Abstract
The secondary cell wall in higher plants consists mainly of cellulose, lignin, and xylan and is the major component of biomass in many species. The Arabidopsis thaliana irregular xylem8 (irx8) mutant is dwarfed and has a significant reduction in secondary cell wall thickness. IRX8 belongs to a subgroup of glycosyltransferase family 8 called the GAUT1-related gene family, whose members include GAUT1, a homogalacturonan galacturonosyltransferase, and GAUT12 (IRX8). Here, we use comparative cell wall analyses to show that the irx8 mutant contains significantly reduced levels of xylan and homogalacturonan. Immunohistochemical analyses confirmed that the level of xylan was significantly reduced in the mutant. Structural fingerprinting of the cell wall polymers further revealed that irx8 is deficient in glucuronoxylan. To explore the biological function of IRX8, we crossed irx8 with irx1 (affecting cellulose synthase 8). The homozygous irx1 irx8 exhibited severely dwarfed phenotypes, suggesting that IRX8 is essential for cell wall integrity during cellulose deficiency. Taken together, the data presented show that IRX8 affects the level of glucuronoxylan and homogalacturonan in higher plants and that IRX8 provides an important link between the xylan polymer and the secondary cell wall matrix and directly affects secondary cell wall integrity.
INTRODUCTION
The vascular bundles, harboring phloem and xylem elements, facilitate nutrient and water distribution in higher plants (Ye, 2002). The xylem elements transport water from the root to the aerial parts of the plant. The rapid movement of water through the xylem elements creates an internal negative pressure. To withstand these forces, the xylem cells deposit a secondary cell wall matrix, mainly consisting of cellulose, lignin, xylan, and a variety of glycoproteins. The deposition of the secondary wall occurs when the cells have reached their terminal morphological state (Turner et al., 2001). This lignified secondary wall is the most abundant element of biomass produced by many plants and also constitutes the major mechanical support for plant growth.
Defects in the constituents of the secondary cell wall may result in an inward collapse of individual xylem cells (Turner and Somerville, 1997; Turner et al., 2001). Genetic screens for such phenotypes in Arabidopsis thaliana have revealed several genes essential for xylem development, referred to as irregular xylem (irx) mutants (Turner and Somerville, 1997) or fragile fiber (fra) mutants (Zhong et al., 2001). Three of the mutants, irx1, irx3, and irx5, have defects in cellulose synthase (CESA) subunits of the secondary cell wall cellulose complex (Turner and Somerville, 1997; Taylor et al., 1999). Several other irx and fra mutants are associated with cellulose and lignin deposition and with signal transduction–associated processes (Jones et al., 2001; Szyjanowicz et al., 2004; Zhong et al., 2005a).
Xylan is the main hemicellulosic component of the secondary cell wall in dicot plants and the second most abundant biopolymer in plants (Ebringerova and Heinze, 2000). Arabidopsis is believed to mainly contain xylan polymers consisting of a β-(1,4)–linked xylose backbone, decorated with α-d-glucuronic acid and/or 4-O-methyl-α-d-glucuronic acid. These residues normally occupy O-2 positions at every 6 to 12 xylose residues. The xylose backbone may also be decorated with l-arabinose and/or acetylated on C-2 or C-3 positions (Ebringerova and Heinze, 2000). The role of xylan in the secondary wall is not clear. However, it has been suggested that xylans coat the cellulose microfibrils (Awano et al., 2002) and may influence the helicoidal orientation of the microfibrils (Reis and Vian, 2004). Biochemical analyses showed that several species, such as wheat (Triticum aestivum; Porchia et al., 2002), pea (Pisum sativum; Waldron and Brett, 1983; Baydoun et al., 1989), poplar (Populus spp), and sycamore (Acer pseudoplatanus; Dalessandro and Northcote, 1981), harbor xylosyltransferase activities thought to be responsible for xylan formation. However, the genes corresponding to these activities remain elusive. The only previously characterized mutations known to have major effects on xylan synthesis are fra8, which encodes a putative glucuronyltransferase associated with glucuronoxylan synthesis during secondary wall formation (Zhong et al., 2005b), and irx9, which encodes a family 43 putative glycosyltransferase of unknown function (Bauer et al., 2006).
Arabidopsis has 40 putative glycosyltransferases that have been assigned to family 8 (GT8) in the CAZY classification system and that fall into at least two evolutionarily distinct groups (Sterling et al., 2006). One of these groups within GT8 is the 25-member GAUT1-related superfamily, named after galacturonosyltransferase1 (GAUT1). GAUT1 is a homogalacturonan (HG) α1,4-galacturonosyltransferase that elongates HG oligosaccharides by catalyzing the transfer of galacturonic acid (GalA) from UDP-GalA onto the HG oligosaccharides. Two mutants with defects in other members of the GAUT1-related superfamily, parvus (Lao et al., 2003; Shao et al., 2004) and quasimodo1 (qua1) (Bouton et al., 2002), have been reported. Mutations in QUA1 (GAUT8) resulted in a dwarfed plant phenotype, and the corresponding gene product was suggested to be involved in HG synthesis (Bouton et al., 2002). However, the qua1 mutant displayed a significant decrease in both HG and xylan synthase activity (Orfila et al., 2005). The parvus mutant also displayed a dwarfed phenotype, which was more pronounced under low humidity, and displayed reduced levels of branching in rhamnogalacturonan I (RG-I) linkages (Lao et al., 2003). The mutant, furthermore, displayed a marked reduction in β-(1,4)–linked xylose linkages.
The majority of the irx mutants have been identified based on genetic screens for collapsed xylem cells (Turner et al., 2001). However, recent reports using extensive microarray data mining and coexpression analyses have identified at least eight novel gene products involved in secondary cell wall deposition (Brown et al., 2005; Persson et al., 2005). Several of these candidates were also implicated in poplar wood formation, monitored through microarray experiments (Aspeborg et al., 2005). Here, we describe the characterization of the irx8 mutant, which carries a defect in a member (At5g54690; GAUT 12) of the GAUT1-related gene family. Biochemical analyses of fractionated cell wall material, together with immunolabeling, suggest that irx8 is deficient in glucuronoxylan and in a subfraction of HG. Genetic crosses of irx8 with other secondary cell wall mutants support this conclusion and provide evidence that IRX8 plays an essential role in secondary cell wall integrity.
RESULTS
irx8 Has Collapsed Xylem Vessels and Displays a Dwarfed Phenotype
IRX8 is coexpressed with the secondary cell wall CESA complex genes (CESA4, 7, and 8) over ∼500 microarray data sets (Persson et al., 2005). To evaluate the function of IRX8, five independent homozygous T-DNA insertion lines with insertions in or near the IRX8 gene were established (Figure 1A). At least two of the lines (irx8-1 and ixr8-2) were complete nulls for the expression of IRX8 as assessed by RT-PCR (data not shown). Significant reduction in IRX8 transcript level was observed in the other three lines (irx8-3, -4, and -5), which harbor T-DNA inserts in the 5′ untranslated region (UTR) and introns of the gene, respectively (Figure 1A). All mutant lines displayed very similar phenotypes, suggesting that the irx8-3, -4, and -5 alleles may have defects in translation of the transcripts produced.
Figure 1.
Phenotypic Features of irx8 Mutant Plants.
(A) Relative positions of T-DNA insertions for the irx8 mutants.
(B) to (E) Alterations in growth of irx8 mutant plants.
(B) Seven-week-old plants showing shorter inflorescence stems in irx8 mutants (right) compared with the wild type (left). Bar = 5 cm.
(C) Seven-week-old rosette leaves from irx8-1 (bottom) are significantly smaller compared with wild-type (top) leaves.
(D) irx8 plants had smaller flowers with short anthers (arrow) and reduced amounts of pollen. The female organs appeared normal.
(E) The siliques from irx8 mutants were significantly smaller and contained no seeds.
(F) Hand-cut transverse sections from wild-type (left) and irx8-1 (right) stems stained with Toluidine blue. Arrows indicate collapsed xylem vessels. Ph, phloem; XE, xylem.
(G) Transverse root sections (1-μm thick) from 7-week-old wild type (left) and irx8 (right) stained with Toluidine blue. Arrows indicate collapsed xylem vessels. Bars = 10 μm.
The mutant plants had stunted growth and smaller leaves, siliques, and flowers compared with the wild type (Figures 1B to 1E). Interestingly, although the leaf size was considerably smaller than the wild-type leaf, the trichomes of the mutants were the same size (data not shown). In accordance with the reduced silique size (Figure 1E), no seeds could be obtained from the homozygous mutant plants. Homozygous mutants were therefore produced as segregants from heterozygous parents for phenotypic analyses. The semisterility of the mutant plants may be explained by a shorter anther filament and reduced number of pollen (Figure 1D; data not shown). The irx8 mutant plants also exhibited collapsed xylem vessels in roots and stems (Figures 1F and 1G), suggesting a decrease in xylem wall strength in the mutants.
The organization of the xylem tissue in irx8 roots was dramatically altered. The xylem files form a closely packed array of cell files in wild-type roots, leading to the regular occurrence of tripartite cell corners between xylem cell files (Figure 1G). By contrast, the central cylinder of irx8 roots was disorganized, and the xylem files did not form a closely packed array. As a consequence, we were unable to detect tripartite xylem cell corners in the mutant (Figure 1G).
The development of the xylem vessels in previously characterized irx mutants (irx1 to 5) appears to be normal until maturation of the xylem elements (Turner and Somerville, 1997). To investigate if the irx8 mutants exhibited similar defects during xylem development, regions of stems with different degrees of xylem maturation were sectioned and analyzed (Figures 2A to 2I). Similar to the irx1 mutants, the mature xylem vessels were deformed in the irx8 mutant. However, in contrast with irx1, the irx8 mutants also exhibited deformations in initiating xylem cells (i.e., the protoxylem cells; cf. Figures 2C, 2F, and 2I). These observations suggest that IRX8 plays an important role for secondary cell wall integrity throughout xylem maturation. In addition, the irx8 mutants contain ∼50% more xylem cells than wild-type stems at a comparable developmental stage. This may reflect a compensatory response in the mutants, presumably to maintain sufficient water-transporting capacity.
Figure 2.
Early Onset of Irregular Xylem Phenotype in irx8.
Hand-cut transverse sections from different developmental points of the stem for the wild type, irx8-1, and irx1 stained with Toluidine blue. Insets show enlarged xylem vessels. Arrows indicate collapsed xylem vessels. Bars = 20 μm.
(A) to (C) Sections from base, middle, and top of wild-type stems, respectively.
(D) to (F) Sections from base, middle, and top of irx8-1 stems, respectively.
(G) to (I) Sections from base, middle, and top of irx1 stems, respectively.
To examine the cell walls surrounding the xylem cells in more detail, 7-week-old basal stem and root sections were examined using transmission electron microscopy. Figure 3 shows that the xylem cells are significantly smaller in the mutant compared with the wild type. The cell walls surrounding the xylem vessels are also severely reduced in thickness in the mutants. Similar to previously characterized irx1, ixr3, and irx5 mutants (Turner and Somerville, 1997), the irx8 mutants show an uneven distribution of cell wall deposition compared with the wild type (Figure 3). Additionally, the region that separates cells, consisting of residual primary cell walls, is wider and exhibits less contrast in the mutant compared with the wild type (Figure 3), possibly reflecting differences in the composition of cell wall polymers that make up this region.
Figure 3.
Reduction in Cell Wall Thickness and Cell Size in irx8 Xylem Cells.
Transmission electron microscopy of cell walls from the metaxylem region of wild-type and irx8-1 mature stems and roots. Bars = 2 μm.
(A) Wild-type (left) and irx8-1 (right) stem sections. PW, residual primary cell walls.
