Abstract
Phosphatidylinositol monophosphate 5-kinase (PIP5K) plays an essential role in coordinating plant growth, especially in response to environmental factors. To explore the physiological function of PIP5K, we characterized Arabidopsis thaliana PIP5K9, which is constitutively expressed. We found that a T-DNA insertion mutant, pip5k9-d, which showed enhanced PIP5K9 transcript levels, had shortened primary roots owing to reduced cell elongation. Transgenic plants overexpressing PIP5K9 displayed a similar root phenotype. Yeast two-hybrid assays identified a cytosolic invertase, CINV1, that interacted with PIP5K9, and the physiological relevance of this interaction was confirmed by coimmunoprecipitation studies using plant extracts. CINV1-deficient plants, cinv1, had reduced activities of both neutral and acid invertases as well as shortened roots. Invertase activities in pip5k9-d seedlings were also reduced, suggesting a negative regulation of CINV1 by PIP5K9. In vitro studies showed that PIP5K9 interaction indeed repressed CINV1 activities. Genome-wide expression studies revealed that genes involved in sugar metabolism and multiple developmental processes were altered in pip5k9-d and cinv1, and the altered sugar metabolism in these mutants was confirmed by metabolite profiling. Together, our results indicate that PIP5K9 interacts with CINV1 to negatively regulate sugar-mediated root cell elongation.
INTRODUCTION
The phosphatidylinositol (PI) signaling pathway is crucial for the complex regulation of eukaryotic cells. Although the importance of phosphoinositides as lipid signaling molecules in eukaryotic cells was first explored in the 1950s (Hokin and Hokin, 1953), investigation of the role of these signaling molecules in higher plants began more recently. Phosphatidylinositol monophosphate 5-kinase (PIP5K), a key enzyme in the PI signaling pathway, catalyzes the synthesis of membrane PI-4,5-bisphosphate [PI(4,5)P2] by phosphorylation of PI-4-phosphate (PI4P) at the 5 position of the inositol ring (Divecha et al., 1995). PIP5Ks play essential roles in multiple cellular processes and may regulate gene transcription, mRNA export, cell cycle progression, and DNA repair (Irvine, 2003). PIP5Ks are present in various cellular compartments, including the plasma membrane, Golgi, vesicles, and even the nuclear matrix (Payrastre et al., 1992). In mammalian cells, localization of PIP5Ks and PI(4,5)P2 to nuclear speckles is associated with pre-mRNA processing factors (Boronenkov et al., 1998) and regulators involved in G1- to S-phase progression (Divecha et al., 2002). In addition, PIP5K isoforms can be dynamically regulated by Src Tyr kinase and Shp-1 Tyr phosphatase (Bairstow et al., 2005) or by phosphorylation and factors including membrane receptors, Rho family GTPases, ARF family GTPase, or by interacting with phospholipase D (Oude Weernink et al., 2004).
In Arabidopsis thaliana, 15 PIP5K isoforms have been identified, and 9 of them are highly similar to identified PIP5Ks (Mueller-Roeber and Pical, 2002). Previous studies showed that At PIP5K1 was rapidly induced upon drought, salt, and abscisic acid treatment (Mikami et al., 1998); however, which phosphoinositide molecules are produced by At PIP5K1 in vivo are still unknown, although this enzyme catalyzes the synthesis of PI(4,5)P2 and PI(3,4,5)P3 in insect cells (Elge et al., 2001) or PI(3,5)P2 and PI(4,5)P2 in Escherichia coli (Westergren et al., 2001). In addition, differential expression of PIP5K isoforms in various tissues and during cellular responses to environmental stimuli suggests that they perform different functions (Lin et al., 2004). In Oryza sativa, Os PIP5K1 negatively regulates rice heading time by controlling floral induction genes (Ma et al., 2004). In mammalian cells, there was an intimate relationship between PI(4,5)P2 and membrane actin cytoskeleton (Lassing and Lindberg, 1985; Janmey and Stossel, 1987). In plants, PI(4,5)P2 regulates actin dynamics in growing and maturing root hairs through interacting with profilin (Braun et al., 1999). Rac-type Rho proteins physically associate with PIPK activity and PI(4,5)P2 to regulate actin organization and polar growth of the pollen tube (Kost et al., 1999). Although PIP5Ks can serve as a regulator of many cellular proteins that are implicated in numerous processes of signal transduction and cytoskeleton organization (Anderson et al., 1999; Oude Weernink et al., 2004), their physiological roles and mechanism of action in plants remain unclear.
Multiple factors are known to affect plant root growth, one of which is sucrose. In addition, recent studies have unraveled the critical role of sugar metabolism in other developmental processes, including germination, hypocotyl elongation, floral transition, and cell expansion (ApRees, 1974; Bhowmik et al., 2001). Sucrose is the major form of translocated sugars from source to sink through phloem in plants (Koch, 1996), and its metabolism supplies the substrate for cellular biosynthesis. Sucrose cannot be used directly; it is irreversibly hydrolyzed to hexoses by invertase for further metabolism and biosynthesis. The invertase, one of the most important enzymes in sugar metabolism, is also critical for regulating many processes in actively growing tissues, such as young seedlings, leaves, and roots. In addition, this enzyme is also involved in the plant's responses to environmental stimuli because sucrose and hexose, substrate and product of invertase, respectively, are not only nutriments but also signaling molecules (Ricardo and ApRees, 1970; Kim et al., 2000; Heyer et al., 2004; Roitsch and Gonzalez, 2004).
Several isoforms of invertase with different biochemical characteristics and distinct subcellular localizations have been identified (Sturm, 1999; Tang et al., 1999). The cell wall–bound form (cwINV) is insoluble, and the vacuolar form (vaINV) is soluble. Both cwINV and vaINV are acidic, whereas the cytosolic form (cytINV) is neutral/alkaline. Compared with cwINV and vaINV, which have been characterized extensively, the cytINV is less studied as a result of its low enzymatic activities (Sturm, 1999; Vargas et al., 2003). The neutral invertase has a conserved C terminus and is not affected by heavy metals, but it is strongly inhibited by the cleavage products (Roitsch and Gonzalez, 2004). Recent studies showed that in response to stimuli, invertase transcript levels are upregulated. On the other hand, silencing of invertase activities depends on posttranslational mechanisms, including pH and protein inhibition (Rausch and Greiner, 2004). Based on its functions in supplying carbohydrates to the sink, regulating source–sink transitions, amplifying signals that control source–sink relations, and integrating signals and defense responses (Roitsch, 1999), invertase is considered a central modulator of assimilation partitioning and plant defense responses.
Here, we report the physiological functions and mechanism of action of an Arabidopsis PIP5K, PIP5K9, in vivo. We show that PIP5K9 interacts with CINV1 to negatively regulate invertase activity. This negative regulation affects sugar signaling/metabolism and inhibits root cell elongation, resulting in shortened primary roots.
RESULTS
Isolation and Structural Analysis of PIP5K9, Which Is Constitutively Expressed in Various Tissues and Repressed by Low Temperature
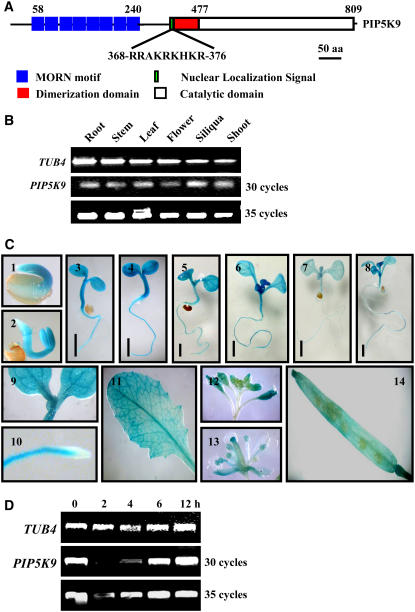
To study the physiological role of plant PIP5Ks, full-length cDNA of an Arabidopsis PIP5K, PIP5K9, was amplified by PCR. The coding sequence of PIP5K9 contains 2448 bp and encodes an 815–amino acid polypeptide (∼92.1 kD) with high similarity to other PIP5Ks. Structural organization analysis reveals the presence of multiple MORN (for membrane occupation and recognition nexus) motifs (eight times) at the N terminus, a catalytic domain at the C terminus, and a dimerization domain at the middle region of the protein (Figure 1A). This organization of different motifs is similar to that found in other plant PIP5Ks. Interestingly, PIP5K9 has a unique nuclear localization signal just C terminal of the dimerization domain (Figure 1A) that is not present in other Arabidopsis PIP5Ks, suggesting that PIP5K9 may have a nuclear function.
Figure 1.
Structural and Expression Patterns of PIP5K9.
(A) Structural organization of PIP5K9. MORN motifs and the catalytic domain are located at the N or C terminus, respectively. The dimerization domain and unique nuclear localization signals are located in the middle region. aa, amino acids.
(B) Semiquantitative RT-PCR analysis revealed the constitutive expression of PIP5K9 in various tissues, including silique, stem, root, leaf, and flower. The Arabidopsis TUB4 gene was used as a positive internal control.
(C) Promoter–reporter gene fusion studies showed expression of PIP5K9 in Arabidopsis seedlings (panels 1 to 8), cotyledon (panel 9), root elongation zone (panel 10), leaf (panel 11), flower (panels 12 and 13), and silique (panel 14). Bars = 2 mm.
(D) Semiquantitative RT-PCR analysis revealed the upregulation of PIP5K9 by low temperature. Twelve-day-old Arabidopsis seedlings were treated with 4°C for 2, 4, 6, and 12 h. The Arabidopsis TUB4 gene was used as a positive internal control.
PIP5K9 is expressed in various tissues, including roots, stems, flowers, siliques, shoots, and leaves (Figure 1B), as determined by semiquantitative RT-PCR. The expression pattern of PIP5K9 was further examined by promoter–reporter gene fusion studies. Analysis of >20 independent Arabidopsis transgenic lines showed that PIP5K9 was expressed in seedlings at young stages (1 to 8 d; Figure 1C, panels 1 to 8), including cotyledons (Figure 1C, panel 9) and the root elongation zone but not in the meristem zone (Figure 1C, panel 10). Transcripts of PIP5K9 were also detected in leaves (Figure 1C, panel 11), flowers (mainly in anther; Figure 1C, panels 12 and 13), and siliques (Figure 1C, panel 14).
In contrast with PIP5K1, PIP5K9 transcript levels were not altered by abscisic acid and NaCl treatment (data not shown). PIP5K9 expression, however, was repressed under low temperature (4°C) for 2 or 4 h but recovered at 6 h after treatment (Figure 1D). This result was confirmed by DNA microarray analysis (Lin et al., 2004).
pip5k9-d Has Enhanced Expression and Results in Shortened Roots as a Result of Repressed Cell Elongation
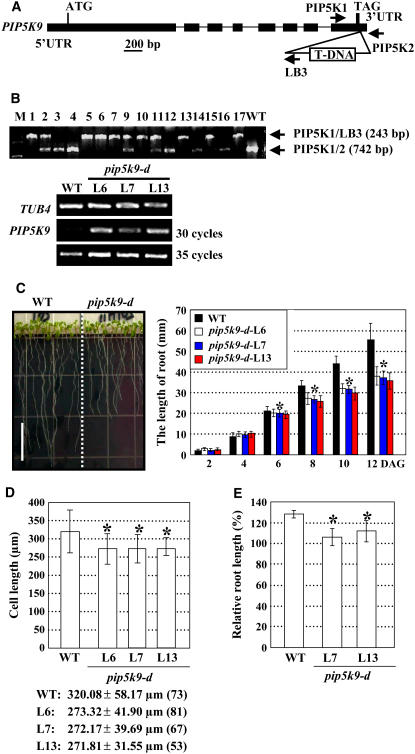
To dissect the physiological roles of PIP5K9, a T-DNA insertion line, Garlic_210_H11, was identified from the Syngenta Arabidopsis Insertion Library (Torrey Mesa Research Institute). This line contained a single T-DNA (data not shown) inserted into the 3′ untranslated region of PIP5K9 (Figures 2A and 2B). RT-PCR analysis of homozygous mutant lines (Figure 2B, top panel) showed enhanced expression of PIP5K9 (Figure 2B, bottom panel), indicating that the T-DNA insertion mutant, Garlic_210_H11, is a gain-of-function mutant. We named this mutant pip5k9-d. Microarray analysis further confirmed the increased expression of PIP5K9 in this mutant.
