Abstract
The pentatricopeptide repeat (PPR) family represents one of the largest gene families in plants, with >440 members annotated in Arabidopsis thaliana. PPR proteins are thought to have a major role in the regulation of posttranscriptional processes in organelles. Recent studies have shown that Arabidopsis PPR proteins play an essential, nonredundant role during embryogenesis. Here, we demonstrate that mutations in empty pericarp4 (emp4), a maize (Zea mays) PPR-encoding gene, confer a seed-lethal phenotype. Mutant endosperms are severely impaired, with highly irregular differentiation of transfer cells in the nutrient-importing basal endosperm. Analysis of homozygous mutant plants generated from embryo-rescue experiments indicated that emp4 also affects general plant growth. The emp4-1 mutation was identified in an active Mutator (Mu) population, and cosegregation analysis revealed that it arose from a Mu3 element insertion. Evidence of emp4 molecular cloning was provided by the isolation of four additional emp4 alleles obtained by a reverse genetics approach. emp4 encodes a novel type of PPR protein of 614 amino acids. EMP4 contains nine 35–amino acid PPR motifs and an N-terminal mitochondrion-targeted sequence peptide, which was confirmed by a translational EMP4–green fluorescent protein fusion that localized to mitochondria. Molecular analyses further suggest that EMP4 is necessary to regulate the correct expression of a small subset of mitochondrial transcripts in the endosperm.
INTRODUCTION
The maize (Zea mays) seed comprises two major compartments, the embryo and the endosperm, whose developments are simultaneously initiated by a double fertilization event. The embryo consists of an embryonic axis surrounded by a single massive cotyledon, the scutellum. The embryonic axis comprises all of the tissues that will give rise to the seedling structure, which represents the beginning of the new sporophytic generation. In maize, as in most cereals, the endosperm represents the main storage component of the seed and persists at maturity, during which it provides nutrients to the seedling upon germination. Detailed studies of endosperm development have been reported (Kiesselbach, 1949). The existence of four distinct domains in the differentiated endosperm was determined from a combination of cytological and molecular studies (Bommert and Werr, 2001; Costa et al., 2004; Olsen, 2004). The embryo-surrounding region (ESR) is found at the endosperm–embryo interface. This tissue is thought to play a role in embryo nutrition and/or in establishing a physical barrier between the embryo and the endosperm during seed development. The identification of three small Cys-rich peptides in the ESR is suggestive of an additional signaling role for this tissue (Bonello et al., 2002). The basal region of the endosperm is characterized by the early presence of transfer cells that facilitate nutrient import into the maize kernel. The expression of several basal endosperm transfer layer (BETL)–specific genes has been detected in this tissue (Hueros et al., 1995, 1999b; Gomez et al., 2002; Gutierrez-Marcos et al., 2004), although their functions are not well understood. The bulk of the endosperm is composed of the central starchy endosperm, in which starch and proteins are synthesized and stored. Storage product accumulation begins in this region at ∼12 d after pollination (DAP) and continues until seed maturity. The fourth domain consists of the aleurone, the outermost epidermal cell layer that accumulates proteins and lipids.
For many years, mutant analysis has served as a powerful tool for the identification of genes required for seed development (Neuffer and Sheridan, 1980; Sheridan and Neuffer, 1980; Scanlon et al., 1994). We have focused our attention on a group of mutants, termed empty pericarp (emp), which define a subset of defective kernel (dek) mutants with the most severe reduction in endosperm size. These mutants are easily recognizable in segregating mature ears because they have a flattened appearance as a result of compression by the surrounding normal seeds (Scanlon et al., 1994, 1997). Of the emp genes isolated to date, emp2 is the only one to have been molecularly characterized and was found to encode a heat-shock response regulator protein (Fu et al., 2002). Here, we describe the molecular and genetic characterization of emp4-1, a recessive lethal emp mutation that originated from an active Mutator (Mu) population. Developing emp4-1 seeds were retarded in their growth compared with wild-type seeds and displayed severe morphological abnormalities as well as altered patterns of gene expression within the endosperm transfer tissue. Cosegregation analysis revealed that the mutation was attributable to the insertion of a Mu3 element in the emp4 locus. The isolation of the emp4 gene was achieved through a gene-tagging approach, whereas proof of its molecular cloning was obtained after a reverse genetics–based screen, which identified four additional independent emp4 alleles. emp4 is a single-copy gene, which encodes a novel type of pentatricopeptide repeat (PPR) protein. Our data further suggest that EMP4 is a mitochondrion-targeted PPR protein that is necessary for the correct regulation of mitochondrial gene expression in the endosperm.
RESULTS
Phenotypic and Genetic Characterization of emp4-1
The emp4-1 mutation, previously designated aborted seed7065 (Giulini et al., 2000), was isolated in the F2 generation of a cross between a Mu line and a maize inbred line. The recessive seed-lethal mutation was maintained by selfing heterozygous plants and propagated by outcrossing emp4-1/+ plants to different inbred lines (A188, W64A, and H99).
The emp4-1 mutation was mapped to chromosome 1L after crossing heterozygous mutant plants to the complete set of B-A translocations. Because the emp4-1 seed phenotype resembles that of the dek22 mutant (Clark and Sheridan, 1986), which also resides on the same chromosomal arm, individuals heterozygous for each mutant were intercrossed to test for genetic allelism (for details, see Methods). The resulting ears contained only wild-type seeds, indicating that dek22 and emp4-1 are not allelic.
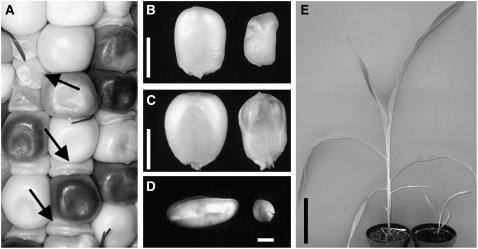
emp4-1 mutants are macroscopically recognizable as early as 10 DAP, because of the pale, translucent, and collapsed appearance of mutant caryopses (Figure 1A). A comparison of wild-type and mutant sibling kernels at 20 and 30 DAP showed a reduction in the size of the mutant caryopsis, which was surrounded by a normal maternal pericarp (Figures 1B and 1C). At 20 DAP, emp4-1/emp4-1 embryos appeared much smaller than wild-type sibling embryos (Figure 1D), and at maturity, homozygous emp4-1 seeds were lethal and were not able to germinate. However, we were able to rescue emp4-1/emp4-1 embryos at 20 DAP by culturing them on a basal medium and then transferring the seedlings to soil. Although not altered significantly in their vegetative architecture, homozygous mutant plants exhibited delayed growth (Figure 1E) but nonetheless were able to reach reproductive maturity.
Figure 1.
Mutant emp4-1 Seed and Plant Phenotypes.
(A) Ear segregating for wild-type and emp4-1 mutant kernels (arrows).
(B) Twenty-DAP wild-type (left) and mutant (right) kernels.
(C) Thirty-DAP wild-type (left) and mutant (right) kernels.
(D) Isolated 20-DAP wild-type (left) and mutant (right) embryos.
(E) Comparison of wild type (left) and homozygous emp4-1 mutant (right) plants obtained from 20-DAP rescued and cultured embryos.
Bars = 5 mm in (B) and (C), 1 mm in (D), and 5 cm in (E).
Endosperm Domains Are Altered in emp4-1 Mutant Kernels
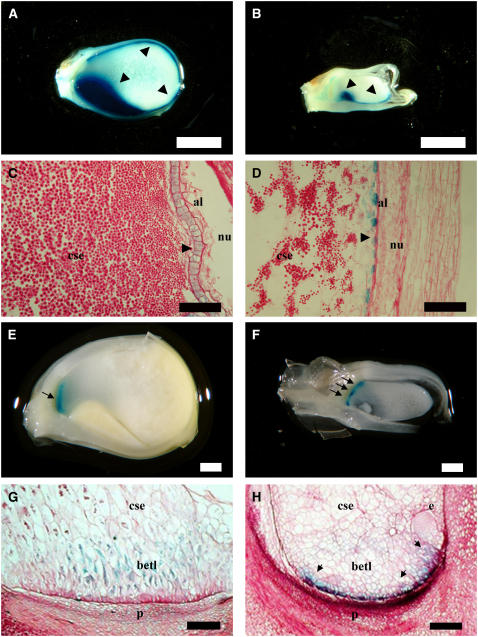
Macroscopically, emp4-1 mutants exhibit more profound phenotypic defects on the endosperm than on the embryo. Therefore, we decided to compare the development of wild-type and mutant sibling kernels to determine which cell types of the endosperm were perturbed in emp4-1 seeds. To analyze the aleurone tissue in emp4-1 seeds, stable maize lines carrying a Vp1 promoter transcriptional fusion to β-glucuronidase (GUS) (Costa et al., 2003) were introgressed in the emp4-1 background. GUS staining was performed on longitudinally cut hand sections of 20-DAP wild-type and mutant kernels (Figures 2A and 2B). In wild-type seeds, GUS staining was present throughout the aleurone, extending from the germinal to the abgerminal face of the seed (Figure 2A), whereas GUS staining was detected only on the germinal face of mutant kernels (Figure 2B). Wild-type aleurone usually appeared as a single layer of isodiametric cells (Figure 2C), whereas in mutants, it was discontinuous and contained many irregularly shaped cells (Figure 2D). Frequently, the presence of ectopic GUS staining was observed in discrete portions of the basal endosperm.