(B) Wild-type (left) and irx8-1 (right) root sections.
Expression of IRX8
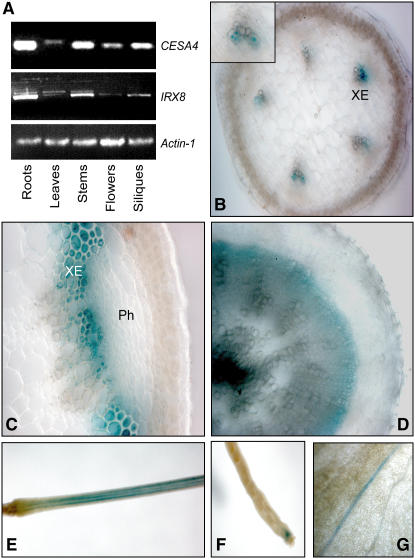
To examine where and when IRX8 is expressed, we isolated RNA from roots, leaves, stems, flowers, and siliques from 7-week-old plants and assessed the relative expression levels using semiquantitative RT-PCR (Figure 4A). Roots, stems, and siliques contained significantly higher levels of IRX8 transcripts than the other tissues, indicating that IRX8 is highly expressed where secondary walls are deposited. CESA4 showed a very similar transcript distribution, corroborating coexpression of the IRX8 and CESA4 genes (Figure 4A).
Figure 4.
Expression of IRX8.
(A) Expression of IRX8 and one of the secondary wall cellulose synthase subunits, CESA4, in different tissues assessed by RT-PCR. Actin was used as control. All tissues are from 7-week-old plants.
(B) to (G) Expression patterns of the IRX8 gene assessed through an IRX8 promoter:GUS construct. The GUS expression is indicated by blue color.
(B) Cross section of a 7-week-old stem at an expanding internode showing GUS staining at the developing metaxylem. Inset shows enlarged xylem vessels. XE, xylem vessels.
(C) Cross section of a 7-week-old stem at the base showing GUS staining in both interfascicular fibers and xylem. Ph, phloem.
(D) Cross section of 7-week-old root showing GUS staining associated with developing secondary xylem.
(E) Part of inflorescence stem carrying siliques showing strong GUS expression.
(F) Tip of silique.
(G) Magnification of rosette leaf showing GUS staining associated with vascular strands.
To investigate IRX8 expression in more detail, we produced transgenic plants in which the β-glucuronidase (GUS) reporter gene was placed under the control of the 1.4-kb promoter region upstream of IRX8. GUS activity was observed in the vascular tissue of stems, roots, leaves, and floral tissues (Figures 4B to 4G), corroborating the hypothesis that IRX8 is involved in secondary cell wall synthesis. In elongating internodes, GUS signals were only present in developing xylem cells, where secondary wall material is being deposited (Figure 4B). In stems where rapid elongation was no longer occurring, GUS staining was present in both xylem cells and interfascicular fiber cells (Figure 4C). These cells are surrounded by thickened secondary cell wall deposits, consistent with a role for IRX8 in secondary wall formation. A similar GUS pattern was observed in root sections (Figure 4D), where GUS expression was prevalent in tissues undergoing secondary growth. Strong GUS activity was also found in inflorescence stems carrying siliques (Figure 4E).
To confirm that all essential elements were present in the promoter:GUS construct, we replaced the GUS gene with an IRX8 cDNA and transformed it into the irx8-1 and irx8-2 mutants. There were no significant differences in growth between transformed mutant plants and wild-type plants, indicating that the promoter targeted IRX8 expression to the correct locations. In addition, the irregular xylem phenotype was completely rescued in the transformed plants (see Supplemental Figures 1A to 1D online). These observations suggest that the residual IRX8 transcript levels in the 5′ UTR and intron-located irx8 T-DNA mutants most likely do not result in correctly translated proteins.
Compositional Analysis of Total Cell Wall Material in irx8
The collapsed xylem phenotype suggests an alteration in the composition of the cell walls associated with the xylem elements. Fourier transform infrared (FTIR) spectroscopy and cellulose and sugar analyses were therefore initially undertaken to analyze different aspects of the cell wall composition. FTIR spectra of total cell wall material from 7-week-old mutants and wild-type stems and roots were collected and analyzed using principal component analysis. The spectrum for irx8 showed a clear separation from wild-type spectra based on principal component 1, indicating clear alterations in the composition of the mutant cell walls (data not shown). The corresponding loadings showed that spectra from both irx8 roots and stems have a negative correlation with peaks at 985, 1043, and 1173 cm−1, corresponding to potential alterations in noncellulosic polymers compared with the wild type (data not shown; Mouille et al., 2003; Persson et al., 2005). Previous studies have shown that irx8 contains ∼25% less cellulose compared with the wild type (Brown et al., 2005; Persson et al., 2005). However, irx1 and irx3 plants typically have an 80% reduction in cellulose compared with the wild type but exhibit rather minor growth defects (Turner and Somerville, 1997). Thus, these data suggest that the severity of the irx8 phenotype can only partially be attributed to the reduction in cellulose deposition.
To estimate whether the irx8 mutations affect synthesis of specific polysaccharides, we analyzed the neutral sugar and uronic acid composition of cell walls from stems and roots. Table 1 shows that the irx8-1 and irx8-2 mutants have slightly higher levels of arabinose, rhamnose, mannose, and galactose compared with the wild type in both stems and roots. However, the main difference was a >50% reduction of xylose in the cell walls isolated from mutant stems and roots compared with the wild type (Table 1).
Table 1.
Neutral Monosaccharide Composition of Cell Walls from irx8 Stems and Roots
| Rha | Fuc | Ara | Xyl | Man | Gal | Glu | Uronic Acids | ||
|---|---|---|---|---|---|---|---|---|---|
| Stem | Col-0 | 6.72 ± 0.56 | 0.89 ± 0.43 | 8.24 ± 0.16 | 93.53 ± 5.25 | 14.32 ± 0.81 | 20.61 ± 1.20 | 55.83 ± 0.82 | 131.81 ± 10.19 |
| irx8-1 | 9.20 ± 0.54 | 0.85 ± 0.11 | 19.09 ± 0.08 | 40.02 ± 1.84 | 27.67 ± 1.05 | 38.04 ± 1.27 | 69.72 ± 1.66 | 142.46 ± 9.45 | |
| irx8-2 | 10.52 ± 0.25 | 1.58 ± 0.35 | 19.14 ± 0.01 | 36.58 ± 1.19 | 22.62 ± 0.61 | 36.38 ± 2.11 | 62.53 ± 1.75 | 123.63 ± 14.18 | |
| Root | Col-0 | 4.27 ± 0.24 | 0.85 ± 0.05 | 10.85 ± 0.56 | 42.37 ± 2.43 | 4.99 ± 0.35 | 22.55 ± 2.41 | 40.32 ± 3.33 | 102.57 ± 4.44 |
| irx8-1 | 4.68 ± 0.24 | 0.96 ± 0.07 | 14.93 ± 0.99 | 18.42 ± 1.23 | 6.13 ± 0.40 | 24.79 ± 1.33 | 50.01 ± 2.81 | 123.49 ± 3.15 | |
| irx8-2 | 5.00 ± 0.27 | 1.09 ± 0.05 | 14.33 ± 0.89 | 13.49 ± 0.94 | 5.96 ± 0.45 | 26.20 ± 1.35 | 53.83 ± 1.50 | 115.38 ± 14.00 |
Neutral sugar composition from 7-week-old roots and stems of irx8-1, irx8-2, and wild-type plants. The amounts of sugar are presented as mean values of micrograms of sugar/milligrams of dry weight (n = 5) ± se.
Compositional Analysis of Fractionated Cell Wall Material from irx8 Stems
To determine if the changes in polysaccharide composition were associated with a specific type of wall polymer, we treated total cell walls from 7-week-old stems sequentially with enzymes and solvents to preferentially enrich for specific classes of polymers. The recovered fractions were analyzed for glycosyl residue composition by gas chromatography–mass spectrometry (GC-MS) of trimethylsilyl derivatives, a method that allows quantification of specific uronic acids and neutral sugars (York et al., 1985).
Endopolygalacturonase (EPG) and pectin methylesterase (PME)/EPG treatment of cell walls released glycans containing primarily galactose, GalA, xylose, and mannose (Table 2). Both treatments resulted in an increased amount of released galactose, arabinose, and rhamnose in the irx8 mutant alleles compared with the wild type. The increases in galactose content persisted in all of the fractions (Table 2). Interestingly, the released amount of GalA in both EPG and PME/EPG fractions was significantly reduced for the irx8 mutant alleles compared with the wild type. These results show that irx8 contains reduced levels of enzyme accessible nonesterified and methylesterified HG, indicating a change in wall structure. It is interesting to note that the EGTA/Na2CO3-extracted wall fraction does not show as dramatic a reduction in GalA, suggesting that a subfraction of HG was affected in irx8 walls.
Table 2.
Glycosyl Residue Composition of Total and Fractionated Cell Walls from 7-Week-Old Wild-Type (Col-0), irx8-1, and irx8-5 Stems
| Residue | Ara | Rha | Fuc | Xyl | GalA | Man | Gal | Glc | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Cell Wall Fractiona | mass %b | mol % | mol % | mol % | mol % | mol % | mol % | mol % | mol % | |
| EPG | Col-0 | 7.8 | 2.5 | 1.5 | 0.0 | 18.6 | 27.2 | 10.3 | 40.0 | –c |
| irx8-1 | 20.6 | 9.9 | 3.0 | 0.1 | 3.5 | 1.5 | 9.6 | 72.0 | – | |
| irx8-5 | 19.9 | 15.4 | 6.4 | 0.4 | 3.1 | 1.5 | 8.4 | 64.8 | – | |
| PME/EPG | Col-0 | 8.3 | 0.5 | 1.5 | 0.0 | 3.9 | 65.3 | 1.5 | 27.4 | – |
| irx8-1 | 7.5 | 6.7 | 5.1 | 0.2 | 8.8 | 3.9 | 9.5 | 65.5 | – | |
| irx8-5 | 8.7 | 7.4 | 8.5 | 0.4 | 11.7 | 6.5 | 8.8 | 56.4 | – | |
| EGTA/Na2CO3 sol | Col-0 | 33.3 | 6.6 | 6.6 | 0.3 | 1.6 | 81.0 | 1.4 | 2.3 | 0.0 |
| irx8-1 | 42.1 | 9.0 | 6.6 | 0.1 | 2.6 | 75.5 | 0.2 | 5.7 | 0.0 | |
| irx8-5 | 48.2 | 14.2 | 10.4 | 0.0 | 4.7 | 67.3 | 0.0 | 3.1 | 0.0 | |
| 4 n KOH sol | Col-0 | 25.1 | 2.1 | 2.0 | 0.3 | 72.8 | 7.8 | 0.0 | 7.8 | 6.7 |
| irx8-1 | 15.6 | 4.4 | 2.7 | 0.7 | 30.9 | 12.2 | 6.5 | 25.8 | 16.4 | |
| irx8-5 | 9.9 | 4.7 | 2.7 | 0.7 | 30.5 | 11.0 | 5.9 | 29.8 | 14.3 | |
| 4 n KOH insol | Col-0 | 25.5 | 6.8 | 4.2 | 0.2 | 14.4 | 21.0 | 3.6 | 15.4 | 34.1 |
| irx8-1 | 15.6 | 4.3 | 4.3 | 0.2 | 14.8 | 13.2 | 4.9 | 25.0 | 32.9 | |
| irx8-5 | 9.9 | 2.6 | 3.4 | 0.0 | 8.8 | 21.5 | 3.6 | 21.1 | 38.8 | |
| 4 n KOH insol-SHd | Col-0 | 2.7 | 0.8 | 0.3 | 3.6 | 1.6 | 10.0 | 2.7 | 78.0 | |
| irx8-1 | 3.3 | 0.7 | 0.1 | 3.7 | 2.8 | 8.8 | 5.7 | 74.5 | ||
| irx8-5 | 3.5 | 1.5 | 0.1 | 3.8 | 2.9 | 6.7 | 5.7 | 75.3 |
Total alcohol insoluble walls were extracted from stems of Arabidopsis wild type (Col-0), irx8-1, and irx8-5 and sequentially fractionated first by wall-specific enzymes (Aspergillus niger), EPG, and then a combination of PME (A. niger) and EPG (PME/EPG). Remaining walls were subjected to EGTA-sodium carbonate (EGTA/Na2CO3) and finally 4 n potassium hydroxide (4 n KOH sol/4 n KOH insol) extraction. Soluble components were neutralized and dialyzed against water, and the tetramethylsilane (TMS) derivatives were analyzed by GC-MS for glycosyl residue composition.