Figure 2.
Identification and Phenotypic Analysis of the PIP5K9 T-DNA Insertion Mutant.
(A) Scheme of the PIP5K9 gene. Exons (boxes) and introns (lines) are indicated. The T-DNA is inserted at the 3′ end of PIP5K9. LB, left border of the T-DNA. Positions of primers used for identification of the T-DNA insertion (PIP5K1 and LB3) or confirmation of the presence of the T-DNA (PIP5K1 and PIP5K2) are indicated. UTR, untranslated region.
(B) The T-DNA insertion in PIP5K9 was confirmed by PCR with genomic DNA as template. Independent lines L1, L2, L5 to L7, L9 to L11, L13, L15, and L17 contain the T-DNA insertion. Homozygous lines (with a single amplified fragment of 243 bp; L1, L5 to L7, L10, L13, L15, and L17) and heterozygous lines (with two amplified fragments of 243 and 742 bp; L2, L9, and L11) were confirmed by PCR. M, DNA ladder. Transcript levels of PIP5K9 were examined, and the results showed that, compared with the wild type, expression of PIP5K9 was much enhanced in the homozygous lines L6, L7, and L13 (bottom panel). RT-PCR was performed using the Arabidopsis TUB4 gene as a positive internal control.
(C) pip5k9-d mutant plants had shortened roots in 12-d-old seedlings (left panel; bar = 10 mm). Detailed calculation and statistical analysis of root length showed that 6 d after germination (DAG), root of pip5k9-d is significantly shorter than that of wild-type plants (right panel). Heteroscedastic t test analysis demonstrated that the difference was significant (*P < 0.01). Error bars represent se (n > 80).
(D) Calculation of cell length of primary roots indicates decreased cell elongation in pip5k9-d mutant plants. Heteroscedastic t test analysis showed that the difference was significant (*P < 0.01). Error bars represent sd (n > 50). Cell lengths are indicated, and numbers in parentheses show the calculated cell numbers.
(E) Relative root length of seedlings grown at 14°C (shown as percentage; the root length at 22°C was set as 100%), based on the premise that all seedlings had produced and partially expanded the first two leaves before the third rosette leaf primordia were established. Compared with the elongated roots of wild-type seedlings, pip5k9-d plants almost maintained their state at 14 or 22°C. n > 30 for each line and each treatment. Heteroscedastic t test analysis confirmed that the difference was significant (*P < 0.01). Error bars represent se.
Under normal growth conditions, pip5k9-d seedlings displayed shortened primary roots (∼30% reduction) compared with wild-type seedlings. The root shortening was more pronounced at 6 d after germination (Figure 2C). Further observation and statistical analysis of cell lengths at the root elongation zone indicated that the shortened root phenotype was attributable to reduced cell elongation (Figure 2D). The hypocotyl length of pip5k9-d seedlings was not altered significantly under light conditions.
As PIP5K9 expression was repressed by low temperature, we compared seedling root lengths at 14 or 22°C at the same growth stage (when young seedlings had produced the first two leaves and before the third rosette leaf primordia were established) (Kim et al., 1998). Statistical analysis showed no obvious difference in root growth at 14 or 22°C in pip5k9-d seedlings, whereas roots of wild-type seedlings were obviously elongated at 14°C (Figure 2E), indicating that pip5k9-d was resistant to chilling.
Relationship between PIP5K9 Expression Levels and Root Phenotype
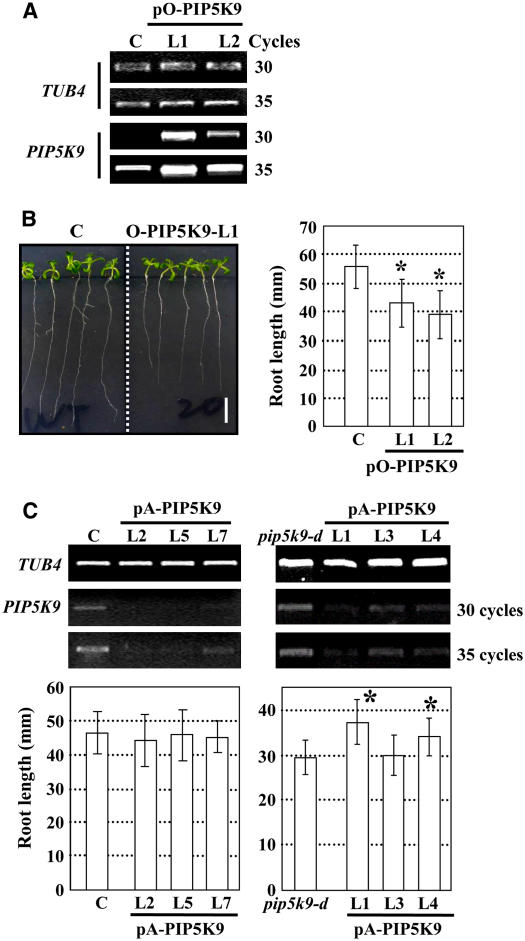
Transgenic approaches were used to further elucidate the physiological role of PIP5K9. An overexpression construct (pO-PIP5K9) or an antisense construct (pA-PIP5K9) were transformed into wild-type (pO-PIP5K9) or wild-type and pip5k9-d (pA-AtPIP5K9) plants, respectively. RT-PCR analysis confirmed the enhanced expression of PIP5K9 in wild-type plants harboring pO-PIP5K9 (Figure 3A) or the reduced expression of PIP5K9 in both wild-type and pip5k9-d plants harboring pA-PIP5K9 (Figure 3C). Statistical analysis of seedling root growth showed that enhanced PIP5K9 expression indeed resulted in shortened roots, similar to that found in pip5k9-d seedlings (Figure 3B), whereas repressed PIP5K9 expression in pip5k9-d rescued the shortened root phenotype to a certain extent (Figure 3C, right panel), although there was no significant change in wild-type seedlings (Figure 3C, left panel). All of these results suggest that PIP5K9 negatively regulates primary root growth.
Figure 3.
Analysis of Transgenic Plants Overexpressing or Deficient in PIP5K9.
(A) RT-PCR analysis showed increased transcript levels of PIP5K9 in transgenic lines 5K9-O-L1 and -L2, transformed with pO-PIP5K9. Arabidopsis TUB4 was used as a positive internal control.
(B) Transgenic plants overexpressing PIP5K9 showed shorter roots compared with control plants (C) (left panel; bar = 10 mm). Calculation and statistical heteroscedastic t test analysis showed a significant decrease of root lengths of the PIP5K9-overexpressing plants (*P < 0.01). Twelve-day-old seedlings were used for root length measurement. Error bars represent se (n > 30).
(C) RT-PCR analysis of PIP5K9 transcript levels in control (C) or pip5k9-d mutant plants transformed with pA-PIP5K9 (top panel). Expression of PIP5K9 was repressed in both control and pip5k9-d plants. The At TUB4 gene was used as a positive internal control. Further analysis showed that repression of PIP5K9 in control plants exhibited no obvious difference in root growth, whereas pip5k9-d plants recovered the root length to some extent. Twelve-day-old plants were measured and analyzed. Error bars represent se (n > 30). Heteroscedastic t test analysis showed a significant difference (*P < 0.01).
PIP5K9 Interacts with a Cytosolic Invertase, CINV1, in Vivo
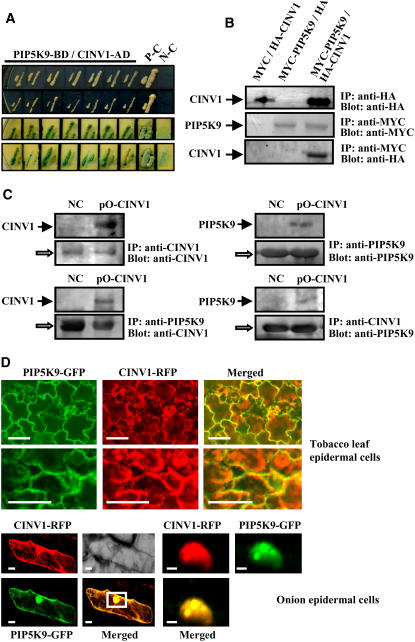
To further explore the potential components of PIP5K9-mediated signaling pathways, proteins that can interact with PIP5K9 were identified using yeast two-hybrid screenings. Truncated derivatives of PIP5K9 consisting of the MORN motif and the dimerization domain, or the dimerization and catalytic domains, were used as baits to screen an Arabidopsis cDNA library. We did not obtain any positive clone after screening of >4.5 × 106 transformants with a bait containing the MORN motif and the dimerization domain. On the other hand, we obtained several positive clones using the PIP5K9 dimerization and catalytic domains after screening of >3 × 106 transformants. Sequence analysis of the confirmed clones revealed one gene encoding a putative cytosolic invertase, which was assigned as CINV1. Two-hybrid analysis showed that the interaction with CINV1 activated the reporter HIS3 gene (even in the presence of 12 mM 3-amino-1,2,4-triazole, a competitive inhibitor of the HIS3 protein) and α-galactosidase (with whole CINV1 protein) (Figure 4A), indicating a strong interaction between PIP5K9 and CINV1.
Figure 4.
PIP5K9 Interacts with Arabidopsis Cytosolic Invertase, CINV1.
(A) Yeast-based two-hybrid analysis showed that yeast cells cotransformed with PIP5K9-BD (dimerization and catalytic domains of PIP5K9 fused with the binding domain of Gal4) and CINV1-AD (whole CINV1 fused with the activation domain of Gal4) could grow on synthetic dropout (SD) medium lacking Leu, Trp, His, and adenine hemisulfate salt (top panel), and the cells could turn blue on the same medium supplemented with X-α-gal (bottom panel). Yeast cells cotransformed with pGADT7-T and pGBKT7-53 were used as positive controls (P-C); those cotransformed with pGADT7-T and pGBK7-Lam were used as negative controls (N-C).
(B) Interaction of PIP5K9 and CINV1 was further confirmed through coimmunoprecipitation measurement. Expression of the HA-tagged CINV1 protein (HA-CINV1) in yeast cells was monitored by immunoprecipitation and protein gel blot analysis with an anti-HA antibody (top panel). The anti-MYC antibody was used to detect the expression of MYC-tagged PIP5K9 fusion proteins by immunoprecipitation and protein gel blot analysis (middle panel). The MYC-PIP5K9 immunoprecipitates were analyzed by anti-HA, confirming that CINV1 can be coimmunoprecipitated with PIP5K9 (bottom panel).
(C) Coimmunoprecipitation of PIP5K9 and CINV1 proteins in vivo. cinv1 mutant plants transformed with pA-PIP5K9 were used as a negative control (NC). Expressions of CINV1 and PIP5K9 were monitored by immunoprecipitation and protein gel blot analysis with anti-CINV1 or anti-PIP5K9 polyclonal antibody, respectively (top panel). Protein extracts were immunoprecipitated (IP) with a polyclonal antibody to PIP5K9, and protein gel blots were analyzed with a polyclonal antibody to CINV1 (bottom panel, left). In a parallel experiment, immunoprecipitation was done using a polyclonal antibody to CINV1, and protein gel blots were analyzed with a polyclonal antibody to PIP5K9 to detect coimmunoprecipitated PIP5K9 protein (bottom panel, right). The open arrows indicate cross reactions with the heavy chain of the protein A–conjugated antibody.
(D) Transient expression of fusion proteins PIP5K9-GFP and CINV1-RFP in tobacco mesophyll cells and onion epidermal cells indicates the colocalization of PIP5K9 and CINV1 to membranes and the nucleus. Bars = 50 μm (top panel and bottom panel, left) or 10 μm (bottom panel, right).
