Abstract
In Arabidopsis thaliana, silencing of hydroxycinnamoyl-CoA shikimate/quinate hydroxycinnamoyl transferase (HCT), a lignin biosynthetic gene, results in a strong reduction of plant growth. We show that, in HCT-silenced plants, lignin synthesis repression leads to the redirection of the metabolic flux into flavonoids through chalcone synthase activity. Several flavonol glycosides and acylated anthocyanin were shown to accumulate in higher amounts in silenced plants. By contrast, sinapoylmalate levels were barely affected, suggesting that the synthesis of that phenylpropanoid compound might be HCT-independent. The growth phenotype of HCT-silenced plants was shown to be controlled by light and to depend on chalcone synthase expression. Histochemical analysis of silenced stem tissues demonstrated altered tracheary elements. The level of plant growth reduction of HCT-deficient plants was correlated with the inhibition of auxin transport. Suppression of flavonoid accumulation by chalcone synthase repression in HCT-deficient plants restored normal auxin transport and wild-type plant growth. By contrast, the lignin structure of the plants simultaneously repressed for HCT and chalcone synthase remained as severely altered as in HCT-silenced plants, with a large predominance of nonmethoxylated H units. These data demonstrate that the reduced size phenotype of HCT-silenced plants is not due to the alteration of lignin synthesis but to flavonoid accumulation.
INTRODUCTION
In the large array of plant natural products, compounds issuing from the phenylpropanoid pathway fulfill important functions, being involved in development and interaction of the plant with its environment (Petersen et al., 1999). For example, coumarins, isoflavonoids, and stilbenes are antimicrobial metabolites produced by plants while defending themselves against pathogen infections. Flavonoids protect plants against UV irradiation and act as signals in plant–symbiont interactions, while phenolic compounds, such as salicylic acid and acetosyringone, are signals implicated in plant–pathogen interactions. In addition, the phenylpropanoid pathway is responsible for the production of the three lignin monomers called monolignols. Lignin is embedded in the plant cell walls, rigidifying them and rendering them impermeable to water. Thus, lignin plays important roles in mechanical support and water transport in plants.
In the phenylpropanoid pathway (Figure 1), p-coumaroyl CoA is situated at the junction of the metabolic routes leading to flavonoids or to phenylpropanoid compounds sensu stricto. Indeed, p-coumaroyl CoA is the common substrate of two enzymes: (1) chalcone synthase (CHS), which catalyzes the formation of the flavonoid skeleton by condensation of p-coumaroyl CoA with three malonyl CoA molecules, and (2) hydroxycinnamoyl-CoA shikimate/quinate hydroxycinnamoyl transferase (HCT), which leads to the biosynthesis of two major lignin building units, namely, the guaiacyl and syringyl units (Figure 1). HCT has been characterized recently (Hoffmann et al., 2003) and shown to catalyze the synthesis of the shikimate and quinate esters of p-coumaric acid, which are the substrates of the cytochrome P450 3-hydroxylase (CYP98A3) (Schoch et al., 2001; Franke et al., 2002). Repression of HCT in Nicotiana benthamiana resulted in marked changes in the concentration of caffeoylquinate isomers and in the amount and composition of lignin, thus demonstrating that HCT functions in phenylpropanoid metabolism in planta (Hoffmann et al., 2004).
Figure 1.
The Phenylpropanoid Pathway.
P-coumaroyl CoA is at the crossroad of metabolic routes leading either to flavonoids or to monolignols and sinapoylmalate. CHS and HCT activities control the metabolic flux entering the two routes. ALDH, aldehyde dehydrogenase; C3H, C3-hydroxylase (CYP98A3); C4H, C4-hydroxylase (CYP73A5); F5H, ferulate 5-hydroxylase (CYP84A1); CAD, cinnamyl alcohol dehydrogenase; CCoAOMT, caffeoyl-CoA O-methyltransferase; CCR, cinnamoyl-CoA reductase; CHI, chalcone isomerase; 4CL, 4-coumaroyl-CoA ligase; COMT I, caffeic acid O-methyltransferase of class I; F3H, flavanone 3-hydroxylase; F3′H, flavonoid 3′-hydroxylase (CYP75B1); FLS, flavonol synthase; PAL, phenylalanine ammonia lyase; SGT, sinapate UDP-glucose sinapoyltransferase; SMT, sinapoylglucose malate sinapoyltransferase; UGTs, UDP sugar glycosyltransferases.
In Arabidopsis thaliana, RNA-mediated posttranscriptional gene silencing of HCT was achieved by plant transformation with a hairpin repeat of a portion of the HCT sequence. Among the primary transformants produced, a large majority displayed severely reduced growth and dark-green/purple coloration of leaves (Hoffmann et al., 2004). A few were less affected, developed a floral stem, and produced seeds. Here, the progeny of the latter was examined for HCT expression, and HCT-silenced lines were selected that displayed varying severity of the growth phenotype. The purple coloration of leaves of silenced plants suggested that they were accumulating anthocyanins. Since some flavonoids are considered as endogenous auxin transport regulators that affect plant development (Jacobs and Rubery, 1988; Brown et al., 2001; Taylor and Grotewold, 2005), we investigated whether growth inhibition of HCT-silenced plants is mediated by the inhibition of auxin transport resulting from flavonoid accumulation.