Figure 2.
Effects of emp4-1 on Aleurone and Basal Transfer Layer Development.
(A) to (D) Longitudinal section of 20-DAP wild-type ([A] and [C]) and mutant ([B] and [D]) kernels carrying the pVP1-GUS transgene and showing GUS precipitate in embryo and aleurone cells.
(E) to (H) Longitudinal section of 14-DAP wild-type ([E] and [G]) and emp4-1 ([F] and [H]) kernels carrying the pBET1-GUS transgene and showing GUS precipitate in basal endosperm transfer layer cells.
(C), (D), (G), and (H) are 10-μm wax sections stained with periodic acid Schiff's stain. Arrows and arrowheads highlight areas of interest. al, aleurone; betl, basal endosperm transfer layer; cse, central starchy endosperm; e, embryo; esr, embryo-surrounding region; nu, nucellus; p, placento-chalazal region. Bars = 2 mm in (A) and (B), 100 μm in (C) and (D), 1 mm in (E) and (F), and 200 μm in (G) and (H).
In addition, sections were counterstained with periodic acid Schiff's stain, which allowed an evaluation of starch and carbohydrate composition in the central endosperm of mutant and sibling wild-type kernels. Starch accumulation appeared notably reduced in the central starchy endosperm of emp4-1 mutants compared with that of their wild-type siblings (Figures 2C and 2D).
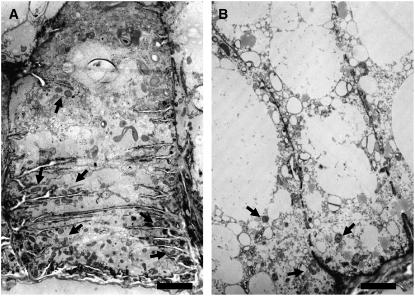
To investigate the development of the BETL, we introgressed the emp4-1 mutation in plants carrying a BETL1 promoter transcriptional fusion to GUS (Hueros et al., 1999a). Mutant and wild-type sibling seeds were analyzed at 14 DAP after staining for GUS (Figures 2E to 2H). In wild-type seeds, expression of the transgene was confined to the BETL (Figure 2E), where it appeared evenly distributed in two or three adjacent cell layers of the transfer region, decreasing progressively toward the center of the endosperm (Figure 2G). By contrast, reporter gene expression was irregular in mutant seeds (Figure 2F); GUS staining was often observed in the peripheral-most cells of the basal endosperm (Figure 2H) but in some cases was absent altogether. In addition, we noticed that some sectors within the basal endosperm of emp4-1 mutants were also devoid of cell wall ingrowths, as indicated by the absence of periodic acid Schiff's stain in these areas (cf. Figures 2G and 2H). To further investigate the morphological abnormalities of the basal endosperm transfer cells in emp4-1 mutants, we sectioned wild-type and mutant kernels and examined them by electron microscopy. In the wild type, the BETL consisted of relatively large, elongated cells with extensive cell wall ingrowths and containing many mitochondria (Figure 3A). By contrast, cells in basal emp4-1 mutant endosperms were smaller, vacuolated, and lacked defined cell wall ingrowths (Figure 3B). In addition, mitochondrial populations were less abundant in mutant transfer cells than in the wild type.
Figure 3.
Transfer Cell Morphology in Wild-Type and emp4-1 Mutant Endosperms.
Electron microscopy images of a typical 9-DAP wild-type basal endosperm transfer cell containing abundant mitochondria (arrows) associated with extensive cell wall ingrowths (A) and a typical transfer cell found in 9-DAP emp4-1 mutant endosperm lacking cell wall ingrowth structures and containing fewer mitochondria (B). Note that the transfer cells are more vacuolated in emp4-1 endosperms than in the wild type. Bars = 2 μm.
Transfer Cell Layer Gene Expression Is Altered in emp4-1 Endosperms
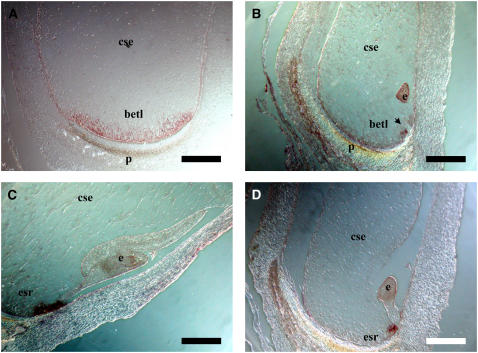
Because of the striking defects found in the basal regions of emp4-1 endosperms, we decided to investigate the expression patterns of genes specifically expressed in tissues of the basal endosperm—the BETL and the ESR—in mutant and wild-type sibling kernels. Therefore, we performed mRNA in situ hybridization analysis on sections of 11-DAP emp4-1 and wild-type kernels using an antisense BETL-specific meg1 probe (Gutierrez-Marcos et al., 2004). As expected, meg1 transcripts were distributed uniformly throughout the BETL of wild-type kernels (Figure 4A), whereas they were distributed irregularly and limited to small sectors in mutant endosperms (Figure 4B). The Zm ESR2 antisense probe was used to detect all three ESR-specific transcripts (Zm ESR1, Zm ESR2, and Zm ESR3) (Opsahl-Ferstad et al., 1997). No differences in expression were detected between wild-type and sibling emp4-1 kernels; in both instances, ESR transcripts were confined to the ESR (Figures 4C and 4D).
Figure 4.
Effects of emp4-1 on Basal Endosperm Gene Expression.
mRNA in situ analysis in wild-type ([A] and [C]) and mutant ([B] and [D]) kernels at 11 DAP with Zm Meg1 antisense probe ([A] and [B]) and Zm Esr2 antisense probe ([C] and [D]). betl, basal endosperm transfer layer; cse, central starchy endosperm; e, embryo; esr, embryo-surrounding region; p, placento-chalazal region. Bars = 500 μm.
Cosegregation Analysis and Cloning of emp4
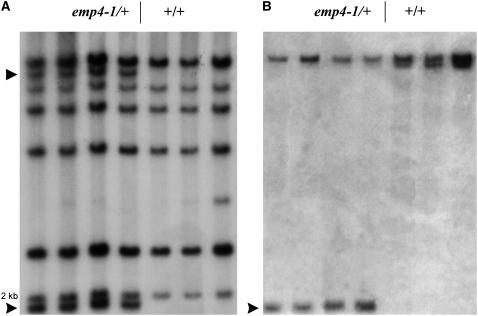
Because the emp4-1 mutant was isolated from an active Mu population, we performed a cosegregation analysis in an attempt to identify whether insertion of a Mu element in the Emp4 locus was responsible for the mutant phenotype. To facilitate the molecular analysis, plants bearing the mutation were outcrossed to W64A, a low Mu copy inbred line, and the F1 plants were selfed to obtain segregating families.
Genomic DNA extracted from leaves of emp4-1/+ and +/+ individuals whose genotype had been ascertained by selfing was compared by DNA gel blot analysis. For this procedure, genomic DNA was digested with PstI, a methylation-sensitive endonuclease that cuts once inside the Mu3 element. Hybridization with a Mu3-specific probe revealed two fragments of ∼9 and 2 kb cosegregating with the mutant phenotype (Figure 5A). Subsequent hybridization with a different Mu3 probe revealed that the 5′ end of the Mu3 element was included in the 2-kb fragment. The 2-kb Mu3 PstI restriction fragment was cloned from a subgenomic library prepared from size-fractionated PstI fragments of emp4-1/+ genomic DNA. The cloned DNA comprised 1115 bp of genomic sequence flanking the Mu3 insertion and was mapped to the long arm of chromosome 1, between markers umc140 and umc106 (see Methods). To verify the identity of the cloned DNA, a 401-bp PstI-MluI sequence (probe 1) was used to reprobe the same PstI-digested DNA gel blot. In emp4-1/+ individuals, the same 2-kb PstI fragment and an additional 11-kb fragment corresponding to the W64A Emp4-1 wild-type allele were identified (Figure 5B). In wild-type individuals, the same 11-kb W64A band was identified, plus a second 10-kb restriction fragment length polymorphism band, corresponding to the Mu line wild-type allele (Figure 5B). Cosegregation analysis was performed on a total of 100 plants, including individuals from F1 and F2 segregating families, and the data obtained were concordant, indicating that the Mu3 element was inserted either in the emp4 gene itself or adjacent to it. DNA gel blot analysis of genomic DNA from a wide range of inbred lines indicated that emp4 is a single-copy gene in maize.
Figure 5.
Cosegregation Analysis of emp4-1 with a Mu3 Element.
(A) Mu3 transposon-tagged PstI fragments (arrowheads) cosegregating with the emp4-1 mutation.
(B) DNA gel blot hybridized with probe 1. A 2-kb PstI band and an 11.2-kb BamHI band show cosegregation with the emp4-1 mutation (arrowhead).
