Abstract
PEROXIN11 (PEX11) is a peroxisomal membrane protein in fungi and mammals and was proposed to play a major role in peroxisome proliferation. To begin understanding how peroxisomes proliferate in plants and how changes in peroxisome abundance affect plant development, we characterized the extended Arabidopsis thaliana PEX11 protein family, consisting of the three phylogenetically distinct subfamilies PEX11a, PEX11b, and PEX11c to PEX11e. All five Arabidopsis PEX11 proteins target to peroxisomes, as demonstrated for endogenous and cyan fluorescent protein fusion proteins by fluorescence microscopy and immunobiochemical analysis using highly purified leaf peroxisomes. PEX11a and PEX11c to PEX11e behave as integral proteins of the peroxisome membrane. Overexpression of At PEX11 genes in Arabidopsis induced peroxisome proliferation, whereas reduction in gene expression decreased peroxisome abundance. PEX11c and PEX11e, but not PEX11a, PEX11b, and PEX11d, complemented to significant degrees the growth phenotype of the Saccharomyces cerevisiae pex11 null mutant on oleic acid. Heterologous expression of PEX11e in the yeast mutant increased the number and reduced the size of the peroxisomes. We conclude that all five Arabidopsis PEX11 proteins promote peroxisome proliferation and that individual family members play specific roles in distinct peroxisomal subtypes and environmental conditions and possibly in different steps of peroxisome proliferation.
INTRODUCTION
Peroxisomes are single membrane–bound organelles that facilitate numerous essential biochemical reactions in nearly all eukaryotic organisms. The significance of peroxisomes is underscored by the human genetic diseases and the lethal plant phenotypes caused by peroxisomal deficiencies (Lin et al., 1999; Gould and Valle, 2000; Hu et al., 2002; Rylott et al., 2003; Schumann et al., 2003; Sparkes et al., 2003; Wanders, 2004; Fan et al., 2005). Plants have a large number of structurally similar but metabolically specialized peroxisomes, namely, leaf peroxisomes, glyoxysomes in germinating seedlings, nodule-specific peroxisomes, glyoxysome-related gerontosomes in senescent tissue, and unspecialized peroxisomes. They mediate photorespiration, fatty acid β-oxidation, the glyoxylate cycle, nitrogen metabolism, the synthesis of plant hormones, and the metabolism of hydrogen peroxide (Beevers, 1979; Hayashi and Nishimura, 2003). Recent findings have also revealed a role for peroxisomes in photomorphogenesis (Hu et al., 2002) and in plant–pathogen interactions (Lipka et al., 2005; McCartney et al., 2005). Thus, plant peroxisomes exert their functions in a variety of plant-specific processes of agricultural and economical significance.
Peroxisomes are highly dynamic and versatile. In eukaryotes, the abundance of peroxisomes is controlled by multiple pathways that are incompletely understood. First, peroxisomes can originate in the endoplasmic reticulum (ER) through a unique budding system in yeast cells, whereby ER-derived vesicles containing at least two of the essential peroxisome biogenesis factors (Pex3p and Pex19p) merge and recruit additional proteins before maturing into a functional peroxisome (Hoepfner et al., 2005; Schekman, 2005). The presence of peroxisomal proteins in ER or ER-like structures has also been reported in plant cells (Mullen et al., 1999, 2001; Karnik and Trelease, 2005). Second, preexisting peroxisomes undergo constitutive divisions in normal dividing cells and induce divisions (proliferations) under certain metabolic and environmental conditions (Guo et al., 2003; Thoms and Erdmann, 2005; Yan et al., 2005). For simplicity, peroxisome division and proliferation will be used interchangeably in this report. Finally, under specific environmental conditions, the entire peroxisome organelle is degraded through vacuole-mediated pexophagy, an autophagy-related process, to maintain cellular homeostasis (Farre and Subramani, 2004).
Various environmental, metabolic, and developmental cues affect the number and size of peroxisomes (Yan et al., 2005). For example, yeast peroxisomes rapidly proliferate when glucose-rich media are replaced by limited carbon and nitrogen sources such as methanol and oleic acid (Chang et al., 1999), and mammalian peroxisome numbers are upregulated by a wide variety of fatty acids and hypolipidemic drugs (Lock et al., 1989). Under these conditions, transcription factors and transcription factor complexes, such as Adr1p and Oaf1p-Pip2p from yeast and the mammalian peroxisome proliferator-activated receptor-α–retinoid X receptor complex, activate a suite of genes in peroxisome biogenesis and function (Karpichev et al., 1997; Desvergne and Wahli, 1999; Gurvitz et al., 2000, 2001; Smith et al., 2002; Rottensteiner et al., 2003b). In plants, peroxisomes are enlarged and clustered at the onset of seed germination and tend to elongate in dark-grown hypocotyls, whereas peroxisomes in other tissues are generally spherical (Mano et al., 2002). In addition, electron microscopy of plant cells revealed an increase of peroxisome number after environmental or metabolic stimuli such as ozone, the herbicide isoproturon, the hypolipidemic drug clofibrate, and high-light irradiance, whereby higher frequencies of peroxisome budding and fission were observed (de Felipe et al., 1988; Ferreira et al., 1989; Palma et al., 1991; Oksanen et al., 2003). Interestingly, expression of the Xenopus peroxisome proliferator-activated receptor-α in tobacco (Nicotiana tabacum) plants altered fatty acid metabolism and increased acyl-CoA oxidase activity and the number of peroxisomes (Nila et al., 2006), suggesting that peroxisome biogenesis and function in plants and animals may share certain regulatory circuitry. Oxidative stress also induced the expression of several Arabidopsis thaliana PEX genes putatively involved with peroxisome biogenesis (Lopez-Huertas et al., 2000). Lastly, it has been suggested that in plants, accumulation of long-chain fatty acids inside the peroxisome may trigger an increase in size and a decrease in number of this organelle, because plants defective in core β-oxidation enzymes contained fewer but larger peroxisomes (Hayashi et al., 1998; Pinfield-Wells et al., 2005). Such an increase in size and reduction in number of peroxisomes were also reported in human fibroblasts from patients with β-oxidation deficiencies (Funato et al., 2006). However, the molecular basis underlying these interesting observations has not been determined.
Extensive research in yeast has yielded the identification of a group of peroxisomal proteins called the peroxins (PEXs), which mediate various aspects of peroxisome biogenesis and maintenance, including the assembly of new membrane structures, peroxisome membrane protein (PMP) targeting, matrix protein import, and peroxisome division/proliferation. There are at least 32 PEX proteins in the yeast Saccharomyces cerevisiae, with ∼20 mammalian and 15 plant homologs identified (Purdue and Lazarow, 2001; Charlton and Lopez-Huertas, 2002; Heiland and Erdmann, 2005). Of these PEX proteins, four groups of PMPs are primarily involved with peroxisome division and proliferation, among which Pex11p was the first to be isolated and is the best characterized. Lack of Pex11p led to the presence of one or two so-called giant peroxisomes in S. cerevisiae cells and caused the cells to cease growth on oleate-containing medium owing to their inability to metabolize oleate. Conversely, when overexpressed, the gene caused an increase in the number of peroxisomes per cell (Erdmann and Blobel, 1995; Marshall et al., 1995). Although this protein was discovered >10 years ago, how Pex11p acts to regulate the proliferation of peroxisomes in yeast is still poorly understood. Recently, three additional families of PMPs, Pex25p/Pex27p, Pex28p/Pex29p, and Pex30p/Pex31p/Pex32p, were also discovered to play roles in peroxisome proliferation by unknown mechanisms (Rottensteiner et al., 2003a; Tam et al., 2003; Vizeacoumar et al., 2003, 2004).
Given the vital and unique role of peroxisomes in plant development and plant responses to abiotic and biotic stresses, we are interested in elucidating the molecular mechanisms controlling peroxisome proliferation in plants by identifying the signals and nuclear/peroxisomal proteins involved in this enigmatic process. The Arabidopsis genome contains five sequence homologs to the yeast Pex11p protein (Charlton and Lopez-Huertas, 2002; Lingard and Trelease, 2006) but no obvious homologous sequences to Pex25p/Pex27p, Pex28p/Pex29p, Pex30p/Pex31p/Pex32p and the above-mentioned transcriptional factors. The lack of apparent plant homologs to most proteins that operate in yeast and mammalian systems to control peroxisome proliferation suggests that it is essential to study this fundamental cell biological process in plants. This line of research will also help us determine how the signals and molecular machinery that control peroxisome proliferation have evolved in different organisms. As a first step toward building a mechanistic model of plant peroxisome division and proliferation and understanding how changes in peroxisome proliferation may affect development, we characterized the Arabidopsis PEX11 protein family, the only obvious sequence homologs to known yeast peroxisomal proteins specifically involved with peroxisome proliferation.
A recent study using suspension cultured Arabidopsis and tobacco (BY2) cells (Lingard and Trelease, 2006) demonstrated that myc-tagged Arabidopsis PEX11 homologs sorted directly from the cytosol to peroxisomes after being transformed into the cells by biolistic bombardments. All five At PEX11 isoforms behaved as peroxisomal membrane proteins, which caused distinct morphological changes to the organelles when overexpressed. Here, we addressed the role of the Arabidopsis PEX11 homologs in peroxisome proliferation and plant development using whole Arabidopsis plants. In addition to testing the subcellular targeting and membrane association of At PEX11 proteins using cell biological and biochemical approaches, we also addressed the evolutionary aspect of eukaryotic PEX11 genes, performed an extended expression analysis of the five Arabidopsis isoforms, conducted cell biological and physiological assays with whole plants containing increased or decreased levels of the At PEX11 proteins, and complemented the yeast pex11 mutant with some At PEX11 isoforms through heterologous expression.
RESULTS
Phylogenetic Analysis of the Arabidopsis PEX11 Protein Family
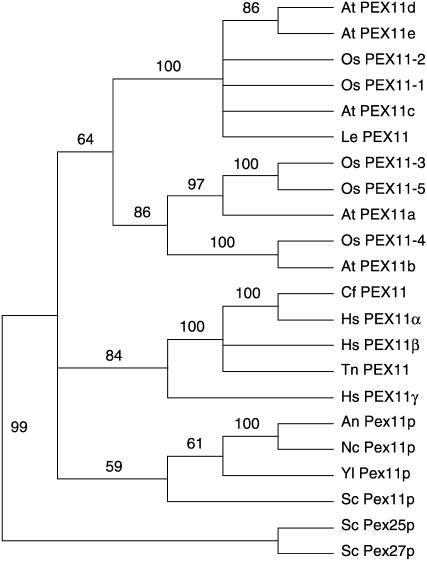
The presence of five PEX11 isoforms in Arabidopsis—PEX11a (At1g47750), PEX11b (At3g47430), PEX11c (At1g01820), PEX11d (At2g45740), and PEX11e (At3g61070)—prompted us to conduct a phylogenetic analysis of eukaryotic PEX11 homologs to determine how PEX11 gene sequences evolved (Figure 1; see Supplemental Figure 1 online). Our phylogenetic analysis demonstrates that all PEX11 sequences from various species form a monophyletic group, which can be separated into plant, animal, and fungal subclades, suggesting that PEX11 genes had a single origin and later evolved independently after the separation of these kingdoms. Among the three human PEX11 isoforms, PEX11α is more closely related to the Canis familiaris (dog) PEX11 protein than to PEX11β and PEX11γ; PEX11α and PEX11β are more similar to the Tetraodon nigroviridis (pufferfish) PEX11 homolog than to PEX11γ. These observations indicate an amplification and divergence of the vertebrate PEX11 genes since the early stage of vertebrate evolution. The plant PEX11 proteins can be divided into two groups: one containing At PEX11c to PEX11e, Os PEX11-1 and -2 of rice (Oryza sativa), and Le PEX11 of tomato (Solanum lycopersicum); and the other containing At PEX11a and PEX11b and Os PEX11-3, -4, and -5. The latter group can be further classified into two subclades: one containing At PEX11a and Os PEX11-3 and -5; and the other containing At PEX11b and Os PEX11-4. Together, these results suggest that plant PEX11 genes diversified before the evolutionary split of monocots from dicots and that Arabidopsis PEX11 proteins can be categorized into three subfamilies: PEX11a, PEX11b, and PEX11c to PEX11e. Because the entire Pex11p protein shares weak sequence similarity with the C terminus of Pex25p and Pex27p, it was suggested that the S. cerevisiae Pex11p, Pex25p, and Pex27p proteins form a so-called PEX11 protein family (Rottensteiner et al., 2003a; Thoms and Erdmann, 2005; Yan et al., 2005). The fact that all PEX11 sequences from different kingdoms form a monophyletic group indicates that the functions of the yeast Pex25p and Pex27p proteins may be more distinct from that of Pex11p than proposed previously.
Figure 1.
A Neighbor-Joining Tree of PEX11 Protein Sequences.
Numbers at the nodes are bootstrap values from 1000 trials. An, Aspergillus nidulans; At, Arabidopsis thaliana; Cf, Canis familiaris; Hs, Homo sapiens; Le, Solanum lycopersicum (formerly Lycopersicon esculentum); Nc, Neurospora crassa; Os, Oryza sativa; Sc, Saccharomyces cerevisiae; Tn, Tetraodon nigroviridis; Yl, Yarrowia lipolytica.