Proper regulation of auxin transport is essential for plant growth and development (Lomax et al., 1995; Friml, 2003). Auxin is transported from cell to cell by uptake and efflux carrier proteins with strict directionality resulting in a basipetal transport from the top toward the base of the plant and influencing vascular tissue differentiation, apical development, organ regeneration, tropic growth, and cell elongation (Friml, 2003; Taylor and Grotewold, 2005). Three families of carrier proteins that are products of the AUX, PIN, or MDR/P-glycoprotein genes have been mainly identified through the characterization of their respective mutants (for reviews, see Muday and DeLong, 2001; Muday and Murphy, 2002; Blakeslee et al., 2005). Among these auxin carriers, PIN proteins constitute a family of efflux carriers located on the basal or apical side of the transporting cell (Galweiler et al., 1998; Gil et al., 2001; Friml et al., 2004; Peer et al., 2004; Wisniewska et al., 2006). Regulatory proteins RCN1 and PID, which mediate protein phosphorylation and PIN polarity, have also been shown to control auxin transport and efflux and influx carriers having asymmetric distributions in transporting cells (Galweiler et al., 1998; Christensen et al., 2000; Benjamins et al., 2001; Muday and DeLong, 2001; Rashotte et al., 2001; Muday and Murphy, 2002; Friml et al., 2004).
Plant flavonoids are considered natural regulators of cellular auxin efflux and consequent auxin polar transport (Jacobs and Rubery, 1988; Brown et al., 2001; Murphy et al., 2002; Buer and Muday, 2004; Peer et al., 2004; Taylor and Grotewold, 2005). Likely sites of flavonoid action have been identified as plasma membrane proteins known as the naphtylphthalamic acid (NPA) binding protein (NBP) complex (Jacobs and Rubery, 1988; Lomax et al., 1995; Murphy et al., 2000, 2002). The tir3/doc1 mutant that is affected both in light responses and in auxin transport defines a calossin-like gene named BIG and displays a reduction in the number of NPA binding sites in the microsomal fraction (Ruegger et al., 1997; Gil et al., 2001). A role of intracellular trafficking has been proposed because treatment with brefeldin A, an inhibitor of vesicular transport, prevents normal membrane localization of PIN1 and blocks auxin efflux (Delbarre et al., 1998; Geldner et al., 2001; Muday and Murphy, 2002; Geldner et al., 2003). Finally, several NBPs occur that differ in their ligand affinity (Lomax et al., 1995; Murphy et al., 2000, 2002; Muday and Murphy, 2002), suggesting multiple physiological functions for the recognition of the NBP complex.
Some studies indicate that NBPs are associated with the cytoplasmic face of the plasma membrane and are distinct from efflux carriers (Muday and DeLong, 2001). Recently, MDR/P-glycoproteins have been uncovered as important players in polar auxin transport when different members of the family were purified from NPA binding complexes and implicated in the stabilization of the membrane efflux systems (Noh et al., 2001, 2003; Murphy et al., 2002; Geisler et al., 2003; Blakeslee et al., 2005). It has been proposed that MDR/P-glycoproteins function as ATP-dependent auxin transporters whose interactions with PIN proteins confer directionality and substrate specificity to the auxin efflux machinery (Blakeslee et al., 2005; Geisler et al., 2005). Auxin efflux activities of Arabidopsis MDR/P-glycoproteins are inhibited by quercetin (Geisler et al., 2005; Bouchard et al., 2006). Recently, PIN expression in yeast and mammalian cells demonstrated a direct catalytic role for PINs in auxin transport, independently of the MDR/P-glycoprotein family (Petrasek et al., 2006). AUX1 has also been functionnally characterized and shown to have influx carrier activity in Xenopus laevis oocytes (Yang et al., 2006). Auxin transport is elevated in plants with the transparent testa4 (tt4) mutation in the CHS gene, consistent with the absence of flavonoids, the putative endogenous negative auxin transport regulators (Buer and Muday, 2004; Peer et al., 2004). Conversely, accumulation of flavonols in tt3 and tt7 mutants or in wild-type and tt4 plants fed with a flavonoid precursor provoked auxin transport inhibition (Brown et al., 2001; Peer et al., 2004). Flavonoid synthesis is tightly controlled by environmental cues (Feinbaum and Ausubel, 1988; Kubasek et al., 1992; Graham, 1998; Winkel-Shirley, 2002), indicating that flavonoid accumulation may be regulated under conditions when auxin transport is modulated.
HCT gene repression has been shown to lead to profound changes in phenylpropanoid metabolism. In HCT-silenced plants, lignin biosynthesis was inhibited and syringyl lignin unit content was decreased (Hoffmann et al., 2004). In Arabidopsis, the impact of HCT repression on lignin biosynthesis has been detected by histochemical staining of HCT-silenced stems (Hoffmann et al., 2004). In this work, we analyze lignin of HCT-deficient Arabidopsis by thioacidolysis and demonstrate that the H nonmethoxylated unit that is present in trace amounts in wild-type plants represents 85% of total lignin monomers. Moreover, we show that many flavonoids, including flavonols and anthocyanins, accumulate in higher amounts in HCT-silenced plants compared with wild-type plants. Flavonoid accumulation was dependent on the light conditions and controlled by the expression of CHS, the first enzyme of the flavonoid pathway. Reduction of plant growth and development was correlated to the extent of flavonoid accumulation and to the level of auxin transport inhibition. Finally, flavonoid accumulation, auxin transport inhibition, and reduced size phenotype were abolished in plants where both HCT and CHS genes were repressed simultaneously, thus demonstrating the role of flavonoids in the developmental alterations of HCT-silenced plants.
