Abstract
Wood is formed by the successive addition of secondary xylem, which consists of cells with a conspicuously thickened secondary wall composed mainly of lignin and cellulose. Several genes involved in lignin and cellulose biosynthesis have been characterized, but the factors that regulate the formation of secondary walls in woody tissues remain to be identified. In this study, we show that plant-specific transcription factors, designated NAC SECONDARY WALL THICKENING PROMOTING FACTOR1 (NST1) and NST3, are key regulators of the formation of secondary walls in woody tissues of Arabidopsis thaliana. In nst1-1 nst3-1 double knockout plants, the secondary wall thickenings in interfascicular fibers and secondary xylem, except for vascular vessels, were completely suppressed without affecting formation of cells destined to be woody tissues. Conversely, as shown previously for NST1, overexpression of NST3 induced ectopic secondary wall thickenings in various aboveground tissues. Furthermore, the expression of chimeric repressors derived from NST1 and NST3 suppressed secondary wall thickenings in the presumptive interfascicular fibers. Because putative orthologs of NST1 and NST3 are present in the genome of poplar, our results suggest that they are also key regulators of the formation of secondary walls in woody plants and could be used as a tool for the genetic engineering of wood and its derivatives.
INTRODUCTION
Wood is a major terrestrial biomass and one of our most important natural materials (Plomion et al., 2001). In the history of plant evolution, acquisition of a mechanism for the formation of woody tissues is considered a particularly important event with regard to successful propagation of vascular plants, making it possible for plants to support taller growth and enabling easier dispersion of pollen and seeds. Wood is formed by the successive addition of secondary xylem, which originates from vascular cambium. The secondary xylem, in both herbaceous and woody plants, consists of cells with a conspicuously thickened secondary wall that develops beneath the primary cell wall and is composed mainly of lignin and cellulose. Various genes involved in the biosynthesis of lignin and cellulose have been characterized (Taylor et al., 1999, 2003; Jones et al., 2001; Sibout et al., 2005), but factors that regulate the formation of secondary walls in woody tissues remain relatively unknown.
VASCULAR-RELATED NAC-DOMAIN6 (VND6) and VND7 transcription factors have been shown to be regulators of the formation of vascular vessels (Kubo et al., 2005). However, the transcription factors that regulate the formation of other woody tissues, including fibers and secondary xylem, are unknown. One of the allelic mutants of REVOLUTA (REV)/INTERFASCICULAR FIBERLESS1 (IFL1) encoding the HD-ZIPIII class transcription factor is defective in differentiation of interfascicular fibers at the bottom of the inflorescence stem (Zhong et al., 1997; Zhong and Ye, 1999). However, the fiberless phenotype of ifl1 may be due to the secondary effect of impaired basipetal transport of auxin (Zhong and Ye, 2001).
Secondary wall thickenings are found not only in xylem but also in seedpods and anther endothecium and are required for the dehiscence of seedpods and anthers (Keijzer, 1987; Spence et al., 1996). We have shown previously that two plant-specific transcription factors, namely, NAC SECONDARY WALL THICKENING PROMOTING FACTOR1 (NST1) and NST2, are redundantly responsible for secondary wall thickenings in anther endothecium and induce ectopic secondary wall thickenings in various tissues when expressed ectopically (Mitsuda et al., 2005). This finding prompted us to use Arabidopsis thaliana as a model to identify the factor(s) involved in regulating the formation of secondary walls in woody tissues because recent molecular genetics analyses suggest that the genes regulating woody growth are not unique to woody plants (Groover, 2005) and, moreover, because cambium-mediated secondary growth can be studied in this model plant (Chaffey et al., 2002).
In this study, we show evidence that the plant-specific transcription factors NST1 and NST3 (At1g32770) redundantly regulate the secondary wall thickenings in interfascicular fiber of inflorescence stems and secondary xylem of hypocotyls in Arabidopsis without affecting the formation of cells destined to be woody tissues. Furthermore, we suppressed the formation of secondary walls in the stem by genetic manipulation using the chimeric repressor for NST1 and NST3. Because putative orthologs of NST1 and NST3 are present in the genome of poplar, our results suggest that they are also key regulators of the formation of secondary walls in woody plants and could provide tools for the genetic engineering of wood and its derivatives.
RESULTS
NST1 and NST3 Are Possible Regulators of the Formation of Secondary Walls in Woody Tissues
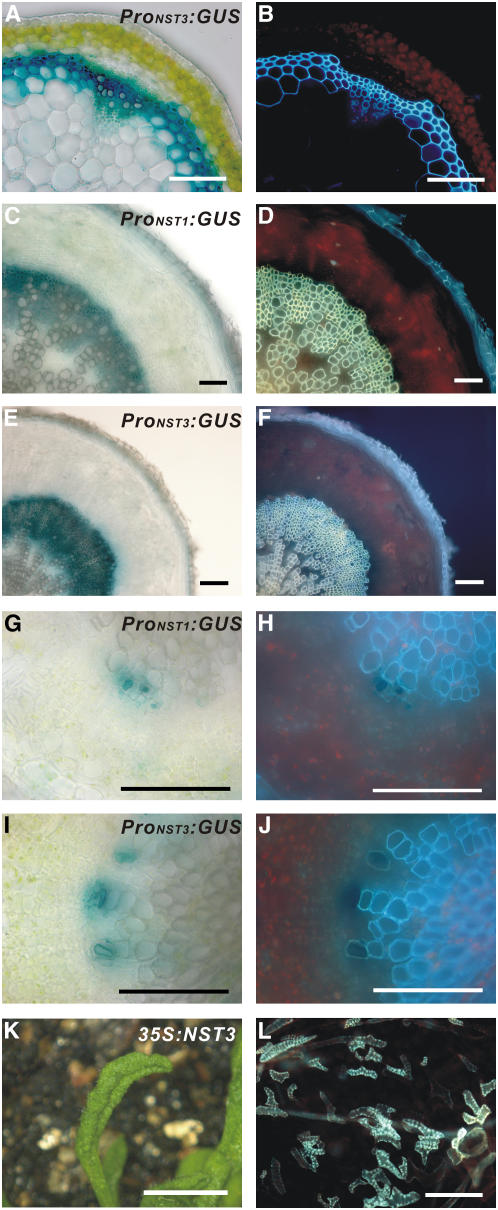
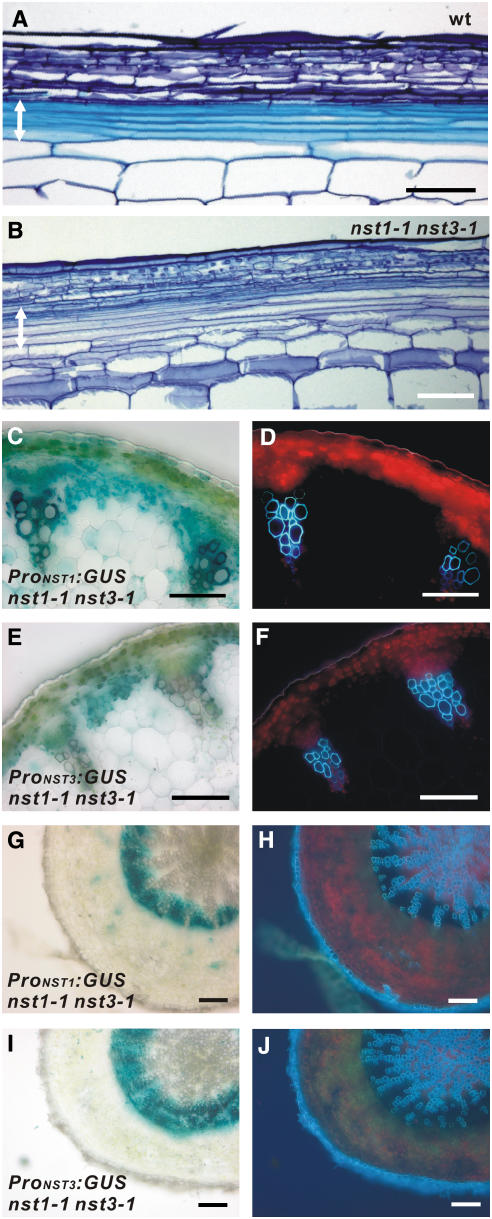