The percentage of total wall mass recovered for a fraction.
Glucose values were not included in EPG and PME/EPG fractions as a result of α-amylase digestion immediately prior to cell wall degrading enzyme digestions. Glucose was included or not detected in all other fractions.
An aliquot of the 4 n KOH insol fraction was subjected to Saeman hydrolysis followed by TMS composition. The Saeman hydrolysis procedure hydrolyzes crystalline cellulose, thereby allowing the detection of glucose attributed to cellulose, whereas standard TMS hydrolysis does not.
Treatment of cell walls with 4 n KOH released glycans containing primarily xylose with lesser amounts of GalA, mannose, galactose, glucose, arabinose, and rhamnose (Table 2). We found a major reduction in the amount of xylose solubilized by 4 n KOH treatment in the mutant walls. This suggests that a xylan or xyloglucan polymer is affected in irx8.
The insoluble material remaining after 4 n KOH treatment from both wild-type and mutant walls consisted mainly of glucose, with small amounts of additional glycosyl residues (Table 2). Presumably, the <25% of nonglucose sugars in the 4 n KOH pellet represent noncellulosic carbohydrates that remain bound to the crystalline cellulose during wall fractionation. No major differences in the composition of the noncellulosic carbohydrate in the pellet fraction from wild-type versus mutant walls were observed.
Glycosyl Linkage Analysis of Fractionated Cell Wall Material from irx8 Stems
To determine structural changes in polysaccharides in irx8 mutant walls, glycosyl residue linkage analyses were performed on the fractionated material generated above (Table 3). The mutant had reduced levels of 4-GalAp and 3,4-GalAp, while terminal-GalAp was increased compared with the wild type in the EPG released fraction. The increase in N-terminal GalAp does not, however, make up for losses in 4-linked or 3,4-linked GalAp. Therefore, it is likely that HG is the wall component reduced in this fraction. Terminal Xylp, as well as 4-Xylp, was also reduced in the EPG released fraction, compared with the wild type, confirming that less xylan was solubilized by EPG treatment of mutant walls compared with the wild type. The 2-linked Rhap, a component of RG-I, was also reduced in the EPG released fraction for the mutants compared with the wild type. There were also notable increases in T-Galp, 2,6-Galp, 3,6-Galp, 3,4,6-Galp, and 4-Manp in irx8 mutants compared with the wild type, corroborating the results from the glycosyl compositional analysis.
Table 3.
Glycosyl Linkage of Fractionated Cell Walls from the Wild Type (Col-0), irx8-1, and irx8-5
| Fractionsa
|
EPG Released
|
EGTA/Na2CO3 Soluble
|
4 n KOH Soluble
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| Residueb | Col-0 | irx8-1 | irx8-5 | Col-0 | irx8-1 | irx8-5 | Col-0 | irx8-1 | irx8-5 |
| T-GalAp | NDc | 7.2 | 1.6 | ND | ND | ND | ND | ND | ND |
| 4-GalAp | 17.5 | ND | ND | 11.2 | 10.5 | 9.7 | 5.5 | 1.5 | 3.5 |
| 2,4-GalAp | ND | ND | ND | 3.1 | 3.2 | 1.0 | 1.5 | 0.9 | 0.9 |
| 3,4-GalAp | 5.3 | ND | ND | 4.2 | 5.0 | 2.2 | 2.1 | 1.2 | 1.4 |
| 2,3,4-GalAp | ND | ND | ND | 5.4 | 6.6 | 1.7 | 1.5 | 1.4 | 1.1 |
| T-Rhap | 2.8 | 1.7 | 2.2 | 1.0 | 1.2 | 1.0 | 0.5 | 0.6 | 0.2 |
| 2-Rhap | 6.8 | 3.2 | 3.7 | 7.8 | 8.4 | 11.3 | 3.2 | 1.7 | 1.9 |
| 3-Rhap | ND | ND | ND | 2.2 | 1.8 | 2.2 | 1.3 | 1.1 | 0.3 |
| 2,3-Rhap | 2.6 | 2.1 | ND | 3.2 | 3.2 | 2.5 | 1.8 | 0.9 | 0.5 |
| 3,4-Rhap | 8.0 | 3.6 | 1.5 | 1.9 | 1.2 | 1.5 | 0.8 | 0.6 | 0.2 |
| 2,4-Rhap | ND | ND | ND | 5.0 | 4.2 | 6.1 | ND | ND | ND |
| 2,3,4-Rhap | ND | ND | ND | 6.8 | 5.0 | 5.6 | 2.1 | 2.5 | 1.5 |
| T-Fucp | ND | ND | ND | 0.8 | 0.6 | 0.8 | 0.6 | 0.6 | 0.4 |
| 3,4-Fucp | 1.7 | 2.1 | 1.6 | 1.8 | 1.0 | 1.4 | 1.4 | 0.9 | 0.5 |
| 2,4-Fucp | ND | ND | ND | 1.7 | ND | ND | ND | 0.2 | 0.1 |
| T-Araf | 4.3 | 6.6 | 10.6 | 3.4 | 7.4 | 5.5 | 2.2 | 4.9 | 2.4 |
| T-Arap | 3.0 | 1.6 | 1.4 | 1.3 | 1.2 | 0.8 | 0.6 | 0.7 | 0.5 |
| 3-Araf | 1.6 | 2.5 | 2.8 | 0.9 | 1.0 | 1.0 | 0.7 | 1.0 | 0.4 |
| 5-Araf | 5.0 | 7.3 | 6.4 | 5.5 | 10.6 | 6.5 | 2.6 | 6.1 | 3.2 |
| 2,4-Arap or 2,5-Araf | ND | ND | ND | 3.2 | 3.7 | 4.0 | ND | ND | ND |
| 3,4-Arap or 3,5-Araf | ND | ND | ND | ND | 1.8 | 1.2 | ND | 2.8 | 2.6 |
| 2,3,4-Arap or 2,3,5-Araf | ND | ND | ND | 6.1 | 4.2 | 8.1 | ND | 2.5 | 3.8 |
| T-Xylp | 3.0 | 1.3 | 2.2 | 0.8 | 1.3 | 0.9 | 2.7 | 2.0 | 1.4 |
| 4-Xylp | 10.8 | 6.2 | 9.2 | 1.0 | 1.1 | 1.1 | 32.0 | 19.6 | 15.5 |
| 2,4-Xylp | ND | ND | ND | ND | ND | ND | 5.5 | 6.5 | 3.4 |
| 3,4-Xylp | ND | ND | ND | 0.7 | 0.7 | 0.6 | 3.3 | 5.7 | 1.8 |
| T-Galp | 3.5 | 6.0 | 14.6 | 4.4 | 4.3 | 7.2 | 4.3 | 3.5 | 4.1 |
| 2-Galp | ND | ND | 0.8 | 0.5 | 0.5 | ND | 0.9 | 1.1 | 1.6 |
| 3-Galp | ND | ND | ND | 2.4 | 2.1 | 3.4 | ND | ND | ND |
| 4-Galp | ND | ND | 3.6 | 6.6 | 3.8 | 7.3 | 3.2 | 2.9 | 4.6 |
| 6-Galp | ND | ND | ND | 3.1 | 2.6 | 4.3 | 1.9 | 2.0 | 8.9 |
| 2,6-Galp | ND | 2.2 | 1.4 | 0.4 | ND | ND | ND | ND | ND |
| 3,6-Galp | ND | 8.4 | 5.7 | 0.6 | 0.4 | ND | 0.8 | 0.8 | 1.4 |
| 3,4,6-Galp | ND | 1.5 | 1.7 | ND | ND | ND | ND | ND | ND |
| T-Manp | 3.2 | 4.8 | 6.0 | ND | ND | ND | 0.3 | 0.5 | 0.4 |
| 4-Manp | 6.9 | 16.4 | 13.8 | ND | ND | ND | 5.5 | 5.3 | 11.4 |
| 2,4-Manp | 14.1 | 13.7 | 8.2 | ND | ND | ND | 0.1 | 1.2 | 0.3 |
| 4,6-Manp | ND | 1.5 | 1.1 | ND | ND | ND | 0.7 | 1.1 | 1.9 |
| T-Glcp | –d | – | – | 0.4 | ND | ND | 0.4 | 0.7 | 0.1 |
| 4-Glcp | – | – | – | ND | ND | ND | 4.1 | 5.7 | 10.1 |
| 2,4-Glcp | – | – | – | 1.9 | 1.4 | 1.2 | 1.2 | 1.6 | 1.6 |
| 3,4-Glcp | – | – | – | ND | ND | ND | 0.8 | 1.4 | 1.3 |
| 4,6-Glcp | – | – | – | ND | ND | ND | 4.3 | 6.4 | 5.3 |
The fractions correspond to the fractions analyzed in Table 2 and described in the Table 2 footnotes. Fractions were analyzed by a modified Hakamori method for glycosyl residue linkage.
Sugar residues are expressed as percentage of total peak areas.
ND, not detected.
Glucose was omitted from EPG-released fractions since they contained large amounts of starch breakdown products.
Linkage analysis of the EGTA/Na2CO3 fractions revealed only modest differences between the wild type and irx8 (Table 3). A slight reduction in 4-linked GalAp confirmed that compositional changes in GalA were associated with HG. The amount of 5-Araf was increased in this fraction of the irx8 walls, possibly reflecting changes in arabinan pectic sidechain abundance and distribution in the walls of these mutants.
The irx8 mutants contained strikingly reduced amounts of 4-Xylp in the 4 n KOH fraction. This result corroborates conclusions from above that the reduced xylose detected by glycosyl residue composition analyses was due to reduced levels of xylans in the mutant. In addition, 4-GalAp and 2-Rhap were reduced in the mutant, suggesting reduced amounts of HG and RG-I backbone polysaccharides (Table 3).
