the translated β-GMCSF mRNA (Fig. 7A, panel 6). Thus, an untranslated ARE-mRNA can nucleate PB formation in the presence of cycloheximide when hDcp2 is knocked down. We also tested whether overexpressed TTP can nucleate PB formation in the presence of cycloheximide. As seen in Figure 7A (panels 10,12), overexpression of TTP triggers efficient PB formation. This is particularly evident for the translationally repressed βHP-GMCSF mRNA (Fig. 7A, panel 12), but also was observed on the translated β-GMCSF mRNA (Fig. 7A, panel 10), suggesting that overexpressed TTP can localize ARE-mRNAs in PBs even before their association with polysomes. When hDcp2 was depleted in combination with TTP overexpression, the accumulation of the β-GMCSF mRNAs in PBs was further enhanced (Fig. 7A, panels 14,16).
Figure 7.
TTP can nucleate PB formation on substrate mRNAs in the presence of cycloheximide. (A) In situ hybridization assays showing localization of β-GMCSF (left panels) and βHP-GMCSF (right panels) mRNAs in cycloheximidetreated HeLa cells in the presence of hDcp2 siRNAs (panels 5–8), TTP protein (panels 9–12), a combination of TTP protein and hDcp2 siRNAs (panels 13–16), or no exogenous siRNAs or protein (panels 1–4). PBs were visualized using GFP-hDcp1a. An enlarged cell section (representing the area in the dotted square) is shown in the top left corner for each image. The percentage of cells with reporter mRNAs in PBs is displayed for each experiment with the total number of cells counted indicated in parentheses. (B) In situ hybridization assays showing localization of β-6bs (top left panels), β-GMCSF (bottom left panels), βHP-6bs (top right panels), or βHP-GMCSF (bottom right panels) mRNAs in cycloheximide-treated HeLa cells in the presence of hDcp2 siRNAs (panels 5–8), MS2-TTP-F126N protein (panels 9–12,17–20), or a combination of both MS2-TTP-F126N protein and hDcp2 siRNAs (panels 13–16), or no exogenous siRNAs or protein (panels 1–4). PBs were visualized using GFP-hDcp1a. The percentage of cells with reporter mRNAs in PBs is displayed for each experiment with the total number of cells counted indicated in parentheses.
We next tested the ability of tethered TTP to nucleate PB formation. As seen in Figure 7B (panel 10), in the presence of cycloheximide the ARE-binding-deficient mutant TTP-F126N protein stimulates formation of PBs when tethered via the MS2 coat protein to the β-6bs mRNA. This is strongly enhanced when polysome formation on the β-6bs mRNA is inhibited by insertion of the 5′ UTR hairpin (Fig. 7B, panel 12) and when hDcp2 is simultaneously knocked down (Fig. 7B, panels 14,16). In contrast, hDcp2 knockdown was insufficient to stimulate PB localization of the β-6bs mRNAs (Fig. 7B, panels 6,8). Importantly, the MS2-TTP-F126N protein likely has no cellular mRNA targets other than the β-6bs mRNAs, since it is mutant for ARE binding (Lai et al. 2002; Lykke-Andersen and Wagner 2005) and does not stimulate PB localization of translated or untranslated β-GMCSF ARE-mRNA (Fig. 7B, panels 18,20). Thus, the PBs observed in Figure 7B are likely highly homogeneous, containing primarily the β-6bs reporter mRNAs. We conclude that TTP can nucleate PB formation on substrate mRNAs under conditions in which other mRNAs are trapped in polysomes. This only occurs when the mRNA decay machinery is rendered limiting by hDcp2 depletion or TTP overexpression.
Discussion
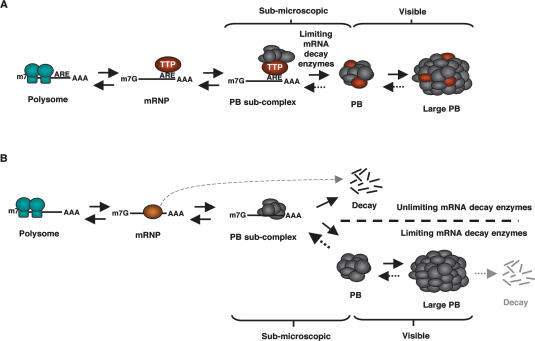
AREs are key elements in post-transcriptional gene regulation that target mRNAs in which they reside for translational silencing and decay, thereby regulating the expression of multiple human genes (Chen and Shyu 1995; Wilusz et al. 2001; Shim and Karin 2002; Zhang et al. 2002). It was observed recently that a number of factors involved in mRNA decay and translational silencing concentrate in cytoplasmic foci called PBs (van Dijk et al. 2002; Eystathioy et al. 2003; Sheth and Parker 2003; Cougot et al. 2004). Here we show that the ARE-binding proteins TTP and BRF-1 nucleate PB formation on ARE-mRNAs. This silences the ARE-mRNAs by sequestering them from the translation machinery until they undergo mRNA decay (Fig. 8A).
Figure 8.
Model for mRNA silencing by PB localization in human cells. (A) TTP and BRF proteins nucleate the formation of a submicroscopic PB subcomplex on ARE-mRNAs. When decay enzymes are limiting, this PB subcomplex aggregates with other mRNA-PB subcomplexes to form PBs, which can grow large enough to be microscopically visible. (B) Under conditions in which mRNA decay enzymes are nonlimiting, mRNAs are efficiently turned over before they can aggregate into a PB, and thus PBs remain submicroscopic (above the dotted line). When mRNA decay enzymes are limiting, PB subcomplexes form larger aggregates (below the dotted line) visible to the microscope that serve to silence translation until mRNA degradation (see Discussion).
TTP and BRF proteins mediate the localization of ARE-mRNAs in PBs
The mechanism by which AREs repress protein expression is poorly understood, but involves both translational silencing and mRNA decay (Chen and Shyu 1995; Gueydan et al. 1999; Piecyk et al. 2000; Wilusz et al. 2001; Shim and Karin 2002; Lopez de Silanes et al. 2005; Mazan-Mamczarz et al. 2006). Here, we show evidence that ARE-mRNA silencing involves delivery of ARE-mRNAs to PBs by the proteins TTP and BRF-1. First, β-GMCSF and β-TNF-α ARE-mRNAs are observed in PBs (Fig. 1). This localization is inhibited upon depletion of TTP/BRF proteins using siRNAs or upon overexpression of dominant-negative mutant TTP proteins (Figs. 2, 4). In contrast, localization of β-GMCSF and β-TNF-α mRNAs in PBs is enhanced upon TTP or BRF-1 overexpression (Fig. 2; Supplementary Fig. S2). Second, TTP and BRF-1, as well as the NTD and CTD of TTP, can target a tethered non-ARE mRNA to PBs (Figs. 3, 4). However, it is important to note that targeting to PBs of β-globin mRNA by tethered TTP and BRF-1 is not as efficient as by AREs. This suggests that the function of the ARE is not completely recapitulated by tethered TTP and BRF-1. Nevertheless, our experiments demonstrate that the TTP family of ARE-binding proteins deliver target ARE-mRNAs to PBs and, in the case of TTP, this can be mediated by both the NTD and CTD.
TTP and BRF proteins target polysome-released ARE-mRNAs to PBs
How do TTP/BRF proteins shift the equilibrium of ARE-mRNAs from polysomes to PBs? The following observations suggest that TTP/BRF proteins act, at least in part, to target ARE-mRNAs to PBs subsequent to their polysome release (Fig. 8A). First, the presence of a hairpin that efficiently inhibits polysome formation does not target an mRNA to PBs unless the mRNA contains an ARE or is tethered to TTP or BRF-1 (Fig. 5). Second, TTP can nucleate the formation of PBs on an ARE-mRNA or a tethered mRNA even when other PB substrates are trapped in polysomes by cycloheximide treatment (Fig. 7). Thus, polysome release is insufficient for mRNA PB localization, and TTP and BRF-1 efficiently localize nontranslated mRNAs to PBs.