In a previous report, we showed that NST1 has strong promoter activity not only in anther endothecium but also in interfascicular fibers of inflorescence stems and cells differentiating into vascular vessels where secondary walls develop (Mitsuda et al., 2005). Therefore, we postulated that NST1 might also regulate the development of secondary wall thickening in xylem. However, in two NST1 T-DNA–tagged lines (Alonso et al., 2003), nst1-1 (SALK_120377) and nst1-2 (SALK_149993), secondary wall thickening in inflorescence stems was not dramatically different from the wild type, even though a slight reduction in secondary wall thickening was occasionally observed in some nst1-1 plants (see Supplemental Figures 1A to 1D online). The observation that disruption of NST1 does not affect xylem formation suggests the presence of a factor that acts redundantly with NST1, as in the case of NST2 in anther endothecium (Mitsuda et al., 2005). Analysis of publicly available microarray data for Arabidopsis revealed that the expression of NST3, a homolog of NST1, is enhanced in stems, as with expression of NST1 (Schmid et al., 2005). Examination of the promoter activities of NST1 and NST3, using promoter-reporter gene constructs (ProNST1:GUS and ProNST3:GUS), revealed that they are observed in the interfascicular fibers of inflorescence stems and the secondary xylem of hypocotyls as well as cells differentiating into vascular vessels, in which secondary walls develop (Figures 1A to 1J; Mitsuda et al., 2005). As was also the case with NST1 and NST2, ectopic expression of NST3 driven by the cauliflower mosaic virus 35S promoter induced ectopic secondary wall thickening displaying a similar appearance to the tracheary element (TE) in various aboveground tissues (Figures 1K and 1L; Mitsuda et al., 2005), indicating that the ability of NST3 to induce secondary wall thickening is similar to that of NST1 and NST2. However, neither of the two NST3 T-DNA–tagged lines (SALK_149909 nor SALK_131657) had any obvious phenotypic abnormalities in the xylem (see Supplemental Figures 1E to 1H online). These observations suggest that NST1 and NST3 might be involved, redundantly, in the formation of secondary walls in the stem and hypocotyl.
Figure 1.
NST3 and Its Promoter Showed Similar Activities to NST1 in Woody Tissues.
(A) and (B) Cross section of an inflorescence stem of Arabidopsis carrying the ProNST3:GUS construct (A) and the same sections under UV illumination (B). Secondary walls containing lignin emitted blue autofluorescence.
(C) to (F) Cross sections of a mature root hypocotyl of transgenic Arabidopsis carrying ProNST1:GUS (C) or ProNST3:GUS (E) and the same sections under UV illumination ([D] and [F]).
(G) to (J) Cross sections of a young root hypocotyl of transgenic Arabidopsis carrying ProNST1:GUS (G) or ProNST3:GUS (I) and the same sections under UV illumination ([H] and [J]).
(K) Upward curling rosette leaf of a 35S:NST3 plant.
(L) Ectopic secondary wall thickening in epidermal cells of rosette leaves, as visualized under UV illumination.
Bars = 100 μm, except in (K), where the bar = 5 mm.
Double T-DNA–Tagged Lines of NST1 and NST3 Show Loss of Secondary Walls in Woody Tissues
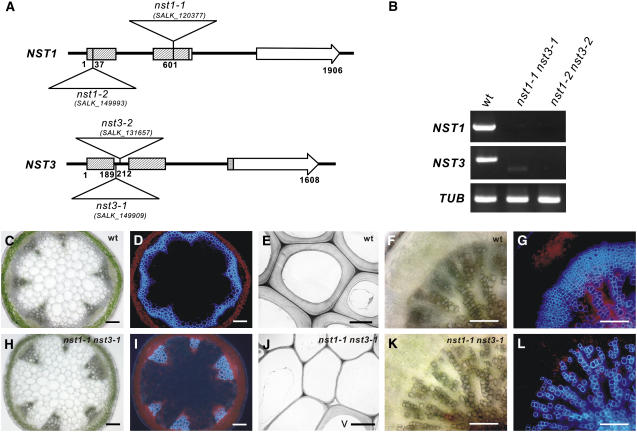
To examine whether NST1 and NST3 redundantly regulate the formation of secondary walls in woody tissues, we prepared homozygous double knockout NST1 and NST3 lines (referred to as nst1-1 nst3-1 hereafter; Figure 2A). We confirmed by RT-PCR analysis that no transcript corresponding to the correct size of NST1 or NST3 mRNA is present in the mutant lines of nst1-1 nst3-1 or nst1-2 nst3-2 (Figure 2B). Although a smaller aberrant transcript amplified with the NST3 primer was faintly detected in the nst1-1 nst3-1 line, sequence analysis revealed that the putative protein encoded by the fragment terminated soon after the first 62 amino acids due to internal deletion of mRNA with frame shift (data not shown). This indicates that nst1-1 nst3-1 and nst1-2 nst3-2 are null mutants. We found that nst1-1 nst3-1 plants completely lost their lignified materials as represented by blue autofluorescence in the region where interfascicular fibers should be formed in wild-type plants (Figures 2C, 2D, 2H, and 2I). This was the case even when plants grew taller than 25 cm, which is a sufficient height for the production of interfascicular fibers with secondary walls in wild-type plants (Ko et al., 2004). Ultrastructural observations using transmission electron microscopy revealed that conspicuously thickened secondary walls were clearly evident in the interfascicular regions of the wild type but not nst1-1 nst3-1 plants, except for vascular vessels (Figures 2E and 2J). Hypocotyls of Arabidopsis are known to form secondary xylem from a relatively early stage and have a similar structure to the trunk of a tree (Chaffey et al., 2002). Observation of a cross section of the hypocotyl of the nst1-1 nst3-1 plant revealed that lignified materials represented by autofluorescence were completely lost in the presumptive secondary xylem, but the secondary walls of vascular vessels did not seem to be affected as in inflorescence stems (Figures 2F, 2G, 2K, and 2L).
Figure 2.
Double T-DNA–Tagged Lines of NST1 and NST3 Showed Loss of Secondary Walls in Woody Tissues.