Immunohistochemistry of irx8
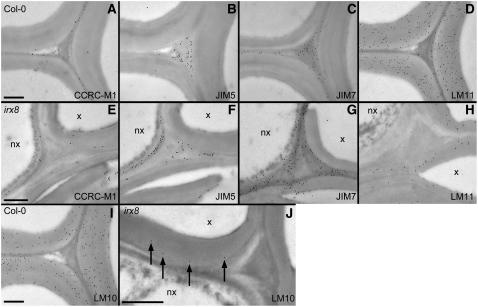
The glycosyl composition and linkage analyses suggest that xylan polymers, and possibly polymers associated with the xylan (specifically, HG polymers), are affected in the irx8 mutant. To localize and examine structural features of the affected polymers, we probed stem and root sections with a series of antibodies that recognize plant cell wall glycans. A total of 26 and 47 different antibodies that bind epitopes in most major classes of wall polysaccharides, including pectins (HG and RG-I), hemicelluloses (xyloglucan and xylan), and galactans and arabinogalactans (including arabinogalactan proteins), were used for immunolocalization studies of stem and root tissue, respectively. No difference in labeling intensity nor patterns could be detected with the majority of the antibodies. For example, no difference in labeling patterns using JIM5 and JIM7, which bind to different patterns of methyl esterified HG (Clausen et al., 2003), and CCRC-M1, which recognizes fucosylated xyloglucan (Puhlmann et al., 1994), were detected in the mutants compared with the wild type (Figures 5A to 5C, 5F to 5H, 5K to 5M, and 5P to 5R). However, the labeling intensity for two monoclonal antibodies, LM10 and LM11, which recognize different epitopes on xylan polymers (McCartney et al., 2005), was significantly reduced in the irx8 mutant (cf. Figures 5D and 5I, 5E and 5J, 5N and 5S, and 5O and 5T). As expected by their epitope specificity, both LM10 and LM11 only bound to regions containing thick secondary cell walls (Figure 5). LM10 has been suggested to be specific to unsubstituted or lower-substituted xylans, whereas LM11 binds to both low and higher substituted xylans (McCartney et al., 2005). The reduction in antibody labeling was more dramatic for LM10 compared with LM11 in the mutants (cf. Figures 5D and 5I, 5E and 5J, 5N and 5S, and 5O and 5T).
Figure 5.
Immunofluorescent Labeling of Transverse Sections of irx8 and Wild-Type Stems and Roots.
Immunofluoscent labeling of 250-μm-thick transverse sections taken from root (rows 1 and 2) and stem (rows 3 and 4) tissues of 7-week-old Col-0 (rows 1 and 3) and irx8-1 (rows 2 and 4) plants. Antibodies used for labeling are indicated in the figure. Bars = 50 μm in (A) to (J) and 20 μm in (K) to (T).
(A) to (E) Wild-type root sections labeled with CCRC-M1 (A), JIM5 (B), JIM7 (C), LM10 (D), and LM11 (E).
(F) to (J) irx8 root sections labeled with CCRC-M1 (F), JIM5 (G), JIM7 (H), LM10 (I), and LM11 (J).
(K) to (O) Wild-type stem sections labeled with CCRC-M1 (K), JIM5 (L), JIM7 (M), LM10 (N), and LM11 (O).
(P) to (T) irx8 stem sections labeled with CCRC-M1 (P), JIM5 (Q), JIM7 (R), LM10 (S), and LM11 (T).
To investigate if the distribution of any wall polymers is altered in the mutant, we visualized different epitopes using immunogold electron microscopy (Figure 6). The majority of antibodies displayed similar patterns in the walls surrounding the xylem vessels in both wild-type and mutant sections, analogous to what was observed for the immunofluorescence above. Both LM10 and LM11 showed significantly less labeling in the walls surrounding the xylem cells in the irx8 sections, corroborating the lower levels in fluorescence for the mutant observed above. Although the immunofluorescence data suggested a complete absence of LM10 labeling (Figures 5I and 5S), the immunogold suggested that some residual LM10 epitopes may be present in irx8 (Figure 6J). These observations support the compositional and linkage analyses above that the irx8 mutants have reduced levels of xylan.
Figure 6.
Immunogold Labeling of Xylem Cell Corners in 95-μm-Thick Transverse Sections Taken from Roots of 7-Week-Old Col-0 and irx8-1 Plants.
(A) to (D) Wild-type sections labeled with CCRC-M1 (A), JIM5 (B), JIM7 (C), and LM11 (D).
(E) to (H) irx8 sections labeled with CCRC-M1 (E), JIM5 (F), JIM7 (G), and LM11 (H).
(I) Wild-type section labeled with LM10.
(J) irx8 section labeled with LM10. The larger plate for irx8 LM10 labeling was included to show the sparse LM10 labeling (indicated by arrows).
x, xylem cells; nx, nonxylem cells. Bars = 1 μm.
Structural Analyses of irx8 Cell Wall Polymers
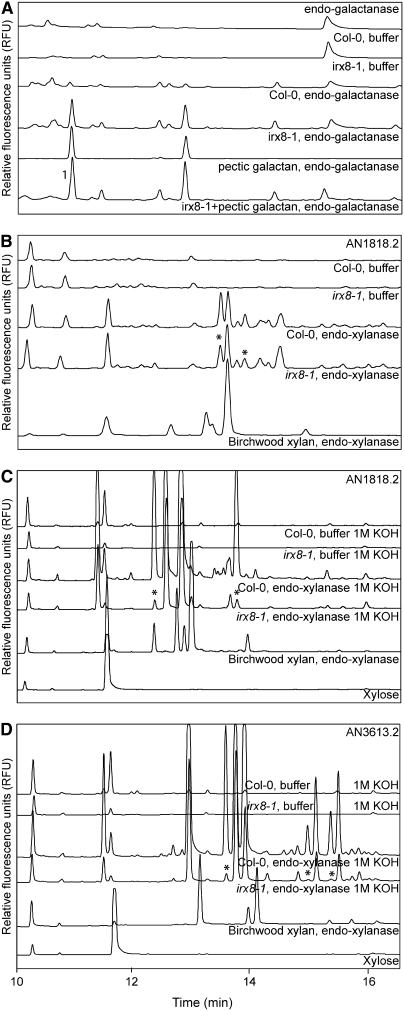
To obtain information about structural differences in irx8 cell wall polymers, we used a series of hydrolytic enzymes (polygalacturonase, xyloglucanase, endomannanase, xylogalacturonase, endogalactanase, cellulase, endoarabinanase, and endoxylanases) to fingerprint polysaccharides by capillary electrophoresis (Bauer et al., 2006) from wild-type and irx8-1 stems and roots. The only differences between the wild type and irx8-1 were observed after incubation with an endogalactanase and the endoxylanase AN1818.2 (Figure 7). Two peaks were significantly more pronounced in the mutant compared with the wild type after digestion with the endogalactanase (Figure 7A). To identify the peaks, we incubated commercially obtained pectic galactan with the endogalactanase. Two peaks, which corresponded to the altered peaks in the irx8 mutant after enzyme treatment, were evident after separation by capillary electrophoresis (Figure 7A). One of the peaks comigrated with galactose (peak 1 in Figure 7A), indicating that more pectic galactan is accessible in irx8 compared with the wild type.
Figure 7.
Capillary Electropherograms of Sugars and Oligosaccharides Released by Hydrolytic Enzyme Digests of Cell Walls from 7-Week-Old Stems.
(A) Endogalactanase treatment. Commercial pectic galactan digested with endogalactanase was used as reference material. The 1 indicates galactose migration. RFU, relative fluorescence units.
(B) Endoxylanase AN1818.2 (Bauer et al., 2006) treatment. Commercial birchwood xylan digested with endoxylanase AN1818.2 was used as reference material.
(C) Endoxylanase AN1818.2 treatment of 1 M KOH fractions.
(D) Endoxylanase AN3613.2 treatment of 1 M KOH fractions. The asterisk indicates peaks that differ between wild-type and irx8 mutants.
The endoxylanase AN1818.2 digestion resulted in a different capillary electrophoresis fingerprint pattern for the mutant compared with the wild type (Figure 7B). At least two peaks were significantly reduced in the irx8 mutant compared with the wild type. However, we were unable to identify the composition of the peaks using external standards, such as digested birchwood xylan (Figure 7B). To increase the enzyme accessibility and to enrich for specific kinds of polymers, we separated wall material into five major fractions. We then applied selected enzymes (polygalacturonase, arabinofuranosidase, and xylanases) to the different cell wall fractions and separated the oligosaccharides labeled with 8-aminopyrene-1,3,6-trisulfonate using capillary electrophoresis. Incubation of material from the 1 and 4 M KOH fractions with the two endoxylanases (AN1818.2 and AN3613.2) generated distinct differences in the fingerprint pattern after capillary electrophoresis separation (Figures 7C and 7D). Two peaks, present in the wild type, were absent in irx8 after treatment with AN1818.2 (Figure 7C). At least two peaks were also significantly reduced in irx8 after treatment with the endoxylanase AN3613.2 (Figure 7D). However, we were unable to identify the structures present in the peaks by capillary electrophoresis using an external standard (Figures 7C and 7D).
To identify the oligosaccharide fragments that differed between the wild type and the irx8 mutant, the peaks were assigned by comparison with matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) mass spectra (Figure 8). Using the endoxylanase AN1818.2, we found abundant peaks corresponding to mass-to-charge (m/z) ratios of 613, 627, 635, and 649 in the wild type (Figure 8A). Unless otherwise mentioned, the ions were observed as monosodium adducts [M+Na]+. The AN1818.2 enzyme cuts xylan polymers adjacent to glucuronic acid, or 4-O-Me glucuronic acid branches, resulting in a fragment consisting of three xylose residues decorated with a glucuronic acid or with a 4-O-Me glucuronic acid residue (P. Vasu, A. Mort, S. Bauer, and C. Somerville, personal communication). The mass of these fragments corresponds to 613 (glucuronic acid branch) and 627 (4-O-Me glucuronic acid branch), respectively. The identity of these peaks has been confirmed by 1H-nuclear magnetic resonance analyses (P. Vasu and A. Mort, personal communication). Whereas the AN1818.2-digested mutant did generate a peak at m/z 627 comparable to the wild type, the peak corresponding to m/z 613 was absent (cf. Figures 8A and 8B). The peak m/z 635, corresponding to an exchange of a proton for an additional sodium ion in the glucuronic acid branch ([M+2Na−H]+), was also absent in the irx8 mutant (cf. Figures 8A and 8B). These data suggest that the irx8 mutant is lacking glucuronic acid branched xylan.
Figure 8.
MALDI-TOF Mass Spectra from Xylanase-Treated Polysaccharides Present in 1 M KOH Fractions.
(A) and (B) MALDI-TOF mass spectra of 1 M KOH fractions from the wild type (A) and irx8 (B) after treatment with endoxylanase AN1818.2.
(C) and (D) MALDI-TOF mass spectra of 1 M KOH fractions from the wild type (C) and irx8 (D) after treatment with endoxylanase AN3613.2.
Arrows indicate absent peaks in irx8 mutant spectra. Symbols indicate the oligosaccharide structure corresponding to the peaks. Closed squares, xylose; open circles, glucuronic acid; Me, methyl. All ions are observed as monosodium adducts [M+Na]+.
The AN3613.2 enzyme produces xylan fragments consisting of four xylose residues decorated with either a glucuronic acid or with a 4-O-Me glucuronic acid residue (P. Vasu and A. Mort, personal communication). We found abundant peaks corresponding to m/z 727, 745, 759, 767, and 781 after AN3613.2 digestion of the wild type (Figure 7C). The m/z 745 corresponds to four xylose residues decorated with a glucuronic acid branch and the m/z 759 to four xylose residues decorated with a 4-O-Me glucuronic acid. The identity of these peaks has been confirmed by 1H-nuclear magnetic resonance analyses (P. Vasu and A. Mort, personal communication). Analogous to AN1818.2, the peak m/z 745, corresponding to the glucuronic acid branched xylan, was absent in the irx8 mutant after AN3613.2 digestion. In addition, the peak m/z 767, corresponding to an exchange of a proton for a sodium ion in the glucuronic acid branch, was absent in the irx8 mutant (cf. Figures 8C and 8D). However, peaks corresponding to 4-O-Me glucuronic acid branched xylan, m/z 759, were comparable to the wild type. Thus, the xylan polymer in irx8 is deficient in glucuronic acid branches but does contain 4-O-Me glucuronic acid branches.