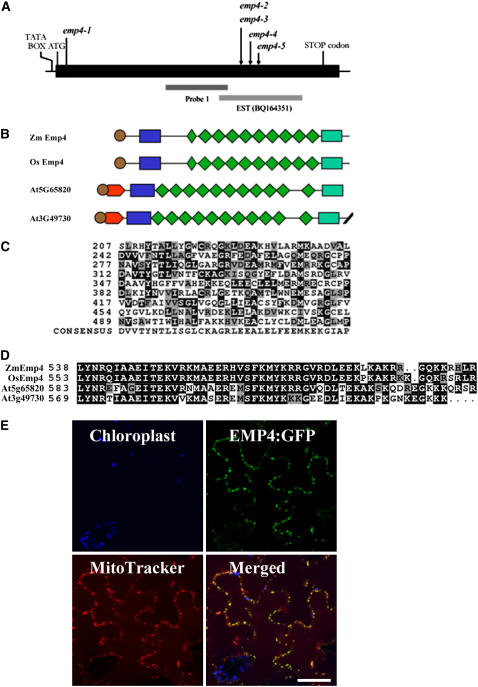
To confirm that the cloned sequence corresponded to the emp4 gene, a reverse genetics approach was used. For this, we screened a Mu gene machine (see Methods) using oligonucleotide primers designed to the emp4 genomic sequence in combination with Mu-specific primers. Four independent germinal Mu insertions in the emp4 locus were identified, each conferring a mutant emp4-1–like seed phenotype. A genetic complementation analysis of emp4-1/+ plants with plants heterozygous for the new insertions later confirmed that all four newly identified Mu insertions were in the same locus and thus defined a new emp4 allelic series: emp4-2, emp4-3, emp4-4, and emp4-5 (see Figure 7A).
Figure 7.
Organization of emp4 Gene and Protein Structure.
(A) Scheme of the emp4 gene. The location of the Mu3 transposon insertion of the emp4-1 allele in the coding sequence and the positions of Mu insertions of known emp4 alleles are indicated with arrows. The putative TATA box, ATG transcriptional start site, and stop codon are shown. Genomic regions comprising probe 1 and EST clone BQ164351 are indicated.
(B) Schematic alignments of maize emp4 (Zm EMP4), rice emp4-like (Os EMP4; AC135956), and Arabidopsis At3g49730 and At5g65820 predicted amino acid products. The PPR motifs and the PPR-like short motif are represented by green rhomboids. The putative signal peptide is shown as brown circles. The novel N-terminal and C-terminal domains are indicated as blue and green boxes, respectively.
(C) Comparison of the nine PPR motifs found in EMP4 with the PPR consensus sequence. Residues identical to the consensus are shaded in black, and similar residues are shaded in gray.
(D) Alignment of the novel EMP4 C-terminal domain present in Zm Emp4, rice Os Emp4 (AC135956), and Arabidopsis At3g49730 and At5g65820.
(E) EMP4 localization in tobacco leaf epidermal cells. Panels show chloroplast autofluorescence (blue), transient expression of EMP4-GFP (green), Mito Tracker Orange localization (red), and merged images. Fluorescent signals were visualized using confocal laser scanning microscopy.
Emp4 Is Expressed in Vegetative and Most Reproductive Plant Tissues
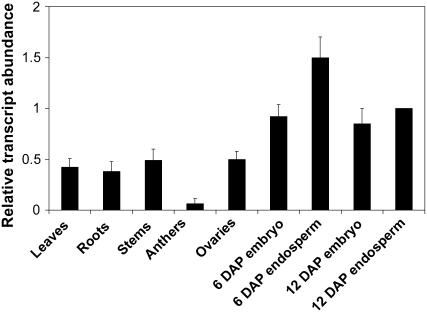
To confirm that the cloned emp4 fragment was indeed expressed, we performed a BLAST search analysis (Altschul et al., 1997), which identified a region with high similarity to an EST from maize immature ears (BQ164351). Gene-specific primers were designed and used in rapid amplification of cDNA ends (RACE) to generate full-length wild-type emp4 cDNA. The sense primer Oest7 and the antisense primer RT5rev were used in combination with the appropriate primers (see Methods and Supplemental Table 1 online) to isolate the emp4 3′ and 5′ regions, respectively. The internal cDNA portion was generated by RT-PCR using a forward primer (RT5) derived from the genomic clone sequence and a reverse primer (Oest3) derived from the EST sequence. emp4 mRNA could not be detected by conventional RNA gel blot analysis, indicating that it is expressed at very low levels. Therefore, we attempted to determine emp4 spatial expression by quantitative RT-PCR (see Methods). Emp4 transcript was detected in a range of tissues, including 6- and 12-DAP embryos and endosperms, leaves, roots, stems, and ovaries, and was found at low levels in anthers (Figure 6).
Figure 6.
emp4 Is Expressed in Vegetative and Reproductive Tissues.
RNA was isolated from leaves, roots, and stems (lanes 1 to 3), anthers (lane 4), ovaries (lane 5), 6-DAP embryos and endosperms (lanes 6 and 7), and 12-DAP embryos and endosperms (lanes 8 and 9). Quantitative RT-PCR analysis was performed using emp4 gene-specific primers, and Actin expression was used to normalize expression values (see Supplemental Table 1 online). Transcript abundance is indicated relative to 12-DAP endosperms, and error bars indicate se (see Methods).
Emp4 Encodes a Mitochondrion-Targeted PPR Protein
To obtain the full-length genomic clone, we screened a maize F2 BAC library (O'Sullivan et al., 2001) with the emp4 PstI-MluI 401-bp probe. Three identical hybridizing BAC clones were isolated, and a 5.2-kb region encompassing the emp4 locus was subcloned and sequenced directly. The alignment between the genomic sequence and the obtained cDNA sequence showed complete homology. The emp4 gene contains a single exon and a putative TATA box at position –38 (Figure 7A). The full-length emp4 cDNA is 1950 bp, excluding the poly(A) tail, and the EMP4 predicted protein contains an open reading frame (ORF) of 614 amino acids. A Pfam search (Bateman et al., 2004) was performed with the putative EMP4 ORF to identify any known amino acid motifs. The results revealed that emp4 encodes a PPR protein that exhibits high homology with one rice (Oryza sativa) and two Arabidopsis thaliana PPR proteins (Figure 7B). The organization of maize EMP4 and rice AC135956 was highly similar, whereas the two Arabidopsis sequences contained an additional domain at the N terminus, located between residues 1 and 61 in At3g49730 and between residues 8 and 69 in At5g65820 (Figure 7B).
The region of the EMP4 protein extending from residue 209 to residue 525 contains nine PPR motifs that show a variable degree of conservation with other PPR proteins (Figure 7C). The PPR motifs are contiguous, except for motifs 7 and 8, which are interrupted by two amino acids. The positions of the nine PPR motifs, therefore, were shifted forward by two amino acids to allow the alignment with the PPR consensus sequence described by Lurin et al. (2004). In addition, a short sequence preceding the PPR tandem repeat motifs was found, showing significant homology with the 31–amino acid long PPR-like short motif (Lurin et al., 2004). Two additional regions were identified in the EMP4 sequence, one located at the N terminus between residues 47 and 102, and the other at the C terminus between amino acids 538 and 590. The C-terminal region had only matches to the Arabidopsis sequences (At3g49730 and At5g65820) and to the rice BAC genomic sequence AC135956 (Figure 7D). By contrast, the N-terminal region, which comprised a stretch of hydrophobic amino acid residues followed by a short sequence of aromatic residues, was conserved in these and other PPR proteins (see Supplemental Figure 1 online). Using TargetP (Emanuelsson et al., 2000), we identified a putative targeting signal peptide at the N terminus of EMP4, which was also predicted for the rice and Arabidopsis homologous PPR proteins. However, a chloroplast subcellular localization signal was predicted for the EMP4 sequence, whereas a mitochondrial localization signal was predicted for rice AC135956 and for the Arabidopsis At3g49730 and At5g65820 putative gene products. The Predotar program (Small et al., 2004) also predicted a chloroplast localization signal for the EMP4 sequence, whereas no prediction was obtained for the other three EMP4-like protein sequences. To experimentally determine the subcellular localization of EMP4, we generated a translational EMP4-GFP fusion construct, which was used for transient expression studies in tobacco (Nicotiana tabacum) leaf epidermal cells. Contrary to the computer predictions, confocal laser scanning microscopy analysis of leaf samples showed in vivo colocalization of the green fluorescent signal of EMP4-GFP with Mito Tracker Orange in mitochondria (Figure 7E).