This raises the question of how TTP and BRF proteins promote localization of ARE-mRNAs in PBs. Our observations suggest that this is mediated through direct interactions between TTP/BRF proteins and PB subunits. First, TTP and BRF-1 stimulate PB localization of mRNA already released from polysomes (Fig. 5). Second, overexpression of the ARE-binding-deficient TTP-F126N mutant protein inhibits ARE-mRNA localization in PBs (Fig. 4), suggesting that titratable trans-acting factors that bind to TTP are important for PB localization. Third, TTP exists in complex with several PB factors (Fenger-Gron et al. 2005). Some of these interactions appear to be direct, as evidenced by interaction assays between bacterially purified TTP and PB components translated in rabbit reticulocyte lysates (C. Egan, T.M. Franks, and J. Lykke-Andersen, unpubl.). Taken together, these observations suggest that TTP nucleates the formation of a PB “subcomplex” on ARE-mRNAs that subsequently mediates the association with the PB (Fig. 8A). Once a PB subcomplex is nucleated by TTP/BRF proteins, how does it mediate the localization of ARE-mRNAs in PBs? Future studies should reveal whether this is a result of active transport or passive diffusion followed by retention in the PB. However, we have not observed any effect on PB formation of depleting actin filaments or microtubules using cytoskeleton inhibitors (J. Dennis and J. Lykke-Andersen, unpubl.).
Do ARE-binding proteins actively release ARE-mRNAs from polysomes?
Experiments using translational inhibitors suggest that mRNAs need to be released from polysomes prior to their localization in PBs (Fig. 8A; Cougot et al. 2004; Teixeira et al. 2005). Our observation that a 5′ UTR hairpin that represses translation initiation does not enhance localization of the β-GMCSF ARE-mRNA in PBs (Fig. 5) suggests that ARE-binding proteins may stimulate polysome release, thus negating the effect of the 5′ UTR hairpin. This is consistent with observations by others that AREs can repress translation (Zhang et al. 2002). Interestingly, tethered TTP and BRF-1 were less efficient than an ARE at localizing an mRNA in PBs (Figs. 1, 3), but regained activity similar to an ARE when translation was inhibited (Fig. 5). This suggests that either TTP and BRF-1 do not efficiently release bound mRNAs from polysomes, or the tethered proteins do not fully recapitulate endogenous protein function. Using sucrose gradient polysome assays, we observed no evidence that an ARE or tethered TTP or BRF-1 stimulates accumulation of polysome-released mRNA (data not shown). However, it is possible that, under these conditions, the released mRNA is rapidly degraded and therefore is undetectable. Candidate factors implicated in releasing ARE-mRNAs from polysomes include (in addition to TTP and BRF proteins) the microRNA miR16 and the associated RNA-induced silencing complex (RISC), which has been implicated in the decay of mRNAs containing the ARE from TNF-α mRNA (Jing et al. 2005), as well as the translation repressor proteins TIA-1 and TIAR, which have affinity for AREs (Dember et al. 1996; Gueydan et al. 1999; Piecyk et al. 2000; Lopez de Silanes et al. 2005; Mazan-Mamczarz et al. 2006). In addition, general translation repressors that are components of PBs may stimulate localization of ARE-mRNAs in PBs by repressing polysome formation. Interestingly, the PB component Rck/p54 exists in complex with TTP (Fenger-Gron et al. 2005), and its yeast ortholog, Dhh1p, is a translational repressor (Coller and Parker 2005). An important goal for future studies is to delineate how the multiple ARE-binding proteins and their associated factors cooperate to silence ARE-mRNAs.
TTP and BRF proteins facilitate the localization of ARE-mRNAs in PBs when mRNA decay enzymes are limiting
Our studies raise the important question of under which conditions TTP and BRF proteins translocate ARE-mRNAs from the cytoplasm to PBs. The following observations suggest that the sequestration of the ARE-mRNAs into microscopically visible PBs takes place under conditions in which mRNA decay enzymes acting on the ARE-mRNA become limiting (Fig. 8A). First, when levels of hDcp2, hXrn1, or PMScl-75 are reduced by knockdown, ARE-mRNAs accumulate at higher concentrations in PBs (Fig. 6). Second, when TTP or BRF-1 proteins are overexpressed, which is predicted to leave endogenous mRNA decay factors limiting for TTP and BRF-1 function, the ARE-mRNAs accumulate at high levels in the PBs (Fig. 2; Supplementary Fig. S2). Third, TTP can nucleate the formation of PBs on substrate mRNAs during cycloheximide treatment only when TTP is overexpressed or hDcp2 is depleted (Fig. 7). Thus, TTP and BRF-1 sequester ARE-mRNAs in PBs when the availability of mRNA decay enzymes becomes limiting for ARE-mRNA decay (Fig. 8A). This is predicted to sequester the ARE-mRNA away from the translation machinery, thereby providing an efficient means of silencing ARE-mRNA function even while the mRNA is awaiting decay.
Can the sequestered ARE-mRNAs degrade in the PB, or do they have to be released from the PB prior to their decay? The observations that the catalytic mRNA decay enzymes hDcp2 and hXrn1 concentrate in PBs (Sheth and Parker 2003; Cougot et al. 2004) and that their knockdown results in enhanced accumulation of ARE-mRNAs in PBs (Fig. 6) suggest that these enzymes function to degrade ARE-mRNAs in the PB. Moreover, any manipulation of TTP and BRF-1 proteins that enhances ARE-mRNA decay also stimulates ARE-mRNA localization in PBs (Fig. 2), whereas those conditions that impair TTP/BRF-1 function result in decreased degradation and PB localization of the ARE-mRNAs (Figs. 2, 4). However, while these observations suggest that ARE-mRNAs can degrade in PBs, it is a formal possibility that mRNA decay enzymes are kept inactive in the PB and only degrade mRNAs when both the mRNA and the mRNA decay enzymes are released from the PB. Even if ARE-mRNA degrades in the PB, it remains to be determined what fraction of the total cellular mRNA degrades there. Moreover, it is unknown whether the exosome assists in ARE-mRNA decay in the general cytoplasm or in conjunction with PBs or PB subunits. Indirect immunofluorescence assays have revealed that several Drosophila exosome subunits are observed in cytoplasmic foci, although it is unclear whether these correspond to PBs (Graham et al. 2006).
A general function of PBs in sequestering mRNAs that undergo delayed decay
The model in Figure 8A can likely be extended to other mRNAs that interface with PBs. In this scenario, the miRISC complex would serve (via GW182) to nucleate PBs on miRNA target mRNAs (Yang et al. 2004; Ding et al. 2005; Jakymiw et al. 2005; Liu et al. 2005b; Pauley et al. 2006), whereas Upf proteins mediate PB formation on NMD substrates (Sheth and Parker 2006)—in human cells, likely via hSmg7 (Unterholzner and Izaurralde 2004). In addition, it can be speculated that, at least in yeast, mRNAs that lack destabilizing cis-elements are targeted after their deadenylation to PBs by the Lsm1-7–Pat1 complex, which localizes in PBs (Sheth and Parker 2003) and specifically associates with and activates decay of deadenylated mRNAs (Tharun et al. 2000; Tharun and Parker 2001).