We next examined whether the full-length PIP5K9 protein was able to interact with CINV1. The entire coding regions of PIP5K9 and CINV1 were fused with those of binding domain (BD) and activating domain (AD), respectively, and the resulting constructs harboring fused MYC-PIP5K9 and HA-CINV1 were cotransformed into yeast cells. Detection of the relevant proteins with antibodies showed that anti-MYC antibody not only immunoprecipitated MYC-PIP5K9 but also coimmunoprecipitated HA-CINV1 from the protein extracts, whereas no HA-CINV1 signals were detected from the immunoprecipitates of yeast cells expressing MYC-PIP5K9 and HA or HA-CINV1 and MYC (two negative controls). These results confirm the interaction of PIP5K9 and CINV1 in yeast cells (Figure 4B).
To further investigate the association between PIP5K9 and CINV1 in vivo, transgenic plants overexpressing both PIP5K9 and CINV1 were generated by transforming pip5k9-d plants with pO-CINV1, and negative control plants were generated by transforming at inv1 plants with pA-PIP5K9. Protein gel blot analysis detected the presence of CINV1 in the immunoprecipitated PIP5K9 complex (Figure 4C, left panel, bottom) and the presence of PIP5K9 in the immunoprecipitated CINV1 complex (Figure 4C, right panel, bottom), indicating a direct interaction between the two proteins in plant cells.
We investigated the subcellular localization of PIP5K9 and CINV1 in vivo by transient expression in tobacco (Nicotiana benthamiana) leaves through agroinfiltration or in onion (Allium cepa) epidermal cells through particle bombardment. Confocal microscopic analysis showed that PIP5K9 was localized mainly in cellular membranes, the nucleus, and the cytosol, and CINV1 was found mainly in the cytosol but was also detected in the nucleus and membranes. When the two proteins were coexpressed in tobacco mesophyll and onion epidermal cells, colocalization of PIP5K9–green fluorescent protein (GFP) and CINV1–red fluorescent protein (RFP) was found mainly in membranes and the nucleus (Figure 4D).
pip5k9-d Is Less Sensitive to Glucose and Sucrose as a Result of Reduced Invertase Activities
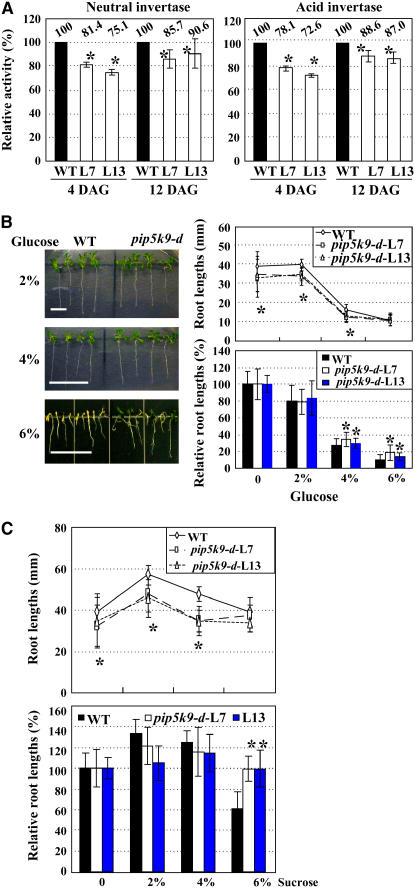
Based on the interactions of PIP5K9 and CINV1, we investigated the effects of PIP5K9 on invertase activity in vivo. Compared with those in wild-type seedlings, neutral invertase activities were reduced by ∼10 to 25% in pip5k9-d seedlings at 4 and 12 d after germination (Figure 5A, left panel). Similar results were obtained with acid invertase activities (Figure 5A, right panel). Together, these results indicate that PIP5K9 negatively regulates invertase activities.
Figure 5.
pip5k9-d Seedlings Were Less Sensitive to High Concentrations of Glucose and Sucrose.
(A) Both total neutral (left panel) and acid invertase (right panel) activities of pip5k9-d mutant plants were reduced compared with those of wild-type plants (which was set as 100%). The measurements were repeated three times, and average data are presented and shown as percentages. Error bars represent se. Heteroscedastic t test analysis showed significant differences (*P < 0.01). DAG, days after germination.
(B) pip5k9-d is less sensitive to high concentrations of supplemented glucose. Growth of pip5k9-d and wild-type plants on the medium supplemented with different concentrations (2, 4, or 6%) of d-glucose was observed (left panel), and root lengths were measured (right panel, top). Statistical calculation on relative root lengths compared with control seedlings without sugar (set as 100%) confirmed the reduced inhibitions of pip5k9-d seedlings (right panel, bottom). Twelve-day-old plants were used for measurements, and the experiments were repeated three times. Bar = 10 mm, and error bars represent se (n > 30). Heteroscedastic t test analysis showed significant differences (*P < 0.01).
(C) pip5k9-d is less sensitive to high concentrations of supplemented sucrose. Root lengths of pip5k9-d and wild-type plants on medium supplemented with different concentrations (2, 4, or 6%) of sucrose were measured (left panel), and relative root lengths compared with control seedlings without sucrose (set as 100%) were statistically calculated (right panel), indicating the repressed sensitivities of pip5k9-d to supplemented sucrose. Twelve-day-old plants were used for measurements, and error bars represent se (n > 30). Heteroscedastic t test analysis showed significant differences (*P < 0.01). The experiments were repeated three times.
The hydrolysis of sucrose to hexoses by invertase is an indispensable reaction in sugar signaling and metabolism, and hence the altered invertase activity may result in abnormal responses of plants to sugar. Therefore, we examined the growth responses of pip5k9-d to different concentrations of sugars. Compared with wild-type seedlings, some pip5k9-d seedlings developed green and expanded cotyledons in the presence of 6% d-glucose and were able to grow continuously (Figure 5B, left panel). In addition, statistical analysis of the primary root growth and relative root lengths confirmed the reduced responses of pip5k9-d to high concentrations of glucose (Figure 5B, right panel) and sucrose (Figure 5C). However, we found no obvious developmental alteration when pip5k9-d seedlings were grown in the presence of mannitol or sorbitol, perhaps because, in contrast with glucose, mannitol and sorbitol are not efficiently metabolized by plants and they mainly cause a constant osmotic stress to affect plant growth, suggesting that the reduced response of pip5k9-d to glucose and sucrose is specific and not related to osmotic stress.
CINV1 Is a Cytosolic (Alkaline/Neutral) Invertase
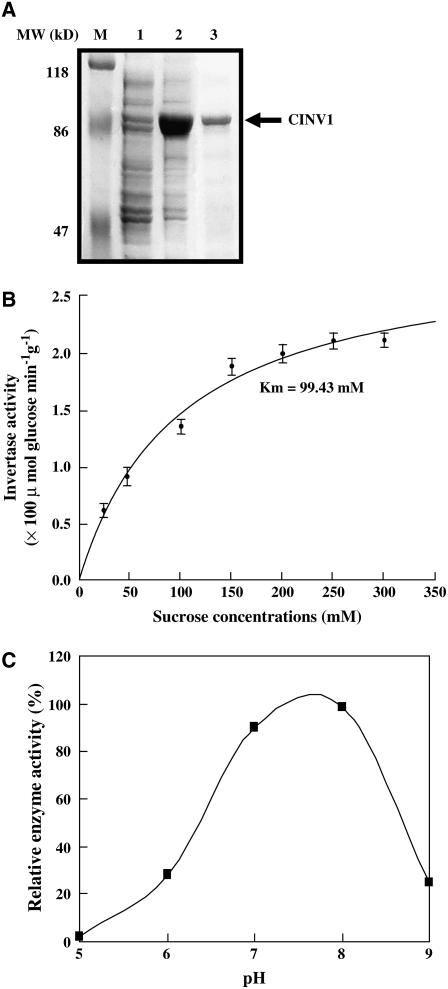
Full-length cDNA of CINV1 (At1g35580) was isolated and confirmed by sequencing. CINV1 contains 1656 bp and encodes a 551–amino acid polypeptide (∼62.8 kD). Structural analysis reveals that CINV1 is a putative cytosolic invertase and classified as an alkaline/neutral invertase based on pH optima. Enzymatic analysis of purified recombinant CINV1 protein, expressed in E. coli, indeed showed the sucrose hydrolase activity of CINV1 (Figure 6). The Km for sucrose is 99.43 mM at pH 7.0, and the pH optimum of CINV1 was in the range ∼7.0 to 8.0, confirming CINV1 as a cytosolic (alkaline/neutral) invertase.
Figure 6.
CINV1 Hydrolyzes Sucrose in Vitro.
(A) Expression and purification of recombinant CINV1. M, protein molecular mass marker; lanes 1 and 2, crude extracts in the absence or presence of isopropylthio-β-galactoside (in a final concentration of 0.5 mM, for 6 h); lane 3, purified recombinant CINV1. The arrow indicates the position of recombinant CINV1.
(B) Invertase activity of purified CINV1 (pH 7.0) under a gradient of sucrose concentrations (25 to 300 mM). The Km was calculated from data of three independent experiments, and error bars represent se.
(C) Invertase activity of purified CINV1 over a pH range from ∼5.0 to 9.0 at the sucrose concentration of 100 mM. Purified CINV1 showed highest activities in the range from ∼7.0 to 8.0.
Deficiency of CINV1 Results in Shortened Roots
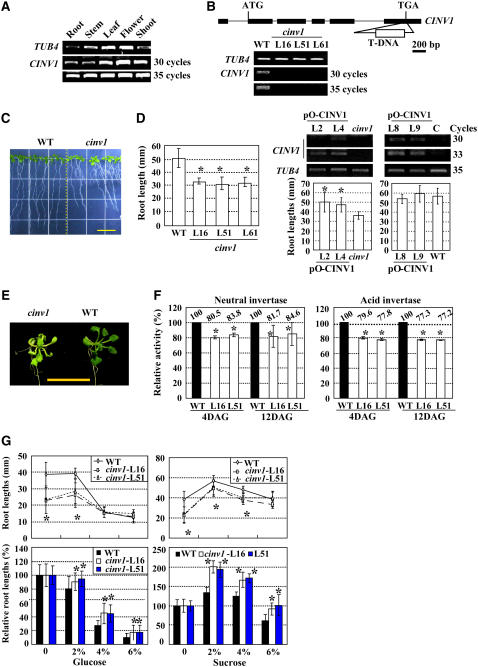
Semiquantitative RT-PCR analysis showed that CINV1 was constitutively expressed in various tissues (Figure 7A), similar to the expression profile of PIP5K9. A putative T-DNA insertion knockout mutant, cinv1, was identified and characterized. The T-DNA is inserted into the 3′ end of the gene (Figure 7B, top panel), causing a deficiency of CINV1 (Figure 7B, bottom panel).
Figure 7.
Identification and Phenotypic Analysis of cinv1 Mutant Plants.
(A) Semiquantitative RT-PCR analysis reveals the constitutive expression of CINV1 in various tissues, including shoot, stem, root, leaf, and flower. The Arabidopsis TUB4 gene was used as a positive internal control.
(B) Scheme of the Arabidopsis CINV1 gene. Exons (boxes) and introns (lines) are indicated. The T-DNA insertion was located at the 3′ end of CINV1. Further RT-PCR analysis showed reduced transcript levels of CINV1 (bottom panel). The Arabidopsis TUB4 gene was used as a positive internal control.
(C) cinv1 plants had shortened roots compared with wild-type plants. Root lengths of 12-d-old plants were measured and calculated. Heteroscedastic t test analysis showed significant differences (right panel; *P < 0.01). Bar = 10 mm (left panel), and error bars represent se (n > 30). The experiments were repeated three times.