(A) Schematic diagram of the structure of the NST1 and NST3 genes and the positions of T-DNA tags in nst1-1 (SALK_120377), nst1-2 (SALK_149993), nst3-1 (SALK_149909), and nst3-2 (SALK_131657) lines. Numbers indicate nucleotide positions from the site of initiation of translation. Boxes and arrows represent exons. Shaded boxes represent coding regions of conserved NAC domains.
(B) RT-PCR analysis of the transcripts of the NST gene in the NST T-DNA–tagged lines. No transcript corresponding to full-length NST1 or NST3 mRNA was detected either in nst1-1 nst3-1 or nst1-2 nst3-2 plants. TUB represents the gene for β-tubulin used as a positive control.
(C), (D), (F), and (G) Cross sections of an inflorescence stem (C) and root-hypocotyl (F) of wild-type plants and the same sections under UV illumination ([D] and [G]).
(H), (I), (K), and (L) Cross sections of an inflorescence stem (H) and root-hypocotyl (K) of nst1-1 nst3-1 plants and the same sections under UV illumination ([I] and [L]).
(E) and (J) Ultrastructural views of the tissue corresponding to interfascicular fibers of inflorescence stems of a wild-type (E) and nst1-1 nst3-1 (J) plants taken by transmission electron microscopy. A thickened secondary wall is visible in the wild type but not the nst1-1 nst3-1 plant except for vascular vessels (V).
Bars = 100 μm, except in (E) and (J), where bars = 5 μm.
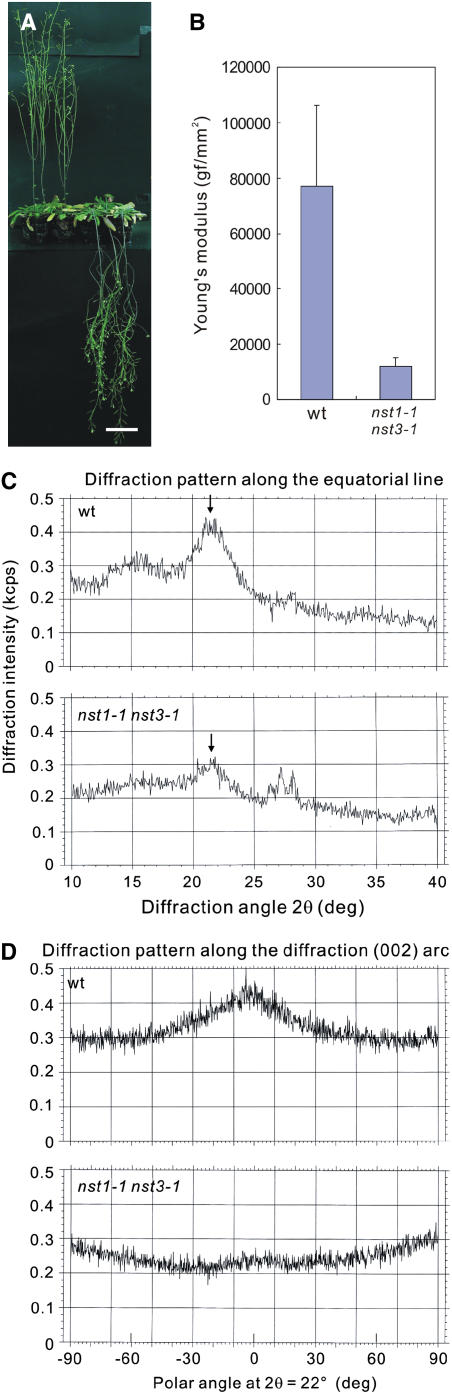
Under short-day conditions, the nst1-1 nst3-1 plants were no longer able to remain upright when they reached >15 cm in height as a result of the loss of secondary walls in the stem cells (Figure 3A). Stems of nst1-1 nst3-1 plants were easily bent and broken. Indeed, the physical strength of inflorescence stems of nst1-1 nst3-1 plants, as represented by Young's modulus, was much lower than that of stems of wild-type plants (Figure 3B). In addition, x-ray diffraction analysis suggested that the nst1-1 nst3-1 plants had no cellulose microfibrils constituting the secondary wall (Figures 3C and 3D). These findings indicate that neither lignin nor cellulose, which constitute secondary walls, was produced in inflorescence stems of nst1-1 nst3-1 plants, with the exception of the vascular vessels. However, the growth rate and overall size of nst1-1 nst3-1 plants were similar to those of wild-type plants, suggesting that the development of vascular vessels was not affected. These results indicate that NST1 and NST3 redundantly regulate the formation of secondary walls in interfascicular fibers and secondary xylem in Arabidopsis.
Figure 3.
Double T-DNA–Tagged Lines of NST1 and NST3 Showed Reduced Stem Strength and Loss of Cellulose Microfibrils.
(A) Wild-type (left) and nst1-1 nst3-1 (right) plants. The nst1-1 nst3-1 plants were unable to stand erect. Bar = 5 cm.
(B) Young's modulus of inflorescence stems of wild-type and nst1-1 nst3-1 plants. Stems of nst1-1 nst3-1 plants were much weaker than wild-type stems (values are means + SD; n = 5).
(C) The x-ray diffraction patterns of wild-type plants (top panel) and nst1-1 nst3-1 plants (bottom panel) along the equatorial line.
(D) The x-ray diffraction patterns of wild-type plants (top panel) and nst1-1 nst3-1 plants (bottom panel) along the (002) arc at 2θ = 22°, which corresponds to the position indicated by the arrows in (C).
A completely separate T-DNA–tagged line, nst1-2 nst3-2, had the same defective phenotype as that of nst1-1 nst3-1 plants (data not shown). We were able to almost entirely reverse the defective phenotype of nst1-1 nst3-1 plants by introducing a genomic fragment containing either the NST1 or NST3 gene (see Supplemental Figure 2 online). Similar restoration of an almost wild-type phenotype occurred when ProNST1:NST1 or ProNST3:NST3 was introduced into nst1-1 nst3-1 plants (data not shown). These observations indicate that the phenotype of nst1-1 nst3-1 plants was due to loss of the activities of the NST1 and NST3 genes.