If the model in Figure 8A provides a general paradigm for how mRNAs interface with PBs, it is predicted that the size of PBs in the cell directly correlates with the availability of mRNA substrates and inversely correlates with the availability of mRNA decay enzymes (Fig. 8B). These predictions are consistent with our observations, as well as with observations by others, of the dynamics of PBs. For example, PBs grow large when the mRNA decay machinery is rendered limiting by an increase in the level of polysome-free cytoplasmic mRNA by puromycin treatment (Teixeira et al. 2005), or by a reduction in cellular Dcp2 and Xrn1 activity (Fig. 6; Sheth and Parker 2003; Cougot et al. 2004). In contrast, PBs disappear when mRNA substrates become limiting upon entrapment of translated mRNAs in polysomes by cycloheximide treatment (Fig. 7; Cougot et al. 2004; Teixeira et al. 2005), or when decapping is rendered nonlimiting by hDcp2 overexpression (Fenger-Gron et al. 2005). These considerations provide a possible explanation for the observation that PBs disappear when miRNAs are depleted from human HeLa cells through depletion of Drosha (Pauley et al. 2006), or when miRNA function is inhibited through depletion of the PB factor GW182 (Yang et al. 2004; Jakymiw et al. 2005; Liu et al. 2005a; Rehwinkel et al. 2005; Pauley et al. 2006). In this case, depletion of a major cellular pathway that employs mRNA decay enzymes is likely to render the mRNA decay enzymes nonlimiting for other mRNA decay pathways, thus degrading them prior to their aggregation into (microscopically visible) PBs (Fig. 8B). In a similar manner, PBs disappear when deadenylases are depleted in yeast cells (Sheth and Parker 2003), even though a subset of PB-associated mRNAs, such as NMD substrates (Sheth and Parker 2006), can undergo mRNA decay independently of deadenylation (Muhlrad and Parker 1994). The concept that PB factors are not inactive when PBs are invisible is underscored by our observation that PBs can form on TTP-associated mRNAs even when other mRNAs are trapped in polysomes by cycloheximide treatment, which normally results in the disappearance of PBs (Fig. 7).
Thus, PB formation may provide a mechanism for the cell to deal with situations in which mRNAs targeted for decay become overabundant. A key purpose for the PB may therefore be to sequester mRNAs targeted for decay away from the translational machinery when the mRNA decay machinery is limiting, thus effectively silencing protein production even before the mRNA is degraded. Interestingly, some mRNAs sequestered in PBs can return to the cytoplasm for translation rather than being degraded (Brengues et al. 2005; Bhattacharyya et al. 2006). For example, Bhattacharyya et al. (2006) demonstrated that the CAT-1 mRNA, which has both miRNA targets and an ARE in its 3′ UTR, can be relieved of miRNA-mediated suppression in PBs by the ARE-binding protein HuR. An interesting question for future studies will be to determine if mRNAs that are targeted to PBs by an ARE can also be removed from PBs by HuR or other ARE-binding proteins. This could conceivably be regulated for those mRNAs by an inability of the PB localizing factors to recruit catalytic mRNA decay enzymes. Alternatively, mRNA decay enzyme function could be inhibited globally under certain conditions. It can also be speculated that aggregation into PBs provides a mechanism to effectively increase the local concentration of mRNA decay enzymes. If mRNA decay can occur in the PB, it is thus predicted that the rate of mRNA decay is higher in the PB than it is in the general cytoplasm. Finally, the PB may act to sequester mRNA decay enzymes to prevent promiscuous mRNA decay in the cytoplasm. Thus, the aggregation of mRNAs into PBs may provide an efficient mechanism to silence mRNAs by simultaneously shutting down translation and stimulating mRNA decay. The PB could therefore be thought of as a “buffer” for the cellular mRNA decay machinery, which ensures that mRNAs targeted for degradation are functionally repressed even when the cellular mRNA decay machinery has become limiting.
Materials and methods
Plasmids
Plasmids encoding the β-wt (pPCBwt), β-GMCSF (pPCBwtATGMCSF), β-Gap (pcB-Gap), and β-6bs (pcTet2Bwt-3MS2) reporter mRNAs have been described previously (Lykke-Andersen and Wagner 2005). The plasmid pPCBwtTNF-α, which encodes the β-TNF-α mRNA, contains the core TNF-α ARE (Carrick et al. 2004) inserted into an ApaI site of the pPCBwt plasmid (Fenger-Gron et al. 2005). To create the constructs for βHP-wt (pcTet2BHP-wt), βHP-MS2 (pcTet2BHP-3MS2), and βHP-GMCSF (pcTet2BHP-GMCSF) mRNA expression, a previously described hairpin sequence (Kozak 1989) was inserted into the HindIII and NheI sites of the corresponding β-globin mRNA expression vectors upstream of the β-globin coding region. A plasmid encoding a tetracycline-responsive activator protein was used to activate transcription of reporter mRNAs (pTet-TTA; Clontech). Plasmids encoding TTP, BRF-1, or derivatives thereof have been described previously (Lykke-Andersen and Wagner 2005). The plasmid pcNEGFP-hDcp1a, which was used as a PB marker, has two tandem copies of the Enhanced GFP (EGFP) inserted into the HindIII site of pcDNA3 and the ORF of hDcp1a inserted between EcoRI and NotI sites. Plasmids used to knock down hDcp2, hXrn1, and PMScl-75 expression were created by cloning an siRNA precursor sequence (see Supplemental Material) into the BseRI and BamHI sites of the pSHAG vector (Paddison et al. 2004). Plasmid sequences are available upon request.
Indirect immunofluorescence and in situ hybridization assays
HeLa cells in DMEM/10% fetal bovine serum (FBS) at ∼50% confluency in 12-well plates were transfected using TransIT HeLaMonster reagent according to the manufacturer’s protocols (Mirus), with a total of 1 μg of plasmid. Cells were split to chamber slides 24 h later. For indirect immunofluorescence experiments, cells were transfected with 50 ng of pcDNA3-Myc-TTP/BRF-1 and 0.95 μg of empty pcDNA3 vector. Forty-eight hours after transfection, cells were fixed in 4% paraformaldehyde for 15 min, and permeabilized and blocked with PBS/1% goat serum/0.1% Triton X-100 for 30 min. Cells were then incubated with rabbit anti-hDcp1a (Lykke-Andersen and Wagner 2005) and mouse anti-myc (9B11; Cell Signaling) antibodies at 1:200 and 1:1000 dilutions, respectively, for 2 h. Following removal of the primary antibody, cells were incubated for 1 h with 4 μg/mL secondary anti-rabbit antibodies labeled with Alexa 488 fluorophore and anti-mouse antibodies labeled with Texas-red fluorophore (Molecular Probes).
For in situ hybridization experiments, cells were transfected in the presence of 50 ng/mL tetracycline with 300 ng of reporter mRNA expression plasmid, 300 ng of pTet-TTA, and 100 ng of pcNEGFP-hDcp1a. Plasmids expressing the following proteins were cotransfected in specific experiments: 50 ng (Figs. 2, 7) or 150 ng (Figs. 3, 5) of pcDNA3-Myc-TTP, 150 ng of pcNMS2-TTP/BRF-1 (Figs. 3, 5), 50 ng of pcDNA3-Myc-TTP-ΔNTD/ΔCTD/F126N (Fig. 4), 200 ng of pcDNA3-Myc-TTP-RBD (Fig. 4), and 150 ng of pcNMS2-TTP-F126N (Fig. 7). pcDNA3 was added to a total of 1 μg of plasmid in each experiment. Forty hours after transfection, transcription of reporter mRNAs was initiated by washing cells in phosphate-buffered saline (PBS) and placing them in DMEM/10% FBS, containing no tetracycline. After a transcriptional pulse of 0–12 h (see Pulsed Expression Experiments and Cell Phenotype Quantification), cells were fixed in 4% paraformaldehyde for 15 min and permeabilized overnight in 70% ethanol. Cells were then rehydrated for 10 min in 50% formamide and 2× SSC. Next, cells were incubated overnight at 37°C in a solution containing 50% formamide, 2× SSC, 0.02% bovine serum albumin (BSA), 2 mM vanadyl–ribonucleoside complexes, 1 μg/mL total yeast RNA, and 0.1 mg/mL dextran sulfate. In order to detect the localization of the β-globin mRNA, four Texas-red labeled 50-nucleotide DNA oligo probes (Invitrogen) complementary to sequences in exons 1, 2, and 3 were also added to the mixture at a concentration of 20 ng/mL each (sequences of oligos are given in the Supplemental Material). Cells were washed twice for 30 min at 37°C in 50% formamide and 2× SSC prior to visualization.