Expression Profiles of Arabidopsis PEX11 Genes
The five Arabidopsis PEX11 genes were shown previously to be expressed in siliques; with the exception of At PEX11a, transcripts of At PEX11b to PEX11e were also detected in roots, leaves, and suspension cultured cells (Lingard and Trelease, 2006). To gain a more comprehensive view of PEX11 gene expression, we conducted a search of web-based microarray databases (https://www.genevestigator.ethz.ch/) and used RT-PCR to analyze PEX11 gene expression in response to stresses. Given that genes functioning in similar processes often display similar expression patterns, we also searched the Arabidopsis coresponse database for genes that are coexpressed with PEX11 family members (http://csbdb.mpimp-golm.mpg.de/csbdb/dbcor/ath/ath_tsgq.html).
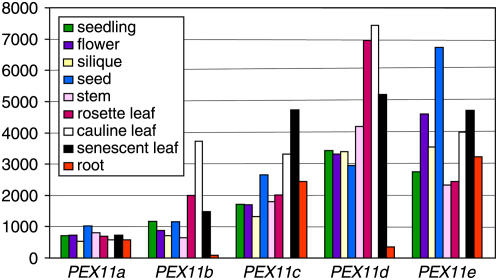
PEX11a is expressed at a constitutively low level in all tissues examined (Figure 2). PEX11b also has fairly low levels of expression in most tissues, especially in root, but exhibits higher expression in cauline leaves (Figure 2). Additionally, we found that PEX11b was consistently upregulated during dark-to-light transitions in young seedlings (see Supplemental Figure 2A online). PEX11d and PEX11e have the highest levels of expression among all family members in leaf and seed tissue, respectively (Figure 2). Lending support to these expression patterns was the finding that ∼15% of the genes coexpressed with PEX11d are involved with photosynthesis, whereas ∼20% of the genes coexpressed with PEX11e belong to the category of gluconeogenesis and the glyoxylate cycle, processes with which peroxisomes are known to be primarily associated during seed germination (see Supplemental Figure 2B online). Peroxisomes are possibly involved in plant senescence, with roles in membrane lipid catabolism into carbohydrate, nitric oxide signaling, and the proteolytic cleavage of proteins (Distefano et al., 1999; Corpas et al., 2001). Consistent with this notion, online microarray data revealed high-level PEX11c to PEX11e expression in natural senescent tissue (Figure 2), and our RT-PCR analysis showed upregulation of PEX11 genes, especially PEX11a and PEX11e, during induced senescence of rosette leaves (see Supplemental Figure 2A online). These data suggest that PEX11a, PEX11c, and PEX11e are constitutively expressed in all tissues and that PEX11d and PEX11e are major PEX11 isoforms in leaf peroxisomes and seed glyoxysomes, respectively. In addition, PEX11a and PEX11e may be strongly involved in glyoxysomal function in senescent tissues, and PEX11b may play a crucial role in leaf peroxisome photorespiration in young seedlings.
Figure 2.
Expression Patterns of the Arabidopsis PEX11 Genes Revealed by Online Microarray Data.
Expression (y axis) is displayed as a signal expression value assigned by GENEVESTIGATOR (https://www.genevestigator.ethz.ch; Zimmermann et al., 2004), in which data used for the analysis were gathered.
Subcellular Localization and Biochemical Analysis of the At PEX11 Proteins
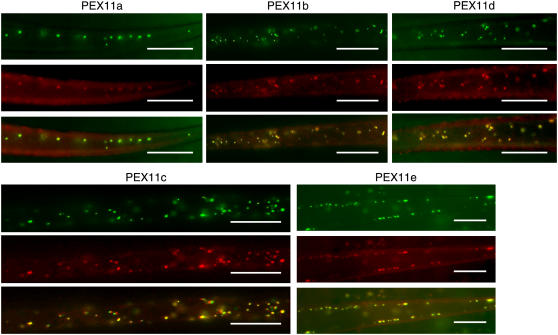
As an initial step to study the biochemical function of At PEX11 proteins, we expressed cyan fluorescent protein (CFP) fusions of the At PEX11 isoforms and tested their subcellular targeting in plants. We fused CFP in-frame to the 5′ end of each Arabidopsis PEX11, cloned the constructs into a binary vector containing the 35S constitutive promoter, and transformed the constructs into plants expressing a peroxisomal marker. This marker is a yellow fluorescent protein (YFP) fused with the peroxisomal targeting signal type 1 (PTS1; composed of Ser-Lys-Leu) at its C-terminal end (Fan et al., 2005). CFP fusion of each Arabidopsis PEX11 protein displayed colocalization with YFP-PTS1 in transgenic seedlings (Figure 3), supporting the previous results that all five At PEX11 proteins were peroxisome-targeted in suspension cultured plant cells in a transient expression assay (Lingard and Trelease, 2006). To show protein colocalization unequivocally, we chose areas in which peroxisomes were mostly spherical for imaging. In fact, overexpression of CFP-PEX11 in the transgenic seedlings conferred abnormal peroxisomal morphology in many cells, a phenotype that became more pronounced as the plants matured and will be discussed in detail in later sections.
Figure 3.
Fluorescence Microscopic Analysis of the Subcellular Localization of At CFP-PEX11 Proteins.
Epifluorescence micrographs of rosette leaf trichomes were taken from 3-week-old T3 plants coexpressing CFP-PEX11 and YFP-SKL proteins. Bars = 20 μm.
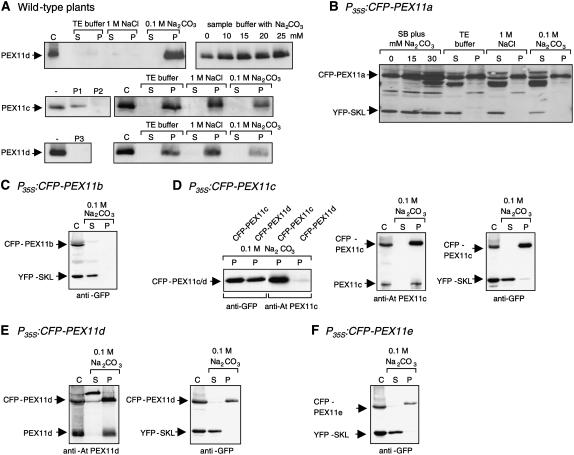
Because gene expression from a strong constitutive promoter and the addition of terminal tags to membrane proteins, as described above and used in a previous study (Lingard and Trelease, 2006), may both potentially alter subcellular targeting, we analyzed the subcellular localization of endogenous PEX11 homologs in wild-type Arabidopsis plants by immunobiochemistry. Polyclonal peptide–specific antibodies were raised against one or two peptides that contain the highest predicted antigenicity from PEX11c and PEX11d (see Methods for peptide sequences). Highly purified leaf peroxisomes were isolated from wild-type plants by two successive density gradients (Ma et al., 2006). When the peroxisomal proteins were separated by SDS-PAGE and analyzed by immunoblotting, two proteins of ∼26 kD each were specifically recognized by the antisera raised against PEX11c and PEX11d, and the cross-reactivity was partially (P1 of PEX11c) or completely (P2 of PEX11c, P3 of PEX11d) blocked by preincubation of the antisera with the peptides (Figure 4A, middle and bottom panels). Because PEX11c and PEX11d share high sequence similarity (Lingard and Trelease, 2006) and are both expressed in rosette leaves (Figure 2), we investigated whether the antisera were able to discriminate between PEX11c and PEX11d using transgenic lines overexpressing each gene as CFP fusions. A polyclonal antiserum against green fluorescent protein (GFP) detected the CFP fusion proteins with nearly the same intensity in isolated peroxisomes from both transgenic lines, indicating comparable protein levels of CFP-PEX11c and PEX11d (Figure 4D, left panel, lanes 1 and 2). When the same protein fractions were incubated with the antiserum against the two peptides (P1 and P2) of At PEX11c, CFP-At PEX11c but not CFP-At PEX11d was recognized in the corresponding transgenic lines (Figure 4D, left panel, lanes 3 and 4). Conversely, the antiserum against the single peptide (P3) of At PEX11d recognized CFP-At PEX11d specifically (data not shown). These data allowed the conclusion that the polyclonal antisera are able to discriminate between At PEX11c and At PEX11d and that both endogenous PEX11 isoforms are targeted to leaf peroxisomes in wild-type plants.
Figure 4.
Biochemical Analyses of Arabidopsis PEX11 Proteins in Isolated Leaf Peroxisomes.
Leaf peroxisomes isolated from wild-type (A) or transgenic plants overexpressing both YFP-SKL and CFP-PEX11a (B), CFP-PEX11b (C), CFP-PEX11c (D), CFP-PEX11d (E), or CFP-PEX11e (F) were subjected to membrane association analysis of the At PEX11 proteins. Leaf peroxisomal proteins (75 μg) were either precipitated directly by chloroform/methanol (control [C]) or diluted and incubated in different salt solutions. After membrane sedimentation, proteins from the supernatant were precipitated by chloroform/methanol and solubilized in standard sample buffer (SB) or in SB supplemented with 15 mM Na2CO3. Endogenous PEX11c and PEX11d were detected with peptide-specific antisera, and YFP-SKL and CFP-PEX11 proteins were detected with a commercial antiserum against GFP. For blocking of the antiserum with peptides, 100 μL of the antiserum was incubated overnight with 5 μg of peptide. P, pellet; S, supernatant.
To date, plant PEX11 homologs have not been characterized biochemically. To investigate the membrane association of the At PEX11 proteins, leaf peroxisomes of wild-type plants harvested from a sucrose density gradient were diluted and incubated in TE buffer (see Methods), 1 M NaCl, or 0.1 M Na2CO3, pH 11. The membranes were sedimented by centrifugation at 100,000g, and proteins from the supernatant were precipitated by chloroform/methanol. Preliminary data showed that PEX11c (data not shown) and PEX11d were not efficiently solubilized in standard SDS sample buffer but were quantitatively recovered in the pellet fraction after Na2CO3 treatment (Figure 4A, top panel), suggesting that the solubility of At PEX11 can be enhanced by residual traces of Na2CO3. To promote protein solubilization and recovery, At PEX11 homologs were subsequently dissolved in SDS sample buffer supplemented with 15 mM Na2CO3 (Figure 4A, middle and bottom panels). After subfractionation of leaf peroxisomes isolated from wild-type plants into soluble and membrane-associated proteins, both PEX11c and PEX11d were detected in the fraction of leaf peroxisomal membranes and remained associated with the membrane in the presence of high salt concentration (1 M NaCl) and at alkaline pH (0.1 M Na2CO3 [pH 11]), demonstrating that the proteins are integral proteins of the peroxisomal membrane (Figure 4A).
Because antisera were available only for PEX11c and PEX11d, we used transgenic plants overexpressing both CFP-At PEX11 and YFP-SKL fusion proteins to investigate the membrane association of the remaining PEX11 homologs. All five CFP-PEX11 fusions, in addition to YFP-PTS1, were immunologically detected in isolated leaf peroxisomes by polyclonal antiserum against GFP (Figures 4B to 4F). Similar to the native PEX11c and PEX11d, recovery of CFP-PEX11 was also enhanced by the addition of Na2CO3 to the SDS sample buffer (Figure 4B; data not shown). In contrast with YFP-SKL, which was recovered in the soluble fraction after sodium carbonate treatment, CFP-PEX11a, CFP-PEX11c, CFP-PEX11d, and CFP-PEX11e fusions behaved as integral membrane proteins (Figures 4B and 4D to 4F). Because of a low recovery of leaf peroxisomes from the Arabidopsis line overexpressing CFP-PEX11b, the level of CFP-PEX11b was below detection in the membrane fraction after further dilution (∼1:4) (Figure 4C). In transgenic plants overexpressing CFP-PEX11d, the fusion protein and endogenous PEX11d were detected by the peptide-specific antiserum with similar cross-reactivity (Figure 4E, left panel), indicating that the endogenous PEX11d gene is expressed at high levels in leaves (Figure 2; see Supplemental Figure 2B online) and presumably represents the most important isoform of PEX11 in leaf peroxisomes.
With these data, we demonstrate by fluorescence microscopy and immunobiochemical methods that (1) all five CFP fusion proteins of PEX11a to PEX11e are targeted to peroxisomes, confirming previous microscopic data obtained from suspension cultured plant cells (Lingard and Trelease, 2006); (2) two endogenous PEX11 isoforms (PEX11c and PEX11d) of wild-type plants are leaf peroxisomal proteins; and (3) the endogenous PEX11c and PEX11d and the CFP fusions of PEX11a and PEX11c to PEX11e are integral proteins of peroxisomal membranes.