NST1 and NST3 Regulate the Expression of Genes Involved in Biosynthesis of Secondary Walls
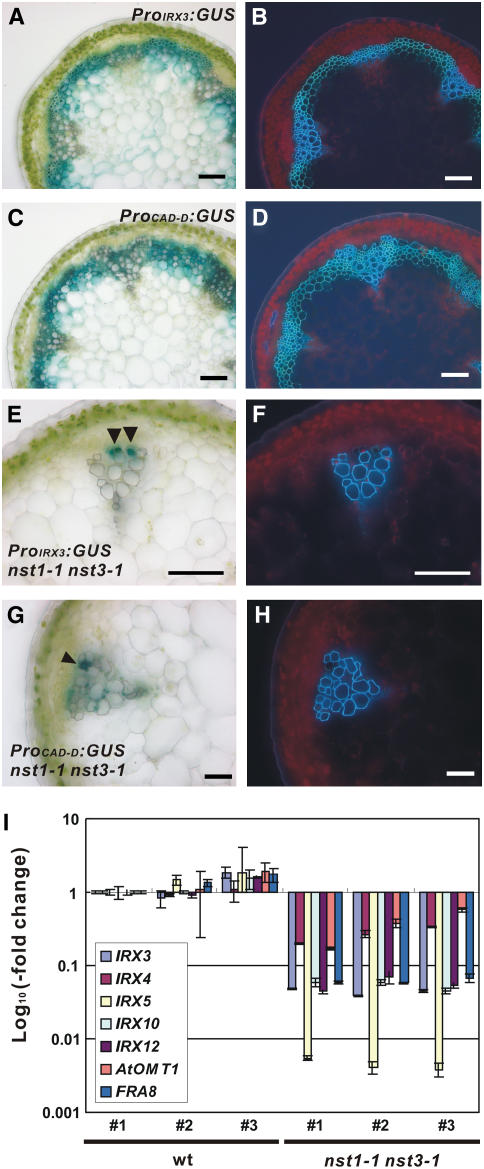
We also analyzed promoter activities of IRREGULAR XYLEM3 (IRX3) and CINNAMYL ALCOHOL DEHYDROGENASE-D (CAD-D), which encode cellulose synthase (Taylor et al., 1999) and an enzyme involved in lignin biosynthesis (Sibout et al., 2005), respectively, to examine whether NST1 and NST3 regulate the expression of genes involved in biosynthesis of secondary walls. The promoter activities of both genes were evident in interfascicular fibers of wild-type background plants (Figures 4A to 4D) but were detected only in cells differentiating into vascular vessels and, in the case of CAD-D, in cells adjacent to vascular vessels, not in cells of interfascicular regions in nst1-1 nst3-1 background plants (Figures 4E to 4H). These findings suggest that neither cellulose nor lignin is produced in the interfascicular regions of nst1-1 nst3-1 plants.
Figure 4.
Expression of Genes Related to Secondary Wall Synthesis Was Suppressed in nst1-1 nst3-1 Plants.
(A) to (D) Cross sections of the inflorescence stem of transgenic plants harboring ProIRX3:GUS (A) or ProCAD-D:GUS (C) and the same sections under UV illumination ([B] and [D]).
(E) to (H) Cross sections of an inflorescence stem ([E] and [G]) and the same sections under UV illumination ([F] and [H]) of an nst1-1 nst3-1 plant transformed with ProIRX3:GUS ([E] and [F]) and ProCAD-D:GUS ([G] and [H]), respectively. The cells indicated by arrowheads are those differentiating into vascular vessels ([E] and [G]). Bars = 100 μm.
(I) The levels of gene expression involved in the biosynthesis of secondary walls. The IRX3 and IRX5; IRX4, IRX12, and At OMT1; and IRX10 and FRA8 genes encode enzymes for cellulose biosynthesis, lignin biosynthesis, and the biosynthesis and modification of xyloglucan, respectively. The y axis represents the log10 ratio of the level of expression relative to that in wild-type plant #1. Error bars represent sd of results from three replicates.
To determine the entire transcriptome of the nst1-1 nst3-1 plants, we performed microarray experiments. A total of 17,514 genes passed the filtering test (see Methods). The expression of 391 genes was suppressed significantly (Q-value < 0.1) with levels of transcripts being 50% or less than those of wild-type plants (see Supplemental Table 1 online). This group of genes significantly overlapped with the group of genes related to the synthesis of secondary walls (Table 1) and to the group of genes whose expression was enhanced in 35S:NST1 plants in a previous study (Mitsuda et al., 2005). Analysis by quantitative RT-PCR confirmed that the expression of genes involved in the biosynthesis of secondary walls, namely, IRX3, IRX4, IRX5, IRX10, IRX12, At OMT1, and FRAGILE FIBER8 (FRA8) (Taylor et al., 1999, 2003; Muzac et al., 2000; Jones et al., 2001; Brown et al., 2005; Sawa et al., 2005; Zhong et al., 2005), was indeed suppressed in nst1-1 nst3-1 plants (Figure 4I). IRX3 and IRX5 encode cellulose synthase (Taylor et al., 1999, 2003), IRX4, IRX12, and At OMT1 are related to lignin biosynthesis (Muzac et al., 2000; Jones et al., 2001; Brown et al., 2005; Sawa et al., 2005), and FRA8 is considered to function in the biosynthesis of xylan (Zhong et al., 2005). These observations support the hypothesis that NST1 and NST3 regulate the formation of secondary walls.
Table 1.
Groups of Genes That Overlapped Significantly with Those Genes Whose Expression Was Suppressed in nst1-1 nst3-1 Plants
| Groups of Genes | Number of Genes | Overlapping Genes | P Value | Odds Ratio | Reference or Resource |
|---|---|---|---|---|---|
| Upregulated genes in 35S:NST1 plants | 636 | 75 | 2.20E-16 | 11.25747 | Mitsuda et al. (2005) |
| Xylem-biased genes | 254 | 33 | 2.20E-16 | 10.58139 | Zhao et al. (2005) |
| Secondary cell wall biosynthesis genes (GO: 0009834) | 6 | 5 | 5.26E-09 | 317.756 | http://www.arabidopsis.org/ |
| Genes involved in the cinnamate- monolignol pathway | 60 | 10 | 2.73E-08 | 13.06842 | Tokimatsu et al. (2005) |
| Genes involved in lignin biosynthesis | 32 | 7 | 4.99E-07 | 18.10865 | http://www.arabidopsis.org/ |
| Genes involved in xyloglucan biosynthesis and modification | 98 | 9 | 2.23E-05 | 6.568023 | Tokimatsu et al. (2005) |
| Lignin biosynthesis genes (GO: 0009809) | 21 | 4 | 0.00028 | 15.0599 | http://www.arabidopsis.org/ |
| Cellulose biosynthesis genes (GO: 0030244) | 34 | 4 | 0.001853 | 8.527619 | http://www.arabidopsis.org/ |
Selected groups of genes that significantly overlapped with genes whose expression was suppressed (0.5-fold or below, with a Q-value < 0.1) in nst1-1 nst3-1 plants are listed. The number of genes in each group examined by microarray analysis is given in the second column, and, of these, the number of genes that is the same as those suppressed in the nst1-1 nst3-1 plants is listed in the third column. P values and odds ratios (= number of genes that actually overlapped/number of genes expected by chance) from Fisher's exact test are listed in the fourth and fifth columns, respectively. The data resource or reference is listed in sixth column. GO and 2.2E-016 represent gene ontology, as defined by the consortium and values below 2.2E-016, respectively.
Cells Destined to Be Woody Tissues Form in nst1-1 nst3-1 Plants
To investigate whether cells destined to be fibers form in nst1-1 nst3-1 plants, we examined longitudinal sections of interfascicular regions. In the wild-type plants, we observed clearly differentiated long and narrow fibrous cells stained sky-blue by Toluidine blue O (Figure 5A). Such staining indicates the accumulation of lignin. In nst1-1 nst3-1 plants, long and narrow fiber-like cells similar to those in wild-type plants were also observed (Figure 5B). However, these cells were not stained similarly as in wild-type plants by Toluidine blue O, probably because of the absence of secondary walls.
Figure 5.
Investigation of Cell Differentiation into Fibers and Secondary Xylem in nst1-1 nst3-1 Plants.
(A) and (B) Longitudinal sections of the inflorescence stems of wild-type (A) and nst1-1 nst3-1 (B) plants after staining with Toluidine blue O. Arrows indicate the tissue corresponding to interfascicular fibers.