Pulsed expression experiments and cell phenotype quantification
Transcription was pulsed for 8 h prior to the observation of the localization of β-wt, β-GMCSF, and β-TNF-α reporter mRNAs (Figs. 1, 2, 4–7). To observe the localization of reporter mRNAs over time, transcription of individual cell samples was induced for the indicated time prior to fixation (Figs. 2, 4). For tethering experiments, transcription of reporter mRNAs was pulsed for 12 h (Figs. 3, 5, 7).
To quantify the number of cells with reporter mRNAs concentrated in PBs, transfected cells expressing detectable quantities of reporter mRNA were scored for reporter mRNA colocalization with GFP-hDcp1a. The cell counts from at least three experiments were averaged to produce a final percentage and standard deviation measurement.
Quantification of RNA concentration in the PB versus the cytoplasm
The intensity of the in situ hybridization signal in the background (outside the cell), the cytoplasm, and the five most intensely staining PBs were calculated for each of three randomly chosen cells using OpenLab software. After subtraction of the background intensity, the average PB intensity was divided by the cytoplasmic intensity to determine the concentration of reporter mRNA in the PB versus the cytoplasm.
RNAi-mediated knockdown
For TTP/BRF knockdown, HeLa (Fig. 2) or HeLa tet-off (Supplementary Fig. S1) cells were seeded onto 3.5-cm wells at 75% density 20 h prior to siRNA transfection. On the following day, each cell sample was incubated for 6 h in a 1-mL transfection mixture containing 10 μL of Lipofectamine 2000 reagent (Invitrogen) and siRNAs at a concentration of 200 nM, along with the following plasmids: 200 ng pcNEGFP-hDcp1a, 600 ng reporter mRNA, and 600 ng pTet-TTA (Clontech) in DMEM/10% FBS containing tetracycline at a concentration of 50 ng/mL. On the following day, cells were split to chamber slides or 12-well plates. For in situ hybridization experiments and mRNA decay assays, expression of mRNA reporters was induced 40 h after siRNA transfection. For hDcp2, hXrn1, and PMScl-75 knockdowns (Fig. 6), HeLa cells were transfected as previously described under “immunofluorescence and in situ hybridization”; however, in this case, cells were also transfected with pSHAG plasmids encoding a precursor siRNA targeted against hDcp2, hXrn1, or PMScl-75 mRNAs.
Polysome profiles
HeLa tet-off cells were seeded onto 10-cm plates at ∼50% density 1 d prior to transfection. On the following day, cells were transfected in the presence of 50 ng/mL tetracyline with 3.75 μg of reporter plasmid, 1.5 μg of a plasmid encoding a desired MS2 coat protein (if relevant), and 500 ng of pcβ-Gap transfection control reporter and pcDNA3 to a total of 10 μg plasmid. Forty hours after transfection, transcription of reporter mRNAs was pulsed for 8 h. Polysome profile experiments were then conducted as described by Johannes and Sarnow (1998).
PB nucleation experiments
Expression of mRNA reporters was pulsed for 8 h. Cycloheximide was added to cells at a concentration of 10 μg/mL for the duration of the pulse, and cells were visualized as described above.
Acknowledgments
We thank Min Han for use of fluorescence microscope, Paula Grissom and the laboratory of Dick McIntosh for supplies used to conduct polysome profiles, and Christian Damgaard for in situ hybridization protocol and TIA-1 clones. This research was supported by grants to J.L. from the National Institutes of Health (grant GM 066811) and the Pew Scholars Program in the Biological Sciences, sponsored by the Pew Scholar Charitable Trusts. T.F. was supported by National Institutes of Health NRSA Institutional Training Grant no. GM-07135 from the National Institute of General Medical Sciences.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1494707
References
- Anderson P., Kedersha N., Kedersha N. RNA granules. J. Cell Biol. 2006;172:803–808. doi: 10.1083/jcb.200512082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrei M.A., Ingelfinger D., Heintzmann R., Achsel T., Rivera-Pomar R., Luhrmann R., Ingelfinger D., Heintzmann R., Achsel T., Rivera-Pomar R., Luhrmann R., Heintzmann R., Achsel T., Rivera-Pomar R., Luhrmann R., Achsel T., Rivera-Pomar R., Luhrmann R., Rivera-Pomar R., Luhrmann R., Luhrmann R. A role for eIF4E and eIF4E-transporter in targeting mRNPs to mammalian processing bodies. RNA. 2005;11:717–727. doi: 10.1261/rna.2340405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya S.N., Habermacher R., Martine U., Closs E.I., Filipowicz W., Habermacher R., Martine U., Closs E.I., Filipowicz W., Martine U., Closs E.I., Filipowicz W., Closs E.I., Filipowicz W., Filipowicz W. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell. 2006;125:1111–1124. doi: 10.1016/j.cell.2006.04.031. [DOI] [PubMed] [Google Scholar]
- Blackshear P.J. Tristetraprolin and other CCCH tandem zinc-finger proteins in the regulation of mRNA turnover. Biochem. Soc. Trans. 2002;30:945–952. doi: 10.1042/bst0300945. [DOI] [PubMed] [Google Scholar]
- Brengues M., Teixeira D., Parker R., Teixeira D., Parker R., Parker R. Movement of eukaryotic mRNAs between polysomes and cytoplasmic processing bodies. Science. 2005;310:486–489. doi: 10.1126/science.1115791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carballo E., Lai W.S., Blackshear P.J., Lai W.S., Blackshear P.J., Blackshear P.J. Feedback inhibition of macrophage tumor necrosis factor-α production by tristetraprolin. Science. 1998;281:1001–1005. doi: 10.1126/science.281.5379.1001. [DOI] [PubMed] [Google Scholar]
- Carballo E., Lai W.S., Blackshear P.J., Lai W.S., Blackshear P.J., Blackshear P.J. Evidence that tristetraprolin is a physiological regulator of granulocyte-macrophage colony-stimulating factor messenger RNA deadenylation and stability. Blood. 2000;95:1891–1899. [PubMed] [Google Scholar]
- Carrick D.M., Lai W.S., Blackshear P.J., Lai W.S., Blackshear P.J., Blackshear P.J. The tandem CCCH zinc finger protein tristetraprolin and its relevance to cytokine mRNA turnover and arthritis. Arthritis Res. Ther. 2004;6:248–264. doi: 10.1186/ar1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.Y., Shyu A.B., Shyu A.B. AU-rich elements: Characterization and importance in mRNA degradation. Trends Biochem. Sci. 1995;20:465–470. doi: 10.1016/s0968-0004(00)89102-1. [DOI] [PubMed] [Google Scholar]