(D) RT-PCR analysis of CINV1 transcripts in control (C) or cinv1 mutant plants transformed with pO-CINV1 (top panel). Expression of CINV1 was enhanced in both control and cinv1 plants, and statistical analysis on root lengths showed that overexpressing CINV1 had no obvious effect on root growth of control plants, whereas in cinv1 plants the short-root phenotype was rescued. Twelve-day-old plants were used for measurements and analysis. Error bars represent se (n > 30). Heteroscedastic t test analysis indicated significant differences (*P < 0.01). The At TUB4 gene was used as a positive internal control.
(E) cinv1 plants showed light green leaves, which may be attributable to inhibition of photosynthesis. Bar = 1 cm.
(F) Both total neutral (left panel) and acid (right panel) invertase activities of cinv1 plants were reduced compared with those of wild-type plants, especially at 4 d after germination (DAG). Activities of seedlings grown at days 4 or 12 were measured and presented as percentages of those of wild-type seedlings. The measurements were repeated three times, and average data are presented. Heteroscedastic t test analysis indicated significant differences (*P < 0.01), and error bars represent se.
(G) cinv1 is less sensitive to high concentrations of supplemented glucose (left panel) or sucrose (right panel). Root lengths of cinv1 and wild-type plants on medium supplemented with different concentrations (2, 4, or 6%) of glucose (left panel) or sucrose (right panel) were measured, and relative root lengths compared with control seedlings without supplemented sugars (set as 100%) were statistically calculated. cinv1 was less inhibited by glucose (left panel, bottom) and high concentration of sucrose (6%; right panel, bottom). Twelve-day-old plants were used for measurements, and error bars represent se (n > 30). Heteroscedastic t test analysis showed significant differences (*P < 0.01). The experiments were repeated three times.
Subsequent phenotypic observation and statistical analysis showed that cinv1 seedlings had shortened roots (∼70% of wild-type seedlings), similar to those of pip5k9-d (Figure 7C). Complemented expression of CINV1 in cinv1 rescued the short-root phenotype (Figure 7D, left panel), whereas wild-type plants with overexpressed CINV1 had no significant change in root growth (Figure 7D, right panel), suggesting the critical role of CINV1 in root growth.
In addition to the root phenotype, cinv1 plants have light green leaves (Figure 7E; more obviously under weak light or in medium without sugar), which may be attributable to the inhibition of photosynthesis and the feedback regulation of photosynthesis by sugars (Foyer, 1987). In source organs, invertase hydrolyzes sucrose to limit carbohydrate export and sugar signaling, and the resulting changes in hexose levels can affect many processes, including photosynthesis.
Further analysis revealed the obviously repressed activities of both neutral and acid invertase in cinv1 seedlings (Figure 7F), similar to those in pip5k9-d seedlings. The positive correlation between invertase activities and shortened roots of cinv1 and pip5k9-d plants suggest that these phenotypes can be attributed to the negative regulation of CINV1 by PIP5K9.
Similar to pip5k9-d seedlings, cinv1 seedlings were more tolerant to high concentrations of glucose and sucrose compared with wild-type seedlings (Figure 7G).
Altered Sugar Metabolism of pip5k9-d and cinv1
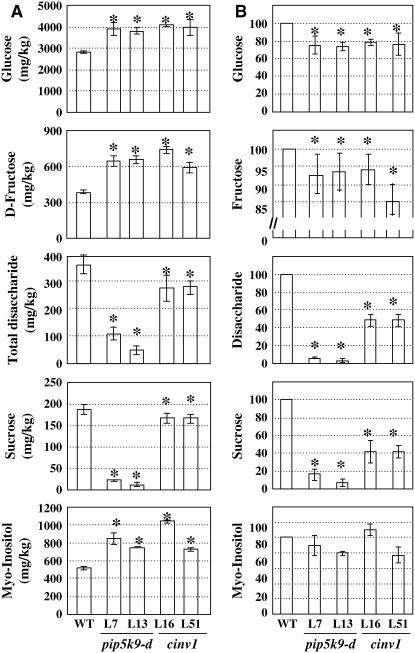
We performed gas chromatography–mass spectrometry (GC-MS) analysis to determine changes in sugar metabolites in pip5k9-d and cinv1 roots (Figure 8). Figure 8A shows that the concentration of sucrose in mutant roots was much lower than that in wild-type roots, as was that of total disaccharide. Although the concentrations of glucose and fructose were higher in mutant roots, the total amounts in roots were reduced. Notably, the concentration of myo-inositol was obviously increased, which may be attributable to the altered PI signaling pathway (i.e., feedback of enhanced PIP5K levels). The similar changes in sugar metabolites in pip5k9-d and cinv1 roots reflected the regulatory effects of PIP5K9 (negative role) and CINV1 in sugar metabolism. Repression of invertase activity results in reduced amounts of hexoses in mutants roots (Figure 8B). Moreover, amounts of several amino acids (e.g., Leu, Val, Thr, Gly, Ser, Pro, and Asn) were undetectable in pip5k9 and cinv1 seedlings (data not shown), indicating repressed amino acid synthesis as well.
Figure 8.
Altered Metabolites in pip5k9-d and cinv1 Roots.
(A) Metabolites in roots of pip5k9-d, cinv1, and wild-type plants grown for 12 d were profiled using GC-MS. The concentrations of glucose, d-fructose, and myo-inositol were increased in both mutant plants, whereas those of sucrose and total disaccharide were obviously reduced compared with those of wild-type plants. Heteroscedastic t test analysis showed significant differences (*P < 0.01), and error bars represent se. The measurement was performed in triplicate (n > 80).
(B) Relative amounts of glucose, d-fructose, sucrose, total disaccharide, and myo-inositol in seedling roots of pip5k9-d and cinv1 were compared with those of wild-type plants (regarded as 100%).
Altered Expression of Multiple Genes in pip5k9-d and cinv1
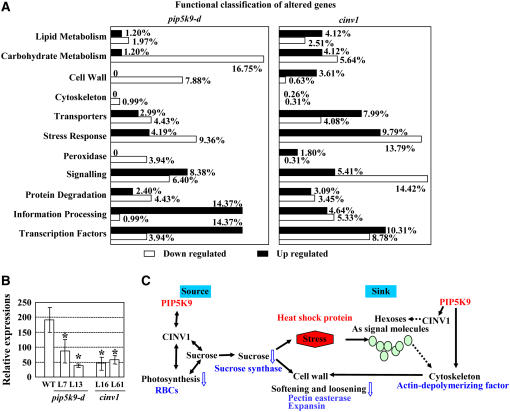
To dissect the molecular mechanism underlying the shortened root phenotype, we performed genome-wide expression studies using ATH1 Arabidopsis whole genome microarray chips (see Supplemental Materials online). Roots of seedlings at the same growth stage were used as materials. Compared with the wild type, expression levels of ∼20 genes were increased or repressed in both pip5k9-d and cinv1. Most of the increased genes encode transcription factors, heat shock proteins, etc. Among the repressed genes are those involved in carbohydrate metabolism (e.g., sugar transport) and genes associated with cell wall metabolism, such as polygalacturonase and pectinesterase (Figure 9A, Table 1; see Supplemental Table 1 online). Besides the common regulated genes, many genes were altered especially in pip5k9-d or cinv1, including those regulating cell wall status, actin dynamics, and jasmonate response (Table 1; see Supplemental Tables 2 and 3 online).
Figure 9.
Expression of Genes Involved in Multiple Developmental Processes Was Altered in pip5k9-d and cinv1.
(A) Altered expression of multiple genes with enhanced PIP5K9 transcript levels or reduced CINV1 transcript levels. Seedling roots of pip5k9-d, cinv1, and wild-type plants were used for RNA extraction and further hybridization. Genes with significant differences in expression (Wilcoxon rank test, P < 0.01) were collected and functionally classified between wild-type and pip5k9-d plants (left panel) and between wild-type and cinv1 plants (right panel).
(B) Real-time RT-PCR analysis showed that RBCs was repressed in pip5k9-d and cinv1 seedlings (grown for 4 d). Transcript levels were normalized with respect to TUB4 transcript levels. Mean values were obtained from three independent PCR amplifications, and error bars indicate se. Heteroscedastic t test analysis showed significant differences (*P < 0.01).
(C) Summary of genes with altered transcript levels through microarray analysis, as well as changes in relevant metabolites and developmental processes. Open arrows indicate the regulatory tendency of the physiological process under enhanced PIP5K9, and the levels of relevant metabolites are highlighted (blue; decreased). The gene regulatory tendencies are highlighted in blue (downregulation) and red (upregulation).
Table 1.
Altered Genes in pip5k9-d and/or cinv1
| Altered in both pip5k9-d and cinv1
| |||||
|---|---|---|---|---|---|
|
pip5k9-d
|
cinv1
|
||||
| L7 | L13 | L16 | L51 | Description | |
| At2g26560 | 1.8 | 3.3 | 2.1 | 1.8 | Similar to latex allergen |
| At5g37260 | 1 | 1.1 | 2 | 1.8 | Putative CCA1, myb family transcription factor |
| At4g31970 | 4.6 | 8.4 | 5.2 | 3.3 | Cytochrome P450 family protein |
| At3g24520 | 0.5 | 0.9 | 0.6 | 0.6 | Heat shock transcription factor family protein |
| At3g01970 | 0.7 | 1.1 | 1.1 | 0.8 | WRKY family transcription factor |
| At1g75830 | 5.4 | 4.5 | 4.6 | 3.6 | Putative plant defensin fusion protein |
| At2g30540 | 0.5 | 1.4 | 1.4 | 3.1 | Glutaredoxin family protein |
| At2g45220 | −1.2 | −0.8 | −0.3 | −0.7 | Pectinesterase family protein |
| At1g05650 | −2.3 | −0.6 | −2.3 | −1.3 | Polygalacturonase |
| At1g08900 | −1.5 | −1 | −0.5 | −0.6 | Sugar transporter–related |
| Altered in pip5k9-d
| |||
|---|---|---|---|
| L7 | L13 | ||
| At1g26590 | 5.2 | 3.3 | CAF protein similar to C2H2 zinc finger protein |
| At4g12400 | 0.6 | 1.9 | Stress-induced protein, sti1-like protein |
| At4g00680 | −1.4 | −3.1 | Putative actin-depolymerizing factor |
| At1g01750 | −1.2 | −4.2 | Similar to actin-depolymerizing factor4 |
| At1g62980 | −2.8 | −1.8 | Expansin At-EXP6 |
| At5g57530 | −1.1 | −3.4 | Xyloglucan endotransglycosylase |
| At1g52050 | −1.9 | −4.4 | Jasmonate-inducible protein |
| At2g23630 | −1.3 | −2.7 | Putative pectinesterase |
| Altered in cinv1
| |||
|---|---|---|---|
| L16 | L51 | ||
| At2g23170 | 2.3 | 2 | Auxin-responsive GH3 family protein |
| At5g35190 | 1 | 1.1 | Extensin-like protein extensin |
| At1g54970 | 0.9 | 0.8 | Similar to Pro-rich protein |
| At3g45960 | −0.8 | −0.7 | Expansin family protein (EXPL3) |
| At1g70880 | −2.3 | −2.7 | Similar to ripening-induced protein |
| At5g58690 | −0.8 | −0.7 | Phosphoinositide-specific phospholipase-like protein |
The roots of wild-type, pip5k9-d (lines 7 and 13), and cinv1 (lines 16 and 51) seedlings at the same developmental stage were used for the microarray analysis using Arabidopsis ATH1 chips. Numbers indicate the regulation ratios (Log2).