IRX8 Affects Secondary Cell Wall Assembly and Integrity
Compositional, linkage, and structural analyses suggest, together with immunohistochemistry, that IRX8 is deficient in glucuronoxylan synthesis and possibly affects scaffolding of pectic polymers during secondary wall deposition. A recent study reported that another irx mutant, irx9, is also involved in xylan synthesis (Bauer et al., 2006). Since IRX8 and IRX9 are tightly coexpressed and have very similar phenotypes and glycosyl compositions, we anticipated that the two genes affect the same polymer. To test this, we generated irx8 irx9 double mutants. As expected, the phenotypic traits were not additive, and the double mutant plants looked similar to the single mutant parent plants (Figures 9A and 9B). However, the irx8 irx9 double mutants had significantly less xylose compared with the irx8 and irx9 single mutant plants (Figure 9C). Whereas irx8 and irx9 contain ∼40 to 50% xylose compared with the wild type, the double mutant contained only ∼20% xylose compared with wild-type plants. These data suggest that both IRX8 and IRX9 are involved in the synthesis of a xylan polymer or polymers but may affect synthesis of different components of the polymer(s).
Figure 9.
Analyses of irx8 irx9 Double Mutants.
(A) Whole-plant morphology of wild-type, irx9-2, irx8-2, and irx8-2 irx9-2 plants. Bar = 5 cm.
(B) Agarose gels showing wild-type, irx8-2, irx9-2, and irx8-2 irx9-2 genotypes as assessed by PCR. The irx8 genotyping (top panel) contains both genomic primers and insert primer. The two bottom panels are PCRs with genomic primers (top) and insert primer (bottom) for irx9, respectively.
(C) Neutral sugar composition from 7-week-old stems of wild-type, irx8-2, irx9-2, and irx8-2 irx9-2 plants. The amounts of sugar are presented as mean values (n = 5) of micrograms of sugar/milligrams of dry weight ± se.
The stunted growth and the deformed xylem cells in irx8 mutant plants suggest that the integrity of the secondary wall is significantly reduced. However, this can not be solely attributed to a reduction in cellulose, considered to be responsible for the main tensile strength of the cell wall, since irx8 mutants only show a 20 to 25% reduction in cellulose content compared with the wild type (Brown et al., 2005; Persson et al., 2005). Therefore, it appears that the function of IRX8 has an impact on the integrity of the secondary wall. To assess to what degree the xylan polymer contributes to tensile strength in the absence of secondary wall cellulose, we produced an irx8-2 irx1 double mutant. The irx1 (CESA8) mutant causes disruption of cellulose production during secondary wall formation and results in an 80% reduction in total cellulose content (Turner and Somerville, 1997). The homozygous double mutants were severely dwarfed and sterile (Figures 10A to 10C). The leaves were dark green, indicating an increase in chloroplasts per leaf area of the mutants, which was probably due to the reduced cell size. Several double mutant plants were unable to bolt and only grew rosette leaves (data not shown), which may indicate that the cell walls in the emerging xylem cells were unable to sustain water transport and therefore prohibited stem growth. We generated stem sections in mutant plants, in which stems developed, to assess the xylem cell structure. Figure 10D shows that the xylem cells in the irx8 irx1 mutant plants are severely deformed and are smaller in size compared with the wild type, possibly reflecting a reduction in water transporting ability. To investigate whether the double mutant phenotype is specific for irx8, we also crossed irx9-1 with irx1. The resulting phenotypes were very similar to irx8 irx1 (data not shown), suggesting that the xylan polymer is essential for secondary cell wall integrity.
Figure 10.
Analyses of irx1 irx8 Double Mutants.
(A) Agarose gels showing wild-type, irx1, irx8-2, and irx8-2 irx1 genotypes as assessed by PCR. Top panel shows bands from the wild type and mutants after restriction digest with BpiI, which does not cut in irx1. Bottom panel shows irx8 genotypes after PCR.
(B) Rosette leaves from 4-week-old wild-type, irx1, irx8-2, and irx8-2 irx1 plants. Bar = 5 cm
(C) Whole-plant morphology of wild-type, irx1, irx8-2, and irx8-2 irx1 (arrow) plants. Inset shows magnified irx8-2 irx1 plant. Bars = 5 cm.
(D) Hand-cut transverse sections from 7-week-old stems for wild-type and irx8-2 irx1 plants. Arrows indicate severely deformed xylem cells. XE, xylem; Ph, phloem. Bar = 50 μm.
(E) irx8-2 irx1 mutant phenotype setting flowers and siliques directly on rosette. Bar = 5 cm.
DISCUSSION
The irx8 mutants exhibit a significant loss in secondary cell wall integrity in both roots and stems. Consistent with the phenotype, the IRX8 gene is exclusively expressed in developing fiber and xylem cells where secondary cell walls are deposited. Biochemical analyses of the irx8 mutants show that they are deficient in glucuronoxylan and, to a lesser extent, HG.
Immunohistochemical analyses support a role for IRX8 in xylan production. Only two antibodies, LM10 and LM11, of the >40 antibodies tested showed a significant reduction in labeling of mutant walls compared with the wild type. The LM10 and LM11 antibodies recognize different epitopes on xylan polymers (McCartney et al., 2005). Interestingly, the reduction in the LM10 epitope is more dramatic than that of the LM11 epitope, suggesting that not all xylan polymers are affected in irx8. The structures of the epitopes recognized by the LM10 and LM11 antibodies are distinct but have not been determined in detail. The LM10 epitope has been reported to reside on xylans that are unsubstituted or carry only a low level of substitution (McCartney et al., 2005), but binding studies performed in our laboratory suggest that this antibody also binds to branched xylan preparations (A.G. Swennes and M.G. Hahn, unpublished data). The changes in GalA and galactose contents resulting from the irx8 mutation were not discernible with the antibodies available against polysaccharides containing those sugars.
Another mutant, fra8, is also deficient in glucuronoxylan (Zhong et al., 2005b). Similar to IRX8, FRA8 is expressed exclusively in tissues where secondary cell deposition occurs. Both of the gene products appear to be glycosyltransferases, albeit of different families. In addition, preliminary results indicate that the xylan-deficient irx9 mutant, affecting a family 43 member, is also deficient in glucuronoxylan (S. Persson, S. Bauer, and C. Somerville, unpublished data). This implies that at least three enzymes are involved in glucuronoxylan synthesis. Two hypotheses seem plausible: the gene products of FRA8, IRX8, and IRX9 may work together to catalyze different steps in the overall synthesis of glucuronoxylan, or there are several different types of xylan and the three enzymes are type-specific.
The first hypothesis suggests that the three glycosyltransferases perform different steps in xylan synthesis, for example, in the process of attaching the glucuronic acid branch to the xylan backbone or in synthesis of specific glycans required for xylan production. IRX8 belongs to a family of genes related to GAUT1, a gene shown to encode a galacturonosyltransferase (Sterling et al., 2006). IRX8 is designated GAUT12 in this gene family and, in accordance with the putative galacturonosyltransferase activity suggested from characterization of GAUT1 (Sterling et al., 2006), the irx8 mutant has reduced levels of 4-linked and 3,4-linked GalA in the pectinase-released wall fraction. The irx8 mutant also contains lower levels of GalA and xylose in this fraction (Table 2). These results may indicate either a reduction in xylosylated GalA residues (e.g., xylogalacturonan) or a reduced association of a specific pectin with xylan. The latter may indicate that, in addition to glucuronoxylan, the irx8 mutant also is deficient in particular types of HG or RG-I that are branched with β-1,4-xylans. Duan et al. (2003, 2004) have reported that 4-linked xylose residues are attached to O-4 of rhamnose in RG-I in Diospyros kaki, and similar structures have been reported in other species (Yamada et al., 1987). In addition, Nakamura et al. (2002) have reported that β-1,4-xylans are attached to O-3 of GalA in HG of soybean (Glycine max). The absence of immunolabeling of major pectic polymers in the thick secondary wall may further indicate that the potential link for xylan may be to a very specific pectin polymer, one for which an antibody probe is not currently available. Interestingly, a mutant (qua1) affecting a GAUT1-related gene family member (GAUT8) is also affected in both HG and xylan synthesis (Bouton et al., 2002; Orfila et al., 2005). Arabidopsis qua1 mutant stems have reduced HG:α 1,4-galacturonosyltransferase activity and β-1,4-xylosyltransferase activity as well as reduced amounts of HG and xylan in stem walls (Orfila et al., 2005). Although the enzymatic function of QUA1 (GAUT8) has not yet been described, the results would support a role for at least some of the GAUT1-related gene family members as galacturonosyltransferases that synthesize a glycan (e.g., HG) required for xylan synthesis. Interestingly, another mutant affecting a GAUT1-related gene family member (GATL1), parvus, had reduced levels of RG-I and 4-linked xylose (Lao et al., 2003; Shao et al., 2004), corroborating a role for the gene family in pectin–xylan interfaces. Since completing this work, it has come to our attention that Maria Peña, William York, Zheng-Hua Ye, and coworkers have identified, by mutant analysis, a complex oligosaccharide associated with xylan synthesis (W. York, personal communication). These authors have suggested that IRX8 may be involved in synthesis of a complex glycan primer for xylan synthesis.
A functional role for IRX8 in pectin assembly/distribution was also evident from the distribution of cell wall material after fractionation. Significantly more material was extracted from mutant walls with EPG and Na2CO3, agents that release pectic polymers, than could be released from wild-type walls (Table 2). Similar results were also obtained when cell wall material was extracted with water, which also releases pectic polymers. The irx8 mutant walls released twice as much material compared with wild-type walls (data not shown), suggesting that certain pectic polymers are cross-linked to a lower extent in the irx8 mutant walls. Curiously, the irx8 mutant has a wider and more diffuse residual primary cell wall that is located outside of the thickened secondary walls. Immunogold labeling shows, for example, that JIM5 and JIM7 epitopes are abundant in this region. JIM5 and JIM7 recognize HG with different patterns of methyl esterification (Clausen et al., 2003). Therefore, it is plausible that an increase in the width of the residual primary wall may be due to an increase in accumulation of some pectic polymers or a decreased compactness of the pectic matrix in these residual primary walls. The increase in the residual primary wall in irx8 could therefore be a result of impaired scaffolding between the xylan polymer and specific pectic polymers in the secondary wall.
The second hypothesis, that two or more types of xylan exist, is supported in part by the differences in glucuronoxylan and 4-O-methyl glucuronoxylan in the irx8 and irx9 mutants. MALDI-TOF analyses revealed that both the irx8 and irx9 mutants are essentially completely deficient in glucuronoxylan but appear to contain significant levels of 4-O-methyl glucuronoxylan. Interestingly, the irx9 mutants displayed reverse differences in LM10 and LM11 immunolabeling compared with irx8 (i.e., the levels of LM10 epitopes were similar to the wild type in the irx9 mutant, while the LM11 epitope was absent; Bauer et al., 2006). These data suggest that IRX8 and IRX9 may be involved in the production of different parts, or types, of xylan polymer. The xylose level in the irx8 irx9 double mutant plants was, furthermore, reduced to <20% of the wild-type levels compared with 40 to 50% reductions in the single mutant lines, again indicating that IRX8 and IRX9 affect different parts, or types, of xylan polymers. However, the double mutants did not display any major additive developmental phenotype, corroborating that IRX8 and IRX9 both are involved in xylan production. These data suggest that more than one type of xylan is made in secondary walls and that the FRA8, IRX8, and IXR9 gene products may act on parallel pathways of xylan synthesis rather than as components of a concerted pathway. However, definitive proof of the enzymatic functions of FRA8, IRX8, and IRX9 must await in vitro biochemical evidence. We attempted to express IRX8 in insect cells but were unable to detect significant amounts of the protein (data not shown).