(C) to (F) Cross section of an inflorescence stem ([C] and [E]) and the same sections under UV illumination ([D] and [F]) of an nst1-1 nst3-1 plant transformed with ProNST1:GUS ([C] and [D]) and ProNST3:GUS ([E] and [F]), respectively.
(G) to (J) Cross sections of a root-hypocotyl ([G] and [I]) and the same sections under UV illumination ([H] and [J]) of an nst1-1 nst3-1 plant transformed with ProNST1:GUS ([G] and [H]) and ProNST3:GUS ([I] and [J]), respectively. The presumptive interfascicular fibers and secondary xylem were stained with ProNST1:GUS and ProNST3:GUS, even though secondary wall thickening was completely suppressed. Bars = 100 μm.
In addition, we found that the promoter activities of NST1 and NST3, as represented by the β-glucuronidase (GUS) activities expressed by the ProNST1:GUS and ProNST3:GUS reporter genes, respectively, were clearly evident in the interfascicular regions of nst1-1 nst3-1 plants, in which interfascicular fibers normally form in wild-type plants, even though no secondary walls were ever detected (Figures 5C to 5F). Moreover, in the hypocotyls, the promoter activities of NST1 and NST3 were apparently observed in the presumptive secondary xylem of nst1-1 nst3-1 plants (Figures 5G to 5J). These observations indicate that the cells where NST1 and NST3 should be expressed were formed in nst1-1 nst3-1 plants. Because the promoter activities of NST1 and NST3 were tightly associated with tissues where secondary walls develop, it is considered that cells destined to be woody tissues form even in the absence of NST1 and NST3.
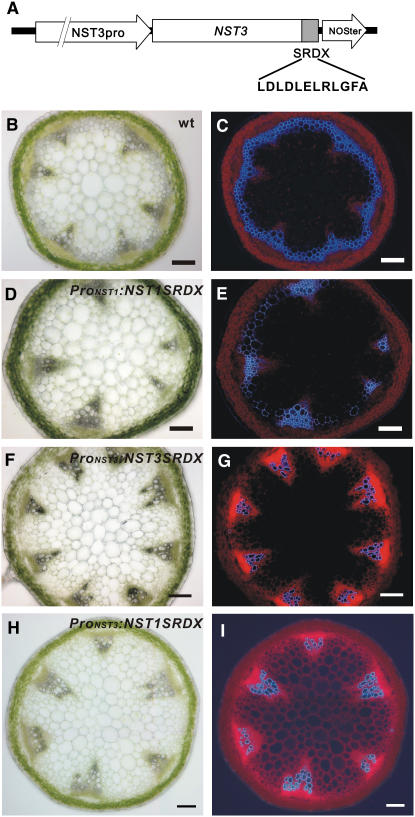
Manipulation of Secondary Walls in Woody Tissues Using a Chimeric Repressor
In an attempt to manipulate the formation of secondary walls, we applied our chimeric repressor gene-silencing technology (CRES-T), in which NST1 or NST3 fused to the EAR motif repression domain (SRDX) dominantly represses the transcription of its target genes (Hiratsu et al., 2003) (Figure 6A). We found that two out of 25 T1 transgenic plants expressing the chimeric NST3 repressor, driven by its own promoter (ProNST3:NST3SRDX), had conspicuously reduced secondary wall thickening in the presumptive interfascicular fibers (Figures 6F and 6G). We also observed a similar but modest effect when ProNST1:NST1SRDX was employed (Figures 6D and 6E) in addition to the previously reported defect in secondary wall thickening in anther endothecium (Mitsuda et al., 2005). By contrast, the expression of the chimeric NST1 repressor driven by the NST3 promoter (ProNST3:NST1SRDX) induced a severe defect in the formation of secondary walls in the presumptive interfascicular fibers in 19 out of 45 T1 plants, without affecting anther dehiscence (Figures 6H and 6I). This was probably due to the synergistic effect of the combination of NST1, which has a strong ability to induce secondary wall thickening (Mitsuda et al., 2005), and the NST3 promoter, which has strong activity in interfascicular fibers and secondary xylem in hypocotyls. These results were consistent with those obtained with T-DNA–tagged lines and suggest the possibility of manipulation of wood formation by genetic engineering.
Figure 6.
Application of CRES-T to NSTs Can Suppress Fiber Formation.
(A) Schematic diagram of one of the introduced constructs. SRDX and NOSter indicate the transcriptional repression domain composed of 12 amino acids and the transcriptional terminator sequence, respectively.
(B) to (I) Cross sections of an inflorescence stem ([B], [D], [F], and [H]) and the same sections under UV illumination ([C], [E], [G], and [I]) of a wild-type, ProNST1:NST1SRDX, ProNST3:NST3SRDX, and ProNST3:NST1SRDX plant, respectively. Secondary wall thickening in the presumptive interfascicular fiber was conspicuously suppressed. Bars = 100 μm.
DISCUSSION
NST1 and NST3 Redundantly Regulate the Formation of Secondary Walls in Woody Tissues Independent from the Formation of Cells Destined to Be Woody Tissues
In this study, we concluded that NST1 and NST3 redundantly regulate the formation of secondary walls in woody tissues, including interfascicular fibers of inflorescence stems and secondary xylem of hypocotyls, except vascular vessels. Our analyses demonstrated that neither lignin nor cellulose, components of secondary walls, is produced in interfascicular fibers and secondary xylem in nst1-1 nst3-1 plants. Furthermore, we showed that the expression of genes involved in the biosynthesis of secondary walls is clearly downregulated in nst1-1 nst3-1 plants and, in addition, that the promoter activities of some of these genes are not detected in the interfascicular regions of nst1-1 nst3-1 plants (Table 1, Figure 4).
However, when grown under short-day conditions for >3 months, we occasionally noted low-level synthesis of secondary walls in presumptive interfascicular fibers in the base of inflorescence stems of nst1-1 nst3-1 plants (see Supplemental Figure 3 online). This observation suggests the presence of residual activity of an NST transcription factor(s), such as NST2 (Mitsuda et al., 2005). In fact, we were able to detect weak promoter activity of NST2 in the interfascicular regions of inflorescence stems (Mitsuda et al., 2005) (data not shown).
We also concluded that cells destined to be fibers and secondary xylem were properly formed independent of the activity of NST1 and NST3. This was confirmed based on the observation that the promoter activities of NST1 and NST3 were evident in the presumptive interfascicular fibers and secondary xylem of nst1-1 nst3-1 plants, and fiber-like cells were evident in the interfascicular region in the nst1-1 nst3-1 plants (Figure 5). Although ectopic expression of NSTs induced ectopic secondary wall thickening with a similar appearance to TE, we previously reported that NSTs do not have the ability to transdifferentiate cells into TE because neither genes related to programmed cell death, the final step of differentiation into TE, nor genes for vascular markers were enhanced, but only genes related to the biosynthesis of secondary walls were upregulated in the plants ectopically expressing NST1 (Mitsuda et al., 2005). Furthermore, the ectopic expression of NSTs induced various patterns of secondary wall thickening depending on the cell type and did not change the shape of cells where ectopic secondary walls develop (data not shown) (Mitsuda et al., 2005). These findings support the scenario that NST1 and NST3 are not responsible for the formation of cells destined to be woody tissue, but rather are involved in the formation of secondary walls after the establishment of the cell identity of woody tissues.