Phenotypic analysis indicated that photosynthesis was affected in cinv1, and the reduced sucrose amounts in both cinv1 and pip5k9-d mutants implied changes in the CO2 fixation/carbohydrate metabolism balance. We examined the expression of genes encoding photosynthesis-related proteins, including RBCs (ribulose-1,5-bisphosphate carboxylase small subunit), PC (plastocyanin), and PAL (phenylalanine ammonia lyase), which were regarded as markers for photosynthesis (Krapp et al., 1993; Ehness et al., 1997; Zhou et al., 1998). Quantitative real-time RT-PCR analysis showed that the transcript level of RBCs was obviously repressed in both pip5k9-d and cinv1 seedlings (Figure 9B), whereas no obvious changes of PC and PAL1 transcript (data not shown) were observed. These results indicate that the effect of enhanced PIP5K9 expression on photosynthesis is likely executed through its negative regulation of CINV1 and suggest that in shoots, reduced photosynthesis led to a repression of sucrose production. In addition, sucrose deficiency results in altered processes, including less flexibility of the cell wall (altered expression of cell wall–related genes) and increased stress responses (enhanced expressions of heat shock proteins) in vivo. Sugar metabolism and signaling are involved in and mediate various biological processes, in which hexoses serve as major signaling molecules (Figure 9C).
DISCUSSION
In this article, we describe the isolation and functional characterization of an Arabidopsis PIP5K, PIP5K9, and its roles in sugar-mediated root development through interacting with a cytosolic invertase. The catalytic domain of PIP5K9 is highly homologous with that of At PIP5K1 (Mueller-Roeber and Pical, 2002). The presence of the MORN motifs, which are not found in animal and yeast PIPKs, suggests that plant PIP5Ks may be regulated by a different mechanism (Ma et al., 2006). We note that PIP5K9 contains a nuclear localization signal (Figure 1A) not found in any other PIP5Ks. PIP5K9-GFP fusion studies indeed confirmed that PIP5K9 is localized to nuclei, in addition to membranes and the cytosol. In mammalian cells, type I and II PIPKs are enriched in nuclei and associated with distinct subnuclear domains (nuclear speckles) containing pre-mRNA processing factors (Boronenkov et al., 1998). The nuclear localization of PIP5K9 suggests that this enzyme may play a role in gene regulation.
PIP5K9 Represses a Cytosolic Invertase through Protein–Protein Interactions
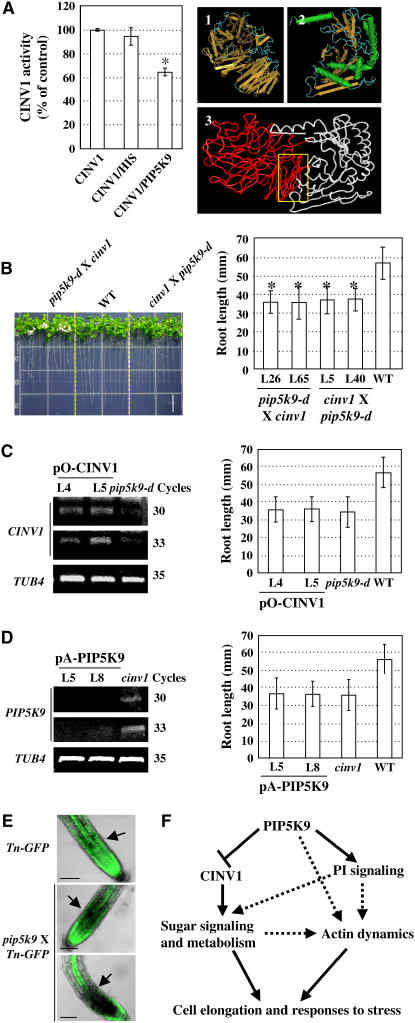
By yeast-based two-hybrid assays and coimmunoprecipitation studies, we demonstrated the interactions of PIP5K9 with a cytosolic invertase, CINV1, in Arabidopsis. Subcellular localization analysis revealed that PIP5K9 and CINV1 are present in overlapping compartments, and indeed, the CINV1 deficiency mutant, cinv1, had shortened roots and was less sensitive to high concentrations of sugar, similar to the PIP5K9-overexpressing mutant, pip5k9-d. This finding suggests a negative relationship between PIP5K9 and CINV1. Further analysis of the impact of PIP5K9 binding to CINV1 on invertase activity in vitro (Figure 10A, left panel), on the phenotypes of transgenic plants (Figures 10C and 10D) and of double mutants (Figure 10B), and on enzymatic activities (Figures 5A and 7F) confirmed the negative regulatory effects of PIP5K9 on CINV1. At present, the precise mechanism by which PIP5K9 represses CINV1 is not known. Stereostructure analysis suggests that PIP5K9 might modulate CINV1 activity or substrate binding capacities through C-terminal interactions (Figure 10A, right panel, panel 3).
Figure 10.
Hypothesized Function Model of PIP5K9.
(A) PIP5K9 represses CINV1 activities in vitro (left panel). Purified HIS peptide (∼32 kD) was used as a control. CINV1 activities were measured in the presence of 100 mM sucrose, pH 7.0, and equal amounts of purified PIP5K9 or HIS peptide were added to test the effects of PIP5K9 on CINV1. The activities were calculated from three independent experiments, and error bars represent se. The hypothesized regulatory mechanism of PIP5K9 on CINV1 through interactions (right panel) was simulated with β-fructosidase from Thermotoga maritime (panel 1) and with the human Hs PIP5K IIβ (panel 2). The dimerization and catalytic domains of PIP5K9 could bind to the C terminus of CINV1 and modulate the CINV1 activity or substrate binding capacities (panel 3).
(B) Growth of heterozygous F2 generation double-cross plants, pip5k9-d × cinv1 and cinv1 × pip5k9-d, for 12 d. Detailed calculation and statistical analysis showed that root lengths of the homozygous F3 generation plants were significantly shorter than those of wild-type plants (*P < 0.01; heteroscedastic t test analysis). Error bars represent se (n > 30). Bar = 10 mm.
(C) RT-PCR analysis indicated enhanced CINV1 transcript levels in pip5k9-d mutant plants transformed with pO-CINV1 (left panel) using the At TUB4 gene as a positive internal control. Measurement and statistical analysis showed that overexpressing CINV1 in pip5k9-d had no obvious effect on root growth. Twelve-day-old plants were measured and analyzed. Error bars represent se (n > 30).
(D) RT-PCR analysis revealed the repressed expression of PIP5K9 in cinv1 mutant plants transformed with pA-PIP5K9 (left panel) using the At TUB4 gene as a positive internal control. Measurement and statistical analysis showed that repression of PIP5K9 had no obvious effect on root growth of cinv1 plants. Twelve-day-old plants were measured and analyzed. Error bars represent se (n > 30).
(E) Actin microfibers were altered under enhanced PIP5K9 expression, as highlighted by arrows. Arabidopsis seedlings harboring talin-GFP were used for genetic crosses.
(F) Working model of the effects of PIP5K9 on cell elongation. PIP5K9 intervenes with sugar metabolism and signaling by interacting with CINV1. Alternatively, PIP5K9 can directly or indirectly regulate cytoskeleton dynamics to coordinate cell elongation and responses to stress.
PIP5K9 and CINV1 Regulate Sugar Metabolism and Signaling Related to Root Cell Elongation
Plants harness light energy to fix carbon dioxide and supply fixed carbon to leaf cells or convert it to sucrose, the major form of translocated sugars from the source to the sink though phloem tissues (Koch, 1996). Therefore, sucrose synthesis is coordinated with the rate of photosynthesis, which in turn is feedback-regulated by sugar accumulation in source leaves (Jang and Sheen, 1994; Roitsch, 1999). Previous work has shown that overexpression or repression of invertases would result in chloroplast damage and photosynthesis inhibition (Sonnewald et al., 1991; Bussis et al., 1997; Tang et al., 1999). The decrease in RBC expression levels in both pip5k9-d and cinv1 seedlings suggests that the repressed CINV1 activity altered the balance of CO2 fixation/carbohydrate metabolism, consistent with the reduced sucrose production in mutant seedlings. Reduced sucrose production in source leaves would lead to a deficiency of sucrose in the sink. Because 30% or more of cellular carbohydrate metabolism is contributed to the synthesis of wall components, it is not surprising that reduced sucrose levels in pip5k9-d and cinv1 roots induce a series of responses, including changes in cell wall state.
Cell wall, which is crucial for cell shape, has two opposing properties: strength and flexibility, both of which are important in cell elongation. Once elongation ceases, the wall becomes tighter and less flexible. Genome-wide expression analysis of roots of pip5k9-d and cinv1 revealed the repression of genes encoding polygalacturonase and pectinesterase (pectin methylesterase). The cell wall is composed mainly of polymers that can be relaxed by enzymes (e.g., polygalacturonases), which degrade pectin or methylated polygalacturonic acid (Collmer and Keen, 1986). Although pectin methylesterase does not relax cell walls directly, it can regulate cell wall degradation through other mechanisms. When pectin methylesterase acts on homogalacturonans, the demethylesterification reaction releases protons to decrease the pH, which further promotes the action of endopolygalacturonases and therefore contributes to cell wall loosening (Nari et al., 1986; Micheli, 2001). Changes in the expression level of these genes indicate that cell wall flexibility in mutant roots is reduced, resulting in an abnormal extension of root cells. In addition, genes encoding expansin and xyloglucan endotransglycosylase or expansin homologs were repressed in roots of pip5k9-d and cinv1, respectively. Together, our genome-wide expression studies indicate that cell walls in both pip5k9-d and cinv1 roots are less flexible, resulting in inhibited root cell elongation and shortened roots.
We note a downregulation of the gene encoding sucrose synthase (SuSy) in pip5k9-d, which catalyzes a readily reversible reaction and is involved in both the degradation and synthesis of sucrose, and of the gene encoding a sugar transport protein. SuSy is present in higher levels in sucrose-using tissues than in sucrose-synthesizing tissues, suggesting that it is involved in sucrose degradation in roots. In addition, SuSy also plays a major role in channeling the substrate UDP-glucose to cellulose synthase. As such, it acts as a molecular switch between survival metabolism and growth and/or differentiation processes involving cellulose synthesis (Haigler et al., 2001). The sucrose deficiency represents abiotic stress for plant cells. This deficiency feedback represses the expression of the gene encoding SuSy and induces genes encoding heat shock proteins or transcription factors.
Cell wall invertase (acid invertase) may supply carbohydrates to the sink and is a central modulator of source–sink transitions through maintaining the sucrose concentration gradient (Dickinson et al., 1991; Roitsch, 1999). Vacuolar invertase (acid invertase) acts on sucrose accumulation in mature sink vacuoles (Yelle et al., 1988, 1991). A sucrose deficiency might cause the feedback repression of acid invertase activities in both pip5k9-d and cinv1 (Figures 5A and 7F).
Metabolite analysis showed increased concentrations of glucose and fructose in both pip5k9-d and cinv1 roots, which may be attributable to altered sugar sensing and signaling. In addition to serving as substrates in carbon and energy metabolism and polymer biosynthesis, sugars also act as signaling molecules to regulate plant growth and development (Sheen et al., 1999). In many cases, sucrose is not the direct signaling molecule, and numerous experiments have documented the role of hexoses in triggering sugar responses and signaling in plants (Sheen et al., 1999). The role of hexoses as signaling molecules, but not substrates, was further confirmed by the observation that hexokinase is a dimeric cytosolic enzyme essential for glycolysis (Sheen et al., 1999), and high levels of sugar repress the expression of genes for sugar production and stimulate those for sugar storage and use (Koch, 1996; Ehness et al., 1997; Godt and Roitsch, 1997; Ismail et al., 1997). These genes were not altered, except for that encoding SuSy, further implying the role of hexoses as signaling molecules in pip5k9-d and cinv1 plants.
PIP5K9 May Function Independently of CINV1
Cell expansion involves the coordinated assembly of the cytoskeleton and the cell wall (Carpita and Gibeaut, 1993). During cell growth, the rate of vesicle delivery to the tip via actin microfilaments and their deposition at the tip determine the growth rate. Decreased actin microfilaments in root elongation zones (Figure 10E), in plants with enhanced PIP5K9 expression, suggests a specific role of PIP5K9 in root development.