Mitochondrial Transcripts Are Depleted in emp4-1 Endosperms
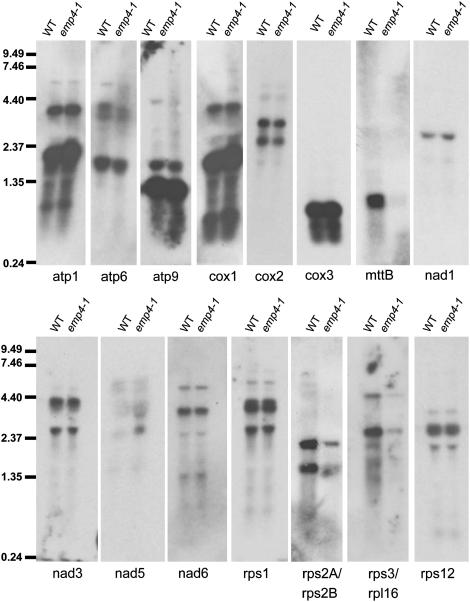
Because EMP4 appeared to target mitochondria, we investigated whether the emp4-1 mutation had any effect on gene expression in this organelle. To this end, we initially performed a microarray analysis of mitochondrial gene expression in 12-DAP wild-type and emp4-1 mutant endosperms (see Methods). This analysis revealed a considerable reduction in gene expression for only a small subset of mitochondrial genes in emp4-1 endosperms (see Supplemental Table 2 online). To confirm these data, we performed RNA gel blot hybridization analysis. We found lower expression of both rps2A/rps2B and rps3/rpl16 and a drastic reduction in mttb (orfX) transcript in emp4-1 mutant endosperms compared with sibling wild-type endosperms (Figure 8). To confirm that the reduced levels of rps2A, rps2B, rps3/rpl16, and mttb (orfX) RNA were not attributable to reduced mitochondrial DNA levels or rearrangements in the mitochondrial genome structure in emp4-1 mutants, we performed DNA gel blot hybridization analysis using specific probes for the corresponding genes and other mitochondrial genes (see Supplemental Figure 2 online). This analysis revealed no differences in DNA levels between wild-type and emp4-1 seeds, suggesting that the observed reduction of some mitochondrial transcripts did not result from DNA defects in mitochondria of emp4-1 seeds.
Figure 8.
RNA Gel Blot Analysis of Mitochondrial Transcript Abundance in Wild-Type and emp4-1 Mutant Endosperms.
Twenty micrograms of total RNA from 12-DAP wild-type and emp4-1 endosperms was analyzed by RNA gel blot hybridization using gene-specific probes. Molecular mass markers are indicated in kilobases at left.
DISCUSSION
Cloning of a Novel emp Gene, emp4, by Transposon Tagging
The phenotype conferred by emp defines a subclass of dek mutants characterized by seeds exhibiting an extreme reduction in endosperm size yet possessing a normal pericarp (Sheridan and Neuffer, 1980; Scanlon et al., 1994, 1997). Because of their dramatic mutant seed phenotype, emp mutants are thought to result from lesions in genes involved in housekeeping functions. To date, only one mutant of this class, emp2, has been molecularly characterized, and the wild-type gene was found to encode a negative regulator of the heat-shock response (Fu et al., 2002). In the absence of EMP2, heat-shock factor genes are upregulated, eventually leading to seed abortion (Fu et al., 2002). Here, we describe the molecular characterization of a second emp gene, termed emp4, which was achieved by transposon tagging. The emp4-1 mutant allele resulted from the insertion of a Mu3 element in the coding region of emp4. Four additional emp4 mutant alleles were identified using a reverse genetics mutagenesis approach. In all cases, a Mu element was found to be inserted within the emp4 coding region and mutant lines exhibited identical emp seed phenotypes. Further molecular evidence revealed that emp4 is a single-copy gene in maize with an essential role in seed development.
emp4 Encodes a PPR Protein Found in Maize and Other Plants
Sequence analysis indicated that emp4 encodes a 614–amino acid protein that is highly homologous with the PPR class of proteins. PPR proteins constitute one of the largest families of conserved proteins in plants and are characterized by multiple repeats of a degenerate 35–amino acid motif containing a distribution of hydrophobic and hydrophilic residues (Small and Peeters, 2000; Hattori et al., 2004). The repeats are usually present as tandem arrays, with an average number of 12 motifs per polypeptide. By contrast, EMP4 contains nine PPR motifs preceded by a short sequence showing partial homology with the 31–amino acid PPR-like short motif described previously by Lurin et al. (2004).
Many PPR proteins contain unrelated sequences either before or after the tandem arrays of PPR motifs. Sequence analyses revealed that EMP4 possesses additional N-terminal and C-terminal regions, which flank the nine PPR repeats. In most PPR proteins, the N terminus contains organelle-targeting signals that show a poor degree of sequence conservation. By contrast, some PPR proteins show a high degree of amino acid conservation at the C terminus, which is used to classify plant PPR proteins into reduced family groups (Lurin et al., 2004). Interestingly, the domain found at the N terminus of EMP4 was conserved in a wide range of plant PPR proteins, whereas the domain identified at the C-terminal region of EMP4 was found in only three other proteins: two Arabidopsis PPR proteins of the P subfamily (Lurin et al., 2004) and a predicted rice PPR protein. Whether the N-terminal region and/or the C-terminal region conserved in EMP4 and the other plant EMP4-like PPR proteins confer a distinct function to these PPRs remains to be determined.
EMP4 Is a Nucleus-Encoded Mitochondrial PPR Protein
PPR genes in plants are thought to encode RNA binding proteins with essential roles in organelles (Fisk et al., 1999; Bentolila et al., 2002; Hashimoto et al., 2003; Kazama and Toriyama, 2003; Koizuka et al., 2003; Meierhoff et al., 2003; Williams and Barkan, 2003; Lurin et al., 2004; Yamazaki et al., 2004; Kotera et al., 2005; Schmitz-Linneweber et al., 2005). PPR proteins are usually targeted to mitochondria or chloroplasts; however, GLUTAMINE-RICH PROTEIN23 is a notable example of a nucleus-localized PPR protein (Ding et al., 2006). Although computer predictions identified a chloroplast-targeted sequence peptide in EMP4, homozygous emp4-1 plants did not display any phenotypes typical of chlorophyll-deficient mutants, such as albinism or lethality. Furthermore, an EMP4-GFP translational fusion construct was found to colocalize with mitochondria, indicating that emp4-1 might affect mitochondrial rather than chloroplast function. Microarray and RNA gel blot analyses further revealed depleted levels of some mitochondrial transcripts, namely rps2A, rps2B, rps3/rpl16, and mttb (orfX), in mutant emp4-1 endosperms compared with their wild-type siblings. rps2A, but not rps2B, is thought to encode the functional version of the ribosomal protein RPS2 (Perrotta et al., 2002). rps3 also encodes a ribosomal protein (Hunt and Newton, 1991), and deletion mutants reported for this gene have resulted in necrotic leaf striations, reduced levels of mitochondrial protein synthesis, severely reduced plant growth, and variable ratios of normal versus mutant mitochondrial DNA (Hunt and Newton, 1991; Newton et al., 1996; Sakamoto et al., 1996). mttb (orfX), on the other hand, encodes a Sec-independent transporter protein (Bogsch et al., 1998; Weiner et al., 1998), which is thought to have a role either in importing folded proteins or in exporting them from the matrix into the intermembrane space (Bogsch et al., 1998). However, no mutants defective in mttb have been reported in plants. Thus, it is unclear to what extent depletion of mttb alone, or in combination with reduced levels of rps2A, rps2B, and rps3/rpl16 transcripts, is responsible for the observed phenotype of emp4-1.
Despite finding irregularities in mitochondrial populations of emp4-1 endosperm transfer cells compared with sibling wild-type cells, we were unable to detect any differences in mitochondrial genome structure and transmission by DNA gel blot analysis. Therefore, it appears that EMP4 is a mitochondrion-targeted protein that regulates transcript levels of a subset of mitochondrial genes that map to distinct regions of the mitochondrial genome (Clifton et al., 2004) and thus are independently transcribed. Consistent with what is predicted for most PPR proteins, EMP4 might function directly as an RNA binding protein. Alternatively, EMP4 might act indirectly as part of a protein complex that regulates mitochondrial gene expression. It is currently unclear how PPRs interact with their target RNAs, although a recent elegant study demonstrated several sites of a maize chloroplast PPR protein, CRP1, to bind specifically to different mRNAs (Schmitz-Linneweber et al., 2005). Furthermore, PPRs are thought to participate in RNA maturation steps, such as editing, stabilization, cleavage, splicing, and translation. At present, it is uncertain in which process EMP4 might be involved. From our molecular analyses, it seems that emp4-1 does not affect precursor RNA processing in the mitochondria. However, transcript stability for a small subset of mitochondrial genes might be affected. It is thus possible that an RNA surveillance mechanism might operate to degrade mRNAs that are not edited in emp4-1 mutants. However, sequence analysis of the mitochondrial transcripts that showed altered expression in emp4-1 endosperms did not reveal any changes in editing compared with the wild type (data not shown). Although technically challenging, future experiments could include run-on transcription analysis of isolated mitochondria from emp4-1 endosperms to assess whether EMP4 affects mitochondrial transcription.