Changes in Peroxisome Morphology and Function in PEX11-Overexpressing Plants
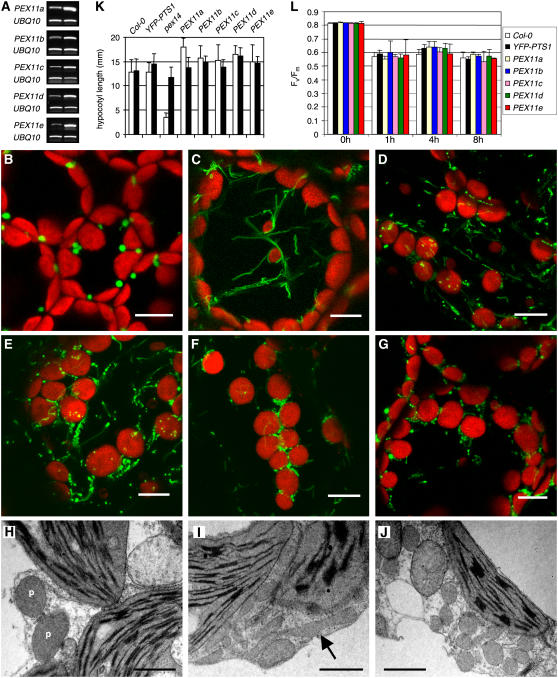
To assess the role that each of the five PEX11 proteins plays in peroxisome proliferation within Arabidopsis, we analyzed YFP-PTS1 plants containing either P35S:PEX11 or P35S:CFP-PEX11 by fluorescence microscopy. Consistent patterns of altered peroxisome morphology were observed in 8 to 15 independent T2 lines overexpressing each PEX11 gene, with or without the CFP tag, as represented in Figure 5 and Supplemental Figures 3A to 3F online. Compared with the typical punctate fluorescence pattern observed in the YFP-PTS1 control lines, both elongation and an increased proliferation of peroxisomes were observed in plants overexpressing each PEX11 gene, and these peroxisomes also had smaller diameters than those of the wild type (Figures 5A to 5G). Overexpression of PEX11a or PEX11b preferentially resulted in peroxisome elongation (Figures 5C and 5D), whereas overexpression of PEX11c to PEX11e mostly led to an increased number of peroxisomes, many of which were still clustered together (Figures 5E to 5G). Unfortunately, the high level of peroxisome elongation and aggregation made quantification of peroxisome abundance unrealistic in these lines. To corroborate observations via confocal microscopy, electron microscopy was performed on leaf mesophyll cells of a PEX11c overexpressor. Indeed, the tubulated and proliferated peroxisomes were narrower than those of the wild type, and many of the proliferated peroxisomes were not completely separated after division (Figures 5H to 5J). Based on these data, we conclude that all five At PEX11 proteins positively affect peroxisome proliferation. This result is somewhat different from the data obtained by Lingard and Trelease (2006), who found that PEX11c and PEX11d led to peroxisome elongation without fission, PEX11e led to duplication without elongation, and PEX11b-transformed cells displayed peroxisome aggregation without changes in peroxisome length or number.
Figure 5.
Peroxisome Phenotypes in Arabidopsis Overexpressing PEX11 Genes.
(A) RT-PCR analysis of PEX11 and UBIQUITIN10 (UBI10) transcripts from YFP-PTS1 plants overexpressing the PEX11 genes. Left lane, YFP-PTS1 control plant; right lane, plant overexpressing each PEX11 gene, whose phenotypes are shown in (C) to (G).
(B) to (G) Confocal micrographs of leaf mesophyll cells from 6-week-old YFP-SKL plants containing P35S:CFP-PEX11a (C), P35S:CFP-PEX11b (D), P35S:CFP-PEX11c (E), P35S:CFP-PEX11d (F), and P35S:CFP-PEX11e (G), compared with the control YFP-SKL plant (B). The larger organelles in the background with red chlorophyll fluorescence are chloroplasts. Bars = 10 μm.
(H) to (J) Electron micrographs of mesophyll cells from 5-week-old wild-type (H) and P35S:CFP-PEX11c ([I] and [J]) plants. (I) and (J) emphasize the induction of peroxisome (P) elongation and multiplication. The arrow in (I) points to an elongated peroxisome. Bars = 1 μm.
(K) Sucrose dependence assay. Hypocotyl lengths of 5-d-old etiolated plants grown on Murashige and Skoog medium plates in the absence (open bars) or presence (closed bars) of 1% sucrose were measured. Error bars indicate sd (n > 34). This experiment was repeated three times, and similar results were obtained.
(L) Fv/Fm measurement. The maximum quantum yield of photosystem II (Fv/Fm) was calculated based on chlorophyll fluorescence measurement of dark-adapted leaves from 4-week-old plants before and after 1, 4, and 8 h of high light treatment. Error bars indicate sd (n > 15).
Obtaining stable transgenic lines allowed us to observe any developmental phenotypes in the PEX11-overexpressing plants. Interestingly, none of the plants overexpressing PEX11 proteins and displaying severely altered peroxisome morphology had any obvious growth and pigmentation phenotypes, indicating that these morphologically aberrant peroxisomes might still be fully functional. To further address this question, we examined the competence of the peroxisomes in these transgenic plants with sugar dependence and chlorophyll fluorescence assays.
Given that peroxisome β-oxidation and the glyoxylate cycle are crucial steps in lipid mobilization, peroxisome mutants tend to develop poorly on media without supplemental sugar, because of a lack of energy availability during germination (Hayashi et al., 1998). This phenotype, which is more pronounced in the dark, can be rescued by exogenous sugar. Thus, a significant difference in hypocotyl length between young seedlings of Arabidopsis mutants germinated on sucrose-free and sucrose-containing media compared with wild-type plants indicates a defect in peroxisome function of the mutant (Zolman et al., 2000, 2001). The peroxisomal protein import mutant pex14 (J. Hu, unpublished data) is shown here as an example (Figure 5K). When grown on sucrose-free medium, the hypocotyls of 5-d-old etiolated PEX11-overexpressing plants were slightly longer than those of the control plants (Figure 5K; see Supplemental Figure 3G online). This difference in hypocotyl length on sugar-free medium is statistically significant, because for each pairwise t test performed between a PEX11 overexpressor and Columbia (Col-0) or between a PEX11 overexpressor and YFP-PTS1, P was consistently <0.0001 (P < 0.0083 after Bonferoni adjustments). This finding suggests that glyoxysomes in the PEX11-overexpressing seedlings function properly and raises the interesting possibility that these glyoxysomes may be able to mobilize lipid more efficiently than those of the wild type.
To determine the activity of photorespiration in the PEX11-overexpressing plants, we measured the maximum quantum yield of photosystem II by calculating the ratio of the variable fluorescence of chlorophyll a to the maximum fluorescence (Fv/Fm) in dark-adapted leaves, a method commonly used in plant peroxisome research (Hayashi et al., 2005). Before chlorophyll fluorescence measurements, 4-week-old plants grown in normal light intensity (80 μmol·m−2·s−1) were transferred to chambers containing high light (1500 μmol·m−2·s−1) to induce plant photorespiration activity. PEX11-overexpressing lines did not exhibit obvious differences in Fv/Fm from wild-type Col-0 or YFP-PTS1 control plants either before or after 1, 4, and 8 h of high-light illumination (Figure 5L), suggesting that the photorespiration activities of the PEX11-overexpressing lines are comparable to those of the wild-type plants under our experimental conditions.
Disrupting Arabidopsis PEX11 Functions through RNA Interference Decreases Peroxisome Abundance
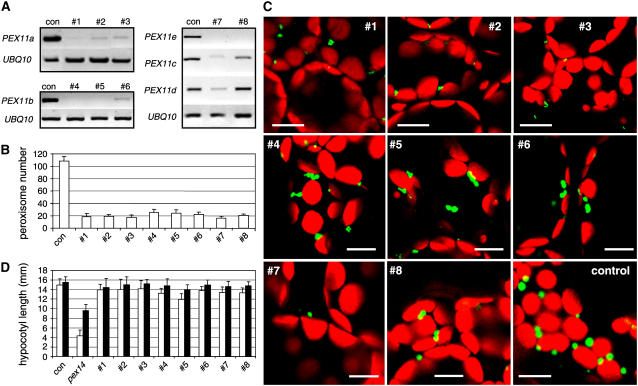
We were unable to identify true T-DNA insertion mutants for any of the Arabidopsis PEX11 genes from publicly available T-DNA insertion collections. To elucidate more clearly the role of each PEX11 gene in the regulation of peroxisome abundance, full-length cDNA fragments of PEX11a, PEX11b, and PEX11e were cloned as inverted repeats into the double-stranded RNA vector pFGC5941 (ABRC). Given the high degree of sequence similarity between PEX11c, PEX11d, and PEX11e, the PEX11e-silencing construct was intended to reduce the expression of all three genes. Plants in a YFP-PTS1 background were transformed with the PEX11-silencing constructs under the control of the 35S promoter. Approximately 58 transgenic lines—24 for PEX11a, 16 for PEX11b, and 18 for PEX11e—were subjected to RT-PCR analysis with gene-specific primers. Eleven, 10, and 9 plants, respectively, showed more than half reduction in the expression level of the corresponding genes. RT-PCR results from representative RNA interference (RNAi) plants are shown in Figure 6A.
Figure 6.
Peroxisome Phenotypes Conferred by Reducing the Expression of PEX11 Genes in Arabidopsis.
(A) RT-PCR analysis of PEX11 and UBI10 transcripts from RNAi plants in which the expression of PEX11a (lines 1 to 3), PEX11b (lines 4 to 6), and PEX11c to PEX11e (lines 7 and 8) is reduced. Controls (con) are YFP-PTS1 plants.
(B) Numerical analysis of peroxisomes in leaf mesophyll cells from 4-week-old T2 RNAi plants. Numbers shown were obtained from epifluorescence images captured from 150 μm × 150 μm of a cell (n > 17). Error bars indicate sd.
(C) Confocal microscopy of cells from (B). Bars = 10 μm.
(D) Sucrose dependence assay of PEX11 RNAi lines. The hypocotyl lengths of 5-d-old etiolated plants grown on sugar-free (open bars) or sugar-containing (closed bars) Murashige and Skoog medium plates were measured. Error bars indicate sd (n > 30).
To investigate the effect of complete or strong silencing of PEX11a, PEX11b, and PEX11c to PEX11e on peroxisome proliferation, the RNAi lines were analyzed by fluorescence microscopy and consistent phenotypes were observed in T1 and T2 generations. Strong silencing of PEX11a, PEX11b, PEX11c to PEX11e, or PEX11e alone resulted in a reduction of the total number of peroxisomes by >75% (Figure 6B) (P < 0.001 in each pairwise t test). Plants in which PEX11a or PEX11c to PEX11e were silenced exhibited a strong reduction in peroxisome number and size (Figures 6B and 6C, lines 1 to 3 and 7). On the other hand, plants with strong reduction of PEX11b expression displayed a weaker phenotype in peroxisome abundance and size (Figures 6B and 6C, lines 4 to 6). PEX11e-silenced plants in which the expression of PEX11c was only slightly reduced contained a smaller number of peroxisomes, but the organelle size remained normal (Figures 6B and 6C, line 8). Many peroxisomes in the RNAi plants were irregularly shaped, such as those shown in lines 1 to 6 and 8 (Figure 6C), and many appeared to have started the division processes but failed to separate, resulting in dumbbell-shaped peroxisomes (Figure 6C, lines 4 to 6 and 8). The RNAi plants did not show any obvious growth or leaf pigmentation defects (data not shown). Sugar dependence and chlorophyll fluorescence assays of T2 plants revealed no significant difference between the RNAi lines and the wild-type plants in hypocotyl length on sugar-free medium (Figure 6D) or in Fv/Fm in dark-adapted leaves (data not shown), suggesting that the reduced peroxisome abundance in these plants did not severely affect glyoxysomal function during germination and leaf peroxisomal function during photorespiration.
In summary, we conclude that the five At PEX11 proteins are at least partially redundant in regulating peroxisome proliferation and that PEX11a and PEX11c to PEX11e appear to play a stronger role than PEX11b in peroxisome proliferation in leaf tissue under normal growth conditions.
Complementation of the S. cerevisiae Pex11p Null Mutant with Arabidopsis PEX11 Proteins
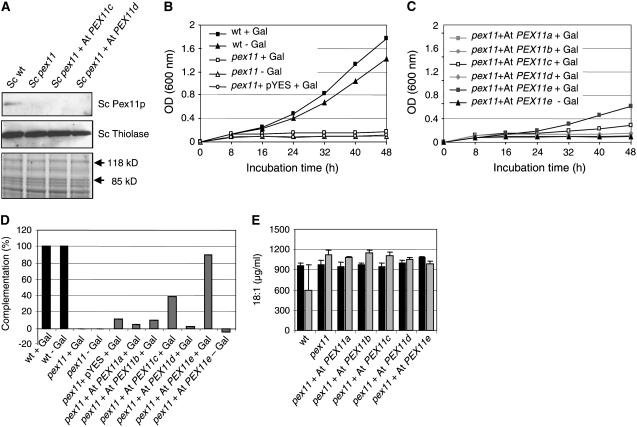
Our phylogenetic, cell biological, biochemical, and genetic analyses together suggested that all of the Arabidopsis PEX11 proteins are able to promote peroxisome proliferation. To provide direct evidence that Arabidopsis PEX11 homologs and Sc Pex11p share a conserved function in peroxisome biogenesis, we tested whether At PEX11 genes could complement the growth phenotype of the Sc pex11 mutant. Peroxisomes are the only subcellular compartment in fungi in which fatty acids can be oxidized, and peroxisome proliferation and gene induction of peroxisomal matrix and PEX proteins are generally upregulated when oleic acid is the sole carbon source (Chang et al., 1999). Thus, the S. cerevisiae Pex11p null mutants are unable to grow on medium with oleic acid as the sole carbon source (Erdmann and Blobel, 1995), indicating a severe peroxisomal dysfunction. Despite the growth deficiencies and reduced peroxisome abundance, import of PTS1- and PTS2-containing matrix proteins (e.g., thiolase; Figure 7A) and membrane proteins, however, was not reported to be affected by Pex11p deficiency (Erdmann and Blobel, 1995).