RESULTS
HCT Silencing Results in Severe Growth Inhibition and Flavonoid Accumulation
Arabidopsis plants transformed with a hairpin repeat of a portion of the HCT sequence driven by the 35S promoter to induce RNA interference (RNAi) against HCT transcripts were severely affected in their growth and development compared with wild-type controls. A majority of primary transformants (T0 population) displayed a severe dwarf phenotype and did not develop a floral stem (Hoffmann et al., 2004). The plants that produced seeds gave rise to T1 progeny that was studied in this work and showed three typical size phenotypes of silenced plants compared with controls (Figure 2A, plants vs, s, and i). Plants with intermediate and small sizes (Figure 2A, plants i and s) were fully fertile and used for further study (see below). HCT expression in plant stems was measured by protein gel blots using a specific antiserum raised against the purified Arabidopsis recombinant protein (see Methods). HCT protein was hardly detectable (Figure 2B, plants i and s) or undetectable at all (Figure 2B, plant vs). Consistently, HCT activity, measured by HPLC analysis of the reaction product, p-coumaroylshikimate (Hoffmann et al., 2004), was considerably reduced in stems of all silenced plants compared with controls (Figure 2C). HCT activity of silenced plants with different reduced size phenotypes was in the range of 1 to 4% of the control value (Figure 2C). The HCT-deficient plants not only had a dwarf phenotype but also displayed a dark-purple coloration on leaves, indicating a strong accumulation of anthocyanins. We addressed the question of HCT silencing specificity by analyzing the expression of the closest members of the Arabidopsis acyltransferase family that includes ∼60 genes (Hoffmann et al., 2003; D'Auria, 2006). Expression studies by RT-PCR have demonstrated that At5g57840 and At2g19070 genes that share 40 and 36% identity with HCT (At5g48930), respectively, were not affected in HCT-deficient plants (data not shown), thus confirming the specificity of HCT silencing.
Figure 2.
HCT Silencing Has a Strong Impact on Plant Growth.
(A) Three typical size phenotypes (i, intermediate; s, small; vs, very small) were observed among T1 progeny of transformants carrying an HCT hairpin repeat (right-hand plants) compared with the wild-type control plant (on the left).
(B) Protein extracts from wild-type stems or stems of plant types i and s or leaves of plant vs were immunoblotted with a specific anti-HCT serum raised against the Arabidopsis protein. The arrow indicates the position of HCT protein. At the right is the position of markers of known molecular mass (given in kilodaltons).
(C) HCT activity was strongly reduced in plants of small size phenotypes (i, s, and vs) compared with the wild type. Mean activity values and standard errors were calculated from four determinations.
To confirm the activation of the flavonoid pathway upon HCT silencing, we extracted leaf tissues of HCT-silenced and wild-type plants and compared their flavonoid content by HPLC analysis. Comparison of HPLC profiles (Figure 3) demonstrated the accumulation of several flavonols (as revealed by their absorbance at 345 nm; Figure 3C) and anthocyanins (as revealed by their absorbance at 530 nm; Figure 3D) in HCT-silenced leaves compared with wild-type tissues (Figures 3A and 3B). Products were identified by the characteristic UV absorbance spectra of their aglycones, and the nature of the glycosides was inferred from their mass spectra (Figures 3E to 3H). In wild-type leaf tissues, only one major peak was detected at 345 nm, which corresponded to sinapoylmalate (Figure 3A). In HCT-deficient plants, several additional peaks corresponding to glycosides of kaempferol and quercetin appeared (Figure 3C). According to their chromatographic and spectral properties and compared with published data (Bloor and Abrahams, 2002; Jones et al., 2003; Tohge et al., 2005), the main flavonol derivatives were kaempferol-3-O-[rhamnosyl (1>2) glucoside]-7-O-rhamnoside (Figure 4, K1), kaempferol-3-O-glucoside-7-O-rhamnoside (Figure 4, K2), and kaempferol-3-O-rhamnoside-7-O-rhamnoside (Figure 4, K3), together with lower amounts of quercetin homologues (Figure 4, Q1 to Q3). At 530 nm, no peak was detectable in wild-type extract (Figure 3B), whereas in HCT-silenced extract, several anthocyanins were identified as cyanidin derivatives based on their UV absorbance spectra (Figure 3D, C1 to C5). Their mass spectra fit published data (Bloor and Abrahams, 2002; Tohge et al., 2005), allowing us to infer the exact nature of these molecules. A few examples of mass spectra are presented in Figures 3E to 3H, and the structures of the different compounds are given in Figure 4. The main ions observed on the mass spectra of the flavonol glycosides were the deprotonated molecules [M-H]− (Figures 3E and 3G) and the [M]+ ion for the cyanidin derivatives (Figures 3F and 3H). In addition, other ions could be observed that correspond to the loss of glycoside moieties. Table 1 presents the physico-chemical properties that allowed the identification of the different products and their amounts measured in the various genotypes. Under standard light conditions (Table 1), flavonoids were found in higher amounts in HCT-silenced 2-month-old leaves compared with controls where kaempferol derivatives were barely detectable and only two anthocyanins (C1 and C2) were present in low amounts. It is noteworthy that quercetin derivatives were only detected in HCT−s plants. Not surprisingly, no flavonoid at all was detectable in CHS− plants used as controls (Wesley et al., 2001; Dunoyer et al., 2004). The highest accumulation of flavonoids was measured in silenced plants of small size (HCT−s) compared with plants of intermediate size (HCT−i), suggesting a role of these metabolites in plant growth inhibition. In contrast with the increase of flavonols and anthocyanins, sinapoylmalate levels remained roughly unchanged in HCT-deficient tissues (also compare SM peaks in Figures 3A and 3C). This result is at odds with the putative involvement of HCT in sinapoylmalate biosynthesis (Figure 1) and suggests the existence of an alternative HCT-independent pathway. It is important to note that sinapoylglucose, the biosynthetic precursor of sinapoylmalate, was also detected in the extracts, but it never accounted for >3% of sinapoylmalate amount. Taken together, the data demonstrate that HCT repression had a major impact on phenylpropanoid metabolism and resulted in the redirection of the main metabolic flux toward the flavonoid pathway.