- Chen C.Y., Gherzi R., Ong S.E., Chan E.L., Raijmakers R., Pruijn G.J., Stoecklin G., Moroni C., Mann M., Karin M., Gherzi R., Ong S.E., Chan E.L., Raijmakers R., Pruijn G.J., Stoecklin G., Moroni C., Mann M., Karin M., Ong S.E., Chan E.L., Raijmakers R., Pruijn G.J., Stoecklin G., Moroni C., Mann M., Karin M., Chan E.L., Raijmakers R., Pruijn G.J., Stoecklin G., Moroni C., Mann M., Karin M., Raijmakers R., Pruijn G.J., Stoecklin G., Moroni C., Mann M., Karin M., Pruijn G.J., Stoecklin G., Moroni C., Mann M., Karin M., Stoecklin G., Moroni C., Mann M., Karin M., Moroni C., Mann M., Karin M., Mann M., Karin M., Karin M. AU binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell. 2001;107:451–464. doi: 10.1016/s0092-8674(01)00578-5. [DOI] [PubMed] [Google Scholar]
- Chou C.F., Mulky A., Maitra S., Lin W.J., Gherzi R., Kappes J., Chen C.Y., Mulky A., Maitra S., Lin W.J., Gherzi R., Kappes J., Chen C.Y., Maitra S., Lin W.J., Gherzi R., Kappes J., Chen C.Y., Lin W.J., Gherzi R., Kappes J., Chen C.Y., Gherzi R., Kappes J., Chen C.Y., Kappes J., Chen C.Y., Chen C.Y. Tethering KSRP, a decay-promoting AU-rich element-binding protein, to mRNAs elicits mRNA decay. Mol. Cell. Biol. 2006;26:3695–3706. doi: 10.1128/MCB.26.10.3695-3706.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coller J., Parker R., Parker R. General translational repression by activators of mRNA decapping. Cell. 2005;122:875–886. doi: 10.1016/j.cell.2005.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cougot N., Babajko S., Seraphin B., Babajko S., Seraphin B., Seraphin B. Cytoplasmic foci are sites of mRNA decay in human cells. J. Cell Biol. 2004;165:31–40. doi: 10.1083/jcb.200309008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMaria C.T., Brewer G., Brewer G. AUF1 binding affinity to A + U-rich elements correlates with rapid mRNA degradation. J. Biol. Chem. 1996;271:12179–12184. doi: 10.1074/jbc.271.21.12179. [DOI] [PubMed] [Google Scholar]
- Dember L.M., Kim N.D., Liu K.Q., Anderson P., Kim N.D., Liu K.Q., Anderson P., Liu K.Q., Anderson P., Anderson P. Individual RNA recognition motifs of TIA-1 and TIAR have different RNA binding specificities. J. Biol. Chem. 1996;271:2783–2788. doi: 10.1074/jbc.271.5.2783. [DOI] [PubMed] [Google Scholar]
- Ding L., Spencer A., Morita K., Han M., Spencer A., Morita K., Han M., Morita K., Han M., Han M. The developmental timing regulator AIN-1 interacts with miRISCs and may target the argonaute protein ALG-1 to cytoplasmic P bodies in C. elegans. Mol. Cell. 2005;19:437–447. doi: 10.1016/j.molcel.2005.07.013. [DOI] [PubMed] [Google Scholar]
- Eystathioy T., Jakymiw A., Chan E.K., Seraphin B., Cougot N., Fritzler M.J., Jakymiw A., Chan E.K., Seraphin B., Cougot N., Fritzler M.J., Chan E.K., Seraphin B., Cougot N., Fritzler M.J., Seraphin B., Cougot N., Fritzler M.J., Cougot N., Fritzler M.J., Fritzler M.J. The GW182 protein colocalizes with mRNA degradation associated proteins hDcp1 and hLSm4 in cytoplasmic GW bodies. RNA. 2003;9:1171–1173. doi: 10.1261/rna.5810203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan X.C., Steitz J.A., Steitz J.A. Overexpression of HuR, a nuclear-cytoplasmic shuttling protein, increases the in vivo stability of ARE-containing mRNAs. EMBO J. 1998;17:3448–3460. doi: 10.1093/emboj/17.12.3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenger-Gron M., Fillman C., Norrild B., Lykke-Andersen J., Fillman C., Norrild B., Lykke-Andersen J., Norrild B., Lykke-Andersen J., Lykke-Andersen J. Multiple processing body factors and the ARE binding protein TTP activate mRNA decapping. Mol. Cell. 2005;20:905–915. doi: 10.1016/j.molcel.2005.10.031. [DOI] [PubMed] [Google Scholar]
- Ferraiuolo M.A., Basak S., Dostie J., Murray E.L., Schoenberg D.R., Sonenberg N., Basak S., Dostie J., Murray E.L., Schoenberg D.R., Sonenberg N., Dostie J., Murray E.L., Schoenberg D.R., Sonenberg N., Murray E.L., Schoenberg D.R., Sonenberg N., Schoenberg D.R., Sonenberg N., Sonenberg N. A role for the eIF4E-binding protein 4E-T in P-body formation and mRNA decay. J. Cell Biol. 2005;170:913–924. doi: 10.1083/jcb.200504039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fillman C., Lykke-Andersen J., Lykke-Andersen J. RNA decapping inside and outside of processing bodies. Curr. Opin. Cell Biol. 2005;17:326–331. doi: 10.1016/j.ceb.2005.04.002. [DOI] [PubMed] [Google Scholar]
- Gherzi R., Lee K.Y., Briata P., Wegmuller D., Moroni C., Karin M., Chen C.Y., Lee K.Y., Briata P., Wegmuller D., Moroni C., Karin M., Chen C.Y., Briata P., Wegmuller D., Moroni C., Karin M., Chen C.Y., Wegmuller D., Moroni C., Karin M., Chen C.Y., Moroni C., Karin M., Chen C.Y., Karin M., Chen C.Y., Chen C.Y. A KH domain RNA binding protein, KSRP, promotes ARE-directed mRNA turnover by recruiting the degradation machinery. Mol. Cell. 2004;14:571–583. doi: 10.1016/j.molcel.2004.05.002. [DOI] [PubMed] [Google Scholar]
- Graham A.C., Kiss D.L., Andrulis E.D., Kiss D.L., Andrulis E.D., Andrulis E.D. Differential distribution of exosome subunits at the nuclear lamina and in cytoplasmic foci. Mol. Biol. Cell. 2006;17:1399–1409. doi: 10.1091/mbc.E05-08-0805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gueydan C., Droogmans L., Chalon P., Huez G., Caput D., Kruys V., Droogmans L., Chalon P., Huez G., Caput D., Kruys V., Chalon P., Huez G., Caput D., Kruys V., Huez G., Caput D., Kruys V., Caput D., Kruys V., Kruys V. Identification of TIAR as a protein binding to the translational regulatory AU-rich element of tumor necrosis factor α mRNA. J. Biol. Chem. 1999;274:2322–2326. doi: 10.1074/jbc.274.4.2322. [DOI] [PubMed] [Google Scholar]
- Ingelfinger D., Arndt-Jovin D.J., Luhrmann R., Achsel T., Arndt-Jovin D.J., Luhrmann R., Achsel T., Luhrmann R., Achsel T., Achsel T. The human LSm1-7 proteins colocalize with the mRNA-degrading enzymes Dcp1/2 and Xrnl in distinct cytoplasmic foci. RNA. 2002;8:1489–1501. [PMC free article] [PubMed] [Google Scholar]
- Jakymiw A., Lian S., Eystathioy T., Li S., Satoh M., Hamel J.C., Fritzler M.J., Chan E.K., Lian S., Eystathioy T., Li S., Satoh M., Hamel J.C., Fritzler M.J., Chan E.K., Eystathioy T., Li S., Satoh M., Hamel J.C., Fritzler M.J., Chan E.K., Li S., Satoh M., Hamel J.C., Fritzler M.J., Chan E.K., Satoh M., Hamel J.C., Fritzler M.J., Chan E.K., Hamel J.C., Fritzler M.J., Chan E.K., Fritzler M.J., Chan E.K., Chan E.K. Disruption of GW bodies impairs mammalian RNA interference. Nat. Cell Biol. 2005;7:1267–1274. doi: 10.1038/ncb1334. [DOI] [PubMed] [Google Scholar]