Figure 7.
Complementation of the S. cerevisiae Pex11p Null Mutant by At PEX11 Proteins.
(A) Immunoblot analysis of the yeast pex11 mutant for Sc Pex11p deficiency (top panel) and thiolase expression (middle panel). The region between 118 and 85 kD of the Coomassie blue–stained gel (bottom panel) was used as the loading control. Expression of At PEX11c and At PEX11d was below the immunological detection level using peptide-specific antibodies against these proteins (data not shown).
(B) and (C) Growth curves of controls (B) and selected transformants with high complementation efficiency (C).
(D) Complementation efficiency of pex11 transformants expressing At PEX11a to At PEX11e, calculated based on the difference in the average rate of cell division between the transformants and pex11 relative to that of the wild type and pex11 (see [B], [C], and Supplemental Figure 4C online).
(E) Oleic acid uptake of yeast cells. The concentration of oleic acid in the growth medium was determined at 12 h (black bars) and 36 h (gray bars) after inoculation. Error bars represent sd (n > 3).
Untagged versions of the five At PEX11 cDNAs were cloned into the yeast expression vector pYES2 under the control of a galactose-inducible promoter and transformed into the Pex11p-deficient strain UTL7A Sc pex11 (Figure 7A). In 0.1% oleic acid supplemented with 0.3% galactose, growth of Sc pex11 generally stagnated after 8 h at an OD of ∼0.2, whereas the wild-type cells continued to grow and reached an OD of ∼0.8 after 24 h (Figure 7B; see Supplemental Figure 4B online). Whereas expression of PEX11a, PEX11b, and PEX11d did not promote the growth of the Pex11p null mutant on oleic acid, the transformants expressing PEX11c and PEX11e grew significantly better, as indicated by increases in OD of ∼20 and 100%, respectively, compared with Sc pex11 (see Supplemental Figure 4B online). The expression level of At PEX11c and At PEX11d, however, was below the detection limit when total yeast membranes were probed with the antiserum against PEX11c or PEX11d (data not shown). On solid medium, a similar growth ability on oleic acid was also observed for yeast transformants expressing At PEX11c and At PEX11e (see Supplemental Figure 4A online).
A large number of transformants were screened for their ability to grow on oleic acid, and those with high complementation efficiency were selected for further analysis. To investigate the complementation of yeast pex11 by Arabidopsis homologs in more detail, we monitored the growth of selected transformants in a time-dependent manner in the presence and absence of 0.3% galactose. The presence of 0.3% galactose did not affect the growth of the wild type and the Sc pex11 mutant to any considerable extent (Figure 7B). Likewise, transformation of Sc pex11 with the empty pYES2 vector did not stimulate the growth of the yeast cells on oleic acid (Figure 7B). After a lag period of ∼24 h, the transformants expressing PEX11c and PEX11e started to grow on oleic acid (Figure 7C). Enhanced growth of the transformant expressing PEX11e was completely abolished in the absence of galactose (Figure 7C), demonstrating its dependence on gene expression from the galactose-inducible promoter. To determine the complementation efficiency of the transformants, the average rates of cell division were calculated in the second growth period between 32 and 48 h. In contrast with the null mutant, which had a cell division rate of ∼0.005, the transformants expressing PEX11c and PEX11e were able to reach 5- and 10-fold higher values of 0.025 and 0.05, respectively (see Supplemental Figure 4C online). PEX11c and PEX11e were able to complement the difference in the rate of division between the wild type and the mutant by ∼40 and 90%, respectively (Figure 7D). Based on these data, we conclude that two of the five At PEX11 homologs are able to confer to the Pex11p null mutant the ability to grow on oleic acid.
To provide a second line of evidence for the hypothesis that Sc pex11 expressing At PEX11e can use oleic acid as a carbon source for cell growth and proliferation, we analyzed the uptake of oleic acid from the medium by yeast cells. In the initial phase of growth during the first 12 h, the concentration of oleate (18:1) in the growth medium was decreased slightly from the starting concentration of 1000 to ∼950 μg/mL for all yeast strains (Figure 7E). In the subsequent growth phase, the concentration of oleate was reduced significantly in the wild-type cultures to ∼600 μg/mL, indicating that the wild-type cells took up significant amounts of 18:1 from the growth medium for subsequent fatty acid metabolization, allowing cell proliferation (Figure 7E; see Supplemental Figure 4D online). By contrast, the concentration of oleate in the medium remained constant or even increased slightly in the cultures of pex11 and the transformants expressing At PEX11a to At PEX11d. Only for the transformants expressing At PEX11e was a significant reduction of oleic acid in the growth medium determined (∼100 μg/mL, ∼30% of the wild type; Figure 7E; see Supplemental Figure 4D online). These data support the idea that the yeast Pex11p null mutants expressing At PEX11e obtained the ability to metabolize externally supplied oleate by peroxisomal β-oxidation.
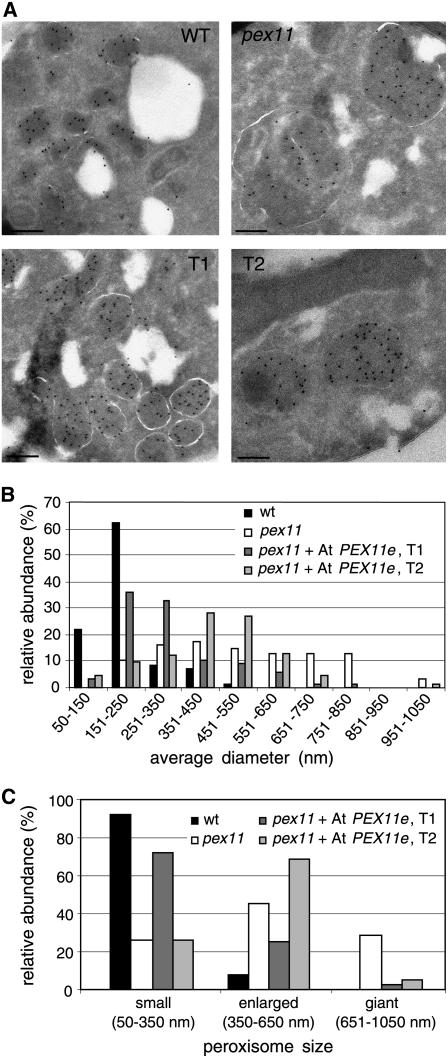
Because At PEX11e conferred partial complementation of the yeast mutant, we analyzed the ultrastructure of the yeast cells by immunoelectron microscopy to determine the effect of the expression of this gene in Sc pex11 on peroxisome size and abundance. Peroxisomes from cells grown for 36 h in oleic acid were labeled by 10-nm gold particles coupled to antibodies against yeast thiolase (Fox3p). As reported previously (Erdmann and Blobel, 1995), the mutant Sc pex11 contained a smaller number of giant peroxisomes, the diameter of which was increased by a factor of 3 to 4 compared with the wild type (Figure 8A). In some cases, these giant peroxisomes seemed to include other subcellular compartments of unknown identity (data not shown). These mutant phenotypes were partially rescued upon expression of At PEX11e (Figure 8A, T1 and T2). Measurement of the average diameter of a large number of peroxisomes and arbitrary classification of peroxisomes into small (50 ≤ diameter ≤ 350 nm), enlarged (350 < diameter ≤ 650 nm), and giant (650 < diameter ≤ 1050 nm) peroxisomes revealed that wild-type cells possessed a large number of small peroxisomes (92%), whereas the Pex11p-deficient mutant Sc pex11 contained a significant percentage of enlarged (45%) and giant (29%) peroxisomes (Figures 8B and 8C). By contrast, upon expression of At PEX11e, only a negligible portion of giant peroxisomes was detectable in the two independent transformants (2 and 5%). Instead, the transformants contained mostly small (transformant T1, 72%) or enlarged (transformant T2, 69%) peroxisomes and resembled the wild type (Figure 8C). These results demonstrate that expression of At PEX11e in Sc pex11 can convert the peroxisome structural phenotype in oleic acid medium from a small number of giant peroxisomes to a large number of small peroxisomes per cell.
Figure 8.
Peroxisome Morphology in the Sc pex11 Mutant Overexpressing At PEX11e.
(A) Electron micrographs of S. cerevisiae cells grown for 36 h in oleic acid medium. T1 and T2 are two pex11 transformants expressing At PEX11e. Peroxisomes were immunolabeled in ultrathin sections by 10-nm gold particles coupled to anti-thiolase serum. Bars = 200 nm.
(B) and (C) Semiquantitative analysis of alterations in peroxisome size conferred by overexpression of At PEX11e in the S. cerevisiae pex11 null mutant.
DISCUSSION
Arabidopsis Possesses a Divergent Family of PEX11 Proteins with Overlapping and Distinct Roles in Peroxisome Proliferation
Although the metabolic reactions mediated by peroxisomes vary tremendously among eukaryotes (Titorenko and Rachubinski, 2004), the general theme of peroxisome biogenesis shares many conserved features among different species. As an example, PEX11 from various yeast species, trypanosoma, and mammals has the ability to promote peroxisome division and proliferation (Thoms and Erdmann, 2005). In this study, we show that overexpression of each individual At PEX11 gene in Arabidopsis results in peroxisome elongation and hyperproliferation. The incomplete separation of many proliferated peroxisomes in these lines may be explained by the fact that the membrane separation machinery was outpaced by such a hyperproliferation of peroxisomes. Data from our overexpression analysis is to some extent inconsistent with the report of Lingard and Trelease (2006). For instance, in suspension cultured cells, PEX11b was shown to cause only peroxisome aggregation, PEX11c and PEX11d led to peroxisome elongation without fission, and PEX11e led to duplication without elongation, yet we found both peroxisomal elongation and duplication in plants overexpressing all of these genes. This discrepancy may have been caused at least in part by the fact that peroxisomes and peroxisomal proteins in cell culture and whole plants are under slightly different regulation and behave somewhat distinctly. It is interesting that the PEX11-overexpressing plants grown on sucrose-free medium for 5 d had longer hypocotyls than wild-type plants, indicating that the glyoxysomal activity in these plants may be slightly higher, possibly as a result of an increase of the overall volume of peroxisomal matrix resulting from the increased peroxisomal length or abundance.
With five members divided into three subfamilies, the plant PEX11 family appears to be the largest among all PEX11 families in various organisms (Figure 1), and it is the largest protein family of all the PEX proteins in Arabidopsis (Charlton and Lopez-Huertas, 2002). Because all five At PEX11 proteins are peroxisomal and capable of inducing peroxisome proliferation, the question arose whether these proteins are functionally overlapping. Our RNAi study suggests that At PEX11 protein functions are at least partially redundant, given that plants with complete or strong silencing of individual PEX11 genes exhibited partial reduction in peroxisome number. Loss of Pex11p in yeast cells led to fewer, but giant, peroxisomes (Erdmann and Blobel, 1995). However, reduction of PEX11 expression in Arabidopsis decreased the size of the organelles in PEX11a and PEX11c to PEX11e RNAi lines, suggesting mechanistic differences between yeast and plants in the regulation of peroxisome size. The PEX11 RNAi plants that displayed apparent reduction of the number of peroxisomes did not show an obvious sugar dependence or photorespiration phenotype, indicating that the peroxisomes in these lines were still functional in the normal range. To this end, it will be necessary to obtain mutants in which all five PEX11 genes are silenced if we are to understand to what extent all five genes are needed for peroxisome proliferation and in what way plant development would be compromised if the functions of all At PEX11 genes are completely disrupted.
Individual members of the At PEX11 family may also play specific roles. Mammals have three PEX11 isoforms, among which PEX11β is constitutively expressed and essential for embryo viability, PEX11α is strongly induced by metabolic cues, and the specific function for PEX11γ is elusive (Li et al., 2002a, 2002b). Overexpression of PEX11β had a greater impact on peroxisome proliferation than that of PEX11α (Schrader et al., 1998). Whether or not members of the mammalian PEX11 protein family are functionally equivalent is still unclear. Plants contain several metabolically distinct subtypes of peroxisomes; hence, the five PEX11 homologs in Arabidopsis may constitute isoforms involved with peroxisome proliferation in specific peroxisome variants. Consistent with this notion is our finding that PEX11d and PEX11e are highly expressed in leaves and seeds, respectively, PEX11b is strongly light-inducible, and PEX11a and PEX11e are upregulated by induced senescence. Furthermore, RNAi lines in which PEX11b was completely silenced displayed a weaker peroxisome phenotype than RNAi lines of PEX11a and PEX11c to PEX11e, suggesting that PEX11b may not play as strong a role in constitutive peroxisome division in the leaf tissue as some other PEX11 proteins. In addition to tissue and peroxisome subtype specificity, the biochemical functions of the At PEX11 proteins may also differ. Two lines of evidence from this study support this view. First, overexpression of PEX11a, PEX11b, and PEX11c to PEX11e resulted in partially distinct peroxisome phenotypes in plants, with PEX11c to PEX11e causing an obviously more complete division/proliferation of the organelles. Thus, PEX11a and PEX11b may be more specialized in membrane elongation, whereas PEX11c to PEX11e are capable of executing the entire peroxisome division process. Second, only PEX11c and PEX11e were able to partially complement the growth phenotype of the yeast pex11 mutant, raising the possibility of functional differences among At PEX11 isoforms.