PI(4,5)P2 is known to bind to several proteins regulating the assembly/disassembly of actin filaments, including profiling, gelsolin, α-actinin, cofilin, filamin, and vinculin (Janmey and Stossel, 1987; Caroni, 2001), and some of these interactions have been documented in plant root hair and pollen tube (Braun et al., 1999; Kost et al., 1999). Recently, A.J. Davis, Y.J. Im, I.Y. Perera, and W.F. Boss (unpublished data) found that At PIP5K1 could be involved directly in microfilament dynamics through interaction with eEF1A. However, whether PI(4,5)P2 production is necessary for PIPK modulation of actin dynamics in vivo is still unknown. Higher levels of myo-inositol in pip5k9-d seedling roots suggest a role of the PI signaling pathway in root development. This result also suggests that PIP5K9 may function independently of CINV1 to regulate root developmental processes, through PI signaling, or to affect actin dynamics or the interaction and crosstalk between PI signaling and cytoskeleton dynamics (Figure 10F).
Reduced sucrose levels, especially in pip5k9-d, suggest that sugar metabolism is more affected in this mutant than in cinv1. Although both cinv1 and pip5k9-d display reduced sucrose sensitivity compared with the wild type, there is a clear difference in their responses; the former is more sensitive to low sucrose concentration than the latter. As exemplified by PIP5K9, other molecules of the PI signaling pathway may regulate developmental processes by interacting with other proteins, independent of their roles in PI metabolism (Toker, 2002).
Auxin is known to play critical roles in root development. Although both pip5k9-d and cinv1 plants are slightly less sensitive to auxin, we found that the shortened root phenotype could not be rescued (data not shown) by auxin, suggesting that auxin is not the main reason for the abnormal root growth.
In summary, we show here that one member of the At PIP5K family, PIP5K9, links PI signaling to sugar metabolism through its interaction with CINV1 and negatively regulates root cell elongation as well as other developmental processes. Given that the Arabidopsis genome contains multiple PIP5Ks that are differentially expressed in different tissues and subcellular locations, we expect future studies of other At PIP5K family members and their interacting proteins to shed light on their complicated roles in plant growth and development.
METHODS
Plant Material, Growth Conditions, and Measurement of Root Length
Arabidopsis thaliana ecotype Columbia was used in these experiments. Plants were transformed using the floral dipping method (Clough and Bent, 1998). Arabidopsis seeds were surface-sterilized and sown on plates containing Murashige and Skoog (MS) medium. Plates were placed vertically and incubated under long-day conditions (20 h of light, 21°C), and seedling root lengths were measured 12 d after germination. MS medium supplemented with 0, 2, 4, and 6% sugar (glucose, sucrose, sorbitol, or mannitol) was used to investigate the effects of sugar on root length. All experiments were performed in triplicate, and the data represent mean values ± se.
Isolation of Full-Length PIP5K9 and CINV1 cDNAs
Primers PIP5K9-1 (5′-GGGTACCTGCTCTGCGGTTTGTCTGAGCGTTTC-3′; added KpnI site underlined) and PIP5K9-2 (5′-CGGATCCCAATCTGTACTCAATTCTCTGTCTA-3′; added BamHI site underlined) were used to amplify the full-length PIP5K9 cDNA from an Arabidopsis cDNA library. The amplified DNA fragment was subcloned into pMD18-T vector (TaKaRa) and sequenced. CINV1 cDNA was amplified using primers CINV1-1 (5′-CCGGAATTCATGGAAGGTGTTGGACTAAGAG-3′; added EcoRI site underlined) and CINV1-2a (5′-CGCGGATCCACCTAAAGCGGTTTTTCAGAGTTG-3′; added BamHI site underlined). Computational analyses, including sequence comparisons and BLAST search, were performed using the GCG SeqWeb program (version 2.0.2; Accelrys). Protein domain analysis was performed at http://swissmodel.expasy.org//SWISS-MODEL.html. Protein three-dimensional structure analysis was performed using Cn3D version 4.1 program (www.ncbi.nlm.nih.gov) and VEGA ZZ 2.0.4 (http://www.ddl.unimi.it/).
Semiquantitative RT-PCR and Quantitative Real-Time RT-PCR Analysis
Semiquantitative RT-PCR analyses were performed to study the expression of PIP5K9 and CINV1 in various tissues of wild-type plants as well as in pip5k9-d and cinv1 mutants. Total RNAs extracted from various tissues of wild-type, transgenic, or mutant seedlings were used to synthesize cDNAs by reverse transcription. Equal amounts of first-strand cDNAs were used as templates for PCR amplification using primers PIP5K9-3 (5′-AGGATGCTAGGTTCAGAGTTAGG-3′) and PIP5K9-4 (5′-GGTTCTGTCTTCTCATATCTGGC-3′) and CINV1-3 (5′-ATCGTTACTGGCTGTGACCC-3′) and CINV1-4 (5′-ACGTACCTTCCGAGCTTACC-3′), respectively. The Arabidopsis tubulin4 gene was amplified using primers tub4-1 (5′-AGAGGTTGACGAGCAGATGA-3′) and tub4-2 (5′-CCTCTTCTTCCTCCTCGTAC-3′) and served as an internal positive control (Marks et al., 1987).
Quantitative real-time RT-PCR analysis was performed with the RotorGene 3000 system (Corbett Research) using the SYBR green detection protocol (TaKaRa). TUB4 (At5g44340) mRNA was used as an internal control, and relative amounts of mRNA were calculated using the comparative threshold cycle method. Primers used were as follows: RBCs (At1g67090; sense, 5′-TTACCTTCCTGACCTTACCGATTCCG-3′, and reverse, 5′-CCACATTGTCCAGTACCGTCCATCAT-3′), PC (sense, 5′-GCTACCGTCACCATCCCTTCTTTC-3′, and reverse, 5′-GTTGCCACTGCGATGACACCGAA-3′), PAL1 (sense, 5′-TGAGTTTAGGAAGCCAGTTGTGAATCT-3′, and reverse, 5′-CGATGAGAAGTAGCACCAAAACCAGTAG-3′).
Promoter–Reporter Gene Fusion Studies
An ∼1.1-kb promoter region of PIP5K9 was amplified by PCR using primers PIP5K9-5 (5′-CAAGCTTAAGAAGGCTGTTATTGTGGCG-3′; added HindIII site underlined) and PIP5K9-6 (5′-GCGTCGACAAGCAGAAACGCTCAGACAAA-3′; added SalI site underlined) using Arabidopsis genomic DNA as template. The 1103-bp DNA fragment was subcloned into pCAMBIA1300 + pBI101.1 (Liu et al., 2003), and the resulting promoter–β-glucuronidase (GUS) construct was transformed into Arabidopsis. Positive transgenic plants were used for detection of GUS activities (Jefferson et al., 1987). Photography was performed using a Nikon microscope with a digital camera.
Identification of pip5k9-d and cinv1 Mutants
The PIP5K9 genomic sequence was used to query the flanking sequence database of the Arabidopsis mutation population (http://www.syngenta.com). One insertion line, Garlic_210_H11, was identified. The Arabidopsis T-DNA insertion mutant cinv1, SALK_095807, was obtained from the SALK collection.
A PCR-based approach was used to identify the mutants. Two pairs of primers, pip5k9-sense (5′-CGTCGTACTCTACCTTGGCATTAT-3′) and pip5k9-anti (5′-TTTTCAGTGGTGTCATTGGATAGG-3′) and pip5k9-sense and LB3 (5′-TAGCATCTGAATTTCATAACCAATCTCGATACAC-3′) were used to confirm the exact insertion position of the T-DNA and to identify homozygous lines. Numbers of T-DNA inserts were examined by DNA gel blot hybridization, and homozygous lines were used for further analysis. Transcript levels in mutant and wild-type plants were analyzed by RT-PCR. For the analysis of the cinv1 mutant, primers used were LBa1 (5′-TGGTTCACGTAGTGGGCCATCG-3′), cinv1-sense (5′-ATCGTTACTGGCTGTGACCC-3′), and cinv1-anti (5′-ACGTACCTTCCGAGCTTACC-3′).
Reciprocal crosses were performed using either pip5k9-d or cinv1 as the female parent. After removal of the petals, sepals, and androecia from large green buds, artificial fecundation was performed at 12:00 to 3:00 pm over the next 2 d.
Generation of PIP5K9 Sense and Antisense and CINV1 Sense Constructs
The PIP5K9 genomic DNA was amplified using primers PIP5K9-1 and PIP5K9-2 and subcloned into pCAMBIA1301 to create pO-PIP5K9 harboring PIP5K9 in the sense orientation. The entire PIP5K9 cDNA fragment was amplified with primers PIP5K9-7 (5′-GCTCTAGATACGAGGAGCAGTTTCTTTT-3′; added XbaI site underlined) and PIP5K9-8 (5′-GGGGTACCCTCCCTTTTGAGAGCTTATTAC-3′; added KpnI site underlined) and then subcloned into pCAMBIA1301 to generate pA-PIP5K9 harboring PIP5K9 in the antisense orientation. The CINV1 cDNA was amplified with primers CINV1-5 (5′-CGCGGATCCGAATGGAAGGTGTTGGACTAAGAG-3′; added BamHI site underlined) and CINV1-6 (5′-ACGCGTCGACACCTAAAGCGGTTTTTCAGAGTTG-3′; added SalI site underlined) and subcloned into pCAMBIA1301 to produce construct pO-CINV1 harboring CINV1 in the sense orientation. The generated constructs were used for plant transformation. Transgenic plants were analyzed by RT-PCR to determine transcript levels of PIP5K9 or CINV1 using external primers PIP5K9-9 (5′-ATGTCTGGCCTTGACGTACG-3′) and PIP5K9-10 (5′-GGTCCAAAGTCTGCTGTCCG-3′) or CINV1-3 and CINV1-4, respectively.
Homozygous T3 lines were used for further analysis.
Longitudinal Section of Root and Measurement of Cell Length
Root sections (0.5 mm) of pip5k9-d and wild-type plants grown on vertically oriented plates for 12 d were sampled, fixed in 2.5% glutaraldehyde (16 to 48 h at 4°C), and dehydrated through a graded ethanol series. Samples were embedded in Epon812 resin (Fluka) and polymerized at 60°C. Longitudinal sections (3 μm) were cut and stained briefly with filtered 1% toluidine blue. Sections were microscopically examined and photographed to measure cell lengths (Liu et al., 2003; Yang et al., 2005).
Yeast Two-Hybrid Screening
Screening for PIP5K9 interacting proteins was performed using the Matchmaker GAL4 Two-Hybrid System 3 (Clontech, TaKaRa). Two cDNA fragments coding for the MORN motif and the dimerization region, or the dimerization region and the catalytic domain, of PIP5K9 were amplified with primers PIP5K9-11 (5′-CCGGAATTCATGTCTGGCCTTGACGTACG-3′; added EcoRI site underlined) and PIP5K9-12 (5′-CGCGGATCCGGTCCAAAGTCTGCTGTCCG-3′; added BamHI site underlined) or PIP5K-13 (5′-CCGGAATTCAGCGATGGAGGTTGCTATGT-3′; added EcoRI site underlined) and PIP5K9-14 (5′-CGCGGATCCCTCTGTCTATGATTTGTTGTTC-3′; added BamHI site underlined), then subcloned into pGBKT7 (BD) to generate the bait constructs pGBKT7-1BD and pGBKT7-2BD.
The yeast strain AH109 was used as the host strain and transformed using a modified lithium acetate method (Clontech user manual) with bait plasmids. The transformants were subsequently transformed with an Arabidopsis cDNA library constructed with pGADT7 vector. More than three million transformants were screened, and positive colonies were subsequently tested for growth on supplemented SD medium lacking Leu, Trp, and His with X-α-gal or on supplemented SD medium lacking Leu, Trp, His, and adenine hemisulfate salt. Positive clones by both selections were further assayed on the same medium supplemented with 2, 4, 6, 8, 10, 12, or 16 mM 3-amino-1,2,4-triazole to test the strength of the interaction.