- Jing Q., Huang S., Guth S., Zarubin T., Motoyama A., Chen J., Di Padova F., Lin S.C., Gram H., Han J., Huang S., Guth S., Zarubin T., Motoyama A., Chen J., Di Padova F., Lin S.C., Gram H., Han J., Guth S., Zarubin T., Motoyama A., Chen J., Di Padova F., Lin S.C., Gram H., Han J., Zarubin T., Motoyama A., Chen J., Di Padova F., Lin S.C., Gram H., Han J., Motoyama A., Chen J., Di Padova F., Lin S.C., Gram H., Han J., Chen J., Di Padova F., Lin S.C., Gram H., Han J., Di Padova F., Lin S.C., Gram H., Han J., Lin S.C., Gram H., Han J., Gram H., Han J., Han J. Involvement of microRNA in AU-rich element-mediated mRNA instability. Cell. 2005;120:623–634. doi: 10.1016/j.cell.2004.12.038. [DOI] [PubMed] [Google Scholar]
- Johannes G., Sarnow P., Sarnow P. Cap-independent polysomal association of natural mRNAs encoding c-myc, BiP, and eIF4G conferred by internal ribosome entry sites. RNA. 1998;4:1500–1513. doi: 10.1017/s1355838298981080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N., Stoecklin G., Ayodele M., Yacono P., Lykke-Andersen J., Fritzler M.J., Scheuner D., Kaufman R.J., Golan D.E., Anderson P., Stoecklin G., Ayodele M., Yacono P., Lykke-Andersen J., Fritzler M.J., Scheuner D., Kaufman R.J., Golan D.E., Anderson P., Ayodele M., Yacono P., Lykke-Andersen J., Fritzler M.J., Scheuner D., Kaufman R.J., Golan D.E., Anderson P., Yacono P., Lykke-Andersen J., Fritzler M.J., Scheuner D., Kaufman R.J., Golan D.E., Anderson P., Lykke-Andersen J., Fritzler M.J., Scheuner D., Kaufman R.J., Golan D.E., Anderson P., Fritzler M.J., Scheuner D., Kaufman R.J., Golan D.E., Anderson P., Scheuner D., Kaufman R.J., Golan D.E., Anderson P., Kaufman R.J., Golan D.E., Anderson P., Golan D.E., Anderson P., Anderson P. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J. Cell Biol. 2005;169:871–884. doi: 10.1083/jcb.200502088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Circumstances and mechanisms of inhibition of translation by secondary structure in eucaryotic mRNAs. Mol. Cell. Biol. 1989;9:5134–5142. doi: 10.1128/mcb.9.11.5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai W.S., Carballo E., Strum J.R., Kennington E.A., Phillips R.S., Blackshear P.J., Carballo E., Strum J.R., Kennington E.A., Phillips R.S., Blackshear P.J., Strum J.R., Kennington E.A., Phillips R.S., Blackshear P.J., Kennington E.A., Phillips R.S., Blackshear P.J., Phillips R.S., Blackshear P.J., Blackshear P.J. Evidence that tristetraprolin binds to AU-rich elements and promotes the deadenylation and destabilization of tumor necrosis factor α mRNA. Mol. Cell. Biol. 1999;19:4311–4323. doi: 10.1128/mcb.19.6.4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai W.S., Carballo E., Thorn J.M., Kennington E.A., Blackshear P.J., Carballo E., Thorn J.M., Kennington E.A., Blackshear P.J., Thorn J.M., Kennington E.A., Blackshear P.J., Kennington E.A., Blackshear P.J., Blackshear P.J. Interactions of CCCH zinc finger proteins with mRNA. Binding of tristetraprolin-related zinc finger proteins to Au-rich elements and destabilization of mRNA. J. Biol. Chem. 2000;275:17827–17837. doi: 10.1074/jbc.M001696200. [DOI] [PubMed] [Google Scholar]
- Lai W.S., Kennington E.A., Blackshear P.J., Kennington E.A., Blackshear P.J., Blackshear P.J. Interactions of CCCH zinc finger proteins with mRNA: Non-binding tristetraprolin mutants exert an inhibitory effect on degradation of AU-rich element-containing mRNAs. J. Biol. Chem. 2002;277:9606–9613. doi: 10.1074/jbc.M110395200. [DOI] [PubMed] [Google Scholar]
- Lai W.S., Parker J.S., Grissom S.F., Stumpo D.J., Blackshear P.J., Parker J.S., Grissom S.F., Stumpo D.J., Blackshear P.J., Grissom S.F., Stumpo D.J., Blackshear P.J., Stumpo D.J., Blackshear P.J., Blackshear P.J. Novel mRNA targets for tristetraprolin (TTP) identified by global analysis of stabilized transcripts in TTP-deficient fibroblasts. Mol. Cell. Biol. 2006;26:9196–9208. doi: 10.1128/MCB.00945-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Rivas F.V., Wohlschlegel J., Yates J.R., III, Parker R., Hannon G.J., Rivas F.V., Wohlschlegel J., Yates J.R., III, Parker R., Hannon G.J., Wohlschlegel J., Yates J.R., III, Parker R., Hannon G.J., Yates J.R., III, Parker R., Hannon G.J., Parker R., Hannon G.J., Hannon G.J. A role for the P-body component GW182 in microRNA function. Nat. Cell Biol. 2005a;7:1261–1266. doi: 10.1038/ncb1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Valencia-Sanchez M.A., Hannon G.J., Parker R., Valencia-Sanchez M.A., Hannon G.J., Parker R., Hannon G.J., Parker R., Parker R. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat. Cell Biol. 2005b;7:719–723. doi: 10.1038/ncb1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lopez Silanes I., Galban S., Martindale J.L., Yang X., Mazan-Mamczarz K., Indig F.E., Falco G., Zhan M., Gorospe M., Galban S., Martindale J.L., Yang X., Mazan-Mamczarz K., Indig F.E., Falco G., Zhan M., Gorospe M., Martindale J.L., Yang X., Mazan-Mamczarz K., Indig F.E., Falco G., Zhan M., Gorospe M., Yang X., Mazan-Mamczarz K., Indig F.E., Falco G., Zhan M., Gorospe M., Mazan-Mamczarz K., Indig F.E., Falco G., Zhan M., Gorospe M., Indig F.E., Falco G., Zhan M., Gorospe M., Falco G., Zhan M., Gorospe M., Zhan M., Gorospe M., Gorospe M. Identification and functional outcome of mRNAs associated with RNA-binding protein TIA-1. Mol. Cell. Biol. 2005;25:9520–9531. doi: 10.1128/MCB.25.21.9520-9531.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykke-Andersen J., Wagner E., Wagner E. Recruitment and activation of mRNA decay enzymes by two ARE-mediated decay activation domains in the proteins TTP and BRF-1. Genes & Dev. 2005;19:351–361. doi: 10.1101/gad.1282305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykke-Andersen J., Shu M.D., Steitz J.A., Shu M.D., Steitz J.A., Steitz J.A. Human Upf proteins target an mRNA for nonsense-mediated decay when bound downstream of a termination codon. Cell. 2000;103:1121–1131. doi: 10.1016/s0092-8674(00)00214-2. [DOI] [PubMed] [Google Scholar]