Targeting and Membrane Association of the Arabidopsis PEX11 Proteins
Gene duplication like that of At PEX11 may often lead to new isoforms targeted to different subcellular compartments. Using a different expression system (constitutive expression in Arabidopsis plants versus transient expression in suspension cultured cells), we confirmed the previous microscopic data that all five PEX11s are targeted to peroxisomes (Lingard and Trelease, 2006). In addition, we analyzed the Arabidopsis PEX11 family members immunobiochemically to verify that endogenous PEX11 isoforms are peroxisome-targeted and to investigate their membrane association. Peptide-specific antisera against PEX11c and PEX11d were able to discriminate between these two isoforms and allowed the detection of both proteins in isolated leaf peroxisomes. Even though the isolated leaf peroxisomes were of high purity and the activities of marker enzymes for mitochondria and chloroplasts were hardly detectable (Ma et al., 2006), dual targeting of PEX11c and PEX11d to a second compartment other than peroxisomes, although unlikely, cannot be fully excluded by this biochemical approach.
In different species, however, conflicting data were reported on the membrane association of PEX11 homologs. S. cerevisiae Pex11p was characterized as a peripheral membrane protein that resides in the inner surface of the peroxisome membrane (Marshall et al., 1995), whereas other studies concluded that the trypanosome PEX11 and mammalian PEX11α are embedded in the peroxisome membrane, presumably by two transmembrane α-helical domains, and exposed at both terminal ends to the cytosol (Lorenz et al., 1998; Schrader et al., 1998). The two Sc Pex11p–related proteins with extended N-terminal extensions, Pex25p and Pex27p, have been characterized as peripheral membrane proteins (Rottensteiner et al., 2003a; Tam et al., 2003). We show in this study that At PEX11a and At PEX11c to At PEX11e behave as integral membrane proteins that remain associated with the peroxisomal membrane in the presence of harsh alkaline conditions such as 0.1 M Na2CO3 (pH 11), under which peripheral membrane proteins are released and recovered in the supernatant after membrane sedimentation by 100,000g (Fujiki et al., 1982). These data led us to the conclusion that the At PEX11 homologs, and presumably plant PEX11 homologs in general, are integral proteins of the peroxisomal membrane.
The tight membrane association of At PEX11 homologs is in agreement with a topology study on plant PEX11 (Lingard and Trelease, 2006). Interestingly, despite their sequence similarity over the entire polypeptide, myc-tagged versions of At PEX11 proteins were reported to adapt to different membrane topologies in Arabidopsis and tobacco BY-2 suspension cultured cells. At PEX11b to At PEX11e were shown to expose both their N and C termini to the cytosol, similar to what was reported for the mammalian PEX11 homologs, but the C terminus of PEX11a was found to locate oppositely to the N terminus, facing the peroxisomal matrix (Lingard and Trelease, 2006). Future studies need to focus on determining which domains of At PEX11 mediate the recruitment of soluble cytosolic and/or matrix proteins involved in the elongation and scission of the peroxisome membrane.
Complementation of the Yeast Pex11p Mutant by Arabidopsis PEX11 Proteins
Phenotypic complementation of a yeast null mutant by putative homologs from higher eukaryotes is an important method to conclude orthology and functional similarity for many, especially nonenzymatic, proteins, and it has been applied to PEX11 homologs from various organisms (Lorenz et al., 1998; Kiel et al., 2005). Taking advantage of the available S. cerevisiae pex11 null mutant (Erdmann and Blobel, 1995), we investigated whether Arabidopsis PEX11 homologs could functionally complement the inability of the mutant to grow on oleic acid and rescue the peroxisome morphology phenotype. We demonstrated that two of five Arabidopsis PEX11 homologs were able to confer such ability, with PEX11e reproducibly giving stronger complementation than PEX11c. However, complementation of yeast pex11 by PEX11d, PEX11a, or PEX11b was not observed in any experiment. In a standard BLAST search with Sc PEX11p as the query, the Arabidopsis PEX11 homologs were retrieved exactly in this order: PEX11e (E value = 0.98), PEX11c (E value = 1.1), and PEX11d (E value = 4.4), whereas PEX11a and PEX11b were too distantly related to be detected. Hence, PEX11a and PEX11b are most likely structurally and functionally too divergent from Sc PEX11 for functional complementation. Regarding the different degrees of complementation efficiency of PEX11c to PEX11e, it is tempting to speculate that sequence similarity between specific domains may be an important criterion for the successful complementation between Sc Pex11p and Arabidopsis homologs. Alternatively, the different complementation ability of At PEX11 homologs may be influenced by different levels of expression and/or different rates of targeting to the peroxisome membrane. Quantitative data on targeting of tagged At PEX proteins to peroxisomes in yeast are required to address these possibilities.
The Mode of Action for PEX11 and Other Proteins in Peroxisome Proliferation—Many Questions Unanswered
Despite its conserved role in diverse species, the biochemical function of PEX11 remains elusive. Several independent lines of evidence suggest that PEX11 proteins are directly involved in peroxisome proliferation either by representing one of the major structural components of peroxisome membranes (Erdmann and Blobel, 1995; Passreiter et al., 1998; Voncken et al., 2003), and thereby specifically shaping the membrane, or by recruiting other proteins to the membrane (Thoms and Erdmann, 2005). However, primary and secondary effects of gene disruptions are difficult to separate, and some evidence also suggests a different physiological role of PEX11 in vivo. van Roermund et al. (2000) provided intriguing evidence that Sc Pex11p is directly involved in the β-oxidation of medium-chain fatty acid (C8) and may transport an essential metabolite across the peroxisome membrane. In partial support of this hypothesis, gene disruption of some essential enzymes of fatty acid β-oxidation was also reported to reduce the number and increase the size of peroxisomes per cell (Hayashi et al., 1998; Pinfield-Wells et al., 2005; Funato et al., 2006). However, Li and Gould (2002) excluded this hypothesis by showing that PEX11 can promote peroxisome division in both mammalian and yeast cells lacking the major receptor (PEX5) for peroxisomal enzymes, suggesting that PEX11's primary role is in peroxisome division and that it can fulfill its function in the absence of peroxisome metabolism.
Based on current evidence, we favor the model that PEX11 proteins recruit additional proteins to the membrane to execute the proliferation process. The only distinctive domain present in the Arabidopsis PEX11 proteins is a dilysine motif at the extreme C terminus of PEX11c to PEX11e (see Supplemental Figure 1 online) as well as in the trypanosome PEX11 and human PEX11α proteins (Yan et al., 2005). The dilysine motif (KXKXX) was shown to contribute to the binding of COPI to peroxisomes (Cosson and Letourneur, 1997), suggesting that some PEX11 proteins may mediate the division of peroxisomes through membrane vesiculation in a coatomer-dependent manner. The importance of this domain in PEX11 is still unclear. The rat PEX11α protein, with its C terminus exposed to the cytosol, was reported to be involved in recruiting ADP-ribosylation factor, a member of the small GTPase family, and COPI to peroxisome membranes (Passreiter et al., 1998; Anton et al., 2000). However, this domain is dispensable for the function of trypanosome PEX11 and Arabidopsis PEX11e in peroxisome proliferation (Maier et al., 2000; Lingard and Trelease, 2006). PEX11 has not been shown to interact with other proteins, except with itself (Marshall et al., 1995, 1996). Dynamin-related proteins (DRPs), including the Arabidopsis DRP3A, have also been shown to play a role in the division of peroxisomes along with the division of other subcellular membranes (Hoepfner et al., 2001; Koch et al., 2003, 2004; Li and Gould, 2003; Mano et al., 2004). Although evidence for direct interaction between PEX11 and DRP is lacking, overexpression of PEX11β was shown to recruit more dynamin-like proteins to the peroxisome (Li and Gould, 2003). To this end, it will be crucial to identify proteins associated with the Arabidopsis PEX11 homologs and adaptors that link the functions of PEX11 and dynamin-related proteins to determine the biochemical function of PEX11.
METHODS
Plant Material, Growth Conditions, and Treatments
Arabidopsis thaliana plants used in this study are of the Col-0 background. Seeds were germinated on 1× Murashige and Skoog medium (Gibco) after a 2-d stratification period, with or without 1% sucrose, and with the addition of appropriate antibiotics when necessary. Unless specified otherwise, plants were grown with a 16-/8-h light/dark photoperiod under 70 to 100 μmol·m−2·s−1 light conditions at 22°C.
To examine light responsiveness, 6-d-old etiolated seedlings were exposed to 2 h of 70 μmol·m−2·s−1 light. For induced senescence treatments, leaves were cut off from 20-d-old plants and placed in water in a Petri dish and incubated at 28°C in the dark for 2 d. Control leaves were incubated at the same temperature in the light for 2 h.
Sequence Alignment and Phylogenetic Analysis
The amino acid sequences of the PEX11 proteins from various organisms were obtained from the National Center for Biotechnology Information website (http://www.ncbi.nlm.nih.gov/entrez/) and the Arabidopsis Information Resource and then aligned by the ClustalW method using the Megalign program from the Lasergene 6 software package (DNASTAR). PAUP* 4.0 (Phylogenetic Analysis using Parsimony; Sinauer Associates) was used to make the neighbor-joining tree and perform the bootstrap analysis, using the distance-analysis function with 1000 replicates. A 50% accuracy value was used as the cutoff for branch reliability.
Generating P35S:CFP-PEX11, P35S:PEX11, and PEX11 RNAi Plants
To clone P35S:CFP-PEX11, we used RT-PCR to amplify the coding region of PEX11a (At1g47750), PEX11b (At3g47430), or PEX11d (At2g45740). First-strand cDNA was made from mRNA of wild-type Col-0 seedlings using primers At1g47750Fw (5′-CGCGGATCCATGGCTACGAAAGCTCCAGA-3′) and At1g47750Rv (5′-CGGGGTACCTCAACAAGAGATCCAGTTCT-3′), At3g47430Fw (5′-CGCGGATCCATGTCTTTGGACACTGTGGA-3′) and At3g47430Rv (5′-CGGGGTACCTCACGATGGCCAGTTCCTAT-3′), or At2g45740Fw (5′-CGCGGATCCATGGGGACGACGTTAGATGT-3′) and At2g45740Rv (5′-CGGGGTACCTCAGGGTGTTTTGATCTTGG-3′), respectively. The coding regions of PEX11c (At1g01820) or PEX11e (At3g61070) were amplified from the cDNA clones 118F11 and 125J9, respectively. Primers used were At1g01820Fw (5′-AAACCCGGGAAATGAGTACCCTTGAGACCAC-3′) and At1g01820Rv (5′-CGAGCTCTCAGACCATCTTGGACTTGG-3′) or At3g61070Fw (5′-CGCGGATCCATGACTACACTAGATTTGAC-3′) and At3g61070Rv (5′-CGGGGTACCTCAAGGTGTCTTCAACTTGG-3′). The resulting RT-PCR fragments were cloned into the BamHI and KpnI sites or the SmaI and SacI sites at the C terminus of CFP in a modified pCAMBIA1300 vector (CAMBIA) containing the 35S promoter.
To clone P35S:PEX11, we amplified the coding regions of PEX11a to PEX11e from the P35S:CFP-PEX11 vector, using the primers At1g47750Fw2 (5′-ACGCGTCGACATGGCTACGAAAGCTCCAGA-3′) and At1g47750Rv, At3g47430Fw2 (5′-GGGGTACCATGTCTTTGGACACTGTGGA-3′) and At3g47430Rv2 (5′-CGGAGCTCTCACGATGGCCAGTTCCTAT-3′), At1g01820Fw2 (5′-GGGGTACCATGAGTACCCTTGAGACCAC-3′) and At1g01820Rv, At2g45740Fw2 (5′-GGGGTACCATGGGGACGACGTTAGATGT-3′) and At2g45740Rv2 (5′-CGGAGCTCTCAGGGTGTTTTGATCTTGG-3′), and At3g61070Fw2 (5′-GGGGTACCATGACTACACTAGATTTGAC-3′) and At3g61070Rv2 (5′-CGGAGCTCTCAAGGTGTCTTCAACTTGG-3′), respectively. The resulting PCR fragment was cloned into the KpnI and SacI sites or the SalI and KpnI sites in the pCAMBIA vector containing the 35S promoter.