Yeast DNA was extracted and transformed into Escherichia coli strain XL1-Blue to amplify the cDNA inserts. The plasmids were then sequenced and analyzed. To confirm the interaction, full-length cDNAs of positive clones were amplified and subcloned into pGADT7 vector and then retransformed into yeast cells. Full-length cDNA of CINV1 was amplified with primers CINV1-1 and CINV1-2a or CINV1-1 and CINV1-2b (5′-CGCGGATCCCTTTTCCCATTTTCTTTTTCCTTAC-3′; added BamHI site underlined). CINV1-pGADT7 and bait (pGBKT7-2BD) were cotransformed into AH109 cells. pGBKT7-PIP5K9 was constructed with the primers PIP5K9-11 and PIP5K9-16 (5′-ACGCGTCGACCTCTGTCTATGATTTGTTGTTC-3′; added SalI site underlined).
Coimmunoprecipitation Analysis
Coimmunoprecipitation experiments using extracts prepared from yeast cells were performed as described by Nam and Li (2002), except that a protease inhibitor cocktail (Sigma-Aldrich), anti-MYC monoclonal antibody (NeoMarkers), anti-HA polyclonal antibody (NeoMarkers), nitrocellulose membrane (Schleicher and Schuell), and a nitroblue tetrazolium/bromochloroindolyl phosphate system were used.
Coimmunoprecipitation experiments using plant extracts were performed essentially according to Li et al. (2002). Transgenic plants overexpressing the relevant proteins were selected by protein gel blot analysis. Shoots of transgenic plants were harvested and ground to a fine powder in liquid nitrogen. The powder was further ground in cold grinding buffer (20 mM Tris-HCl, pH 8.8, 150 mM NaCl, 1 mM EDTA, 20% glycerol, 1 mM phenylmethylsulfonyl fluoride, and 1% protease inhibitor cocktail [Sigma-Aldrich]), and the suspension was spun at 6000g for 15 min at 4°C. The resulting supernatant was further centrifuged at 100,000g for 40 min at 4°C to sediment total membranes. The pellet was resuspended in membrane solubilization buffer (10 mM Tris-HCl, pH 7.3, 150 mM NaCl, 1 mM EDTA, 10% Triton X-100, 1 mM phenylmethylsulfonyl fluoride, and 1% protease inhibitor cocktail [Sigma-Aldrich]), and the suspension was diluted with the membrane solubilization buffer without Triton X-100 to a final Triton X-100 concentration of <2%. The solution was preincubated with anti-PIP5K9 or anti-CINV1 at 4°C for >3 h, followed by the addition of one-tenth volume of protein A–Sepharose (Amersham). Antibody was then added, and the mixture was further incubated with one-tenth volume of protein A–Sepharose at 4°C with gentle shaking (overnight). Beads were washed twice each with five volumes of PBS buffer supplemented with 0.1% Tween 20. Bound proteins were eluted with elution buffer (0.1 M Gly, pH 3.0). The eluted protein were separated by 8 or 10% SDS-PAGE, transferred to a nitrocellulose membrane (Schleicher and Schuell), and analyzed by protein gel blotting with anti-CINV1 or anti-PIP5K9 polyclonal antibody (no cross-reactivity with other PIP5K or invertase). The reactive bands on membranes were visualized by exposure to nitroblue tetrazolium/bromochloroindolyl phosphate.
Agrobacterium-Mediated Transient Expression (Agroinfiltration) of PIP5K9-GFP and CINV1-RFP Fusion Proteins
The coding region of PIP5K9 was amplified with primers PIP5K9-17 (5′-CATGCCATGGCTGGCCTTGACGTACGAG-3′; added NcoI site underlined) and PIP5K9-18 (5′-GGACTAGTTGATTTGTTGTTCTGTGGAAATACTT-3′; added SpeI site underlined) and subcloned into vector pCAMBIA1302. The full-length CINV1 was amplified with primers CINV1-7 (5′-ACGCGTCGACATGGAAGGTGTTGGACTAAGAG-3′; added SalI site underlined) and CINV1-2a and subcloned into vector pGDR (Goodin et al., 2002).
Tobacco (Nicotiana benthamiana) plants grown in a greenhouse were used for agroinfiltration experiments. Binary constructs were transformed into Agrobacterium tumefaciens strain C58 by the freeze-thaw method (An et al., 1988) and used to infiltrate tobacco leaves. Agrobacterial cells were grown at 28°C overnight. Cells were collected by centrifugation and resuspended in MES buffer (10 mM MgCl2, 10 mM MES, pH 5.6, and 150 μM acetosyringone) to a final OD600 of 0.6. The agrobacterial suspension was infiltrated into the abaxial surface of fully expanded leaves (Goodin et al., 2002). Four to six infiltration sites were used per leaf. GFP- and RFP-expressing cells were detected with a confocal laser scanning microscope (Zeiss LSM 510 META; argon laser excitation wavelength, 488 nm [GFP] or 543 nm [RFP]).
Transient expression of GFP or RFP fusion proteins in onion (Allium cepa) epidermal cells was performed with a model PDS-1000/He Biolistic particle delivery system (Bio-Rad). Five micrograms of purified plasmids was coated with 0.6- to 1-μm gold particles, and bombardment was performed with the following parameters: 1100-p.s.i. rupture disc; 27-inch Hg vacuum; 6-cm distance from the stopping screen to the target tissues. After bombardment, onion epidermal tissue was incubated in MS medium at 25°C in darkness for at least 24 h. Cell plasmolysis was achieved by incubating onion skin pieces in 1 M sucrose solution for 10 min.
Gene Expression Analysis Using Affymetrix Microarrays
Seedling roots of pip5k9-d, cinv1, and wild-type plants were harvested and frozen quickly in liquid nitrogen. RNA samples were processed as recommended by Affymetrix, including RNA extraction, converting to double-stranded cDNA, and synthesis of biotin-labeled copy RNA. Fifteen micrograms of biotin-labeled copy RNA was purified and fragmented into 35- to 300-nucleotide fragments and then hybridized with an Arabidopsis genome array (Affymetrix ATH1). The hybridization was performed with two biological replicates for each sample. After hybridization, arrays were scanned using an Agilent GeneArray scanner (Affymetrix G2500A). Microarray data were normalized using the Affymetrix Microarray Suit program (version 5.0). The algorithm absolute call flag was set to indicate the reliability of the data points according to P (present), M (marginal), and A (absent). Significant differences for each gene between the wild-type and pip5k9-d mutant plants, or the wild-type and cinv1 mutant plants, were examined using the Wilcoxon rank test of the Affymetrix Microarray Suit program (P < 0.01) and analysis of variance by fitting a mixed model in the MAANOVA package under R 1.8.0 for multiple testing (false discovery rate–corrected P < 0.05). Genes with the consensus significant difference in two independent samples were selected and listed. Annotations for the selected probe set identification were rechecked at the Affymetrix website.
Enzymatic Assay of Invertase
Invertase activities in plants were determined according to Pelleschi et al. (1997). A microcon (Millipore) was used to desalt supernatants, and the glucose amount was measured with the glucose assay kit (Sigma-Aldrich) during the assay. Enzymatic assay of purified proteins in vitro was performed according to Vargas et al. (2003). The reaction was performed in 0.05 M HEPES-NaOH, pH 7.0, and 0.001 M Na2EDTA, and the results were analyzed with the Enzyme Kinetics Module (SYSTAT software).
Metabolite Profiling by GC-MS
Metabolites were analyzed essentially as described by Fiehn et al. (2000). One hundred milligrams of seedlings or roots (fresh weight) was harvested and ground into a fine powder in liquid nitrogen. Seven hundred microliters of methanol was immediately added to the powder to stop enzymatic activity, and 50 μL of 0.2 ng/mL rabitol was then added. After centrifugation at 10,000g for 2 min, the supernatant was transferred to a tube and dried. For methoximation, 80 μL of methoxyamine hydrochloride in pyridine (20 mg/mL) was used at 30°C for 90 min. Afterward, 50 μL of N-methyl-N-trimethylsilyl-trifluoroacetamide was added, and the mixture was incubated at 37°C for 30 min. GC-MS was performed using an Agilent Network GC system 6890 N/Mass Selective Detector MS 5973.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: PIP5K9 (AJ810853), CINV1 (AM230708), PC (M20937), and PAL1 (L33677).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table 1. Common Altered Genes under Deficiency of CINV1 and Enhanced PIP5K9.
Supplemental Table 2. Altered Genes under Enhanced PIP5K9.
Supplemental Table 3. Altered Genes under Deficiency of CINV1.
Supplementary Material
Acknowledgments
We greatly thank Nam-Hai Chua for critical comments on and revision of the manuscript, Xiao-Ya Chen for careful reading of and helpful comments on the manuscript, Xiao-Yan Gao for performing cross sections of roots, and Shu-Ping Xu for the particle bombardment experiments. This study was supported by the National Nature Science Foundation (Grants 30425029 and 30421001).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Hong-Wei Xue (hwxue@sibs.ac.cn).
Online version contains Web-only data.
References
- An, G., Erbert, P.R., Mitra, A., and Ha, S.B. (1988). Binary vectors. In Plant Molecular Biology Manual A3. (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 1–19.
- Anderson, R.A., Boronenkov, I.V., Doughman, S.D., Kunz, J., and Loijens, J.C. (1999). Phosphatidylinositol phosphate kinases, a multifaceted family of signaling enzymes. J. Biol. Chem. 274 9907–9910. [DOI] [PubMed] [Google Scholar]
- ApRees, T. (1974). Pathways of carbohydrate breakdown in higher plants. In Biochemistry. Series I, Vol. II. Plant Biochemistry, D.H. Northcote, ed (London: Butterworth), pp. 89–127.