To date, the most common mitochondrion-targeted PPR proteins reported in plants are the Restorers of fertility (Rf) described in petunia (Petunia hybrida), radish (Raphanus sativus), rice, and sorghum (Sorghum bicolor) (Bentolila et al., 2002; Brown et al., 2003; Desloire et al., 2003; Koizuka et al., 2003; Klein et al., 2005; Wang et al., 2006). Rf genes are able to restore male fertility in cytoplasmic male-sterile lines, in which some mitochondrial DNA produces unusual ORFs encoding novel proteins that interfere with normal mitochondrial function and pollen development (Schnable and Wise, 1998; Chase and Gabay-Laughnan, 2003). In maize, naturally occurring rf alleles are homozygous-viable, whereas the majority of rf alleles recovered by spontaneous mutations have deleterious, often lethal, effects on seed development (Chase and Gabay-Laughnan, 2003). These homozygous-lethal rf alleles may result from loss-of-function mutations in nuclear genes that are required for the expression of the mitochondrial cytoplasmic male-sterile locus as well as for the expression of mitochondrial genes encoding essential functions (Cushing et al., 2005). At present, no known Rf loci map close to the emp4 locus; nonetheless, it would be interesting to explore whether emp4 has overlapping functions with any Rf gene.
emp4 Is Necessary for Seed Development and Plant Growth
The analysis of knockout mutants for several PPR-encoding genes in Arabidopsis suggests that they play essential roles in plant development (Lurin et al., 2004; Cushing et al., 2005; Prasad et al., 2005; Ding et al., 2006). Furthermore, the phenotypic diversity observed among different ppr mutants affecting embryogenesis implies that each PPR-encoded gene accounts for a distinct morphogenetic program (Cushing et al., 2005; Ding et al., 2006). Our study provides an example of a maize mitochondrion-targeted PPR required for seed development. We show that lesions in emp4 are associated with specific developmental defects, which are first recognizable in the highly metabolic cells of the basal endosperm transfer layer. During normal endosperm development, cells in the BETL develop highly specialized cell wall ingrowths that increase the surface area of the associated plasmalemma to facilitate the uptake of nutrients from the maternal sporophyte to the seed (Thompson et al., 2001). These cells contain dense cytoplasm rich in mitochondria and organelles of the endomembrane secretory system located close to the wall invaginations (Davis et al., 1990). Because transfer cell differentiation is a high-energy-requiring process (Offler et al., 2003), it is hardly surprising that emp4-1 and other seed mutants in maize displaying reduced or abnormal mitochondria populations also possess a poorly differentiated BETL (Charlton et al., 1995).
At later stages of development, emp4-1 endosperms were poorly filled and displayed some abnormalities in the epidermal aleurone layer. It is possible that these defects are a direct consequence of the emp4-1 mutation, or alternatively, that they result directly from the absence of a wild-type BETL. The latter notion is supported by previous studies that highlight the critical role of the BETL in sugar import and metabolism during seed development (Weber et al., 1997; Emes et al., 2003). The initial stages of seed development are influenced by the level of hexose, which is regulated by invertases that are bound to the transfer cell walls (Miller and Chourey, 1992; Weber et al., 1997; Cheng and Chourey, 1999). Mutant analysis has also shown that defects in the BETL are correlated with reduced seed size, occasionally leading to seed abortion (Lowe and Nelson, 1946; Brink and Cooper, 1947; Maitz et al., 2000; Costa et al., 2003).
Homozygous emp4-1 embryos were retarded in their growth and unable to germinate, unless explanted and cultured on a basal medium in vitro, suggesting that the defects observed in developing emp4-1/emp4-1 embryos were attributable to poor nutrient transfer from the endosperm. However, rescued emp4-1/emp4-1 mutant plants also exhibited retarded growth but lacked any discernible developmental abnormalities. This finding could be explained by the fact that emp4-1 might alter the expression of mitochondrial transcripts with nonessential functions during vegetative plant development. Certainly, mitochondria have been shown to perform various specialized functions; consequently, the proteomic composition of this organelle varies in a tissue-dependent manner and in response to diverse developmental cues and genetic factors (Colas des Francs-Small et al., 1992; Bardel et al., 2002; Hochholdinger et al., 2004).
Together, our data suggest that EMP4 regulates the expression of a small group of mitochondrial genes. Further studies of EMP4 function could provide insight into the not well understood occurrence of nucleus–mitochondrion interactions.
METHODS
Mutant Isolation and Propagation
The emp4-1 mutation was first identified in the selfed progeny of a maize (Zea mays) stock crossed as female to a Mu line (Giulini et al., 2000). This F2 progeny segregated for normal and defective seeds in a 3:1 ratio, showing that emp4-1 behaves as a monogenic recessive mutation. Because emp4-1 is a seed-lethal mutation, it has been maintained in heterozygosis and propagated by outcrossing plants carrying the mutation to different maize inbred lines, including the low-copy Mu line W64A.
For embryo-rescue experiments, immature embryos were excised from caryopses and cultivated on a synthetic medium as described by Consonni et al. (2003). Embryo-rescue seedlings at the three-leaf stage were transferred to soil and, after a period of acclimatization, grown in pots under standard greenhouse conditions.
Genetic Allelism Tests
dek22 was kindly provided by the Maize Genetic Cooperation Stock Center (stock 127D). To determine allelism between emp4 and dek22, dek22/+ plants were selfed and outcrossed as males to a minimum of 20 emp4-1/+ plants whose genotype was determined by DNA gel blot analysis. Ears obtained were scored to determine genetic allelism between the two mutants.
To determine genetic allelism between emp4-1 and other emp4-like mutants isolated from Biogemma's Mu population (see below), reciprocal crosses were performed using a minimum of 20 emp4-1/+ plants that were self-pollinated and outcrossed as males to plants heterozygous for each putative allele. Progeny obtained from these crosses were scored to confirm the presence of mutant alleles and to determine genetic complementation for each mutant tested.
GUS Marker Gene Analysis
emp4-1/+ plants were crossed to lines carrying either pBET1-GUS reporter (Hueros et al., 1999a) or pVP1-GUS (Costa et al., 2003). F1 progeny were genotyped for the presence of the transgene by PCR using GUS-specific oligonucleotides (Gutierrez-Marcos et al., 2004), and plants were self-pollinated or sibbed. Wild-type and mutant kernels were cut longitudinally, and GUS was detected histochemically according to the methods described previously (Costa et al., 2003; Gutierrez-Marcos et al., 2004). Wax sections were mounted onto BDH slides and counterstained with periodic acid Schiff's reagent according to the procedure published by Spence (2001). Polysaccharides stained purple-red.
Transmission Electron Microscopy
Wild-type and mutant 9-DAP endosperms were fixed overnight in 100 mM phosphate buffer, pH 7.5, with 4% paraformaldehyde and 1% gluteraldehyde at room temperature. The material was then washed in four 15-min changes of phosphate buffer and treated in 1% aqueous osmium tetroxide buffer, followed by dehydration in a graded acetone series and embedding in epoxy resin. Ultrathin sections were cut using a diamond knife at 80 nm, stained with uranyl acetate and lead citrate on a 2168 Ultrostainer (Carlsberg System), and collected on 2-mm copper grids coated with Butvar B98 support film. Grids were examined with a JEOL 2000EX transmission electron microscope operating at 80 kV accelerating voltage, and negatives were digitally imaged after conventional development.
mRNA in Situ Hybridization
In situ hybridization experiments were performed on developing kernels at 11 DAP according to Costa et al. (2003).
Cosegregation Analysis
For cosegregation analysis, maize genomic DNA was extracted from 7-d-old seedlings and leaf tissues using the urea extraction method (Chen and Dellaporta, 1994). For DNA gel blots, 10 μg of digested genomic DNA was separated on 0.8% agarose gels. DNA fragments were transferred to Hybond N+ membranes (Amersham) with 10× SSC (1× SSC is 0.15 M NaCl and 0.015 M sodium citrate). Membranes were subsequently probed with α-32P–labeled probes prepared from gel-purified DNA fragments labeled by random primer extension (Prime-a-gene labeling system; Promega). Hybridization probes were as follows: Mu3-specific probe corresponds to the internal HindIII-XbaI Mu3 cloned fragment; and probe 1 was the 401-bp PstII-MluI restriction fragment obtained from the genomic DNA flanking the Mu3 insertion.
Cloning of the Genomic Fragments
A subgenomic library of the polymorphic Mu3 fragment was prepared with DNA extracted from emp4-1/+ plants in the pBluescript SK+ vector according to the manufacturer's instructions (Stratagene). By screening this library with a Mu3-specific probe, a single hybridizing clone was isolated, and the presence of a Mu3 element and DNA flanking the insertion was determined after sequencing.
BAC Library Screening
A maize BAC library ZmF2 (O'Sullivan et al., 2001) was screened with probe 1 after labeling using [α-32P]dCTP (Megaprime DNA labeling system; Amersham). The library filters were prehybridized for 5 h in hybridization buffer (PerfectHyb Plus; Sigma-Aldrich) at 68°C. Hybridization was performed overnight. Filters were washed twice for 20 min in 2× SSC and 0.1% SDS and twice for 20 min in 0.2× SSC and 0.1% SDS. Hybridizing BACs were identified after 24 h of exposure with a Storm 860 imaging system (Amersham).
emp4 Mapping
The emp4 probe 1 fragment was genetically mapped by restriction fragment length polymorphism using the LHRF population (Lignées Hautement Recombinantes F2 × F252; Highly Recombinant Inbred Lines F2 × F252) (M. Falque, L. Decousset, A. Murigneux, N. Dautrevaux, D. Dervins, A. Jacob, N. Ribiere, C. Ridel, G. Albini, J. Joets, and A. Charcosset, unpublished data). Mapping data were analyzed with Mapmaker/EXP3.0 software (Lander et al., 1987) with RiSib and Kosambi centimorgan parameters.