The IFL1/REV gene (Zhong and Ye, 1999), which controls the identity of the adaxial side of various organs, including xylem (Talbert et al., 1995; Zhong and Ye, 1999; Emery et al., 2003), is perhaps involved in regulating the identity of xylem because ifl1 mutant plants fail to form interfascicular fibers in inflorescence stems but differentiate ectopic xylem-like sclerified cells in upper regions of inflorescence stems as a result of a reduction of basipetal transport of auxin (Zhong and Ye, 2001). It has also been suggested that auxin might serve as a signal for the secondary growth of inflorescence stems (Ko et al., 2004). Thus, IFL1/REV might promote the basipetal transport of auxin, inducing the expression of NST genes necessary for the promotion of secondary wall thickening in fiber cells.
NAC Transcription Factors Are Master Regulators of Secondary Wall Thickenings in Plants
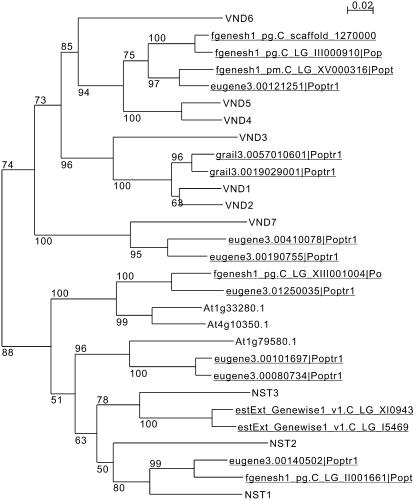
Secondary wall thickenings are formed developmentally in various tissues, including fibers, vascular vessels, secondary xylem, anther endothecium, valve endodermal layer, and the valve margin of siliques. No common factor regulating these secondary wall thickenings has so far been identified, although our studies revealed that the NST1, NST2, and NST3 genes differentially regulate the formation of secondary walls in anther endothecium, interfascicular fibers, and secondary xylem but not vessels. Recently, two NAC domain transcription factors related to NSTs, VND6 and VND7, were shown to regulate the formation of vessel elements (Kubo et al., 2005). These findings suggest that closely related NAC transcription factors, NSTs and VNDs, act as master regulators of secondary wall thickenings in plants.
NSTs and VNDs Are Closely Related but Have Different Functional Roles
It was previously reported that ectopic expression of the VND6 and VND7 genes induces differentiation of xylem vessel elements (Kubo et al., 2005). The NST genes can also induce ectopic secondary wall thickening similar to TE when expressed ectopically (Figure 1L). NSTs and VNDs are phylogenetically classified into separated branches but in the same subfamily (Figure 7). The phenotype of nst1-1 nst3-1 plants could be restored by the expression of VND6 or VND7 under the control of the NST3 promoter (N. Mitsuda and M. Ohme-Takagi, unpublished results). However, microarray analysis showed that the levels of expression of VND genes change dynamically during differentiation into TE, while those of the NST genes do not (Kubo et al., 2005). Moreover, genes related to programmed cell death in the final step of TE differentiation are not induced in 35S:NST1 plants (Mitsuda et al., 2005). These findings suggest that NSTs and VNDs have similar abilities but function differently. VNDs may act as regulators of the formation of vascular vessels, while NSTs act in the formation of secondary walls in other tissues. Interestingly, promoter activities of NSTs were evident in cells differentiating into vascular vessels (Figures 1G to 1J; Mitsuda et al., 2005), suggesting that NSTs may have some function in the formation of secondary walls of vascular vessels even though no defect was observed in vascular vessels of nst1-1 nst3-1 plants. Other factors may also function redundantly with NST1 and NST3 in the formation of secondary walls of vascular vessels.
Figure 7.
Phylogenetic Tree of the NST and VND Genes, Including Putative Homologs in Poplar.
Genes that aren't underlined represent Arabidopsis genes, and those underlined represent poplar genes. Numbers at branches indicate bootstrap values from 100 trials. The NST and VND genes were clearly separated into distinct clusters.
It is curious that the NST gene can induce striated secondary wall thickenings similar to TE in the epidermis when expressed ectopically (Mitsuda et al., 2005) because NSTs are not regulators of vascular vessels but of fibers whose secondary walls are evenly distributed inside the primary cell wall (Turner and Hall, 2000). This might be because epidermal cells have a high potential to differentiate into TE in response to certain stimuli, such as wounding (Cline and Neely, 1983). Further studies are therefore required to reveal the precise functions of NSTs and VNDs in more detail.
NSTs May Be Key Regulators of Secondary Wall Synthesis during Wood Formation in Trees
Wood formation is the sum of several complex processes involving production of xylem mother cells from the cambium, sequential cell divisions, elongation of cells, formation of secondary walls, and cell death. Recent molecular and anatomical studies have suggested that these processes are not unique to woody plants but are shared with herbaceous plants, such as Arabidopsis. For example, secondary xylem of root hypocotyl of Arabidopsis is known to have a similar structure to that of the trunk of a tree (Chaffey et al., 2002). In the root hypocotyl of nst1-1 nst3-1 plants, the formation of secondary walls was completely suppressed in secondary xylem except vascular vessels (Figure 2K), suggesting that the NSTs play a pivotal role in secondary wall synthesis during wood formation. Actually, putative homologs of NST1 and NST3 are present in the genome of poplar, one of the best-characterized woody plants (Figure 7). Thus, it seems likely that a common mechanism for the control of wood formation exists in herbaceous and woody plants. Because we succeeded in manipulating the formation of secondary walls using our CRES-T system (Figure 6), identification of these genes could provide important tools for the manipulation of wood quality and for wood production by genetic engineering.
METHODS
Construction of Plasmids
The protein-coding regions of the NST3 gene were amplified from the Arabidopsis thaliana cDNA library with appropriate primers (see Supplemental Table 2 online). The 5′ upstream region of 3027 bp, which extended from the site of initiation of translation of the NST3 gene, was used for preparation of the ProNST3:GUS, ProNST3:NST3, and ProNST3:NST3SRDX gene constructs. These genes and 35S:NST3 were constructed from modified vectors derived from pGreenII0029 (Hellens et al., 2000) and p35SSRDXG (Mitsuda et al., 2006). For complementation analysis, we used genomic fragments including NST1 (9580 bp) and NST3 (5199 bp), which contained 6523 and 3069 bp of the respective promoter regions. The region corresponding to the transgene of each vector, with the exception of the pGreen-based vectors, was transferred to the pBCKH plant expression vector (Mitsuda et al., 2006) using the Gateway system (Invitrogen).
Conditions for Plant Growth and Transformation
Arabidopsis plants were grown in soil at 22°C with 16 h (long-day condition) or 8 h (short-day condition) of light daily. Unless otherwise stated, plants were grown under the long-day condition. For transformation, a T-DNA vector carrying the appropriate construct was introduced into Agrobacterium tumefaciens strain GV3101 by electroporation, and the resultant Agrobacterium was infiltrated into Arabidopsis using the floral dip method (Clough and Bent, 1998).
Assessment of the Mechanical Strength of Inflorescence Stems
We used the bottom 5 cm of inflorescence stems taller than 25 cm for measurement of Young's modulus according to a previously described method (Kojima and Yamamoto, 2004).