To clone the PEX11 RNAi constructs, the Arabidopsis vector pFGC5941 for double-stranded RNA production was obtained from the ABRC. A fragment of PEX11a cDNA (747 bp) was amplified by PCR using primers pex11aF (5′-GCTCTAGAGGCGCGCCATGGCTACGAAAGCTCCA-3′) and pex11aR (5′-CGGGATCCATTTAAATTCAACAAGAGATCCAGT-3′); primers used to amplify PEX11b cDNA (684 bp) were pex11bF (5′-GCTCTAGAGGCGCGCCATGTCTTTGGACACTGTG-3′) and pex11bR (5′-CGGGATCCATTTAAATTCACGATGGCCAGTTCC-3′); primers designed to amplify PEX11e cDNA (696 bp) were pex11eF (5′-GCTCTAGAGGCGCGCCATGACTACACTAGATTTG-3′) and pex11eR (5′-CGGGATCCATTTAAATTCAAGGTGTCTTCAACTTG-3′). Each fragment was first cloned between the AscI and SwaI sites of pFGC5941 before an inverted repeat of the same fragment was inserted into the BamHI and XbaI sites of pFGC5941 already containing the sense repeat.
All PCR amplifications were performed using the Pfu DNA polymerase (Stratagene) and protocols suggested by the manufacturer. Agrobacterium tumefaciens–mediated transformation of Arabidopsis plants was performed using the floral dip method (Clough and Bent, 1998). Transgenic plants were selected on Murashige and Skoog plates containing 50 ng/μL kanamycin and 25 ng/μL hygromycin for P35S:CFP-PEX11 and P35S:PEX11 and 50 ng/μL kanamycin and 10 ng/μL glufosinate ammonium (BASTA; Crescent Chemical) for PEX11 RNAi selections.
RT-PCR Analysis of PEX11 Transcripts
Total RNA was extracted with TRIzol reagent (Invitrogen) or the RNeasy plant mini kit (Qiagen) and subjected to reverse transcription with Superscript III reverse transcriptase (Invitrogen) or the Omniscript RT kit (Qiagen). The PEX11-specific primers At1g47750F (5′-GCTCGTCTTACTCATAATCGC-3′) and At1g47750R (5′-CATTAGGAGCCGATAACACTCC-3′) were used to amplify a 391-bp product from PEX11a; At3g47430F (5′-CAGTGATCCGTTTCTTGGCG-3′) and At3g47430R (5′-GGCCAGTTCCTATACCAACC-3′) were used to amplify a 432-bp product from PEX11b; At1g01820F (5′-TGCTCTCATTAGCCCTGTTCCC-3′) and At1g01820R (5′-GGACTTGGGATGTGACGGCAAT-3′) were used to amplify a 486-bp product from PEX11c; At2g45740F (5′-TGTCTGGCTTGGGAGATCAGGA-3′) and At2g45740R (5′-TGTCTGGCTTGGGAGATCAGGA-3′) were used to amplify a 272-bp product from PEX11d; At3g61070F (5′-GTCCTTACTCGGGAAGTCGAAG-3′) and At3g61070R (5′-GATAAGTGAGGTGGTAAACC-3′) were used to amplify a 395-bp product from PEX11e; and UBQ10-1 (5′-TCAATTCTCTCTACCGTGATCAAGATGCA-3′) and UBQ10-2 (5′-GGTGTCAGAACTCTCCACCTCAAGAGTA-3′) from the UBI10 gene (At4g05320) were used to amplify a cDNA product of ∼320 bp. PCR conditions were as follows: 94°C for 3 min, followed by 26 cycles at 94°C for 45 s, 57°C for 45 s, and 72°C for 1 min, and a final extension at 72°C for 7 min.
Epifluorescence, Confocal Laser Scanning, and Electronic Microscopy
A Zeiss Axiophot and a Zeiss Axio Imager.M1 microscope (Carl Zeiss) were used to visualize fluorescent proteins. For in vivo detection of YFP and CFP, leaf tissue was mounted in water and viewed using the Axiophot microscope with the YFP filter (excitation, 500 ± 12.5 nm; emission, 540 ± 20 nm) or the CFP filter (excitation, 440 ± 10 nm; emission, 480 ± 15 nm) and the Axio Imager.M1 microscope with the YFP filter (excitation, 500 ± 12 nm; emission, 542 ± 13.5 nm) or the CFP filter (excitation, 438 ± 12 nm; emission, 483 ± 16 nm). A confocal laser scanning microscope (Zeiss LSM5) with a Zeiss PASCAL5.0 system was used to obtain confocal images of YFPs. A 488-nm argon ion laser was used for excitation of YFP and chlorophyll. We used 505- to 530-nm band-pass and 650-nm long-pass emission filters for YFP and chlorophyll, respectively.
For electron microscopy of leaf cells, cauline leaves from 5-week-old YFP-PTS1 plants expressing P35S:CFP-PEX11c were fixed for 2 h at room temperature in 2.5% glutaraldehyde and 0.1 M phosphate buffer, pH 7.2, followed by a secondary fixation in 1% (w/v) OsO4 in the same buffer. Samples were then dehydrated in a graded series of acetone and embedded in Spurr's epoxy resin. Ultrathin sections (70 to 90 nm) were cut by a MT-X ultramicrotome, stained with 2% uranyl acetate and lead citrate, and observed with a JEM-100CX II transmission electron microscope (JEOL).
For electron microscopy of yeast cells, ultrathin cryosections were prepared as described previously (Tokuyasu, 1973, 1980; Kreykenbohm et al., 2002). Yeast cells were fixed with 2% paraformaldehyde (1 volume of growth medium + 1 volume of 4% [w/v] paraformaldehyde) for 30 min at room temperature. After centrifugation, cells were postfixed with 4% paraformaldehyde (in PBS) overnight and for 2 h with 4% paraformaldehyde and 0.1% glutaraldehyde, both steps on ice. After two washings with PBS and 0.02% Gly, cells were embedded in 10% gelatin, cooled on ice, and cut into small blocks. The blocks were infused with 2.1 M sucrose and 0.4% paraformaldehyde overnight. After washing in 2.3 M sucrose and 0.02% Gly, blocks were mounted on metal pins and frozen in liquid nitrogen. Ultrathin sections were cut in an ultracryomicrotome (Leica Microsystems) using a diamond knife (Diatome). For immunolabeling, sections were incubated with a polyclonal rabbit antiserum against thiolase of Saccharomyces cerevisiae (1:150) (Erdmann and Blobel, 1995) for 20 min, followed by incubation with protein A–gold (10 nm) for 20 min. Sections were contrasted with uranyl acetate/methyl cellulose (Liou et al., 1996) for 10 min on ice, embedded in the same solution, and examined with a Philips CM120 electron microscope. Electron micrograph images were taken with a CCD camera (TVIPS). For semiquantitative analysis of peroxisome size, approximately six slices were analyzed for each yeast strain and the average diameter of anti-thiolase–labeled peroxisomes was determined as the mean of two rectangular diameters of maximum length per peroxisome. The relative size of peroxisomes was determined as 74 for the wild type, 87 for pex11, 86 for pex11 + At PEX11e (T1), and 92 for pex11 + At PEX11e (T2).
Sucrose Dependence Assay
Five-day-old etiolated seedlings germinated on 1× Murashige and Skoog medium (Gibco) in the presence or absence of 1% supplemented sucrose were first scanned with an EPSON scanner. Hypocotyl length was then measured on the scanned images with ImageJ (http://rsb.info.nih.gov/ij/). Standard deviations for the data were calculated using the Excel program (Microsoft). Statistical significance was calculated using Student's t test to determine differences between hypocotyl lengths of the control and PEX11-overexpressing/RNAi lines on the unsupplemented medium. The experiment was repeated three times, and similar results were obtained.
Determination of Fv/Fm
Plants were grown for 4 weeks at 22°C with 80 μmol·m−2·s−1 light conditions and under a 12-/12-h light/dark cycle. These plants were then transferred to high-light chambers (1500 μmol·m−2·s−1) for 1, 4, and 8 h. In vivo chlorophyll a fluorescence was measured from three plants for each genotype (five leaf regions from each plant) using a pulse amplitude modulation fluorometer (MAXI IMAGING-PAM; Heins Walz) using protocols suggested by the manufacturer of the device. Attached leaves were dark-adapted for at least 15 min before measurement, and the Fv/Fm was calculated (Maxwell and Johnson, 2000).
Biochemical Analyses of Leaf Peroxisomes
Leaf peroxisomes were isolated from wild-type and CFP-PEX11 overexpression lines as described (Ma et al., 2006) with minor modifications. Equal amounts of protein (75 μg) harvested from a sucrose density gradient (in TE buffer: 10 mM Tricine and 1 mM EDTA, pH 7.5) were diluted fourfold to ∼0.5 M sucrose in TE buffer or in salt solutions to yield a final concentration of 1 M NaCl or 0.1 M Na2CO3. After incubation on ice for 30 min, membranes were sedimented by centrifugation at 100,000g for 1 h. Proteins were precipitated from the supernatant and the pellet fraction by chloroform/methanol (Wessel and Flugge, 1984) and, if not stated otherwise, dissolved in SDS sample buffer supplemented with 15 mM Na2CO3.
Control samples were precipitated without dilution or salt incubation. Isoform-specific peptides of high predicted antigenicity were selected by multiple sequence alignments and antigenicity prediction, and the following peptides were chosen: residues 39 to 55 (P1; SDGQPGTAQNVDKNTSL) and residues 157 to 171 (P2; KEIGNKDKHQNEQYR) from PEX11c and residues 163 to 179 (P3; GNKYQDEDYRAKLKKSN) from PEX11d. The antiserum against At PEX11c was raised against both P1 and P2 of PEX11c, and the antiserum against At PEX11d was raised against P3 of PEX11d. The peptides were coupled to keyhole limpet hemocyanin and injected into rabbit (Bioscience). For antibody blocking, 100 μL of serum was incubated overnight with 5 μg of peptide, then incubated for ∼6 h with a blocked piece of nitrocellulose membrane to reduce the nonspecific binding of the antibodies to nitrocellulose.
Yeast Complementation Analysis
The coding regions of PEX11a to PEX11e were amplified from P35S:CFP-PEX11 vectors using the primers At1g47750Fw3 (5′-GGAATTCATGGCTACGAAAGCTCCAGA-3′) and At1g47750Rv3 (5′-CCGCTCGAGTCAACAAGAGATCCAGTTCT-3′), At3g47430Fw3 (5′-GGAATTCATGTCTTTGGACACTGTGGA-3′) and At3g47430Rv3 (5′-CCGCTCGAGTCACGATGGCCAGTTCCTAT-3′), At1g01820Fw3 (5′-GGAATTCATGAGTACCCTTGAGACCAC-3′) and At1g01820Rv3 (5′-CCGCTCGAGTCAGACCATCTTGGACTTGG-3′), At2g45740Fw3 (5′-GGAATTCATGGGGACGACGTTAGATGT-3′) and At2g45740Rv3 (5′-CCGCTCGAGTCAGGGTGTTTTGATCTTGG-3′), and At3g61070Fw3 (5′-GGAATTCATGACTACACTAGATTTGAC-3′) and At3g61070Rv3 (5′-CCGCTCGAGTCAAGGTGTCTTCAACTTGG-3′), respectively. The resulting fragments were cut with EcoRI and XhoI (underlined above) and cloned into the pYES2 (Invitrogen) yeast expression vector.
Transformation of UTL7A Sc pex11 (Erdmann and Blobel, 1995) was performed according to Woods and Gietz (2001). Transformants were plated on SD medium (0.5% ammonium sulfate, 0.17% yeast nitrogen base, and appropriate amino acids and bases) containing 2% Glc and lacking uracil and Leu. Oleic acid plates were prepared according to a published protocol (Palmieri et al., 2001), which contained 0.17% yeast nitrogen base, 0.5% ammonium sulfate, 1× amino acids as required, 0.1% yeast extract, 0.5% K phosphate buffer (KH2PO4, pH 6.0), 0.1% oleic acid in 0.5% Tween 80, 0.3% galactose (for protein induction), and 2% agar. The oleic acid/0.5% Tween 80 mixture was semisterilized in the 100°C hot block for 5 min. Preculture of yeast cells was grown in 0.3% Glc to the same OD before 10 μL of the culture was spotted onto the oleic acid plates.
For growth analysis on oleic acid, transformants were first grown overnight in liquid SD medium containing 2% Glc. Next, SD medium containing 1% raffinose was inoculated with yeast cells to a starting OD of 0.05 and grown overnight. Finally, 5 or 20 mL of oleic acid medium (0.1% oleic acid, 0.02% Tween 40, and SD medium with 0.1% yeast extract) containing 0.3% Gal was inoculated to a starting OD of 0.05 with washed yeast cells and grown for the times indicated. For OD determination of yeast cells in oleic acid, 0.5-mL cultures were diluted 1:2 in water, the yeast cells were sedimented by centrifugation and washed again in water, and the OD600 was determined. The rate of cell division (rcd) was calculated for specific time periods (t) of growth on oleic acid as follows: rcd(t) = [log10OD(t) − log10OD(t-8)] × [log102 × 8 h]−1. The extent of complementation (c) of a transformant T was calculated relative to the wild type (percentage) based on the mean rates of cell division for 32, 40, and 48 h of growth on oleic acid (t) as follows: c(T) = [rcd av. (T) − rcd av. (pex11)] × [rcd av. (wild type) − rcd av. (pex11)]−1 × 100.