Isolation of Additional emp4 Alleles
Biogemma's Mu population is a collection of 27,500 plants in which endogenous Mu elements have been allowed to transpose at high frequency. Screening for Mu insertions in EMP4 was performed on F1 plant material, and their genotypes were confirmed on F2 plant material. Mutant screens were performed using a PCR-based method with an efficient transposon terminal inverted repeat (TIR)-specific primer (OmuA, 5′-CTTCGTCCATAATGGCAATTATCTC-3′) that hybridizes with both the 5′ and 3′ TIR and antisense CP1 (5′-AGCTGCTCCTTCTTCTCGTG-3′) and sense TSP1 (5′-GCACTTCTTCCACTGGTGCT-3′) emp4-specific primers. Both primers were designed on the sequence of the fragment named probe 1. Four independent insertions were identified: C0232, C1220, B2033, and E2433. The alleles were subsequently renamed emp4-2, emp4-3, emp4-4, and emp4-5 once a lack of genetic complementation was confirmed.
cDNA Isolation and emp4 Transcript Detection
Total RNA was extracted from maize tissues ground in liquid N2 as described previously (Bateman et al., 2004), and poly(A)+ RNA was isolated using Oligotex (Qiagen) according to the manufacturer's instructions. Reverse transcription and RACE were performed according to the recommended protocol provided with the SMART RACE cDNA amplification kit (BD Biosciences Clontech). Reactions for the 3′ RACE were conducted with primer Oest5 and nested primer Oest7, both designed on the EST BQ164351 sequence. For the 5′ RACE, the universal primer provided with the kit was used in combination with primer Oest7rev and with nested primer. The internal region of the cDNA was generated with primers RT5 and Oest3. Oest7rev and Oest3 primers were designed on the EST BQ164351 sequence, whereas RT5 and RT5rev sequences were deduced from the portion of the genomic clone overlapping the 5′ RACE product. Primer sequences are listed in Supplemental Table 1 online.
For quantitative RT-PCR, cDNA from wild-type and emp4-1 plants was amplified on an ABI PRISM 7300 sequence detection system (Applied Biosystems). Primer pairs were designed with Primer Express 2.0 (Applied Biosystems) to obtain a PCR product of 50 to 100 bp. Amplification reactions were prepared with the SYBR-Green PCR Master kit (Applied Biosystems) according to the manufacturer's specifications with 0.4 μM of primers and with 1 μL of cDNA per reaction. Each reaction was made in triplicate, and the real-time experiment was repeated three times. The efficiency of each primer set and the calculation of the level of expression were determined according to Pfaffl (2001). Error bars in the figures represents the se calculated on experiment repetitions. Expression levels were normalized with the values obtained for the housekeeping Actin gene, which was used as an internal reference gene.
Sequence Analysis
The emp4 ORF was predicted using ORF Finder (http://www.ncbi.nlm.nih.gov/gorf/gorf.html). PPR motifs and the C-terminal domain were identified in EMP4 with the protein search software Pfam (http://www.sanger.ac.uk/Software/Pfam/search.shtml) (Bateman et al., 2004). PPR motifs were aligned with the HMMER-derived consensus from 2357 PPR motifs. All of the alignments were calculated using ClustalX (Thompson et al., 1994) and Boxshade (Bioinformatics Group, Swiss Institute for Experimental Cancer Research). The presence of a targeting sequence in the ORF was determined using TargetP (http://www.cbs.dtu.dk/services/TargetP) (Emanuelsson et al., 2000) and GENOPLANTE Predotar version 1.03 (http://genoplante-info.infobiogen.fr/predotar/predotar.html) (Small et al., 2004). The BLAST search against PlantGDB (http://www.plantgdb.org/cgi-bin/PlantGDBblast) (Dong et al., 2004) was performed to identify the C-terminal domain in rice (Oryza sativa) and Arabidopsis thaliana sequences.
Subcellular Localization of EMP4
To generate a translational protein fusion between EMP4 and GFP, the full-length ORF of emp4 was amplified by PCR and introduced into the binary vector pGWB5 (a gift from Tsuyoshi Nakagawa, Shimane University) by GATEWAY in vitro site-specific recombination (Invitrogen). This binary construct contained a cauliflower mosaic virus 35S promoter that allowed the constitutive expression of the EMP4:GFP and was introduced into Agrobacterium tumefaciens strain GV3101 (pMP90) (Koncz and Schell, 1986) by electroporation. Agrobacterium-mediated transient expression was then performed in tobacco (Nicotiana tabacum SR1 cv Petit Havana) as described by Batoko et al. (2000), with some modifications. Briefly, a single colony of the transformed Agrobacterium was used to inoculate 5 mL of YEB medium, supplemented with 100 μg/mL kanamycin and 10 μg/mL gentamycin. The bacterial culture was incubated at 28°C overnight, and the bacteria were pelleted by centrifugation at 2200g for 15 s in a microcentrifuge at room temperature. The pellet was washed three times with 1 mL of the infiltration buffer (50 mM MES, pH 5.6, 2 mM Na3PO4, 0.5% glucose [w/v], and 100 μM acetosyringone) (Sigma-Aldrich) and then resuspended in 1 mL of the same buffer. The bacterial suspension was diluted with infiltration buffer to adjust the inoculum concentration to 0.1 OD600. The inoculum was delivered to the lamina tissues of tobacco leaves by gentle-pressure infiltration through a small needle puncture created in the lower epidermis. The plant was incubated under normal growing conditions for 48 h before confocal microscopy analysis. Leaf samples were harvested at 48 h after infiltration, and protein fusions were visualized using a Zeiss LSM 510 META set to measure an emission band of 475 to 525 nm for GFP and an emission band of 599 nm for Mito Tracker Orange for mitochondrial colocalization. The software LSM dummy (Zeiss) was used for postacquisition image processing.
Mitochondrial Gene Expression Analysis
For RNA isolation, 12-DAP endosperms were harvested in liquid N2 and stored at −80°C. Total RNAs were extracted according to the Trizol–chloroform procedure (Gibco BRL). Two micrograms of total RNA was used for cDNA synthesis with either Cy3 or Cy5 dye molecules, using the 3DNA 900 expression array detection kit for microarrays (Genisphere). Microarray slides containing all maize mitochondrial genes were obtained from D. Stern (Boyce Thompson Institute, Cornell University). cDNA hybridization and washing procedures of the 3DNA 900 expression array detection kit for microarrays were performed according to the manufacturer's instructions. The microarray hybridization was repeated four times to sample the technical variability, with repeats of each dye combination. Microarray slides were scanned using a Scan Array Express HT scanner (Perkin-Elmer), and spot features were identified, quantified, and assessed using BlueFuse (BlueGnome). The complete data set was then addressed using the BlueFuse spot hybridization value score (pON > 0.60). Images were manually flagged, and data from four independent hybridizations was used for statistical analysis.
For RNA gel blot analysis, 20 μg of total RNA was isolated from 12-DAP endosperms fractionated on a 1.5% agarose gel, blotted onto a Hybond-N+ membrane (Amersham), and hybridized with 32P-labeled mitochondrial probes: atp1, atp6, atp9, cox1, cox2, cox3, mttb (orfX), nad1, nad3, nad5, nad6, rps1, rps2A, rps2B, rps3, rpl16, and rps12 (for details, see Supplemental Table 1 online). For DNA gel blot analysis, 10 μg of 12-DAP endosperm genomic DNA was digested with EcoRI and HindIII restriction enzymes, separated on a 0.8% agarose 1× Tris-acetate-EDTA gel, blotted onto a Hybond-N+ membrane (Amersham), and hybridized with the same 32P-labeled mitochondrial probes.
Accession Number
Sequence data for the Emp4 cDNA can be found in the GenBank/EMBL data libraries under accession number DQ291135.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table 1. Details of Primer Names and Sequences Used.
Supplemental Table 2. Microarray Data from Four Independent Experiments Showing Relative Expression of Mitochondrial Transcripts between Wild-Type and emp4-1 Endosperms.
Supplemental Figure 1. Amino Acid Sequence Similarity between the N-Terminal Domain of EMP4 and a Small Group of Rice and Arabidopsis PPR Proteins.
Supplemental Figure 2. DNA Gel Blot Analysis of Mitochondrial Genes in Wild-Type and emp4-1 Endosperms.
Supplementary Material
Acknowledgments
We thank D. Stern (BTI) for providing microarray slides and R. Capper and N. Saunders (University of Oxford) for assistance and advice with the microarray analysis. We also thank K. Newton (University of Missouri) for kindly providing the mitochondrial clones for RNA gel blot analysis, J. Sheldon (Oxford Brookes University) for her help with resin sectioning and electron microscopy, and J. Baker (University of Oxford) for image processing. This work was supported by the Italian Ministero dell'Università e della Ricerca Scientifica e Tecnologica (Project COFIN PRIN 2001 to G.C.) and by European Union Framework V (MAZE) Project QLK3-200-00196.