- Mazan-Mamczarz K., Lal A., Martindale J.L., Kawai T., Gorospe M., Lal A., Martindale J.L., Kawai T., Gorospe M., Martindale J.L., Kawai T., Gorospe M., Kawai T., Gorospe M., Gorospe M. Translational repression by RNA-binding protein TIAR. Mol. Cell. Biol. 2006;26:2716–2727. doi: 10.1128/MCB.26.7.2716-2727.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlrad D., Parker R., Parker R. Premature translational termination triggers mRNA decapping. Nature. 1994;370:578–581. doi: 10.1038/370578a0. [DOI] [PubMed] [Google Scholar]
- Mukherjee D., Gao M., O’Connor J.P., Raijmakers R., Pruijn G., Lutz C.S., Wilusz J., Gao M., O’Connor J.P., Raijmakers R., Pruijn G., Lutz C.S., Wilusz J., O’Connor J.P., Raijmakers R., Pruijn G., Lutz C.S., Wilusz J., Raijmakers R., Pruijn G., Lutz C.S., Wilusz J., Pruijn G., Lutz C.S., Wilusz J., Lutz C.S., Wilusz J., Wilusz J. The mammalian exosome mediates the efficient degradation of mRNAs that contain AU-rich elements. EMBO J. 2002;21:165–174. doi: 10.1093/emboj/21.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paddison P.J., Silva J.M., Conklin D.S., Schlabach M., Li M., Aruleba S., Balija V., O’Shaughnessy A., Gnoj L., Scobie K., Silva J.M., Conklin D.S., Schlabach M., Li M., Aruleba S., Balija V., O’Shaughnessy A., Gnoj L., Scobie K., Conklin D.S., Schlabach M., Li M., Aruleba S., Balija V., O’Shaughnessy A., Gnoj L., Scobie K., Schlabach M., Li M., Aruleba S., Balija V., O’Shaughnessy A., Gnoj L., Scobie K., Li M., Aruleba S., Balija V., O’Shaughnessy A., Gnoj L., Scobie K., Aruleba S., Balija V., O’Shaughnessy A., Gnoj L., Scobie K., Balija V., O’Shaughnessy A., Gnoj L., Scobie K., O’Shaughnessy A., Gnoj L., Scobie K., Gnoj L., Scobie K., Scobie K., et al. A resource for large-scale RNA-interference-based screens in mammals. Nature. 2004;428:427–431. doi: 10.1038/nature02370. [DOI] [PubMed] [Google Scholar]
- Pauley K.M., Eystathioy T., Jakymiw A., Hamel J.C., Fritzler M.J., Chan E.K., Eystathioy T., Jakymiw A., Hamel J.C., Fritzler M.J., Chan E.K., Jakymiw A., Hamel J.C., Fritzler M.J., Chan E.K., Hamel J.C., Fritzler M.J., Chan E.K., Fritzler M.J., Chan E.K., Chan E.K. Formation of GW bodies is a consequence of microRNA genesis. EMBO Rep. 2006;7:904–910. doi: 10.1038/sj.embor.7400783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng S.S., Chen C.Y., Xu N., Shyu A.B., Chen C.Y., Xu N., Shyu A.B., Xu N., Shyu A.B., Shyu A.B. RNA stabilization by the AU-rich element binding protein, HuR, an ELAV protein. EMBO J. 1998;17:3461–3470. doi: 10.1093/emboj/17.12.3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piecyk M., Wax S., Beck A.R., Kedersha N., Gupta M., Maritim B., Chen S., Gueydan C., Kruys V., Streuli M., Wax S., Beck A.R., Kedersha N., Gupta M., Maritim B., Chen S., Gueydan C., Kruys V., Streuli M., Beck A.R., Kedersha N., Gupta M., Maritim B., Chen S., Gueydan C., Kruys V., Streuli M., Kedersha N., Gupta M., Maritim B., Chen S., Gueydan C., Kruys V., Streuli M., Gupta M., Maritim B., Chen S., Gueydan C., Kruys V., Streuli M., Maritim B., Chen S., Gueydan C., Kruys V., Streuli M., Chen S., Gueydan C., Kruys V., Streuli M., Gueydan C., Kruys V., Streuli M., Kruys V., Streuli M., Streuli M., et al. TIA-1 is a translational silencer that selectively regulates the expression of TNF-α. EMBO J. 2000;19:4154–4163. doi: 10.1093/emboj/19.15.4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillai R.S., Bhattacharyya S.N., Artus C.G., Zoller T., Cougot N., Basyuk E., Bertrand E., Filipowicz W., Bhattacharyya S.N., Artus C.G., Zoller T., Cougot N., Basyuk E., Bertrand E., Filipowicz W., Artus C.G., Zoller T., Cougot N., Basyuk E., Bertrand E., Filipowicz W., Zoller T., Cougot N., Basyuk E., Bertrand E., Filipowicz W., Cougot N., Basyuk E., Bertrand E., Filipowicz W., Basyuk E., Bertrand E., Filipowicz W., Bertrand E., Filipowicz W., Filipowicz W. Inhibition of translational initiation by Let-7 MicroRNA in human cells. Science. 2005;309:1573–1576. doi: 10.1126/science.1115079. [DOI] [PubMed] [Google Scholar]
- Ramos S.B., Stumpo D.J., Kennington E.A., Phillips R.S., Bock C.B., Ribeiro-Neto F., Blackshear P.J., Stumpo D.J., Kennington E.A., Phillips R.S., Bock C.B., Ribeiro-Neto F., Blackshear P.J., Kennington E.A., Phillips R.S., Bock C.B., Ribeiro-Neto F., Blackshear P.J., Phillips R.S., Bock C.B., Ribeiro-Neto F., Blackshear P.J., Bock C.B., Ribeiro-Neto F., Blackshear P.J., Ribeiro-Neto F., Blackshear P.J., Blackshear P.J. The CCCH tandem zinc-finger protein Zfp36l2 is crucial for female fertility and early embryonic development. Development. 2004;131:4883–4893. doi: 10.1242/dev.01336. [DOI] [PubMed] [Google Scholar]
- Rehwinkel J., Behm-Ansmant I., Gatfield D., Izaurralde E., Behm-Ansmant I., Gatfield D., Izaurralde E., Gatfield D., Izaurralde E., Izaurralde E. A crucial role for GW182 and the DCP1:DCP2 decapping complex in miRNA-mediated gene silencing. RNA. 2005;11:1640–1647. doi: 10.1261/rna.2191905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen G.L., Blau H.M., Blau H.M. Argonaute 2/RISC resides in sites of mammalian mRNA decay known as cytoplasmic bodies. Nat. Cell Biol. 2005;7:633–636. doi: 10.1038/ncb1265. [DOI] [PubMed] [Google Scholar]
- Sheth U., Parker R., Parker R. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science. 2003;300:805–808. doi: 10.1126/science.1082320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheth U., Parker R., Parker R. Targeting of aberrant mRNAs to cytoplasmic processing bodies. Cell. 2006;125:1095–1109. doi: 10.1016/j.cell.2006.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim J., Karin M., Karin M. The control of mRNA stability in response to extracellular stimuli. Mol. Cells. 2002;14:323–331. [PubMed] [Google Scholar]