Figure 3.
HPLC and Liquid Chromatography–Mass Spectrometry Analysis of Soluble Phenolics Extracted from Wild-Type or HCT-Deficient Leaf Tissues.
(A) and (B) Profiles of wild-type extracts at 345 and 530 nm, respectively. SM, sinapoylmalate.
(C) and (D) Profiles of HCT-silenced extracts at 345 and 530 nm, respectively.
(E) to (H) Examples of electrospray mass spectra (K1, C4, Q2, and C5 peaks, respectively) used for structure determination by comparison with published data (Table 1, Figure 4). Electrospray mass spectra of K1 and Q2 were obtained in the negative mode and C4 and C5 spectra in the positive mode. m/z, mass-to-charge ratio.
Figure 4.
Flavonoids Accumulated in HCT-Deficient Arabidopsis.
Structure of flavonol glycosides and cyanidin derivatives were inferred from their UV-visible and mass spectral properties (Table 1) and from available data (Graham, 1998; Veit and Pauli, 1999; Bloor and Abrahams, 2002; Jones et al., 2003; Abdulrazzak et al., 2005; Tohge et al., 2005).
Table 1.
Liquid Chromatography–Mass Spectrometry Identification and Levels of the Phenolic Compounds Accumulating in Leaves of HCT-Deficient Plants
| Pseudomolecular Ion (m/z) | Standard Light
|
Low Light
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Peak | Rt (min) | λmax (nm) | Fragments (m/z) | CHS− | Wild Type | HCT−i | HCT−s | HCT−s | |
| Q1 | 14.2 | 256–348 | 755 [M-H]− | 609, 446, 299 | ND | ND | ND | 0.005–0.027 | ND |
| Q2 | 17.9 | 257–353 | 609 [M-H]− | 463, 446, 299 | ND | ND | ND | 0–0.017 | ND |
| Q3 | 20.2 | 256–356 | 593 [M-H]− | 446, 299 | ND | ND | ND | 0–0.018 | ND |
| K1 | 15.6 | 264–342 | 739 [M-H]− | 593, 430, 283 | ND | 0–0.005 | 0.006–0.014 | 0.046–0.31 | 0.001–0.011 |
| K2 | 20.0 | 266–347 | 593 [M-H]− | 447, 430, 283 | ND | 0–0.004 | 0.005–0.009 | 0.039–0.22 | 0–0.005 |
| K3 | 22.6 | 266–348 | 577 [M-H]− | 431, 283 | ND | 0–0.010 | 0.012–0.026 | 0.066–0.32 | 0.004–0.18 |
| C1 | 18.3 | 282–527 | 1137 [M]+ | 889, 535, 287 | ND | 0–0.027 | 0–0.39 | 0.72–5.4 | 0–1.00 |
| C2 | 21.6 | 285–536 | 1343 [M]+ | 1095, 535, 287 | ND | 0.042–0.20 | 0.36–0.68 | 0.75–1.7 | 0–0.39 |
| C3 | 23.2 | 280–531 | 889 [M]+ | 727, 449, 287 | ND | ND | 0–0.19 | 0.34–2.0 | 0–0.12 |
| C4 | 25.3 | 282–525 | 975 [M]+ | 727, 535, 287 | ND | ND | 0–0.64 | 2.3–16.2 | 0–0.32 |
| C5 | 28.2 | 318–536 | 1181 [M]+ | 933, 535, 287 | ND | ND | 0–0.56 | 0.89–1.9 | 0–0.34 |
| SM | 21.9 | 329 | 339 [M-H]− | 223 | 4.5 ± 0.2 | 3.9 ± 0.3 | 4.4 ± 0.7 | 3.7 ± 0.9 | 2.7 ± 0.7 |
For each peak separated by HPLC (Figure 3), the physico-chemical properties are given that permitted the identification of the compounds by comparison with published data (Graham, 1998; Veit and Pauli, 1999; Bloor and Abrahams, 2002; Tohge et al., 2005). Products were quantified from five to six determinations based on absorbance peak area at 345 nm for flavonols or 530 nm for anthocyanins or on fluorescence intensity for sinapoylmalate and using kaempferol, cyanidin, or sinapic acid for drawing up the calibration curves. Values are expressed as nmol·mg−1 fresh weight. Mean values and standard errors are given for sinapoylmalate. For flavonol and anthocyanin derivatives, the lowest and highest measurements (out of five to six independent samples) are given to show variability. Phenolic compounds were extracted from the different genotypes grown under standard or low lighting. Rt, retention time; ND, not detected.