The authors responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) are: José F. Gutiérrez-Marcos (jose.gutierrez@plants.ox.ac.uk) and Gabriella Consonni (gabriella.consonni@unimi.it).
Online version contains Web-only data.
References
- Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. (1997). Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardel, J., Louwagie, M., Jaquinod, M., Jourdain, A., Luche, S., Rabilloud, T., Macherel, D., Garin, J., and Bourguignon, J. (2002). A survey of the plant mitochondrial proteome in relation to development. Proteomics 2 880–898. [DOI] [PubMed] [Google Scholar]
- Bateman, A., et al. (2004). The Pfam protein families database. Nucleic Acids Res. 32 D138–D141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batoko, H., Zheng, H.-Q., Hawes, C., and Moore, I. (2000). A Rab1 GTPase is required for transport between the endoplasmic reticulum and Golgi apparatus and for normal Golgi movement in plants. Plant Cell 12 2201–2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentolila, S., Alfonso, A.A., and Hanson, M.R. (2002). A pentatricopeptide repeat-containing gene restores fertility to cytoplasmic male-sterile plants. Proc. Natl. Acad. Sci. USA 99 10887–10892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogsch, E.G., Sargent, F., Stanley, N.R., Berks, B.C., Robinson, C., and Palmer, T. (1998). An essential component of a novel bacterial protein export system with homologues in plastids and mitochondria. J. Biol. Chem. 273 18003–18006. [DOI] [PubMed] [Google Scholar]
- Bommert, P., and Werr, W. (2001). Gene expression patterns in the maize caryopsis: Clues to decisions in embryo and endosperm development. Gene 271 131–142. [DOI] [PubMed] [Google Scholar]
- Bonello, J.F., Sevilla-Lecoq, S., Berne, A., Risueno, M.C., Dumas, C., and Rogowsky, P.M. (2002). Esr proteins are secreted by the cells of the embryo surrounding region. J. Exp. Bot. 53 1559–1568. [DOI] [PubMed] [Google Scholar]
- Brink, R.A., and Cooper, D.C. (1947). Effect of the de17 allele on development of the maize caryopsis. Genetics 32 350–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, G.G., Formanova, N., Jin, H., Wargachuk, R., Dendy, C., Patil, P., Laforest, M., Zhang, J., Cheung, W.Y., and Landry, B.S. (2003). The radish Rfo restorer gene of Ogura cytoplasmic male sterility encodes a protein with multiple pentatricopeptide repeats. Plant J. 35 262–272. [DOI] [PubMed] [Google Scholar]
- Charlton, W.L., Keen, C.L., Merriman, C., Lynch, P., Greenland, A.J., and Dickinson, H.G. (1995). Endosperm development in Zea mays: Implication of gametic imprinting and paternal excess in regulation of transfer layer development. Development 121 3089–3097. [Google Scholar]
- Chase, C.D., and Gabay-Laughnan, S. (2003). Exploring mitochondrial-nuclear genome interactions with S male-sterile maize. Recent Res. Dev. Genet. 3 31–41. [Google Scholar]
- Chen, J., and Dellaporta, S. (1994). Urea-based plant DNA miniprep. In The Maize Handbook, M. Freeling and V. Walbot, eds (New York: Springer-Verlag), pp. 526–527.
- Cheng, W., and Chourey, P.S. (1999). Genetic evidence that invertase-mediated release of hexoses is critical for appropriate carbon partitioning and normal seed development in maize. Theor. Appl. Genet. 98 485–495. [Google Scholar]
- Clark, J.K., and Sheridan, W.F. (1986). Developmental profiles of the maize embryo-lethal mutants dek22 and dek23. J. Hered. 77 83–92. [Google Scholar]
- Clifton, S.W., et al. (2004). Sequence and comparative analysis of the maize NB mitochondrial genome. Plant Physiol. 136 3486–3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colas des Francs-Small, C., Ambard-Bretteville, F., Darpas, A., Salantin, M., Huet, J., Pernollet, J., and Remy, R. (1992). Variation in polypeptide composition of mitochondria isolated from different potato tissues. Plant Physiol. 98 273–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consonni, G., Aspesi, C., Barbante, A., Dolfini, S., Giuliani, C., Giulini, A., Hansen, S., Brettschneider, R., Pilu, R., and Gavazzi, G. (2003). Analysis of four maize mutants arrested in early embryogenesis reveals an irregular pattern of cell division. Sex. Plant Reprod. 15 281–290. [Google Scholar]
- Costa, L.M., Gutierrez-Marcos, J.F., Brutnell, T.P., Greenland, A.J., and Dickinson, H.G. (2003). The globby1 (glo1–1) mutation disrupts nuclear and cell division in the developing maize seed causing alterations in endosperm cell fate and tissue differentiation. Development 130 5009–5017. [DOI] [PubMed] [Google Scholar]
- Costa, L.M., Gutierrez-Marcos, J.F., and Dickinson, H.G. (2004). More than a yolk: The short life and complex times of the plant endosperm. Trends Plant Sci. 9 507–514. [DOI] [PubMed] [Google Scholar]
- Cushing, D.A., Forsthoefel, N.R., Gestaut, D.R., and Vernon, D.M. (2005). Arabidopsis emb175 and other ppr knockout mutants reveal essential roles for pentatricopeptide repeat (PPR) proteins in plant embryogenesis. Planta. 221 424–436. [DOI] [PubMed] [Google Scholar]
- Davis, R.W., Smith, J.D., and Cobb, B.G. (1990). A light and electron microscopic investigation of the transfer cell region of maize caryopsis. Can. J. Bot. 68 471–479. [Google Scholar]
- Desloire, S., et al. (2003). Identification of the fertility restoration locus, Rfo, in radish, as a member of the pentatricopeptide-repeat protein family. EMBO Rep. 4 588–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, Y.H., Liu, N.Y., Tang, Z.S., Liu, J., and Yang, W.C. (2006). Arabidopsis GLUTAMINE-RICH PROTEIN23 is essential for early embryogenesis and encodes a novel nuclear PPR motif protein that interacts with RNA polymerase II subunit III. Plant Cell 18 815–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, Q., Schlueter, S.D., and Brendel, V. (2004). PlantGDB, plant genome database and analysis tools. Nucleic Acids Res. 32 D354–D359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson, O., Nielsen, H., Brunak, S., and von Heijne, G. (2000). Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 300 1005–1016. [DOI] [PubMed] [Google Scholar]
- Emes, M.J., Bowsher, C.G., Hedley, C., Burrell, M.M., Scrase-Field, E.S., and Tetlow, I.J. (2003). Starch synthesis and carbon partitioning in developing endosperm. J. Exp. Bot. 54 569–575. [DOI] [PubMed] [Google Scholar]
- Fisk, D.G., Walker, M.B., and Barkan, A. (1999). Molecular cloning of the maize gene crp1 reveals similarity between regulators of mitochondrial and chloroplast gene expression. EMBO J. 18 2621–2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, S., Meeley, R., and Scanlon, M.J. (2002). empty pericarp2 encodes a negative regulator of the heat shock response and is required for maize embryogenesis. Plant Cell 14 3119–3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giulini, A., Consonni, G., Aspesi, C., and Gavazzi, G. (2000). Molecular analysis of abs7065, a mutant with severe impairment in seed development. Maize Genet. Coop. News Lett. 74 50–51. [Google Scholar]
- Gomez, E., Royo, J., Guo, Y., Thompson, R., and Hueros, G. (2002). Establishment of cereal endosperm expression domains: Identification and properties of a maize transfer cell–specific transcription factor, ZmMRP-1. Plant Cell 14 599–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez-Marcos, J.F., Costa, L.M., Biderre-Petit, C., Khbaya, B., O'Sullivan, D.M., Wormald, M., Perez, P., and Dickinson, H.G. (2004). maternally expressed gene1 is a novel maize endosperm transfer cell–specific gene with a maternal parent-of-origin pattern of expression. Plant Cell 16 1288–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto, M., Endo, T., Peltier, G., Tasaka, M., and Shikanai, T. (2003). A nucleus-encoded factor, CRR2, is essential for the expression of chloroplast ndhB in Arabidopsis. Plant J. 36 541–549. [DOI] [PubMed] [Google Scholar]
- Hattori, M., Hasebe, M., and Sugita, M. (2004). Identification and characterization of cDNAs encoding pentatricopeptide repeat proteins in the basal land plant, the moss Physcomitrella patens. Gene 343 305–311. [DOI] [PubMed] [Google Scholar]
- Hochholdinger, F., Guo, L., and Schnable, P.S. (2004). Cytoplasmic regulation of the accumulation of nuclear-encoded proteins in the mitochondrial proteome of maize. Plant J. 37 199–208. [DOI] [PubMed] [Google Scholar]
- Hueros, G., Gomez, E., Cheikh, N., Edwards, J., Weldon, M., Salamini, F., and Thompson, R.D. (1999. a). Identification of a promoter sequence from the BETL1 gene cluster able to confer transfer-cell-specific expression in transgenic maize. Plant Physiol. 121 1143–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hueros, G., Royo, J., Maitz, M., Salamini, F., and Thompson, R.D. (1999. b). Evidence for factors regulating transfer cell-specific expression in maize endosperm. Plant Mol. Biol. 41 403–414. [DOI] [PubMed] [Google Scholar]
- Hueros, G., Varotto, S., Salamini, F., and Thompson, R.D. (1995). Molecular characterization of BET1, a gene expressed in the endosperm transfer cells of maize. Plant Cell 7 747–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt, M.D., and Newton, K.J. (1991). The NCS3 mutation: Genetic evidence for the expression of ribosomal protein genes in Zea mays mitochondria. EMBO J. 10 1045–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazama, T., and Toriyama, K. (2003). A pentatricopeptide repeat-containing gene that promotes the processing of aberrant atp6 RNA of cytoplasmic male-sterile rice. FEBS Lett. 544 99–102. [DOI] [PubMed] [Google Scholar]
- Kiesselbach, T.A. (1949). The Structure and Reproduction of Corn. (Lincoln, NE: University of Nebraska Press, College of Agriculture, Experimental Station).