It has been hypothesized that xylans coat the cellulose microfibrils to prevent aggregation of the fibrils (Reis and Vian, 2004). Complete disruption of cellulose production in the secondary wall results in a slightly dwarfed phenotype with collapsed xylem vessels (Turner and Somerville, 1997). However, the irx8 mutant plants display a more severe growth phenotype and an earlier onset of the irregular xylem phenotype. In addition, the overall organization of the central cylinder is substantially affected in irx8 roots, and deficiencies in xylem proliferation are evident in stems. These effects may arise from a defect in cell–cell interactions with feedback effects on cell differentiation. It is interesting to note that the qua1 mutation also shows a cell adhesion phenotype. These observations suggest that xylan may play roles that extend beyond coating cellulose. To determine the biological implication of the xylan deficiency in irx8, we generated irx1 irx8-2 double mutants. The double mutants were severely dwarfed, and only grew about an inch tall. Some of the plants were unable to generate any stems at all, in some instances resulting in production of flowers directly on the rosette (Figure 10). Similar phenotypic effects were also evident when irx1 was crossed with irx9 (data not shown). We believe that the extreme effects of the irx8 mutations may either be due to a defect in coating cellulose, which may lead to defects in cell expansion, or that the irx8 mutation interrupts a connection of xylan to a scaffold that connects it to other secondary wall polymers. We suggest that such a connection may be adequate to preserve secondary cell wall integrity in the absence of cellulose.
Phylogenetic analysis of the Arabidopsis GAUT1-related gene family revealed that IRX8 (GAUT12) clusters closely together with three other GAUT proteins in the GAUT-C clade: At5g15470 (GAUT14), At3g01040 (GAUT13), and At3g58790 (GAUT15) (Sterling et al., 2006). These homologs may be relevant targets for further evaluation of the function of IRX8 and other members in the GAUT-C clade. Only one close homolog to the IRX8 homologs in Arabidopsis was evident in rice (Oryza sativa), Os12g0578500, suggesting that this protein may have an IRX8-like function in rice. An IRX8 ortholog, PttGT8D, has been identified in poplar (Aspeborg et al., 2005). Therefore, this protein may play a role in xylan synthesis and cell wall integrity during wood formation in trees. Thus, identification of the IRX8 gene promises to open up new opportunities to study the synthesis and function of xylans in a wide variety of plants.
METHODS
Plant Material and Genetic Analysis
Arabidopsis thaliana (Col-0) plants were germinated on MS medium in an environmental chamber under continuous light (140 to 220 μmol m−2 s−1) at 22°C. Seedlings were then transferred to soil and grown in greenhouse chambers under 16-h-light/8-h-dark conditions at 22°C.
Insertion lines (Alonso et al., 2003) for irx8 were obtained from the ABRC (http://arabidopsis.org). The lines used were SALK_014026 (exon; irx8-1), SAIL_603_G02 (5′ UTR; irx8-2), SAIL_1262_H02 (5′ UTR; irx8-3), SAIL_1214_C12 (5′ UTR; irx8-4), and SALK_044387 (intron; irx8-5). PCR was performed, with primer sequences generated against the genomic regions flanking the insert and a standard primer for the 3′ end of the insertion sequence, to obtain homozygous insertion lines (primers are listed in Supplemental Table 1 online). RT-PCR was used to ensure that the transcript in the insertion lines was not present. RNA was isolated using a Qiagen RNeasy plant mini kit, and a Qiagen One-Step RT-PCR kit was used for first-strand synthesis and subsequent PCR steps (primers are listed in Supplemental Table 1 online).
Antibodies
All primary antibodies used were monoclonal in the form of hybridoma supernatants harvested from cell lines generated either in mice (CCRC series) or in rat (JIM and LM series). The CCRC series of antibodies were from our laboratory stocks and are available from CarboSource (http://cell.ccrc.uga.edu/∼carbosource/CSS_home.html). The JIM series of antibodies were harvested from hybridoma lines obtained from Keith Roberts (John Innes Institute, Norwich, UK) and are available from CarboSource. The LM series of antibodies were in the form of hybridoma supernatants and were purchased from Plant Probes. Hybridoma supernatants were used undiluted. Secondary antibodies (goat anti-mouse or goat anti-rat) conjugated to Alexa-fluor 488 were purchased from Invitrogen and were used diluted 1:100 (v/v). Secondary antibodies conjugated to 18-nm colloidal gold were purchased from Jackson ImmunoResearch Laboratories and were used as directed by the supplier.
Light Microscopy
Hand-cut stem sections (∼200 μm in thickness) were stained for 5 min in 0.02% aqueous Toluidine blue O (Sigma-Aldrich), rinsed, mounted in water, and viewed with a compound microscope (Leitz DMRB; Leica). Cellulose and lignin were visualized by staining sections with either Calcofluor white (Polysciences) or phloroglucinol (Matheson Coleman and Bell), respectively, and viewing with an epifluorescence microscope (Leitz DMRB; Leica).
Tissue Fixation and Embedding
Tissues harvested from 7-week-old plants grown in soil (Freshour et al., 2003) were transferred directly to fixative (1.6% [v/v] paraformaldehyde and 0.2% [w/v] glutaraldehyde in 25 mM sodium phosphate, pH 7.1, containing 0.02% [v/v] Triton X-100). Subsequent fixation and embedding of the tissue was performed as described (Freshour et al., 1996) except that infiltration of resin was performed using only one intermediate step of 50% (v/v) LR White resin:ethanol followed by three incubations with 100% LR White, all for 24 h each. Sectioning was performed using a Reichert-Jung Ultracut E microtome, and sections were handled as described (Freshour et al., 1996).
Immunocytochemistry
Immunolocalizations (immunofluorescence and immunogold) were performed as described (Freshour et al., 1996, 2003). Immunofluorescence images were captured using a Nikon DS-L1 camera control unit and a DS-5M camera head mounted on the microscope. For immunogold localizations, the lead citrate post-stain was omitted.
Transmission Electron Microscopy
Stem sections corresponding to the basal region from 7-week-old irx8 and wild-type plants were prepared and viewed as described by Turner and Somerville (1997).
Expression of IRX8
RNA was isolated using an RNeasy plant mini kit (Qiagen). A Qiagen One-Step RT-PCR kit was used for first-strand synthesis and for subsequent PCR steps using gene-specific primers (forward 5′-CATCCATCTCCTTTCAGAC-3′ and reverse 5′- TCATGATGCTCTAATGTGAC-3′). The primers used for RT-PCR span four introns so that any potential amplification from genomic DNA could easily be identified based on its larger size. The RT-PCR reactions were repeated two times with identical results. The expression of the actin-1 gene was used as an amplification control.
The tissue-level expression pattern of the IRX8 gene was studied using the GUS reporter gene. A genomic DNA fragment extending 1.4 kb upstream of the ATG starting codon for IRX8 was amplified (see Supplemental Table 1 online) and inserted, using KpnI and NcoI, in front of a GUS gene in the pCAMBIA1304 vector to produce plasmid pSP008. The construct was transformed into wild-type plants by Agrobacterium tumefaciens–mediated transformation (Bechtold and Bouchez, 1994). Transgenic plants were selected on hygromycin and used for GUS reporter analyses. Tissues were incubated in GUS staining solution (100 mM sodium phosphate, pH 7.0, 10 mM EDTA, 1 mM ferricyanide, 1 mM ferrocyanide, and 1 mM 5-bromo-4-chloro-3-indolyl β-d-glucuronic acid) at 37°C. After clearing in 70% ethanol, the tissues were observed for GUS staining under a dissecting microscope.
Complementation of irx8
An IRX8 cDNA was obtained by RT-PCR using RNA from stems (see Supplemental Table 1 online). The GUS cDNA in pSP008 was replaced with the IRX8 cDNA to produce pSP008c. The construct was transformed into heterozygous irx8 mutant plants by Agrobacterium-mediated transformation (Bechtold and Bouchez, 1994). Transgenic plants were selected on hygromycin and scored for homozygous irx8 mutant plants by genetic analysis of insertions as described above. Complementation was assessed by comparing plant size and by analyzing xylem vessel shape in transverse stem sections as described above.
FTIR Spectroscopy
Plants were placed in the dark overnight to deplete starch. Stems from at least 10 individual plants for each line were ground in liquid nitrogen using a mortar and pestle. The material was extracted with 80% ethanol overnight at 65°C. After extraction with acetone, the cell wall material was air-dried at room temperature and then ground to a fine powder by ball-milling for 1 to 2 h. Prior to collection of FTIR spectra, the powder was dried at 30°C overnight and then mixed with KBr and pressed into 13-mm pellets.
Fifteen spectra for each line were collected on a Thermo-Nicolet Nexus 470 spectrometer over the range 4000 to 400 cm−1. For each spectrum, 32 scans were co-added at a resolution of 8 cm−1 for Fourier transform processing and absorbance spectrum calculation using Omnic software (Thermo Nicolet). Spectra were corrected for background by automatic subtraction and saved as JCAMP.DX format for further analysis. Using Win-Das software (John Wiley & Sons), spectra were baseline-corrected, area-normalized, and analyzed using a principal components analysis covariance matrix method (Kemsley, 1998).
Glycosyl Residue Composition Analysis of Total Cell Wall Material
Cell wall material from 7-week-old stems and roots was prepared and ball-milled as described above. Alditol acetate derivatives of the neutral sugars were analyzed essentially as described by Blakeney et al. (1983). One milligram of material was mixed with 250 μL 1 M H2SO4, vortexed, autoclaved for 1 h, and neutralized by 100 μL of 9 M NH4OH. One milliliter of 2% NaBH4 in DMSO was added, and tubes were incubated for 90 min at 42°C. Concentrated acetic acid (250 μL) was added, tubes were vortexed, and 250 μL of 1-methylimidazole and 4 mL of acetic anhydride were added. Samples were incubated for at least 10 min at room temperature, and 8 mL of water was added. Samples were cooled to below 30°C, and the alditol acetates were extracted overnight with CH2Cl2 at 4°C. Neutral sugars from the CH2Cl2 phase were analyzed using gas chromatography with myo-inositol (Sigma-Aldrich) as internal standard.
Fractionation of Walls for TMS Glycosyl Residue Composition Analysis
Plants were grown in 14 h of light (140 to 220 μmol m−2 s−1) at 19°C and 10 h of dark at 14°C at 60% constant relative humidity. Stems from 10-week-old irx8 mutants and wild type were harvested on ice, flash frozen in liquid N2, and ground to a fine powder with a mortar and pestle. The ground material was extracted over a sintered glass funnel (Kimax 60M) with vacuum sequentially with 2 volumes of 100 mL of ice cold 80% ethanol, 100% ethanol, chloroform:methanol (1:1), and 100% acetone. Starch was removed from the walls by treatment with Type-I porcine α-amylase (Sigma-Aldrich; 47 units/100 mg cell wall alcohol-insoluble residue or A.I.R.) in 100 mM ammonium formate for 48 h at 25°C with rotation. Destarched walls were centrifuged, washed thrice with sterile water, twice with 100% acetone, air dried, and weighed. The stem A.I.R. was incubated with purified type II Aspergillus niger EPG (0.8 units/100 mg A.I.R.; gift of Carl Bergmann, Complex Carbohydrate Research Center, University of Georgia) (Benen et al., 1999) in 100 mM ammonium formate, pH 6.0, for 24 h with mixing. The pellet was recovered by centrifugation at 3660g for 20 min. The supernatant was saved and the pellet was subjected to EPG treatment a second time. The two supernatants were combined, and the final pellet was washed thrice with sterile water. The combined EPG plus PME treatment was done in a similar manner using a purified A. niger PME (1.0 units/100 mg A.I.R.; gift of Carl Bergmann) and a similar amount of EPG as described above. The supernatants and washes were pooled, lyophilized thrice in 250 mL of sterile water, and weighed. Pellets recovered following EPG/PME treatment were extracted with 5 mM EGTA in 50 mM Na2CO3 overnight at room temperature with mixing. The supernatant was recovered by centrifugation at 3660g for 20 min prior to a second extraction with 5 mM EGTA in 50 mM Na2CO3 and three sequential water washes. The supernatants and corresponding washes were pooled, filtered (Whatman 934-AH glass microfiber filters; 1.5 nm pore), and neutralized to pH 7.0 with acetic acid. The fractions were dialyzed three times with Spectrapor number 6 1000 MWCO tubing in water containing 0.2% thimerosal, lyophilized, and weighed. The pellet recovered from the EGTA and Na2CO3 extracted walls was extracted twice with cold 4 n KOH in 1 mg/mL of sodium borohydride at 25°C overnight with stirring. The pellet was recovered by centrifugation and washed thrice with sterile water. The supernatants were combined, filtered, neutralized, dialyzed, lyophilized, and weighed as described above. The pellet remaining following the 4 n KOH extraction was neutralized, dialyzed, lyophilized, and weighed as described above. All fractions were analyzed by GC-MS of TMS derivatives directly or following Saeman hydrolysis, prior to GC-MS of TMS derivatives.