Examination of the Crystal State of Cellulose Microfibrils of Inflorescence Stems
The bottom region of the inflorescence stems, as described above, was used for x-ray diffraction analysis according to a previously described method (Abe and Yamamoto, 2005). Nickel-filtered Cu Kα radiation (wavelength, 0.154 nm) at 30 kV and 35 mA was used with the reflection technique.
Isolation of RNA, Microarray Experiments, and Analysis
Total RNA was isolated with Trizol as described previously (Fukuda et al., 1991) from the bottom 4 cm of the inflorescence stems of three independent plants grown under the short-day condition and with a height of between 13 and 17 cm. Microarray analyses were performed with the Arabidopsis 2 Oligo Microarray (Agilent Technologies). All microarray experiments and the analysis of data were performed as described previously (Mitsuda et al., 2005) with the exceptions summarized below. P values for differences between nst1-1 nst3-1 and wild-type plants were calculated by Welch's t test, based on a two-tailed distribution (n = 3). To minimize type-I family-wise errors in multiple and simultaneous statistical tests, we adopted a strategy for suppression of false positives. We calculated a Q-value to estimate the false discovery rate from the P value described above using QVALUE software (Storey and Tibshirani, 2003) with the default setting. We considered genes with a Q-value of <0.1 to be genes expressed at different levels in nst1-1 nst3-1 and wild-type plants. Comprehensive gene group analysis using Fisher's exact test was performed with the R program package (http://www.r-project.org/). Quantitative RT-PCR was performed as described previously (Mitsuda et al., 2005). For the analysis of NST transcripts in the mutant lines, RT-PCR was performed with appropriate primers (see Supplemental Table 2 online).
Light and Fluorescence Microscopy
For observations of lignin autofluorescence, we used a filter with the following specifications: glass, 365; dichroic mirror, 395; long-pass, 400. To observe ectopic secondary wall thickening, we cleared tissues by incubating them overnight in 70% lactic acid at 50°C. To prepare 70- to 150-μm sections of inflorescence stems and hypocotyls, we embedded the tissues in 3% agar then sectioned them on a vibrating microtome (HM-650V; Microm). Assays of GUS activity were performed with T1 or T2 transgenic plants. Plant tissues were fixed briefly, in some cases, in solution containing 0.3% formalin, 0.2% MES, pH 5.8, and 0.3 M mannitol before incubation in 100 mM sodium phosphate buffer, pH 7.0, containing 0.1% Triton X-100, 1 mM 5-bromo-4-chloro-3-indolyl-β-d-glucuronide, and 0.5 mM potassium ferricyanide at 37°C for up to 12 h. Stained stems and hypocotyls were embedded in 3% agar and sectioned. All observations by light and fluorescence microscopy were made with the Axioskop2 plus system (Carl Zeiss).
Ultrastructural Observation by Transmission Electron Microscopy
Short pieces of inflorescence stems were fixed in 30 mM HEPES buffer containing 2% paraformaldehyde and 2% glutaraldehyde then fixed in HEPES buffer containing 2% osmium tetroxide. Fixed tissues were embedded in Q651 resin (Nissin EM). Sections of 80 to 90 nm thick were post-stained with uranyl acetate and lead citrate and observed by a JEM1200EX transmission electron microscope (JEOL) at an accelerating voltage of 80 kV.
Identification of NST Homologs in Poplar
Poplar NAC genes resembling the Arabidopsis NST genes were collected using the Advanced Search tool of the Joint Genome Initiative poplar database (http://genome.jgi-psf.org/Poptr1/Poptr1.home.html) with the command, “find by homology to related protein with E-value <1.0e-20”; the database for Populus trichocarpa; and the query “At2g46770.” The 62 extracted sequences and amino acid sequences of subfamily IIb of NAC transcription factors of Arabidopsis, as defined in a previous study (Mitsuda et al., 2005), were aligned using the ClustalW program with default settings (Chenna et al., 2003). The amino acid sequences corresponding to conserved NAC domains were extracted and realigned. A phylogenetic tree was built by neighboring-joining method using ClustalW with default settings (an alignment and the sequences are shown in Supplemental Table 3 online). Bootstrap values were calculated from 100 trials. The subtree including the NST and VND genes is shown in Figure 7.
Accession Numbers and Data Deposition
NST1 and NST3 reported in this study correspond to the Arabidopsis Genome Initiative locus identifiers At2g46770 and At1g32770, respectively. Microarray data performed in this study can be found in the National Center for Biotechnology Information Gene Expression Omnibus data library under accession number GSE5187.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. The Single T-DNA–Tagged Lines of NST1 and NST3 Did Not Show an Obvious Phenotype in Woody Tissues.
Supplemental Figure 2. The Genomic Fragment of NST1 or NST3 Could Restore the Phenotype of nst1-1 nst3-1 Plants.
Supplemental Figure 3. Prolonged Cultivation Induced Slight Formation of Secondary Walls in Interfascicular Regions Even in the nst1-1 nst3-1 Plants.
Supplemental Table 1. Microarray Data from nst1-1 nst3-1 Plants.
Supplemental Table 2. Oligonucleotides Used in This Study.
Supplemental Table 3. Alignment and Sequences Used for the Alignment.
Supplementary Material
Acknowledgments
We thank the Salk Institute and the ABRC for providing seeds of the NST1 and NST3 T-DNA–tagged plants, Junko Ishida for performing the microarray experiment, and Nobuko Kawanami and Yukie Kimura for technical assistance.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Masaru Ohme-Takagi (m-takagi@aist.go.jp).