CHS Expression Controls Flavonoid Synthesis and the Reduced Size Phenotype of HCT-Deficient Plants
The first committed enzyme of flavonoid biosynthesis is CHS, which, like HCT, uses p-coumaroyl CoA as substrate (Figure 1). Light is known to regulate the expression of several genes involved in flavonoid synthesis, including CHS (Feinbaum and Ausubel, 1988; Kubasek et al., 1992); therefore, we studied whether the growth phenotype of HCT-silenced plants was dependent on light irradiation. Figure 5 shows plants grown under standard light intensity (Figure 5, left-hand plants) or low light intensity (Figure 5, right-hand plants). CHS-RNAi plants (CHS−) that do not synthesize flavonoids (Table 1, CHS− plants) were used as controls. When grown under normal light conditions, the two selected HCT-silenced lines accumulated flavonoids (Figure 3, Table 1) and displayed the typical intermediate and small size phenotypes (Figures 5E and 5G) compared with CHS− and wild-type phenotypes (Figures 5A and 5C). When the same genotypes were cultivated under low light intensity, they produced low levels of flavonoids (Table 1, HCT−s plants under low light), and they all grew at the same pace (Figure 5, right-hand plants). This observation suggests that flavonoid accumulation is implicated in the inhibition of growth of HCT-deficient plants cultivated under standard light conditions.
Figure 5.
Light Controls the Plant Growth Phenotypes.
Wild-type, HCT-deficient (HCT−i and HCT−s) and CHS-deficient (CHS−) plants were cultivated under different light intensities.
(A), (C), (E), and (G) Thirty-day-old plants grown under standard light conditions (70 μmol·m2·s−1).
(B), (D), (F), and (H) Forty-five-day-old plants grown under low light conditions (20 μmol·m2·s−1).
To confirm the crucial role of flavonoids in the growth phenotype, we investigated whether HCT gene repression affected CHS gene regulation by light irradiation. Plants grown under low light intensity were submitted to light stress, and their leaves were examined for CHS transcript and flavonoid and sinapoylmalate accumulation 32 and 54 h later (Figure 6). As expected, the CHS-RNAi Arabidopsis line (CHS−) used as negative control did not accumulate either CHS transcripts or flavonoid compounds (Figures 6A and 6B). By contrast, HCT-silenced and wild-type plants displayed a strong induction of CHS expression upon light stress and a consequent increase of their flavonoid content (Figures 6A and 6B). Concerning CHS gene expression, no clear difference in the kinetics of transcript accumulation was recorded between HCT− and wild-type plants, the CHS signal appearing on RNA gel blots at 32 h and increasing later in both genotypes. However, differences were evident in the kinetics of flavonoid accumulation: in HCT-silenced tissues, flavonoid levels detected at time 0 were barely above those in the wild type, but they increased ∼55% higher than in the wild type after 32 h of light stress (Figure 6B). Later on (after 54 h of intense irradiation), flavonoids accumulated in high quantities in both genotypes, indicating a strong stimulation of the overall biosynthetic pathway. These results indicate that, in HCT-silenced plants, the availability of the entire p-coumaroyl CoA pool for CHS activity led to a higher production of flavonoids at the onset of stimulation by light. Upon light stress, sinapoylmalate was also increased in the wild type but not in HCT-deficient plants (Figure 6C), thus indicating a role of HCT in the increased synthesis of sinapoylmalate. Remarkably, its production was most elevated in CHS-RNAi plants, in agreement with the metabolic flux being available in its totality for the synthesis of phenylpropanoids.
Figure 6.
Effect of Light Stress on CHS Expression and Phenolic Content of Wild-Type, CHS−, and HCT− Plants.
Plants were submitted to light stress (190 μmol·m2 ·s−1) after 45 d of cultivation under low light conditions (20 μmol·m2·s−1). The leaves were extracted at 0-, 32-, and 54-h time points and analyzed by RNA gel blot using a CHS cDNA probe (A) or by HPLC to evaluate flavonoid (B) or sinapoylmalate (C) content. In (A), the position of CHS mRNA is indicated at the left. Flavonoid levels were estimated by adding the amounts of flavonol and anthocyanin derivatives separated by HPLC as illustrated in Figure 3. Mean values and standard errors were calculated from five to seven determinations. nd, not detected.
HCT-Deficient Plants Displayed Decreased Auxin Transport
The changes affecting growth and development of HCT-silenced plants that accumulated flavonoids are reminiscent of those provoked by the inhibition of auxin transport by chemicals such as NPA (Ruegger et al., 1997; Mattsson et al., 1999; Brown et al., 2001; Gil et al., 2001). We compared the levels of flavonoid production and the rates of auxin transport in the stems of HCT-repressed, CHS-repressed, and wild-type genotypes as well as after feeding of NPA (Figure 7). First, despite the wild-type growth rate of CHS− plants (Figure 7A), their auxin transport value was slightly higher than that of the wild type (Figure 7B). This result is similar to the behavior of the tt4 mutant, which is mutated in the CHS gene and is therefore also unable to synthesize flavonoids (Brown et al., 2001; Peer et al., 2001, 2004; Buer and Muday, 2004). By contrast, HCT-deficient plants displayed significantly lower auxin transport values compared with those of the wild-type control. Moreover, in the two HCT-deficient lines that were studied, the extent of auxin transport inhibition correlated with the level of growth inhibition. Similarly, the importance of flavonoid accumulation in HCT-silenced plants was correlated with the severity of the phenotype (Figure 7).
Figure 7.
Comparison of Flavonoid Levels and Auxin Transport Values in Wild-Type, CHS−, and HCT− Plants.
(A) Plants used for experiments.