- Klein, R.R., Klein, P.E., Mullet, J.E., Minx, P., Rooney, W.L., and Schertz, K.F. (2005). Fertility restorer locus Rf1 [corrected] of sorghum (Sorghum bicolor L.) encodes a pentatricopeptide repeat protein not present in the colinear region of rice chromosome 12. Theor. Appl. Genet. 111 994–1012. [DOI] [PubMed] [Google Scholar]
- Koizuka, N., Imai, R., Fujimoto, H., Hayakawa, T., Kimura, Y., Kohno-Murase, J., Sakai, T., Kawasaki, S., and Imamura, J. (2003). Genetic characterization of a pentatricopeptide repeat protein gene, orf687, that restores fertility in the cytoplasmic male-sterile Kosena radish. Plant J. 34 407–415. [DOI] [PubMed] [Google Scholar]
- Koncz, C., and Schell, J. (1986). The promoter of TL-DNA gene 5 controls the tissue-specific expression of chimaeric genes carried by a novel type of Agrobacterium binary vector. Mol. Gen. Genet. 204 383–396. [Google Scholar]
- Kotera, E., Tasaka, M., and Shikanai, T. (2005). A pentatricopeptide repeat protein is essential for RNA editing in chloroplasts. Nature 433 326–330. [DOI] [PubMed] [Google Scholar]
- Lander, E.S., Green, P., Abrahamson, J., Barlow, A., Daly, M.J., Lincoln, S.E., and Newburg, L. (1987). MAPMAKER: An interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1 174–181. [DOI] [PubMed] [Google Scholar]
- Lowe, J., and Nelson, O.E. (1946). Miniature seed —A study in the development of a defective caryopsis in maize. Genetics 31 525–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lurin, C., et al. (2004). Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell 16 2089–2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maitz, M., Santandrea, G., Zhang, Z., Lal, S., Hannah, L.C., Salamini, F., and Thompson, R.D. (2000). rgf1, a mutation reducing grain filling in maize through effects on basal endosperm and pedicel development. Plant J. 23 29–42. [DOI] [PubMed] [Google Scholar]
- Meierhoff, K., Felder, S., Nakamura, T., Bechtold, N., and Schuster, G. (2003). HCF152, an Arabidopsis RNA binding pentatricopeptide repeat protein involved in the processing of chloroplast psbB-psbT-psbH-petB-petD RNAs. Plant Cell 15 1480–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, M.E., and Chourey, P.S. (1992). The maize invertase-deficient miniature1 seed mutation is associated with aberrant pedicel and endosperm development. Plant Cell 4 297–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuffer, M.G., and Sheridan, F.W. (1980). Defective kernel mutants of maize. I. Genetic and lethality studies. Genetics 95 929–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton, K.J., Mariano, J.M., Gibson, C.M., Kuzmin, E., and Gabay-Laughnan, S. (1996). Involvement of S2 episomal sequences in the generation of NCS4 deletion mutation in maize mitochondria. Dev. Genet. 19 277–286. [DOI] [PubMed] [Google Scholar]
- Offler, C.E., McCurdy, D.W., Patrick, J.W., and Talbot, M.J. (2003). Transfer cells: Cells specialized for a special purpose. Annu. Rev. Plant Biol. 54 431–454. [DOI] [PubMed] [Google Scholar]
- Olsen, O.A. (2004). Nuclear endosperm development in cereals and Arabidopsis thaliana. Plant Cell 16 (suppl.): S214–S227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opsahl-Ferstad, H.G., Le Deunff, E., Dumas, C., and Rogowsky, P.M. (1997). ZmEsr, a novel endosperm-specific gene expressed in a restricted region around the maize embryo. Plant J. 12 235–246. [DOI] [PubMed] [Google Scholar]
- O'Sullivan, D.M., Ripoll, P.J., Rodgers, M., and Edwards, K.J. (2001). A Zea mays bacterial artificial chromosome (BAC) library from the European flint inbred line F2. Theor. Appl. Genet. 103 425–432. [Google Scholar]
- Perrotta, G., Grienenberger, J.M., and Gualberto, J.M. (2002). Plant mitochondrial rps2 genes code for proteins with a C-terminal extension that is processed. Plant Mol. Biol. 50 523–533. [DOI] [PubMed] [Google Scholar]
- Pfaffl, M.W. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29 e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad, A.M., Sivanandan, C., Resminath, R., Thakare, D.R., Bhat, S.R., and Srinivasan. (2005). Cloning and characterization of a pentatricopeptide protein encoding gene (LOJ) that is specifically expressed in lateral organ junctions in Arabidopsis thaliana. Gene 353 67–79. [DOI] [PubMed] [Google Scholar]
- Sakamoto, W., Kondo, H., Murata, M., and Motoyoshi, F. (1996). Altered mitochondrial gene expression in a maternal distorted leaf mutant of Arabidopsis induced by chloroplast mutator. Plant Cell 8 1377–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scanlon, M.J., Myers, A.M., Schneeberger, R.G., and Freeling, M. (1997). The maize gene empty pericarp2 is required for progression beyond early stages of embryogenesis. Plant J. 12 901–909. [Google Scholar]
- Scanlon, M.J., Stinard, P.S., James, M.G., Myers, A.M., and Robertson, D.S. (1994). Genetic analysis of 63 mutations affecting maize kernel development isolated from Mutator stocks. Genetics 136 281–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz-Linneweber, C., Williams-Carrier, R., and Barkan, A. (2005). RNA immunoprecipitation and microarray analysis show a chloroplast pentatricopeptide repeat protein to be associated with the 5′ region of mRNAs whose translation it activates. Plant Cell 17 2791–2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable, P.S., and Wise, R.P. (1998). The molecular basis of cytoplasmic male sterility and fertility restoration. Trends Plant Sci. 3 175–180. [Google Scholar]
- Sheridan, W.F., and Neuffer, M.G. (1980). Defective kernel mutants of maize. II. Morphological and embryo culture studies. Genetics 95 945–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small, I., Peeters, N., Legeai, F., and Lurin, C. (2004). Predotar: A tool for rapidly screening proteomes for N-terminal targeting sequences. Proteomics 4 1581–1590. [DOI] [PubMed] [Google Scholar]
- Small, I.D., and Peeters, N. (2000). The PPR motif—A TPR-related motif prevalent in plant organellar proteins. Trends Biochem. Sci. 25 46–47. [DOI] [PubMed] [Google Scholar]
- Spence, J. (2001). Plant histology. In Plant Cell Biology: A Practical Approach, C. Hawes and B. Satiat-Jeunemaitre, eds (Oxford, UK: Oxford University Press), pp. 189–206.
- Thompson, J.D., Higgins, D.G., and Gibson, T.J. (1994). CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, R.D., Hueros, G., Becker, H., and Maitz, M. (2001). Development and functions of seed transfer cells. Plant Sci. 160 775–783. [DOI] [PubMed] [Google Scholar]
- Wang, Z., et al. (2006). Cytoplasmic male sterility of rice with boro II cytoplasm is caused by a cytotoxic peptide and is restored by two related PPR motif genes via distinct modes of mRNA silencing. Plant Cell 18 676–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber, H., Wobus, U., and Borisjuk, L. (1997). Sugar import and metabolism during seed development. Trends Plant Sci. 2 169–174. [Google Scholar]
- Weiner, J.H., Bilous, P.T., Shaw, G.M., Lubitz, S.P., Frost, L., Thomas, G.H., Cole, J.A., and Turner, R.J. (1998). A novel and ubiquitous system for membrane targeting and secretion of cofactor-containing proteins. Cell 93 93–101. [DOI] [PubMed] [Google Scholar]
- Williams, P.M., and Barkan, A. (2003). A chloroplast-localized PPR protein required for plastid ribosome accumulation. Plant J. 36 675–686. [DOI] [PubMed] [Google Scholar]
- Yamazaki, H., Tasaka, M., and Shikanai, T. (2004). PPR motifs of the nucleus-encoded factor, PGR3, function in the selective and distinct steps of chloroplast gene expression in Arabidopsis. Plant J. 38 152–163. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.