Online version contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Abe, K., and Yamamoto, H. (2005). Mechanical linkage between cellulose microfibril and the matrix substance in wood cell walls, determined by X-ray diffraction. J. Wood Sci. 51 334–338. [Google Scholar]
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657. [DOI] [PubMed] [Google Scholar]
- Brown, D.M., Zeef, L.A., Ellis, J., Goodacre, R., and Turner, S.R. (2005). Identification of novel genes in Arabidopsis involved in secondary cell wall formation using expression profiling and reverse genetics. Plant Cell 17 2281–2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaffey, N., Cholewa, E., Regan, S., and Sundberg, B. (2002). Secondary xylem development in Arabidopsis: A model for wood formation. Physiol. Plant 114 594–600. [DOI] [PubMed] [Google Scholar]
- Chenna, R., Sugawara, H., Koike, T., Lopez, R., Gibson, T.J., Higgins, D.G., and Thompson, J.D. (2003). Multiple sequence alignment with the clustal series of programs. Nucleic Acids Res. 31 3497–3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline, M.N., and Neely, D. (1983). The histology and histochemistry of the wound-healing process in geranium cuttings. J. Am. Soc. Hortic. Sci. 108 496–502. [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Emery, J.F., Floyd, S.K., Alvarez, J., Eshed, Y., Hawker, N.P., Izhaki, A., Baum, S.F., and Bowman, J.L. (2003). Radial patterning of Arabidopsis shoots by class III HD-ZIP and KANADI genes. Curr. Biol. 13 1768–1774. [DOI] [PubMed] [Google Scholar]
- Fukuda, Y., Ohme, M., and Shinshi, H. (1991). Gene structure and expression of a tobacco endochitinase gene in suspension-cultured tobacco cells. Plant Mol. Biol. 16 1–10. [DOI] [PubMed] [Google Scholar]
- Groover, A.T. (2005). What genes make a tree a tree? Trends Plant Sci. 10 210–214. [DOI] [PubMed] [Google Scholar]
- Hellens, R.P., Edwards, E.A., Leyland, N.R., Bean, S., and Mullineaux, P.M. (2000). pGreen: A versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol. Biol. 42 819–832. [DOI] [PubMed] [Google Scholar]
- Hiratsu, K., Matsui, K., Koyama, T., and Ohme-Takagi, M. (2003). Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis. Plant J. 34 733–739. [DOI] [PubMed] [Google Scholar]
- Jones, L., Ennos, A.R., and Turner, S.R. (2001). Cloning and characterization of irregular xylem4 (irx4): A severely lignin-deficient mutant of Arabidopsis. Plant J. 26 205–216. [DOI] [PubMed] [Google Scholar]
- Keijzer, C.J. (1987). The processes of anther dehiscence and pollen dispersal. The opening mechanism of longitudinally dehiscing anthers. New Phytol. 105 487–498. [DOI] [PubMed] [Google Scholar]
- Ko, J.H., Han, K.H., Park, S., and Yang, J. (2004). Plant body weight-induced secondary growth in Arabidopsis and its transcription phenotype revealed by whole-transcriptome profiling. Plant Physiol. 135 1069–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima, Y., and Yamamoto, H. (2004). Properties of the cell wall constituents in relation to the longitudinal elasticity of wood (part 2). Origin of the moisture dependency of the longitudinal elasticity of wood. Wood Sci. Technol. 37 427–434. [Google Scholar]
- Kubo, M., Udagawa, M., Nishikubo, N., Horiguchi, G., Yamaguchi, M., Ito, J., Mimura, T., Fukuda, H., and Demura, T. (2005). Transcription switches for protoxylem and metaxylem vessel formation. Genes Dev. 19 1855–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuda, N., Hiratsu, K., Todaka, D., Nakashima, K., Yamaguchi-Shinozaki, K., and Ohme-Takagi, M. (2006). Efficient production of male and female sterile plants by expression of a chimeric repressor in Arabidopsis and rice. Plant Biotechnol. J. 4 325–332. [DOI] [PubMed] [Google Scholar]
- Mitsuda, N., Seki, M., Shinozaki, K., and Ohme-Takagi, M. (2005). The NAC transcription factors NST1 and NST2 of Arabidopsis regulate secondary wall thickenings and are required for anther dehiscence. Plant Cell 17 2993–3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muzac, I., Wang, J., Anzellotti, D., Zhang, H., and Ibrahim, R.K. (2000). Functional expression of an Arabidopsis cDNA clone encoding a flavonol 3-O-methyltransferase and characterization of the gene product. Arch. Biochem. Biophys. 375 385–388. [DOI] [PubMed] [Google Scholar]
- Plomion, C., Leprovost, G., and Stokes, A. (2001). Wood formation in trees. Plant Physiol. 127 1513–1523. [PMC free article] [PubMed] [Google Scholar]
- Sawa, S., Demura, T., Horiguchi, G., Kubo, M., and Fukuda, H. (2005). The ATE genes are responsible for repression of transdifferentiation into xylem cells in Arabidopsis. Plant Physiol. 137 141–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid, M., Davison, T.S., Henz, S.R., Pape, U.J., Demar, M., Vingron, M., Scholkopf, B., Weigel, D., and Lohmann, J.U. (2005). A gene expression map of Arabidopsis thaliana development. Nat. Genet. 37 501–506. [DOI] [PubMed] [Google Scholar]
- Sibout, R., Eudes, A., Mouille, G., Pollet, B., Lapierre, C., Jouanin, L., and Seguin, A. (2005). CINNAMYL ALCOHOL DEHYDROGENASE-C and -D are the primary genes involved in lignin biosynthesis in the floral stem of Arabidopsis. Plant Cell 17 2059–2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spence, J., Vercher, Y., Gates, P., and Harris, N. (1996). Pod shatter in Arabidopsis thaliana, Brassica napus and B. juncea. J. Microsc. 181 195–203. [Google Scholar]
- Storey, J.D., and Tibshirani, R. (2003). Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA 100 9440–9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbert, P.B., Adler, H.T., Parks, D.W., and Comai, L. (1995). The REVOLUTA gene is necessary for apical meristem development and for limiting cell divisions in the leaves and stems of Arabidopsis thaliana. Development 121 2723–2735. [DOI] [PubMed] [Google Scholar]
- Taylor, N.G., Howells, R.M., Huttly, A.K., Vickers, K., and Turner, S.R. (2003). Interactions among three distinct CesA proteins essential for cellulose synthesis. Proc. Natl. Acad. Sci. USA 100 1450–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, N.G., Scheible, W.R., Cutler, S., Somerville, C.R., and Turner, S.R. (1999). The irregular xylem3 locus of Arabidopsis encodes a cellulose synthase required for secondary cell wall synthesis. Plant Cell 11 769–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokimatsu, T., Sakurai, N., Suzuki, H., Ohta, H., Nishitani, K., Koyama, T., Umezawa, T., Misawa, N., Saito, K., and Shibata, D. (2005). KaPPA-View. A web-based analysis tool for integration of transcript and metabolite data on plant metabolic pathway maps. Plant Physiol. 138 1289–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner, S.R., and Hall, M. (2000). The gapped xylem mutant identifies a common regulatory step in secondary cell wall deposition. Plant J. 24 477–488. [DOI] [PubMed] [Google Scholar]
- Zhao, C., Craig, J.C., Petzold, H.E., Dickerman, A.W., and Beers, E.P. (2005). The xylem and phloem transcriptomes from secondary tissues of the Arabidopsis root-hypocotyl. Plant Physiol. 138 803–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, R., Pena, M.J., Zhou, G.K., Nairn, C.J., Wood-Jones, A., Richardson, E.A., Morrison, W.H., Darvill, A.G., York, W.S., and Ye, Z.H. (2005). Arabidopsis fragile fiber8, which encodes a putative glucuronyltransferase, is essential for normal secondary wall synthesis. Plant Cell 17 3390–3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, R., Taylor, J.J., and Ye, Z.H. (1997). Disruption of interfascicular fiber differentiation in an Arabidopsis mutant. Plant Cell 9 2159–2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, R., and Ye, Z.H. (1999). IFL1, a gene regulating interfascicular fiber differentiation in Arabidopsis, encodes a homeodomain-leucine zipper protein. Plant Cell 11 2139–2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, R., and Ye, Z.H. (2001). Alteration of auxin polar transport in the Arabidopsis ifl1 mutants. Plant Physiol. 126 549–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
NOTE ADDED IN PROOF
- While this manuscript was under review, Zhong et al. (2006) reported that SND1 plays a significant role in secondary wall synthesis in interfascicular fibers. SND1 is the same gene as NST3 described in our study.
- Zhong, R., Demura, T., and Ye, Z.H. (2006). SND1, a NAC domain transcription factor, is a key regulator of secondary wall synthesis in fibers of Arabidopsis. Plant Cell 18 3158–3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.