Phenol–Sulfuric Acid Assay and Cellulose Quantitation
A phenol sulfuric acid assay (Dubois et al., 1956) was used to quantitate hexoses recovered in all A.I.R. fractions. Weighed samples were mixed with 0.05 mL of 80% phenol and 5 mL of concentrated sulphuric acid. The samples were incubated at 25°C for 30 min, and optical density at 490 nm was measured. Total sugar was calculated based on comparison to a standard curve of known amounts of Ara, Rha, Fuc, Xyl, Gal, GalA, Man, α-d-glucuronic acid, and Glc.
Cellulose contents were measured on ball-milled material described above essentially according to the Updegraff method (Updegraff, 1969). One milligram of material was incubated with 300 μL of acetic:nitric:aqueous reagent (8:1:2), boiled for 30 min, and centrifuged for 5 min at 2500g. The pellet was resuspended in 800 μL of water, centrifuged for 5 min at 2500g, and resuspended in 400 μL acetone. The material was centrifuged for 5 min at 2500g and dried, and 50 μL H2SO4 was added. The material was vortexed for 90 min at room temperature, diluted to 1 M H2SO4 with water, and further diluted 1:4 with water. One milliliter of 0.2% anthrone in H2SO4 was added, and the absorbance was measured at A620.
Uronic Acid Assay
Uronic acid content was determined by the method of Blumenkrantz and Asboe-Hansen (1973). Samples were hydrolyzed in 1.2 mL of 0.5% (w/v) Na2B4O7 in concentrated H2SO4 at 100°C for 5 min. Twenty microliters of 0.15% m-hydroxybiphenyl dissolved in 0.5% NaOH was added, and the samples were mixed and incubated at room temperature for 30 min. Absorbance was measured at 520 nm. GalA was used as a standard.
Glycosyl Residue Composition
Approximately 1 mg of dry cell walls was subjected to methanolysis for 16 h at 80°C in 1 n methanolic HCl. The mixture was evaporated under dry air flow and treated with Pierce TRI-SIL reagent for 20 min at 80°C. The resulting residue was filtered through glass wool to remove particulates. The flow-through was dried under a stream of air and suspended in hexanes. The silylated sugars were analyzed by GC-MS (York et al., 1985).
In some cases (i.e., 4 n KOH insol-SH fractions in Table 2), Saeman hydrolysis (Selvendran et al., 1979; Zhong et al., 2005b) was performed prior to TMS composition analysis. A small aliquot (2 to 4 mg) of dried 4 n KOH insoluble fractions and 20 μg of inositol internal standard was hydrolyzed in concentrated H2SO4 for 3 h with shaking. Samples were diluted to 1 M H2SO4 and heated to 120°C for 1 h before neutralization with BaOH. The insoluble salts were pelleted by centrifugation at 3600g for 20 min. Supernatants were dried down and analyzed as above for TMS glycosyl residue composition.
Glycosyl Residue Linkage Analysis
Total walls or wall fractions were analyzed for glycosyl residue linkage as described by York et al. (1985). In brief, samples were methylated by a modification of the method of Hakomori (1964), depolymerized, reduced, and acetylated, and the resultant partially methylated alditol acetates were analyzed by GC-MS. Specifically, after suspending walls or fractionated walls in DMSO for 2 to 3 weeks at room temperature by sonication and magnetic stirring, 0.4 mL of 2 M potassium dimsylate was added, and the tubes sealed and magnetically stirred for 6 h at room temperature. The reaction mixture was cooled to 0°C, excess 99.5% methyl iodide (0.7 mL) was added, and the tubes were sealed and incubated for an additional 16 h at room temperature. Samples were cooled to 0°C, 0.4 mL of 2 M potassium dimsylate was added, and the tubes incubated for 3 h at room temperature. This was followed by an additional cooling of samples and addition of 0.7 mL of methyl iodide. The samples were applied to a Waters C18 Sep Pak cartridge, washed four times with 10 mL of water, and eluted with acetonitrile. The eluted samples were reduced with 250 μL of superdeuteride (lithium triethylborodeuteride) for 1.5 h, and the reaction neutralized with 20 μL of acetic acid and dried down four times with methanol. Samples were suspended in 1 mL of a 1:1 ethanol:water mix and filtered through a Dionex On-Guard H II column and dried completely. Following sample workup, the permethylated material was hydrolyzed using 2 M trifluoroacetic acid (3 h in sealed tube at 100°C), reduced with NaBD4, and acetylated using acetic anhydride/pyridine. The resulting partially methylated alditol acetates were analyzed on a Hewlett Packard 5890 GC interfaced to a 5970 MSD (mass selective detector; electron impact ionization mode). Separation was performed on a 30 m Supelco 2330 bonded phase fused silica capillary column.
Fractionation of Cell Walls for Capillary Electrophoresis and MALDI-TOF
Cell wall material from 7-week-old stems and roots was prepared and ball-milled as described above. One hundred milligrams of material was extracted with 50 mM CDTA, pH 6.5, at room temperature overnight and pelleted at 2500g for 5 min. The pellet was washed two times with water, subsequently boiled with 50 mM NaHCO3 and 5 mM EGTA for 1 h, and incubated overnight at 4°C. The sample was centrifuged at 2500g for 5 min and washed two times with water. The pellet was extracted two times with 1 M KOH, supplied with 1 mg/mL sodium borohydride, at 25°C, and the extracted material was pooled. The sample was centrifuged at 2500g for 5 min and washed two times with water. The pellet was extracted two times with 4 M KOH, supplied with 1 mg/mL sodium borohydride, at 25°C, and the extracted material was pooled. The sample was centrifuged at 2500g for 5 min and washed two times with water. The pellet was saved for further analyses. The different fractions were neutralized with HCl and dialyzed three times against water containing 0.05% 4-chloro-1-butanol overnight at 4°C and lyophilized.
Capillary Electrophoresis
Capillary electrophoresis was performed as described by Bauer et al. (2005). Ball-milled cell wall material (1 mg) was incubated with 1 μL of enzyme (10 μg/mL) in 0.1 M NaAc buffer, pH 5.0, with shaking at 37°C overnight. Enzymes were inactivated by boiling, and the material was centrifuged (16,000g for 5 min) to remove insoluble material. Aliquots were dried in a speed-vac and resuspended in 5 μL 20 mM sodium phosphate buffer, pH 7.0. Two microliters of 8-aminopyrene-1,3,6-trisulfonate (100 mM in 15% acetic acid) and 2 μL NaBH3CN (1 M in Me2SO) were added, and samples were incubated at 55°C for 2 h. Water (50 μL) was added, and 10 μL of the diluted material was transferred to 190 μL of water. Samples were analyzed using a capillary electrophoresis systems (P/ACE MDQ molecular characterization system; Beckman Coulter) equipped with a 50-cm eCap capillary (75-μm i.d., 375 μm o.d.; Beckman-Coulter) and laser detection (488-nm excitation; 520-nm emission) at an applied voltage of 22 kV using carbohydrate separation buffer (Beckman-Coulter).
Unless indicated, all enzymes used for polysaccharide fingerprinting were recombinant enzymes that were free of contaminating activities (Bauer et al., 2006). The gene numbers for the enzymes were as follows: polygalacturonase (AN4372.2), xyloglucanase (AN0452.2), β-mannanase EC 3.2.1.78 (Megazyme), xylogalacturonase (Afu8g06890), endogalactanase (AN5727.2), cellulase EC 3.2.1.4 (Megazyme), endoarabinanase (Megazyme), and endoxylanases (AN1818.2; AN3613.2).
MALDI-TOF Mass Spectrometry
MALDI-TOF mass spectrometry analyses were performed essentially as described by Bauer et al. (2005).
Expression of Recombinant IRX8 in Drosophila Cells
Vector pMT/V5-HisA (Invitrogen) was modified by replacing the SpeI-ApaI fragment in the polylinker region with a synthetic fragment containing NcoI and BstEII sites to produce plasmid pCRS/V5-HisA (see Supplemental Table 1 online). An IRX8 cDNA was amplified from stem RNA by RT-PCR with primers that introduced NcoI and BstEII sites at the start codon and at the end of the coding sequence, respectively (see Supplemental Table 1 online). The fragment was cloned into the corresponding sites of pCRS/V5-HisA to produce plasmid pSPe54690, which contained an error-free IRX8 cDNA transcribed under control of the Drosophila metallothionein promoter and with a V5 epitope and a 6xHis tag at the C terminus. Plasmid pSPe54690 was introduced into Drosophila cells using CaHPO4 as described by the manufacturer for transient expression assays (Invitrogen). After 3 d, cells were harvested, proteins were extracted in cell lysis buffer (50 mM Tris, pH 7.8, 150 mM NaCl, and 1% Nonidet P-40) as described by the manufacture, and resolved by SDS-PAGE, and a protein gel blot was probed with commercial anti-His antibody (Invitrogen).
Accession Numbers
Arabidopsis Genome Initiative locus identifiers for the genes mentioned in this article are as follows: IRX1, At4g18780; IRX7 (FRA8), At2g28110; IRX8, At5g54690; and IRX9, At2g37090.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table 1. Primer Sequences Used for Analyses.
Supplemental Figure 1. Complementation of irx8.
Supplementary Material
Acknowledgments
S.P. was a recipient of a Swedish Research Council fellowship (No. 623-2004-4254). We thank Debra Skinner and Charles Gass for advice regarding the floral phenotype. We also thank Jennifer Milne for useful suggestions regarding structural and compositional analyses of the irx8 mutant and Carl Bergmann for the gift of purified Aspergillus niger EPG and PME. We also thank Maria Peña, Will York, and Zheng-Hua Ye for communication of results prior to publication. This work was supported, in part, by grants from the U.S. Department of Energy (DOE-FG02-03ER20133 and DOE DE-FG05-93-ER20097), the National Research Initiative, CSREES, USDA (2003-35318-15377), and the National Science Foundation (0090281 and DBI-0421683).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Chris Somerville (crs@stanford.edu).
Online version contains Web-only data.