- Bairstow, S.F., Ling, K., and Anderson, R.A. (2005). Phosphatidylinositol phosphate kinase type Igamma directly associates with and regulates Shp-1 tyrosine phosphatase. J. Biol. Chem. 280 23884–23891. [DOI] [PubMed] [Google Scholar]
- Bhowmik, P.K., Matsui, T., and Ahmed, Z. (2001). Changes in the activity of sucrose metabolizing enzymes during the germination of wheat seeds. Pak. J. Biol. Sci. 4 1264–1266. [Google Scholar]
- Boronenkov, I.V., Loijens, J.C., Umeda, M., and Anderson, R.A. (1998). Phosphoinositide signaling pathways in nuclei are associated with nuclear speckles containing pre-mRNA processing factors. Mol. Biol. Cell 9 3547–3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun, M., Baluska, F., von Witsch, M., and Menzel, D. (1999). Redistribution of actin, profilin and phosphatidylinositol-4,5-bisphosphate in growing and maturing root hairs. Planta 209 435–443. [DOI] [PubMed] [Google Scholar]
- Bussis, D., Heineke, D., Sonnewald, U., Willmitzer, L., Raschke, K., and Heldt, H.W. (1997). Solute accumulation and decreased photosynthesis in leaves of potato plants expressing yeast-derived invertase either in the apoplast, vacuole or cytosol. Planta 202 126–136. [DOI] [PubMed] [Google Scholar]
- Caroni, P. (2001). New EMBO members' review: Actin cytoskeleton regulation through modulation of PI(4,5)P(2) rafts. EMBO J. 20 4332–4336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpita, N.C., and Gibeaut, D.M. (1993). Structural models of primary cell walls in flowering plants: Consistency of molecular structure with the physical properties of the walls during growth. Plant J. 3 1–30. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Collmer, A., and Keen, N.T. (1986). The role of pectic enzymes in plant pathogenesis. Annu. Rev. Phytopathol. 24 383–409. [Google Scholar]
- Dickinson, C.D., Altabella, T., and Chrispeels, M.J. (1991). Slow-growth phenotype of transgenic tomato expressing apoplastic invertase. Plant Physiol. 95 420–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Divecha, N., Roefs, M., Los, A., Halstead, J., Bannister, A., and D'Santos, C. (2002). Type I PIPkinases interact with and are regulated by the retinoblastoma susceptibility gene product-pRB. Curr. Biol. 12 582–587. [DOI] [PubMed] [Google Scholar]
- Divecha, N., Truong, O., Hsuan, J.J., Hinchliffe, K.A., and Irvine, R.F. (1995). The cloning and sequence of the C isoform of PtdIns4P 5-kinase. Biochem. J. 309 715–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehness, R., Ecker, M., Godt, D.E., and Roitsch, T. (1997). Glucose and stress independently regulate source and sink metabolism and defense mechanisms via signal transduction pathways involving protein phosphorylation. Plant Cell 9 1825–1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elge, S., Brearley, C., Xia, H.J., Kehr, J., Xue, H.W., and Mueller-Roeber, B. (2001). An Arabidopsis inositol phospholipid kinase strongly expressed in procambial cells: Synthesis of PtdIns(4,5)P2 and PtdIns(3,4,5)P3 in insect cells by 5-phosphorylation of precursors. Plant J. 26 561–571. [DOI] [PubMed] [Google Scholar]
- Fiehn, O., Kopka, J., Dormann, P., Altmann, T., Trethewey, R.N., and Willmitzer, L. (2000). Metabolite profiling for plant functional genomics. Nat. Biotechnol. 18 1157–1161. [DOI] [PubMed] [Google Scholar]
- Foyer, C.H. (1987). Evidence for different kinases in thylakoid protein phosphorylation. Biochem. J. 248 103–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godt, D.E., and Roitsch, T. (1997). Regulation and tissue-specific distribution of mRNAs for three extracellular invertase isoenzymes of tomato suggests an important function in establishing and maintaining sink metabolism. Plant Physiol. 115 273–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodin, M.M., Dietzgen, R.G., Schichnes, D., Ruzin, S., and Jackson, A.O. (2002). pGD vectors: Versatile tools for the expression of green and red fluorescent protein fusions in agroinfiltrated plant leaves. Plant J. 31 375–383. [DOI] [PubMed] [Google Scholar]
- Haigler, C.H., Ivanova-Datcheva, M., Hogan, P.S., Salnikov, V.V., Hwang, S., Martin, K., and Delmer, D.P. (2001). Carbon partitioning to cellulose synthesis. Plant Mol. Biol. 47 29–51. [PubMed] [Google Scholar]
- Heyer, A.G., Raap, M., Schroeer, B., Marty, B., and Willmitzer, L. (2004). Cell wall invertase expression at the apical meristem alters floral, architectural, and reproductive traits in Arabidopsis thaliana. Plant J. 39 161–169. [DOI] [PubMed] [Google Scholar]
- Hokin, M.R., and Hokin, L.E. (1953). Enzyme secretion and the incorporation of p32 into phospholipides of pancreas. J. Biol. Chem. 203 967–977. [PubMed] [Google Scholar]
- Irvine, R.F. (2003). Nuclear lipid signalling. Nat. Rev. Mol. Cell Biol. 4 349–360. [DOI] [PubMed] [Google Scholar]
- Ismail, I., De Bellis, L., Alpi, A., and Smith, S.M. (1997). Expression of glyoxylate cycle genes in cucumber roots responds to sugar supply and can be activated by shading or defoliation of the shoot. Plant Mol. Biol. 35 633–640. [DOI] [PubMed] [Google Scholar]
- Jang, J.C., and Sheen, J. (1994). Sugar sensing in higher plants. Plant Cell 6 1665–1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janmey, P.A., and Stossel, T.P. (1987). Modulation of gelsolin function by phosphatidylinositol 4,5-bisphosphate. Nature 325 362–364. [DOI] [PubMed] [Google Scholar]
- Jefferson, R.A., Kavanagh, T.A., and Bevan, M.W. (1987). GUS fusions: Beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, G.T., Tsukaya, H., and Uchimiya, H. (1998). The CURLY LEAF gene controls both division and elongation of cells during the expansion of the leaf blade in Arabidopsis thaliana. Planta 206 175–183. [DOI] [PubMed] [Google Scholar]
- Kim, J.Y., Mahe, A., Brangeon, J., and Prioul, J.L. (2000). A maize vacuolar invertase, IVR2, is induced by water stress. Organ/tissue specificity and diurnal modulation of expression. Plant Physiol. 124 71–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch, K.E. (1996). Carbohydrate-modulated gene expression in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47 509–540. [DOI] [PubMed] [Google Scholar]
- Kost, B., Lemichez, E., Spielhofer, P., Hong, Y., Tolias, K., Carpenter, C., and Chua, N.H. (1999). Rac homologues and compartmentalized phosphatidylinositol 4,5-bisphosphate act in a common pathway to regulate polar pollen tube growth. J. Cell Biol. 145 317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krapp, A., Hofmann, B., Shafer, C., and Stitt, M. (1993). Regulation of the expression of rbcS and other photosynthetic genes by carbohydrates: A mechanism for the ‘sink’ regulation of photosynthesis. Plant J. 3 817–828. [Google Scholar]
- Lassing, I., and Lindberg, U. (1985). Specific interaction between phosphatidylinositol 4,5-bisphosphate and profilactin. Nature 314 472–474. [DOI] [PubMed] [Google Scholar]
- Li, J., Wen, J., Lease, K.A., Doke, J.T., Tax, F.E., and Walker, J.C. (2002). BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell 110 213–222. [DOI] [PubMed] [Google Scholar]
- Lin, W.H., Ye, R., Ma, H., Xu, Z.H., and Xue, H.W. (2004). DNA chip-based expression profile analysis indicates involvement of the phosphatidylinositol signaling pathway in multiple plant responses to hormone and abiotic treatments. Cell Res. 14 34–45. [DOI] [PubMed] [Google Scholar]
- Liu, W., Xu, Z.H., Luo, D., and Xue, H.W. (2003). Roles of OsCKI1, a rice casein kinase I, in root development and plant hormone sensitivity. Plant J. 36 189–202. [DOI] [PubMed] [Google Scholar]
- Ma, H., Lou, Y., Lin, W.H., and Xue, H.W. (2006). MORN motifs in plant PIPKs are involved in the regulation of subcellular localization and phospholipid binding. Cell Res. 16 466–478. [DOI] [PubMed] [Google Scholar]
- Ma, H., Xu, S.P., Luo, D., Xu, Z.H., and Xue, H.W. (2004). OsPIPK 1, a rice phosphatidylinositol monophosphate kinase, regulates rice heading by modifying the expression of floral induction genes. Plant Mol. Biol. 54 295–310. [DOI] [PubMed] [Google Scholar]
- Marks, M.D., West, J., and Weeks, D.P. (1987). The relatively large beta-tubulin gene family of Arabidopsis contains a member with an unusual transcribed 5′ noncoding sequence. Plant Mol. Biol. 10 91–104. [DOI] [PubMed] [Google Scholar]
- Micheli, F. (2001). Pectin methylesterases: Cell wall enzymes with important roles in plant physiology. Trends Plant Sci. 6 414–419. [DOI] [PubMed] [Google Scholar]
- Mikami, K., Katagiri, T., Iuchi, S., Yamaguchi-Shinozaki, K., and Shinozaki, K. (1998). A gene encoding phosphatidylinositol-4-phosphate 5-kinase is induced by water stress and abscisic acid in Arabidopsis thaliana. Plant J. 15 563–568. [DOI] [PubMed] [Google Scholar]
- Mueller-Roeber, B., and Pical, C. (2002). Inositol phospholipid metabolism in Arabidopsis. Characterized and putative isoforms of inositol phospholipid kinase and phosphoinositide-specific phospholipase C. Plant Physiol. 130 22–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam, K.H., and Li, J. (2002). BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell 110 203–212. [DOI] [PubMed] [Google Scholar]
- Nari, J., Noat, G., Diamantidis, G., Woudstra, M., and Ricard, J. (1986). Electrostatic effects and the dynamics of enzyme reactions at the surface of plant cells. III. Interplay between limited cell-wall autolysis, pectin methyl esterase activity and electrostatic effects in soybean cell walls. Eur. J. Biochem. 155 199–202. [DOI] [PubMed] [Google Scholar]
- Oude Weernink, P.A., Schmidt, M., and Jakobs, K.H. (2004). Regulation and cellular roles of phosphoinositide 5-kinases. Eur. J. Pharmacol. 500 87–99. [DOI] [PubMed] [Google Scholar]
- Payrastre, B., Nievers, M., Boonstra, J., Breton, M., Verkleij, A.J., and Van Bergen en Henegouwen, P.M. (1992). A differential location of phosphoinositide kinases, diacylglycerol kinase, and phospholipase C in the nuclear matrix. J. Biol. Chem. 267 5078–5084. [PubMed] [Google Scholar]
- Pelleschi, S., Rocher, J.P., and Prioul, J.L. (1997). Effect of water restriction on carbohydrate metabolism and photosynthesis in mature maize leaves. Plant Cell Environ. 20 493–503. [Google Scholar]
- Rausch, T., and Greiner, S. (2004). Plant protein inhibitors of invertases. Biochim. Biophys. Acta 1696 253–261. [DOI] [PubMed] [Google Scholar]
- Ricardo, C.P.P., and ApRees, T. (1970). Invertase activity during the development of carrot roots. Phytochemistry 9 239–247. [Google Scholar]
- Roitsch, T. (1999). Source-sink regulation by sugar and stress. Curr. Opin. Plant Biol. 2 198–206. [DOI] [PubMed] [Google Scholar]
- Roitsch, T., and Gonzalez, M.C. (2004). Function and regulation of plant invertases: Sweet sensations. Trends Plant Sci. 9 606–613. [DOI] [PubMed] [Google Scholar]
- Sheen, J., Zhou, L., and Jang, J.C. (1999). Sugars as signaling molecules. Curr. Opin. Plant Biol. 2 410–418. [DOI] [PubMed] [Google Scholar]
- Sonnewald, U., Brauer, M., von Schaewen, A., Stitt, M., and Willmitzer, L. (1991). Transgenic tobacco plants expressing yeast-derived invertase in either the cytosol, vacuole or apoplast: A powerful tool for studying sucrose metabolism and sink/source interactions. Plant J. 1 95–106. [DOI] [PubMed] [Google Scholar]
- Sturm, A. (1999). Invertases. Primary structures, functions, and roles in plant development and sucrose partitioning. Plant Physiol. 121 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, G.Q., Luscher, M., and Sturm, A. (1999). Antisense repression of vacuolar and cell wall invertase in transgenic carrot alters early plant development and sucrose partitioning. Plant Cell 11 177–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toker, A. (2002). Phosphoinositides and signal transduction. Cell. Mol. Life Sci. 59 761–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargas, W., Cumino, A., and Salerno, G.L. (2003). Cyanobacterial alkaline/neutral invertases. Origin of sucrose hydrolysis in the plant cytosol? Planta 216 951–960. [DOI] [PubMed] [Google Scholar]
- Westergren, T., Dove, S.K., Sommarin, M., and Pical, C. (2001). AtPIP5K1, an Arabidopsis thaliana phosphatidylinositol phosphate kinase, synthesizes PtdIns(3,4)P(2) and PtdIns(4,5)P(2) in vitro and is inhibited by phosphorylation. Biochem. J. 359 583–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, X.H., Xu, Z.H., and Xue, H.W. (2005). Arabidopsis membrane steroid binding protein 1 is involved in inhibition of cell elongation. Plant Cell 17 116–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yelle, S., Chetelat, R.T., Dorais, M., Deverna, J.W., and Bennett, A.B. (1991). Sink metabolism in tomato fruit. IV. Genetic and biochemical analysis of sucrose accumulation. Plant Physiol. 95 1026–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yelle, S., Hewitt, J.D., Robinson, N.L., Damon, S., and Bennett, A.B. (1988). Sink metabolism in tomato fruit. III. Analysis of carbohydrate assimilation in a wild species. Plant Physiol. 87 737–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, L., Jang, J.C., Jones, T.L., and Sheen, J. (1998). Glucose and ethylene signal transduction crosstalk revealed by an Arabidopsis glucose-insensitive mutant. Proc. Natl. Acad. Sci. USA 95 10294–10299. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.