(B) Auxin transport was evaluated by measuring the radioactivity of stem segments of the plants after feeding of tritiated indole-3-acetic acid (see Methods for details) alone or in the presence of 1 μM NPA.
Total flavonol quantities extracted from stems were calculated by adding the amounts of the different flavonol glycosides separated by HPLC as in Figure 3. Mean values and standard errors were calculated from five samples for auxin transport measurements in the absence of NPA and five to seven samples for flavonoid analysis. When auxin transport measurements were performed in the presence of NPA, similar values were obtained in two experiments.
In wild-type plants, the feeding of NPA blocked auxin transport in agreement with published observations (Ruegger et al., 1997; Murphy et al., 2000; Brown et al., 2001). In fact, as shown in Figure 7B, NPA treatment was highly efficient in all genotypes. Taken together, these observations suggested a correlation between flavonoid accumulation, auxin transport inhibition, and plant growth reduction.
HCT-Deficient Plants Displayed Altered Lignification and Collapsed Tracheary Elements in Floral Stems
Histochemical analysis of the stems of different plant genotypes was performed (Figure 8). Wiesner stain revealed lignin in interfascicular fibers and in xylem bundles of all sections. However, in the HCT−s section (Figure 8M), the coloration was noticeably less intense, indicating some changes in lignin content. When stem sections were treated with Mäule reagent to detect S lignin units, interfascicular fibers and vascular tissues of wild-type and CHS− sections stained red (Figures 8B and 8F), whereas both HCT− sections (Figures 8J and 8N) did not stain, indicating that HCT repression decreased S unit biosynthesis. When toluidine blue stain was used, the stem sections of HCT− plants with the small size phenotype (Figures 8O and 8P) displayed the most striking structural alterations compared with wild-type sections (Figures 8C and 8D). Observations at high magnification revealed that the main alterations were in xylem cells, which were small, contorted, and even collapsed (Figure 8P, inset). Pith cell size also appeared strongly reduced, and this is likely at the origin of the reduction of stem diameter in HCT− plants.
Figure 8.
Histochemical Analysis of the Effects of HCT Silencing on Stem Structure and Lignin.
Sections of stems from wild-type, CHS−, HCT−I, and HCT−s plants were stained with Wiesner reagent to detect lignin ([A], [E], [I], and [M]), Mäule reagent to detect S lignin unit ([B], [F], [J], and [N]), or toluidine blue to stain cell walls ([C], [D], [G], [H], [K], [L], [O], and [P]). Arrows show the zones that are scaled up in the insets. cb, cambium; co, cortex; epd, epidermis; if, interfascicular fiber; ph, phloem; pi, pith; xy, xylem. Bars = 100 or 300 μm as indicated.
The impact of HCT silencing on lignin structure was studied by thioacidolysis, which gives rise to H, G, and S thioethylated lignin-derived monomers from H, G, and S lignin units only involved in labile ether bonds. Therefore, the total thioacidolysis yield provides an estimate of the content in these parent lignin structures without any interference from nonlignin phenolics. In agreement with the histochemical analysis (Figures 8A, 8B, 8E, and 8F), the total yield and relative frequency of the H, G, and S thioacidolysis monomers released by 2-month-old floral stems were found to be similar in control and CHS-silenced plants. This result indicates that CHS silencing has no appreciable impact on the lignification of floral stem. By contrast, the dwarf phenotype of the HCT−s line was associated with severe changes in lignification. The relative proportion of lignin-derived H monomers exceeded 85%, whereas H monomers were present in trace amounts in wild-type and CHS− plants (Table 2). The low but nevertheless detectable amount of S lignin units in HCT− stems could be evidenced only by thioacidolysis but not by the less-sensitive Maüle histochemical staining (Figure 8N).
Table 2.
Impact of HCT Silencing on Stem Lignin Structure
| Yield in (H+G+S) Thioacidolysis Monomers (μmol g−1 Dry Sample) | Molar Frequency of Thioacidolysis Monomers (%)
|
|||
|---|---|---|---|---|
| Sample | H | G | S | |
| Wild type | 75.7 ± 8.0 | 0.7 ± 0.0 | 83.5 ± 1.6 | 15.8 ± 1.6 |
| CHS− | 64.8 ± 4.0 | 0.7 ± 0.0 | 86.2 ± 0.3 | 13.1 ± 0.3 |
| HCT−s | 1.0 ± 0.3 | 86.3 ± 1.3 | 11.7 ± 1.1 | 2.0 ± 0.2 |
| CHS−/HCT− | 14.8 ± 1.4 | 84.4 ± 0.8 | 10.6 ± 0.5 | 5.0 ± 0.3 |
The p-hydroxyphenyl (H), guaiacyl (G), and syringyl (S) monomers of lignin were measured after thioacidolysis of dry stems from 2-month-old plants. Mean value and standard error of duplicate analyses are presented.