For electron microscopy and immunoblotting, the oleic acid cultures were inoculated to higher OD to obtain higher yields of cells (OD of 0.2 and 0.5, respectively). For immunoblotting, yeast cells were sedimented, washed, and disrupted mechanically by glass beads in 25 mM Tris-HCl, pH 7.5, and 5 mM EDTA supplemented with protease inhibitors and subfractionated into soluble and membrane proteins by ultracentrifugation. For SDS-PAGE, 20 μg of soluble proteins (anti-thiolase) or 100 μg of membrane proteins (anti-Pex11p) was loaded.
Oleic Acid Determination
For oleic acid analysis, three transformants of the highest complementation efficiency were grown on oleic acid. Aliquots of the medium were removed after 12, 24, and 36 h, and the yeast cells were separated by centrifugation. The fatty acids of the supernatant were first converted to their corresponding methyl esters. Free fatty acids were methylated in methanol with 1-ethyl-3(3-dimethylaminopropyl) carbodiimide hydrochcloride (Stumpe et al., 2001). Fatty acid methyl esters were analyzed by gas chromatography with an Agilent GC 6890 system coupled with a flame ionization detector equipped with a capillary HP INNOWAX column (Hornung et al., 2005).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: NP_014494 (Sc Pex11p), NP_015213 (Sc Pex25p), NP_014836 (Sc Pex27p), NP_564514 (At PEX11a), NP_190327 (At PEX11b), NP_563636 (At PEX11c), NP_850441 (At PEX11d), NP_191666 (At PEX11e), XP_545854 (Cf PEX11), NP_003838 (Hs PEX11α), O96011 (Hs PEX11β), BAD01558 (Hs PEX11γ), CAG13099 (Tn PEX11), EAA65086 (An Pex11p), EAA31192 (Nc Pex11p), CAG81724 (Yl Pex11p), ABF93659 (Os PEX11-1), BAD67925 (Os PEX11-2), ABF95493 (Os PEX11-3), CAD41517 (Os PEX11-4), ABF95492 (Os PEX11-5), and AAF75750 (Le PEX11). Full species names can be found in Figure 1 legend.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Alignment of PEX11 Protein Sequences.
Supplemental Figure 2. Expression Patterns of PEX11 Genes.
Supplemental Figure 3. Peroxisome Phenotype in P35S:PEX11 Plants and Sugar Dependence Assay of P35S:CFP-PEX11 Seedlings.
Supplemental Figure 4. Yeast Complementation by At PEX11 Proteins.
Supplementary Material
Acknowledgments
We thank Andreas Weber, John Froehlich, and Ralf Erdmann for comments on the manuscript; Karen Bird for editing; Chie Awai for technical assistance; Rich Triemer for help with the phylogenetic analysis; Melissa Dawn Lehti-Shiu for help with statistical analysis; Hiroshi Maeda for help with the photorespiration assay; Michael Scharnewski for introduction to fatty acid analysis; Ralf Erdmann for the yeast strains and the antisera against S. cerevisiae thiolase and Pex11p; and the ABRC for the double-stranded RNA vector pFGC5941 and cDNA clones of PEX11c and PEX11e. This work was supported by awards from the U.S. Department of Energy, Michigan State University start-up funds, and the National Science Foundation (Grant MCB 0618335) to J.H. and by grants from the Deutsche Forschungsgemeinschaft (He 565/20-3, Re 1304/2, and Re 1304/4) to H.W. Heldt and to S.R.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Jianping Hu (huji@msu.edu).
Online version contains Web-only data.
References
- Anton, M., Passreiter, M., Lay, D., Thai, T.P., Gorgas, K., and Just, W.W. (2000). ARF- and coatomer-mediated peroxisomal vesiculation. Cell Biochem. Biophys. 32 27–36. [DOI] [PubMed] [Google Scholar]
- Beevers, H. (1979). Microbodies in higher plants. Annu. Rev. Plant Physiol. 30 159–193. [Google Scholar]
- Chang, C.C., South, S., Warren, D., Jones, J., Moser, A.B., Moser, H.W., and Gould, S.J. (1999). Metabolic control of peroxisome abundance. J. Cell Sci. 112 1579–1590. [DOI] [PubMed] [Google Scholar]
- Charlton, W., and Lopez-Huertas, E. (2002). PEX genes in plants and other organisms. In Plant Peroxisomes, A. Baker, and I.A. Graham, eds (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 385–426.
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Corpas, F.J., Barroso, J.B., and del Rio, L.A. (2001). Peroxisomes as a source of reactive oxygen species and nitric oxide signal molecules in plant cells. Trends Plant Sci. 6 145–150. [DOI] [PubMed] [Google Scholar]
- Cosson, P., and Letourneur, F. (1997). Coatomer (COPI)-coated vesicles: Role in intracellular transport and protein sorting. Curr. Opin. Cell Biol. 9 484–487. [DOI] [PubMed] [Google Scholar]
- de Felipe, M., Lucas, M.M., and Pozuelo, J.M. (1988). Cytochemical study of catalase and peroxidase in the mesophyll of Lolium rigidum plants treated with isoproturon. J. Plant Physiol. 132 67–73. [Google Scholar]
- Desvergne, B., and Wahli, W. (1999). Peroxisome proliferator-activated receptors: Nuclear control of metabolism. Endocr. Rev. 20 649–688. [DOI] [PubMed] [Google Scholar]
- Distefano, S., Palma, J.M., McCarthy, I.I., and del Rio, L.A. (1999). Proteolytic cleavage of plant proteins by peroxisomal endoproteases from senescent pea leaves. Planta 209 308–313. [DOI] [PubMed] [Google Scholar]
- Erdmann, R., and Blobel, G. (1995). Giant peroxisomes in oleic acid-induced Saccharomyces cerevisiae lacking the peroxisomal membrane protein Pmp27p. J. Cell Biol. 128 509–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan, J., Quan, S., Orth, T., Awai, C., Chory, J., and Hu, J. (2005). The Arabidopsis PEX12 gene is required for peroxisome biogenesis and is essential for development. Plant Physiol. 139 231–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farre, J.C., and Subramani, S. (2004). Peroxisome turnover by micropexophagy: An autophagy-related process. Trends Cell Biol. 14 515–523. [DOI] [PubMed] [Google Scholar]
- Ferreira, M., Bird, B., and Davies, D.D. (1989). The effect of light on the structure and organization of Leman peroxisomes. J. Exp. Bot. 40 1029–1035. [Google Scholar]
- Fujiki, Y., Hubbard, A.L., Fowler, S., and Lazarow, P.B. (1982). Isolation of intracellular membranes by means of sodium carbonate treatment: Application to endoplasmic reticulum. J. Cell Biol. 93 97–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funato, M., Shimozawa, N., Nagase, T., Takemoto, Y., Suzuki, Y., Imamura, Y., Matsumoto, T., Tsukamoto, T., Kojidani, T., Osumi, T., Fukao, T., and Kondo, N. (2006). Aberrant peroxisome morphology in peroxisomal beta-oxidation enzyme deficiencies. Brain Dev. 28 287–292. [DOI] [PubMed] [Google Scholar]
- Gould, S.J., and Valle, D. (2000). Peroxisome biogenesis disorders: Genetics and cell biology. Trends Genet. 16 340–345. [DOI] [PubMed] [Google Scholar]
- Guo, T., Kit, Y.Y., Nicaud, J.M., Le Dall, M.T., Sears, S.K., Vali, H., Chan, H., Rachubinski, R.A., and Titorenko, V.I. (2003). Peroxisome division in the yeast Yarrowia lipolytica is regulated by a signal from inside the peroxisome. J. Cell Biol. 162 1255–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurvitz, A., Hiltunen, J.K., Erdmann, R., Hamilton, B., Hartig, A., Ruis, H., and Rottensteiner, H. (2001). Saccharomyces cerevisiae Adr1p governs fatty acid beta-oxidation and peroxisome proliferation by regulating POX1 and PEX11. J. Biol. Chem. 276 31825–31830. [DOI] [PubMed] [Google Scholar]
- Gurvitz, A., Wabnegger, L., Rottensteiner, H., Dawes, I.W., Hartig, A., Ruis, H., and Hamilton, B. (2000). Adr1p-dependent regulation of the oleic acid-inducible yeast gene SPS19 encoding the peroxisomal beta-oxidation auxiliary enzyme 2,4-dienoyl-CoA reductase. Mol. Cell Biol. Res. Commun. 4 81–89. [DOI] [PubMed] [Google Scholar]
- Hayashi, M., and Nishimura, M. (2003). Entering a new era of research on plant peroxisomes. Curr. Opin. Plant Biol. 6 577–582. [DOI] [PubMed] [Google Scholar]
- Hayashi, M., Toriyama, K., Kondo, M., and Nishimura, M. (1998). 2,4-Dichlorophenoxybutyric acid-resistant mutants of Arabidopsis have defects in glyoxysomal fatty acid beta-oxidation. Plant Cell 10 183–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi, M., Yagi, M., Nito, K., Kamada, T., and Nishimura, M. (2005). Differential contribution of two peroxisomal protein receptors to the maintenance of peroxisomal functions in Arabidopsis. J. Biol. Chem. 280 14829–14835. [DOI] [PubMed] [Google Scholar]
- Heiland, I., and Erdmann, R. (2005). Biogenesis of peroxisomes. Topogenesis of the peroxisomal membrane and matrix proteins. FEBS J. 272 2362–2372. [DOI] [PubMed] [Google Scholar]
- Hoepfner, D., Schildknegt, D., Braakman, I., Philippsen, P., and Tabak, H.F. (2005). Contribution of the endoplasmic reticulum to peroxisome formation. Cell 122 85–95. [DOI] [PubMed] [Google Scholar]
- Hoepfner, D., van den Berg, M., Philippsen, P., Tabak, H.F., and Hettema, E.H. (2001). A role for Vps1p, actin, and the Myo2p motor in peroxisome abundance and inheritance in Saccharomyces cerevisiae. J. Cell Biol. 155 979–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornung, E., Krueger, C., Pernstich, C., Gipmans, M., Porzel, A., and Feussner, I. (2005). Production of (10E,12Z)-conjugated linoleic acid in yeast and tobacco seeds. Biochim. Biophys. Acta 1738 105–114. [DOI] [PubMed] [Google Scholar]
- Hu, J., Aguirre, M., Peto, C., Alonso, J., Ecker, J., and Chory, J. (2002). A role for peroxisomes in photomorphogenesis and development of Arabidopsis. Science 297 405–409. [DOI] [PubMed] [Google Scholar]
- Karnik, S.K., and Trelease, R.N. (2005). Arabidopsis peroxin 16 coexists at steady state in peroxisomes and endoplasmic reticulum. Plant Physiol. 138 1967–1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpichev, I.V., Luo, Y., Marians, R.C., and Small, G.M. (1997). A complex containing two transcription factors regulates peroxisome proliferation and the coordinate induction of beta-oxidation enzymes in Saccharomyces cerevisiae. Mol. Cell. Biol. 17 69–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiel, J.A., van der Klei, I.J., van den Berg, M.A., Bovenberg, R.A., and Veenhuis, M. (2005). Overproduction of a single protein, Pc-Pex11p, results in 2-fold enhanced penicillin production by Penicillium chrysogenum. Fungal Genet. Biol. 42 154–164. [DOI] [PubMed] [Google Scholar]
- Koch, A., Schneider, G., Luers, G.H., and Schrader, M. (2004). Peroxisome elongation and constriction but not fission can occur independently of dynamin-like protein 1. J. Cell Sci. 117 3995–4006. [DOI] [PubMed] [Google Scholar]
- Koch, A., Thiemann, M., Grabenbauer, M., Yoon, Y., McNiven, M.A., and Schrader, M. (2003). Dynamin-like protein 1 is involved in peroxisomal fission. J. Biol. Chem. 278 8597–8605. [DOI] [PubMed] [Google Scholar]
- Kreykenbohm, V., Wenzel, D., Antonin, W., Atlachkine, V., and von Mollard, G.F. (2002). The SNAREs vti1a and vti1b have distinct localization and SNARE complex partners. Eur. J. Cell Biol. 81 273–280. [DOI] [PubMed] [Google Scholar]
- Li, X., Baumgart, E., Dong, G.X., Morrell, J.C., Jimenez-Sanchez, G., Valle, D., Smith, K.D., and Gould, S.J. (2002. a). PEX11alpha is required for peroxisome proliferation in response to 4-phenylbutyrate but is dispensable for peroxisome proliferator-activated receptor alpha-mediated peroxisome proliferation. Mol. Cell. Biol. 22 8226–8240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X., Baumgart, E., Morrell, J.C., Jimenez-Sanchez, G., Valle, D., and Gould, S.J. (2002. b). PEX11beta deficiency is lethal and impairs neuronal migration but does not abrogate peroxisome function. Mol. Cell. Biol. 22 4358–4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X., and Gould, S.J. (2002). PEX11 promotes peroxisome division independently of peroxisome metabolism. J. Cell Biol. 156 643–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X., and Gould, S.J. (2003). The dynamin-like GTPase DLP1 is essential for peroxisome division and is recruited to peroxisomes in part by PEX11. J. Biol. Chem. 278 17012–17020. [DOI] [PubMed] [Google Scholar]