References
- Alonso, J., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657. [DOI] [PubMed] [Google Scholar]
- Aspeborg, H., et al. (2005). Carbohydrate-active enzymes involved in the secondary cell wall biogenesis in hybrid aspen. Plant Physiol. 137 983–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awano, T., Takabe, K., and Fujita, M. (2002). Xylan deposition on secondary wall of Fagus crenata fiber. Protoplasma 219 106–115. [DOI] [PubMed] [Google Scholar]
- Bauer, S., Vasu, P., Mort, A.J., and Somerville, C.R. (2005). Cloning, expression, and characterization of an oligoxyloglucan reducing end-specific xyloglucanobiohydrolase from Aspergillus nidulans. Carbohydr. Res. 340 2590–2597. [DOI] [PubMed] [Google Scholar]
- Bauer, S., Vasu, P., Persson, S., Mort, A.J., and Somerville, C.R. (2006). Development and application of a suite of polysaccharide degrading enzymes for analyzing plant cell walls. Proc. Natl. Acad. Sci. USA 103 11417–11422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baydoun, E.A., Waldron, K.W., and Brett, C.T. (1989). The interaction of xylosyltransferase and glucuronyltransferase involved in glucuronoxylan synthesis in pea (Pisum sativum) epicotyls. Biochem. J. 257 853–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechtold, N., and Bouchez, D. (1994). In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. In Gene Transfer to Plants, I. Potrykus and G. Spangenberg, eds (Berlin: Springer-Verlag), pp. 19–23.
- Benen, J.A.E., Kester, H.C.M., and Visser, J. (1999). Kinetic characterization of Aspergillus niger N400 endopolygalacturonases I, II and C. Eur. J. Biochem. 259 577–585. [DOI] [PubMed] [Google Scholar]
- Blakeney, A.B., Harris, P.J., Henry, R.J., and Stone, B.A. (1983). A simple and rapid preparation of alditol acetates for monosacharide analysis. Carbohydr. Res. 113 291–299. [Google Scholar]
- Blumenkrantz, N., and Asboe-Hansen, G. (1973). New method for quantitative determination of uronic acids. Anal. Biochem. 54 484–489. [DOI] [PubMed] [Google Scholar]
- Bouton, S., Leboeuf, E., Mouille, G., Leydecker, M.-T., Talbotec, J., Granier, F., Lahaye, M., Hofte, H., and Truong, H.-N. (2002). QUASIMODO 1 encodes a putative membrane-bound glycosyltransferase required for normal pectin synthesis and cell adhesion in Arabidopsis. Plant Cell 14 2577–2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, D.M., Zeef, L.A., Ellis, J., Goodacre, R., and Turner, S.R. (2005). Identification of novel genes in Arabidopsis involved in secondary cell wall formation using expression profiling and reverse genetics. Plant Cell 17 2281–2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clausen, M.H., Willats, W.G.T., and Knox, J.P. (2003). Synthetic methyl hexagalacturonate hapten inhibitors of anti-homogalacturonan monoclonal antibodies LM7, JIM5 and JIM7. Carbohydr. Res. 338 1797–1800. [DOI] [PubMed] [Google Scholar]
- Dalessandro, G., and Northcote, D.H. (1981). Xylan synthetase activity in differentiated xylem cells of sycamore trees (Acer pseudoplatanus). Planta 151 53–60. [DOI] [PubMed] [Google Scholar]
- Duan, J., Wang, X., Dong, Q., Fang, J., and Li, X. (2003). Structural features of a pectic arabinogalactan with immunological activity from the leaves of Diospyros kaki. Carbohydr. Res. 338 1291–1297. [DOI] [PubMed] [Google Scholar]
- Duan, J., Zheng, Y., Dong, Q., and Fang, J. (2004). Structural analysis of a pectic polysaccharide from the leaves of Diospyros kaki. Phytochemistry 65 609–615. [DOI] [PubMed] [Google Scholar]
- Dubois, M., Gilles, K.A., Hamilton, J.K., Rebers, P.A., and Fred, S. (1956). Colorimetric method for determination of sugars and related substances. Anal. Chem. 28 350–356. [Google Scholar]
- Ebringerova, A., and Heinze, T. (2000). Naturally occurring xylans: Structures, isolation procedures and properties. Macromol. Rapid Commun. 21 542–556. [Google Scholar]
- Freshour, G., Bonin, C.P., Reiter, W.D., Albersheim, P., Darvill, A.G., and Hahn, M.G. (2003). Distribution of fucose-containing xyloglucans in cell walls of the mur1 mutant of Arabidopsis. Plant Physiol. 131 1602–1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freshour, G., Clay, R.P., Fuller, M.S., Albersheim, P., Darvill, A.G., and Hahn, M.G. (1996). Developmental and tissue-specific structural alterations of the cell-wall polysaccharides of Arabidopsis thaliana roots. Plant Physiol. 110 1413–1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakomori, S.A. (1964). A rapid permethylation of glycolipid and polysaccharide catalyzed by methylsulfinyl carbanion in dimethyl sulfoxide. J. Biochem. (Tokyo) 55 205–208. [PubMed] [Google Scholar]
- Jones, L., Ennos, A.R., and Turner, S.R. (2001). Cloning and characterization of irregular xylem 4 (irx4): A severely lignin-deficient mutant of Arabidopsis. Plant J. 26 205–216. [DOI] [PubMed] [Google Scholar]
- Kemsley, E.K. (1998). Discriminant Analysis and Class Modelling of Spectroscopic Data. (New York: John Wiley & Sons).
- Lao, N.T., Long, D., Kiang, S., Coupland, G., Shoue, D.A., Carpita, N.C., and Kavanagh, T.A. (2003). Mutation of a family 8 glycosyltransferase gene alters cell wall carbohydrate composition and causes a humidity-sensitive semi-sterile dwarf phenotype in Arabidopsis. Plant Mol. Biol. 53 647–661. [DOI] [PubMed] [Google Scholar]
- McCartney, L., Marcus, S.E., and Knox, J.P. (2005). Monoclonal antibodies to plant cell wall xylans and arabinoxylans. J. Histochem. Cytochem. 53 543–546. [DOI] [PubMed] [Google Scholar]
- Mouille, G., Robin, S., Lecomte, M., Pagant, S., and Hofte, H. (2003). Classification and identification of Arabidopsis cell wall mutants using Fourier-Transform InfraRed (FT-IR) microspectroscopy. Plant J. 35 393–404. [DOI] [PubMed] [Google Scholar]
- Nakamura, A., Furuta, H., Maeda, H., Takao, T., and Nagamatsu, Y. (2002). Analysis of the molecular construction of xylogalacturonan isolated from soluble soybean polysaccharides. Biosci. Biotechnol. Biochem. 66 1155–1158. [DOI] [PubMed] [Google Scholar]
- Orfila, C., Sorensen, S.O., Harholt, J., Geshi, N., Crombie, H., Truong, H.-N., Reid, J.S.G., Knox, J.P., and Sheller, H.V. (2005). QUASIMODO1 is expressed in vascular tissue of Arabidopsis thaliana inflorescence stems, and affects homogalacturonan and xylan biosynthesis. Planta 222 613–622. [DOI] [PubMed] [Google Scholar]
- Persson, S., Wei, H., Milne, J., Page, G.P., and Somerville, C.R. (2005). Identification of genes required for cellulose synthesis by regression analysis of public microarray data sets. Proc. Natl. Acad. Sci. USA 102 8633–8638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porchia, A.C., Sorensen, S.O., and Scheller, H.V. (2002). Arabinoxylan biosynthesis in wheat. Characterization of arabinosyltransferase activity in Golgi membranes. Plant Physiol. 130 432–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puhlmann, J., Bucheli, E., Swain, M.J., Dunning, N., Albersheim, P., Darvill, A.G., and Hahn, M.G. (1994). Generation of monoclonal antibodies against plant cell wall polysaccharides. I. Characterization of a monoclonal antibody to a terminal α-(1→2)-linked fucosyl-containing epitope. Plant Physiol. 104 699–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis, D., and Vian, B. (2004). Helicoidal pattern in secondary cell walls and possible role of xylans in their construction. C. R. Biol. 327 785–790. [DOI] [PubMed] [Google Scholar]
- Selvendran, R.R., March, J.F., and Ring, S.G. (1979). Determination of aldoses and uronic acid content of vegetable fiber. Anal. Biochem. 96 282–292. [DOI] [PubMed] [Google Scholar]
- Shao, M., Zheng, H., Hu, Y., Liu, D., Jang, J.-C., Ma, H., and Huang, H. (2004). The GAOLAOZHUANGREN1 gene encodes a putative glycosyltransferase that is critical for normal development and carbohydrate metabolism. Plant Cell Physiol. 45 1453–1460. [DOI] [PubMed] [Google Scholar]
- Sterling, J.D., Atmodjo, M.A., Inwood, S.E., Kumar Kolli, V.S., Quigley, H.F., Hahn, M.G., and Mohnen, D. (2006). Functional identification of an Arabidopsis pectin biosynthetic homogalacturonan galacturonosyltransferase. Proc. Natl. Acad. Sci. USA 103 5236–5241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szyjanowicz, P.M., McKinnon, I., Taylor, N.G., Gardiner, J., Jarvis, M.C., and Turner, S.R. (2004). The irregular xylem 2 mutant is an allele of korrigan that affects the secondary cell wall of Arabidopsis thaliana. Plant J. 37 730–740. [DOI] [PubMed] [Google Scholar]
- Taylor, N.G., Scheible, W.R., Cutler, S., Somerville, C.R., and Turner, S.R. (1999). The irregular xylem3 locus of Arabidopsis encodes a cellulose synthase required for secondary cell wall synthesis. Plant Cell 11 769–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner, S.R., and Somerville, C.R. (1997). Collapsed xylem phenotype of Arabidopsis identifies mutants deficient in cellulose deposition in the secondary cell wall. Plant Cell 9 689–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner, S.R., Taylor, N., and Jones, L. (2001). Mutations of the secondary cell wall. Plant Mol. Biol. 47 209–219. [PubMed] [Google Scholar]
- Updegraff, D.M. (1969). Semimicro determination of cellulose in biological materials. Anal. Biochem. 32 420–424. [DOI] [PubMed] [Google Scholar]
- Waldron, K.W., and Brett, C.T. (1983). A glucuronyltransferase involved in glucuronoxylan synthesis in pea (Pisum sativum) epicotyls. Biochem. J. 213 115–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada, H., Yanahira, S., Kiyohara, H., Cyong, J.-C., and Otsuka, Y. (1987). Characterization of anti-complementary acidic heteroglycans from the seed of Coix lacryma-jobi var. ma-yuen. Phytochemistry 26 3269–3275. [Google Scholar]
- Ye, Z.H. (2002). Vascular tissue differentiation and pattern formation in plants. Annu. Rev. Plant Biol. 53 183–202. [DOI] [PubMed] [Google Scholar]
- York, W.S., Darvill, A.G., McNeil, M., Stevenson, T.T., and Albersheim, P. (1985). Isolation and characterization of plant cell walls and cell wall components. Methods Enzymol. 118 3–40. [Google Scholar]
- Zhong, R., Burk, D.H., Nairn, C.J., Wood-Jones, A., Morrison, W.H., and Ye, Z.H. (2005. a). Mutation of SAC1, an Arabidopsis SAC domain phosphoinositide phosphatase, causes alterations in cell morphogenesis, cell wall synthesis, and actin organization. Plant Cell 17 1449–1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, R., Burk, D.H., and Ye, Z.H. (2001). Fibers. A model for studying cell differentiation, cell elongation, and cell wall biosynthesis. Plant Physiol. 126 477–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, R., Pena, M.J., Zhou, G.K., Nairn, C.J., Wood-Jones, A., Richardson, E.A., Morrison, W.H., Darvill, A.G., York, W.S., and Ye, Z.H. (2005. b). Arabidopsis fragile fiber8, which encodes a putative glucuronyltransferase, is essential for normal secondary wall synthesis. Plant Cell 17 3390–3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.