Suppression of Flavonoid Production by CHS Silencing Restored Auxin Transport and Normal Development of HCT-Deficient Plants while the Altered Lignification Was Maintained
To univocally demonstrate the implication of the increased synthesis of flavonoids in the growth phenotype of HCT-RNAi plants, the plants were crossed with CHS-RNAi plants that were homozygous for a single locus (Wesley et al., 2001; Dunoyer et al., 2004) and were unable to synthesize flavonoids (Table 1). In parallel, a control cross was performed between HCT−s and wild-type plants (Figure 9A). As expected (since the HCT-silenced parent was hemizygous for the transgene), heterogeneous F1 populations were obtained in both crosses in which approximately one-half of the siblings were HCT-deficient as shown by immunodetection of the HCT protein in stems (Figures 9B and 9D). In the case of the control cross between HCT− and wild-type parents (Figure 9A), the progeny that did not express HCT displayed a growth phenotype similar to that of the HCT-deficient parent, thus confirming that growth inhibition was inheritable. HPLC analysis confirmed that the flavonoid content of the progeny with a reduced size phenotype was high and mirrored that of the HCT-deficient parent (data not shown). In the case of the cross between CHS− and HCT− parents (Figure 9C), all the progeny was silenced for the CHS gene since the CHS− parent was homozygous for the transgene and, consequently, did not accumulate flavonoids (Figure 10A). Although, as mentioned above, half of the progeny was silenced for HCT (Figure 9D), all the progeny grew as wild-type plants (Figure 9C). This means that plants that were simultaneously repressed for HCT and CHS gene expression grew at the same rate as the wild-type controls. Auxin transport rates and flavonol quantities were measured in the double-repressed plants (Figure 10A, CHS−/HCT−) and confirmed that the inhibition of flavonoid production resulting from CHS gene silencing restored a high level of auxin transport and a normal growth rate even though the HCT gene is silenced. Lignification was dramatically altered in CHS−/HCT− progeny as evidenced by the lack of Mäule staining observed in CHS−/HCT− sections as for HCT− parent sections (Figure 10B). Thioacidolysis analysis confirmed the profound changes in lignin structure of both the HCT− parent and progeny (Table 2). Similar to the HCT-silenced parent, lignins in the stems of the double-repressed plants mainly comprised H units. In addition, the yield of the thioacidolysis monomers recovered from the stems of the double-repressed plants was also severely reduced (20% of the control level, Table 2), albeit to a lower extent than in the HCT−s parent (∼1% of the control level, Table 2). The increase of lignin yield in HCT−/CHS− stems compared with HCT− tissues could originate from the absence of flavonoids, such as tannins in the cell walls. Taken together, these results demonstrate that flavonoid accumulation in HCT− plants was responsible for the inhibition of auxin transport and for the reduction of plant growth.
Figure 9.
Analysis of the Crosses between HCT− Plants and Wild-Type or CHS− Plants.
(A) Progeny and parent phenotypes of HCT− × wild type cross; half of the progeny displayed the same growth phenotype as the HCT− parent.
(B) HCT expression in the progeny of HCT− × wild type cross was analyzed by immunoblotting stem extracts with anti-HCT antibodies; progeny of small size did not express HCT, whereas progeny of the wild-type size did. Four representative samples are shown in each case.
(C) Progeny and parent phenotypes of the HCT− × CHS− cross; all the progeny was of the wild-type size.
(D) HCT expression analysis in the progeny of HCT− × CHS− cross by immunoblotting of stem extracts; four extracts (out of seven) that displayed no signal are shown together with four typical positive signals (out of six).
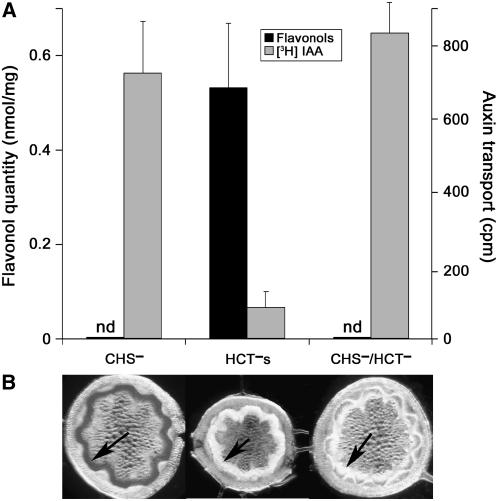
Figure 10.
Suppression of Flavonoid Accumulation in CHS−/HCT− Plants Restored Normal Auxin Transport and Growth.
(A) Auxin transport and flavonol content in stems of CHS−/HCT− progeny were compared with those of CHS− and HCT− parents. Mean values and standard errors were calculated from five to seven determinations. nd, not detected.
(B) The lack of Mäule staining observed in stem sections of CHS−/HCT− progeny and the HCT− parent points to a low content in S lignin unit. Arrows indicate tissues that stained red only in the CHS− section.
DISCUSSION
We have demonstrated that HCT silencing not only resulted in the alteration of the synthesis of lignin but also in the accumulation of various flavonoids, including flavonols and anthocyanins. In particular, the amounts of flavonol derivatives, namely kaempferol and quercetin glycosides, strongly increased in the leaves of HCT-repressed plants. These conjugated forms are synthesized by specific glycosyltransferases from their aglycone precursors (Jones et al., 2003).
Phenolics have long been suspected to interfere with auxin transport (Marigo and Boudet, 1977; Jacobs and Rubery, 1988). When tested in vitro for their capacity to displace NPA from plasma membranes, quercetin and kaempferol proved among the most active phenolic compounds and were shown to perturb auxin transport in hypocotyls of various plants in a manner closely paralleling the inhibitory activity of NPA itself (Jacobs and Rubery, 1988; Lomax et al., 1995; Murphy et al., 2000). These measurements were technically difficult to carry out because exogenously applied flavonols stick to cellulose and do not move away from their application sites. In HCT-silenced plants, that difficulty is not encountered since flavonols are produced in high amounts in situ.