- Lin, Y., Sun, L., Nguyen, L.V., Rachubinski, R.A., and Goodman, H.M. (1999). The Pex16p homolog SSE1 and storage organelle formation in Arabidopsis seeds. Science 284 328–330. [DOI] [PubMed] [Google Scholar]
- Lingard, M.J., and Trelease, R.N. (2006). Five Arabidopsis peroxin 11 homologs individually promote peroxisome elongation, duplication or aggregation. J. Cell Sci. 119 1961–1972. [DOI] [PubMed] [Google Scholar]
- Liou, W., Geuze, H.J., and Slot, J.W. (1996). Improving structural integrity of cryosections for immunogold labeling. Histochem. Cell Biol. 106 41–58. [DOI] [PubMed] [Google Scholar]
- Lipka, V., et al. (2005). Pre- and postinvasion defenses both contribute to nonhost resistance in Arabidopsis. Science 310 1180–1183. [DOI] [PubMed] [Google Scholar]
- Lock, E.A., Mitchell, A.M., and Elcombe, C.R. (1989). Biochemical mechanisms of induction of hepatic peroxisome proliferation. Annu. Rev. Pharmacol. Toxicol. 29 145–163. [DOI] [PubMed] [Google Scholar]
- Lopez-Huertas, E., Charlton, W.L., Johnson, B., Graham, I.A., and Baker, A. (2000). Stress induces peroxisome biogenesis genes. EMBO J. 19 6770–6777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz, P., Maier, A.G., Baumgart, E., Erdmann, R., and Clayton, C. (1998). Elongation and clustering of glycosomes in Trypanosoma brucei overexpressing the glycosomal Pex11p. EMBO J. 17 3542–3555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, C., Haslbeck, M., Babujee, L., Jahn, O., and Reumann, S. (2006). Identification and characterization of a stress-inducible and a constitutive small heat-shock protein targeted to the matrix of plant peroxisomes. Plant Physiol. 141 47–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier, A.G., Schulreich, S., Bremser, M., and Clayton, C. (2000). Binding of coatomer by the PEX11 C-terminus is not required for function. FEBS Lett. 484 82–86. [DOI] [PubMed] [Google Scholar]
- Mano, S., Nakamori, C., Hayashi, M., Kato, A., Kondo, M., and Nishimura, M. (2002). Distribution and characterization of peroxisomes in Arabidopsis by visualization with GFP: Dynamic morphology and actin-dependent movement. Plant Cell Physiol. 43 331–341. [DOI] [PubMed] [Google Scholar]
- Mano, S., Nakamori, C., Kondo, M., Hayashi, M., and Nishimura, M. (2004). An Arabidopsis dynamin-related protein, DRP3A, controls both peroxisomal and mitochondrial division. Plant J. 38 487–498. [DOI] [PubMed] [Google Scholar]
- Marshall, P.A., Dyer, J.M., Quick, M.E., and Goodman, J.M. (1996). Redox-sensitive homodimerization of Pex11p: A proposed mechanism to regulate peroxisomal division. J. Cell Biol. 135 123–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall, P.A., Krimkevich, Y.I., Lark, R.H., Dyer, J.M., Veenhuis, M., and Goodman, J.M. (1995). Pmp27 promotes peroxisomal proliferation. J. Cell Biol. 129 345–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell, K., and Johnson, G.N. (2000). Chlorophyll fluorescence—A practical guide. J. Exp. Bot. 51 659–668. [DOI] [PubMed] [Google Scholar]
- McCartney, A.W., Greenwood, J.S., Fabian, M.R., White, K.A., and Mullen, R.T. (2005). Localization of the tomato bushy stunt virus replication protein p33 reveals a peroxisome-to-endoplasmic reticulum sorting pathway. Plant Cell 17 3513–3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen, R.T., Flynn, C.R., and Trelease, R.N. (2001). How are peroxisomes formed? The role of the endoplasmic reticulum and peroxins. Trends Plant Sci. 6 256–261. [DOI] [PubMed] [Google Scholar]
- Mullen, R.T., Lisenbee, C.S., Miernyk, J.A., and Trelease, R.N. (1999). Peroxisomal membrane ascorbate peroxidase is sorted to a membranous network that resembles a subdomain of the endoplasmic reticulum. Plant Cell 11 2167–2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nila, A.G., Sandalio, L.M., Lopez, M.G., Gomez, M., Del Rio, L.A., and Gomez-Lim, M.A. (2006). Expression of a peroxisome proliferator-activated receptor gene (xPPARalpha) from Xenopus laevis in tobacco (Nicotiana tabacum) plants. Planta 224 569–581. [DOI] [PubMed] [Google Scholar]
- Oksanen, E., Haikio, E., Sober, J., and Karnosky, D.F. (2003). Ozone-induced H2O2 accumulation in field-grown aspen and birch is linked to foliar ultrastructure and peroxisomal activity. New Phytol. 161 791–799. [DOI] [PubMed] [Google Scholar]
- Palma, J.M., Garrido, M., Rodriguez-Garcia, M.I., and del Rio, L.A. (1991). Peroxisome proliferation and oxidative stress mediated by activated oxygen species in plant peroxisomes. Arch. Biochem. Biophys. 287 68–74. [DOI] [PubMed] [Google Scholar]
- Palmieri, L., Rottensteiner, H., Girzalsky, W., Scarcia, P., Palmieri, F., and Erdmann, R. (2001). Identification and functional reconstitution of the yeast peroxisomal adenine nucleotide transporter. EMBO J. 20 5049–5059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passreiter, M., Anton, M., Lay, D., Frank, R., Harter, C., Wieland, F.T., Gorgas, K., and Just, W.W. (1998). Peroxisome biogenesis: Involvement of ARF and coatomer. J. Cell Biol. 141 373–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinfield-Wells, H., Rylott, E.L., Gilday, A.D., Graham, S., Job, K., Larson, T.R., and Graham, I.A. (2005). Sucrose rescues seedling establishment but not germination of Arabidopsis mutants disrupted in peroxisomal fatty acid catabolism. Plant J. 43 861–872. [DOI] [PubMed] [Google Scholar]
- Purdue, P.E., and Lazarow, P.B. (2001). Peroxisome biogenesis. Annu. Rev. Cell Dev. Biol. 17 701–752. [DOI] [PubMed] [Google Scholar]
- Rottensteiner, H., Stein, K., Sonnenhol, E., and Erdmann, R. (2003. a). Conserved function of pex11p and the novel pex25p and pex27p in peroxisome biogenesis. Mol. Biol. Cell 14 4316–4328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rottensteiner, H., Wabnegger, L., Erdmann, R., Hamilton, B., Ruis, H., Hartig, A., and Gurvitz, A. (2003. b). Saccharomyces cerevisiae PIP2 mediating oleic acid induction and peroxisome proliferation is regulated by Adr1p and Pip2p-Oaf1p. J. Biol. Chem. 278 27605–27611. [DOI] [PubMed] [Google Scholar]
- Rylott, E.L., Rogers, C.A., Gilday, A.D., Edgell, T., Larson, T.R., and Graham, I.A. (2003). Arabidopsis mutants in short- and medium-chain acyl-CoA oxidase activities accumulate acyl-CoAs and reveal that fatty acid beta-oxidation is essential for embryo development. J. Biol. Chem. 278 21370–21377. [DOI] [PubMed] [Google Scholar]
- Schekman, R. (2005). Peroxisomes: Another branch of the secretory pathway? Cell 122 1–2. [DOI] [PubMed] [Google Scholar]
- Schrader, M., Reuber, B.E., Morrell, J.C., Jimenez-Sanchez, G., Obie, C., Stroh, T.A., Valle, D., Schroer, T.A., and Gould, S.J. (1998). Expression of PEX11beta mediates peroxisome proliferation in the absence of extracellular stimuli. J. Biol. Chem. 273 29607–29614. [DOI] [PubMed] [Google Scholar]
- Schumann, U., Wanner, G., Veenhuis, M., Schmid, M., and Gietl, C. (2003). AthPEX10, a nuclear gene essential for peroxisome and storage organelle formation during Arabidopsis embryogenesis. Proc. Natl. Acad. Sci. USA 100 9626–9631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, J.J., Marelli, M., Christmas, R.H., Vizeacoumar, F.J., Dilworth, D.J., Ideker, T., Galitski, T., Dimitrov, K., Rachubinski, R.A., and Aitchison, J.D. (2002). Transcriptome profiling to identify genes involved in peroxisome assembly and function. J. Cell Biol. 158 259–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparkes, I.A., Brandizzi, F., Slocombe, S.P., El-Shami, M., Hawes, C., and Baker, A. (2003). An Arabidopsis pex10 null mutant is embryo lethal, implicating peroxisomes in an essential role during plant embryogenesis. Plant Physiol. 133 1809–1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumpe, M., Kandzia, R., Gobel, C., Rosahl, S., and Feussner, I. (2001). A pathogen-inducible divinyl ether synthase (CYP74D) from elicitor-treated potato suspension cells. FEBS Lett. 507 371–376. [DOI] [PubMed] [Google Scholar]
- Tam, Y.Y., Torres-Guzman, J.C., Vizeacoumar, F.J., Smith, J.J., Marelli, M., Aitchison, J.D., and Rachubinski, R.A. (2003). Pex11-related proteins in peroxisome dynamics: A role for the novel peroxin Pex27p in controlling peroxisome size and number in Saccharomyces cerevisiae. Mol. Biol. Cell 14 4089–4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoms, S., and Erdmann, R. (2005). Dynamin-related proteins and Pex11 proteins in peroxisome division and proliferation. FEBS J. 272 5169–5181. [DOI] [PubMed] [Google Scholar]
- Titorenko, V.I., and Rachubinski, R.A. (2004). The peroxisome: Orchestrating important developmental decisions from inside the cell. J. Cell Biol. 164 641–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokuyasu, K.T. (1973). A technique for ultracryotomy of cell suspensions and tissues. J. Cell Biol. 57 551–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokuyasu, K.T. (1980). Immunochemistry on ultrathin frozen sections. Histochem. J. 12 381–403. [DOI] [PubMed] [Google Scholar]
- van Roermund, C.W., Tabak, H.F., van Den Berg, M., Wanders, R.J., and Hettema, E.H. (2000). Pex11p plays a primary role in medium-chain fatty acid oxidation, a process that affects peroxisome number and size in Saccharomyces cerevisiae. J. Cell Biol. 150 489–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizeacoumar, F.J., Torres-Guzman, J.C., Bouard, D., Aitchison, J.D., and Rachubinski, R.A. (2004). Pex30p, Pex31p, and Pex32p form a family of peroxisomal integral membrane proteins regulating peroxisome size and number in Saccharomyces cerevisiae. Mol. Biol. Cell 15 665–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizeacoumar, F.J., Torres-Guzman, J.C., Tam, Y.Y., Aitchison, J.D., and Rachubinski, R.A. (2003). YHR150w and YDR479c encode peroxisomal integral membrane proteins involved in the regulation of peroxisome number, size, and distribution in Saccharomyces cerevisiae. J. Cell Biol. 161 321–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voncken, F., van Hellemond, J.J., Pfisterer, I., Maier, A., Hillmer, S., and Clayton, C. (2003). Depletion of GIM5 causes cellular fragility, a decreased glycosome number, and reduced levels of ether-linked phospholipids in trypanosomes. J. Biol. Chem. 278 35299–35310. [DOI] [PubMed] [Google Scholar]
- Wanders, R.J. (2004). Metabolic and molecular basis of peroxisomal disorders: A review. Am. J. Med. Genet. A. 126 355–375. [DOI] [PubMed] [Google Scholar]
- Wessel, D., and Flugge, U.I. (1984). A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal. Biochem. 138 141–143. [DOI] [PubMed] [Google Scholar]
- Woods, R.A., and Gietz, R.D. (2001). High-efficiency transformation of plasmid DNA into yeast. Methods Mol. Biol. 177 85–97. [DOI] [PubMed] [Google Scholar]
- Yan, M., Rayapuram, N., and Subramani, S. (2005). The control of peroxisome number and size during division and proliferation. Curr. Opin. Cell Biol. 17 376–383. [DOI] [PubMed] [Google Scholar]
- Zimmermann, P., Hirsch-Hoffmann, M., Hennig, L., and Gruissem, W. (2004). GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 136 2621–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolman, B.K., Silva, I.D., and Bartel, B. (2001). The Arabidopsis pxa1 mutant is defective in an ATP-binding cassette transporter-like protein required for peroxisomal fatty acid beta-oxidation. Plant Physiol. 127 1266–1278. [PMC free article] [PubMed] [Google Scholar]
- Zolman, B.K., Yoder, A., and Bartel, B. (2000). Genetic analysis of indole-3-butyric acid responses in Arabidopsis thaliana reveals four mutant classes. Genetics 156 1323–1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.