- Stoecklin G., Ming X.F., Looser R., Moroni C., Ming X.F., Looser R., Moroni C., Looser R., Moroni C., Moroni C. Somatic mRNA turnover mutants implicate tristetraprolin in the interleukin-3 mRNA degradation pathway. Mol. Cell. Biol. 2000;20:3753–3763. doi: 10.1128/mcb.20.11.3753-3763.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoecklin G., Colombi M., Raineri I., Leuenberger S., Mallaun M., Schmidlin M., Gross B., Lu M., Kitamura T., Moroni C., Colombi M., Raineri I., Leuenberger S., Mallaun M., Schmidlin M., Gross B., Lu M., Kitamura T., Moroni C., Raineri I., Leuenberger S., Mallaun M., Schmidlin M., Gross B., Lu M., Kitamura T., Moroni C., Leuenberger S., Mallaun M., Schmidlin M., Gross B., Lu M., Kitamura T., Moroni C., Mallaun M., Schmidlin M., Gross B., Lu M., Kitamura T., Moroni C., Schmidlin M., Gross B., Lu M., Kitamura T., Moroni C., Gross B., Lu M., Kitamura T., Moroni C., Lu M., Kitamura T., Moroni C., Kitamura T., Moroni C., Moroni C. Functional cloning of BRF1, a regulator of ARE-dependent mRNA turnover. EMBO J. 2002;21:4709–4718. doi: 10.1093/emboj/cdf444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoecklin G., Mayo T., Anderson P., Mayo T., Anderson P., Anderson P. ARE-mRNA degradation requires the 5′–3′ decay pathway. EMBO Rep. 2006;7:72–77. doi: 10.1038/sj.embor.7400572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumpo D.J., Byrd N.A., Phillips R.S., Ghosh S., Maronpot R.R., Castranio T., Meyers E.N., Mishina Y., Blackshear P.J., Byrd N.A., Phillips R.S., Ghosh S., Maronpot R.R., Castranio T., Meyers E.N., Mishina Y., Blackshear P.J., Phillips R.S., Ghosh S., Maronpot R.R., Castranio T., Meyers E.N., Mishina Y., Blackshear P.J., Ghosh S., Maronpot R.R., Castranio T., Meyers E.N., Mishina Y., Blackshear P.J., Maronpot R.R., Castranio T., Meyers E.N., Mishina Y., Blackshear P.J., Castranio T., Meyers E.N., Mishina Y., Blackshear P.J., Meyers E.N., Mishina Y., Blackshear P.J., Mishina Y., Blackshear P.J., Blackshear P.J. Chorioallantoic fusion defects and embryonic lethality resulting from disruption of Zfp36L1, a gene encoding a CCCH tandem zinc finger protein of the Tristetraprolin family. Mol. Cell. Biol. 2004;24:6445–6455. doi: 10.1128/MCB.24.14.6445-6455.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor G.A., Carballo E., Lee D.M., Lai W.S., Thompson M.J., Patel D.D., Schenkman D.I., Gilkeson G.S., Broxmeyer H.E., Haynes B.F., Carballo E., Lee D.M., Lai W.S., Thompson M.J., Patel D.D., Schenkman D.I., Gilkeson G.S., Broxmeyer H.E., Haynes B.F., Lee D.M., Lai W.S., Thompson M.J., Patel D.D., Schenkman D.I., Gilkeson G.S., Broxmeyer H.E., Haynes B.F., Lai W.S., Thompson M.J., Patel D.D., Schenkman D.I., Gilkeson G.S., Broxmeyer H.E., Haynes B.F., Thompson M.J., Patel D.D., Schenkman D.I., Gilkeson G.S., Broxmeyer H.E., Haynes B.F., Patel D.D., Schenkman D.I., Gilkeson G.S., Broxmeyer H.E., Haynes B.F., Schenkman D.I., Gilkeson G.S., Broxmeyer H.E., Haynes B.F., Gilkeson G.S., Broxmeyer H.E., Haynes B.F., Broxmeyer H.E., Haynes B.F., Haynes B.F., et al. A pathogenetic role for TNF α in the syndrome of cachexia, arthritis, and autoimmunity resulting from tristetraprolin (TTP) deficiency. Immunity. 1996;4:445–454. doi: 10.1016/s1074-7613(00)80411-2. [DOI] [PubMed] [Google Scholar]
- Teixeira D., Sheth U., Valencia-Sanchez M.A., Brengues M., Parker R., Sheth U., Valencia-Sanchez M.A., Brengues M., Parker R., Valencia-Sanchez M.A., Brengues M., Parker R., Brengues M., Parker R., Parker R. Processing bodies require RNA for assembly and contain nontranslating mRNAs. RNA. 2005;11:371–382. doi: 10.1261/rna.7258505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tharun S., Parker R., Parker R. Targeting an mRNA for decapping: Displacement of translation factors and association of the Lsm1p-7p complex on deadenylated yeast mRNAs. Mol. Cell. 2001;8:1075–1083. doi: 10.1016/s1097-2765(01)00395-1. [DOI] [PubMed] [Google Scholar]
- Tharun S., He W., Mayes A.E., Lennertz P., Beggs J.D., Parker R., He W., Mayes A.E., Lennertz P., Beggs J.D., Parker R., Mayes A.E., Lennertz P., Beggs J.D., Parker R., Lennertz P., Beggs J.D., Parker R., Beggs J.D., Parker R., Parker R. Yeast Sm-like proteins function in mRNA decapping and decay. Nature. 2000;404:515–518. doi: 10.1038/35006676. [DOI] [PubMed] [Google Scholar]
- Unterholzner L., Izaurralde E., Izaurralde E. SMG7 acts as a molecular link between mRNA surveillance and mRNA decay. Mol. Cell. 2004;16:587–596. doi: 10.1016/j.molcel.2004.10.013. [DOI] [PubMed] [Google Scholar]
- van Dijk E., Cougot N., Meyer S., Babajko S., Wahle E., Seraphin B., Cougot N., Meyer S., Babajko S., Wahle E., Seraphin B., Meyer S., Babajko S., Wahle E., Seraphin B., Babajko S., Wahle E., Seraphin B., Wahle E., Seraphin B., Seraphin B. Human Dcp2: A catalytically active mRNA decapping enzyme located in specific cytoplasmic structures. EMBO J. 2002;21:6915–6924. doi: 10.1093/emboj/cdf678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner E., Clement S.L., Lykke-Andersen J., Clement S.L., Lykke-Andersen J., Lykke-Andersen J. An unconventional human Ccr4-Caf1 deadenylase complex in nuclear Cajal bodies. Mol. Cell. Biol. 2006 doi: 10.1128/MCB.01483-06. [Epub December 18, 2006; DOI: http://mcb.asm.org/cgi/content/abstract/MCB.01483-06v1]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz C.J., Wormington M., Peltz S.W., Wormington M., Peltz S.W., Peltz S.W. The cap-to-tail guide to mRNA turnover. Nat. Rev. Mol. Cell Biol. 2001;2:237–246. doi: 10.1038/35067025. [DOI] [PubMed] [Google Scholar]
- Xu N., Chen C.Y., Shyu A.B., Chen C.Y., Shyu A.B., Shyu A.B. Versatile role for hnRNP D isoforms in the differential regulation of cytoplasmic mRNA turnover. Mol. Cell. Biol. 2001;21:6960–6971. doi: 10.1128/MCB.21.20.6960-6971.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita A., Chang T.C., Yamashita Y., Zhu W., Zhong Z., Chen C.Y., Shyu A.B., Chang T.C., Yamashita Y., Zhu W., Zhong Z., Chen C.Y., Shyu A.B., Yamashita Y., Zhu W., Zhong Z., Chen C.Y., Shyu A.B., Zhu W., Zhong Z., Chen C.Y., Shyu A.B., Zhong Z., Chen C.Y., Shyu A.B., Chen C.Y., Shyu A.B., Shyu A.B. Concerted action of poly(A) nucleases and decapping enzyme in mammalian mRNA turnover. Nat. Struct. Mol. Biol. 2005;12:1054–1063. doi: 10.1038/nsmb1016. [DOI] [PubMed] [Google Scholar]
- Yang Z., Jakymiw A., Wood M.R., Eystathioy T., Rubin R.L., Fritzler M.J., Chan E.K., Jakymiw A., Wood M.R., Eystathioy T., Rubin R.L., Fritzler M.J., Chan E.K., Wood M.R., Eystathioy T., Rubin R.L., Fritzler M.J., Chan E.K., Eystathioy T., Rubin R.L., Fritzler M.J., Chan E.K., Rubin R.L., Fritzler M.J., Chan E.K., Fritzler M.J., Chan E.K., Chan E.K. GW182 is critical for the stability of GW bodies expressed during the cell cycle and cell proliferation. J. Cell Sci. 2004;117:5567–5578. doi: 10.1242/jcs.01477. [DOI] [PubMed] [Google Scholar]
- Zhang T., Kruys V., Huez G., Gueydan C., Kruys V., Huez G., Gueydan C., Huez G., Gueydan C., Gueydan C. AU-rich element-mediated translational control: Complexity and multiple activities of trans-activating factors. Biochem. Soc. Trans. 2002;30:952–958. doi: 10.1042/bst0300952. [DOI] [PubMed] [Google Scholar]