Flavonoid biosynthesis is highly regulated by developmental and environmental factors (Feinbaum and Ausubel, 1988; Kubasek et al., 1992; Winkel-Shirley, 2002), and flavonoids are localized to the tissues that transport auxin (Murphy et al., 2000; Peer et al., 2001, 2004; Buer and Muday, 2004), consistent with a role as endogenous regulators of auxin transport. In transgenic plants inhibited in the expression of one lignin biosynthetic gene (by gene knockout or RNAi technology), a wide range of abnormal growth phenotypes was observed, but no clear relationship could be established between the extent of lignin decrease or modification and the growth phenotypes (Boerjan et al., 2003). Here, HCT repression in Arabidopsis resulted in profound changes in lignification as evidenced by the lack of Mäule staining, the reduced Wiesner staining, and the data from thioacidolysis analysis of HCT-deficient tissues. As shown in Table 2, thioacidolysis of wild-type and CHS-deficient stems essentially released the G and S monomers (almost 85 and 15% of lignin-derived monomers, respectively), whereas the H monomer was recovered as a trace component. By contrast, the latter H monomer constituted 85% of the lignin-derived monomers released from small size phenotype plants that displayed in parallel a strong decrease in the proportions in G and S units. These findings are in agreement with the function of HCT in the synthesis of both G and S lignin units and with the role of p-coumaroyl CoA as the precursor of H units (Figure 1). Interestingly, these alterations in lignin structure are much more important than those observed in HCT-silenced N. benthamiana plants submitted to virus-induced gene silencing at later stages of development (Hoffmann et al., 2004). In agreement with the results obtained with a C3H mutant (Abdulrazzak et al., 2005), G and S lignin units could be unequivocally evidenced in HCT-silenced stems, from the specific G and S thioacidolysis monomers. The remanence of some G and S lignin units may be due to the low level of HCT activity measured in HCT-silenced stems (Figure 2). It is interesting to note that at the initial stage of this study, our attempts to isolate knockout mutants from available collections were unsuccessful. This might indicate that such mutants are not viable, but we did not study this issue further.
It is noteworthy that, in addition to HCT itself, genes directly downstream of HCT, namely C3H, CCR, and CCoAOMT, are those whose repression has been shown to have the strongest impact on plant growth (Piquemal et al., 1998; Jones et al., 2001; Pinçon et al., 2001a, 2001b; Franke et al., 2002; Goujon et al., 2003; Abdulrazzak et al., 2005; Patten et al., 2005). HCT repression had a particularly strong impact on growth, likely due to its double role in lignin biosynthesis upstream and downstream of the C3H step (see Figure 1). Like HCT, CCR has CoA ester as substrate, and its inhibition could substantially increase the p-coumaroyl CoA pool and flavonoid production through CHS activity as demonstrated here for HCT-silenced plants. In view of the results presented here, it would be of high interest to evaluate the function of flavonoids in the inhibition of growth observed for various plant genotypes affected in lignification.
The comparison of phenolic profiles of wild-type and HCT-deficient plants provides new clues concerning the synthesis of sinapoylmalate, the major phenylpropanoid of Arabidopsis leaves. First of all, sinapoylmalate levels did not change significantly in HCT-deficient plants that only synthesized trace amounts of S lignin unit. Similar observations have been made recently in the study of C3H-deficient plants (Abdulrazzak et al., 2005). These data suggest that sinapoylmalate and S lignin unit are produced through distinct biosynthetic routes, a hypothesis strengthened by the unusual accumulation of sinapoylated cyanidin derivatives in the leaves of HCT-deficient plants (Table 1, C2 and C5). However, HCT appears to be involved in the increased accumulation of sinapoylmalate under conditions of light stress (Figure 5C), thus indicating that distinct biosynthetic pathways may operate under particular conditions.
Flavonoid-deficient plants with mutations in genes encoding various flavonoid biosynthetic enzymes have provided useful tools for studying the roles of flavonoids in auxin transport, normal growth and development, and responses to gravity and light (Shirley et al., 1995; Brown et al., 2001; Buer and Muday, 2004; Peer et al., 2004). The tt4 mutant affected in the CHS gene has elevated auxin transport in Arabidopsis inflorescences as is the case for the CHS-RNAi line used here and consistent with the absence of negative auxin transport regulators. tt3 and tt7 mutants, which accumulate kaempferol and quercetin derivatives, display reduced auxin transport (Peer et al., 2001, 2004) in accordance with in vitro experiments that have shown the ability of flavonols to decrease polar auxin transport (Jacobs and Rubery, 1988; Murphy et al., 2000). In HCT-deficient plants, the phenylpropanoid metabolic flux is rerouted into the flavonoid biosynthetic pathway, resulting in a higher accumulation of a whole set of flavonoids. The deregulation of flavonoid synthesis results in the inhibition of auxin transport and has severe consequences for plant growth and development. When plants are grown under low lighting, CHS is not expressed, flavonoids do not accumulate, and the detrimental effects on plant development are undetectable (Figure 5). It is noteworthy that auxin biosynthesis is also controlled by light and is increased in plants exposed to intense light (Buer and Muday, 2004). Thus, light simultaneously induces auxin and flavonoid synthesis, with the fine-tuning of auxin transport and concentration ultimately controlling plant development (Friml, 2003). Recently, quercetin has been shown to inhibit auxin efflux activity of MDR/P-glycoproteins (Geisler et al., 2005; Bouchard et al., 2006). The study of the localization and activity of auxin efflux carriers in HCT-deficient plants accumulating flavonoids should shed new light on the molecular mechanisms underlying the inhibition of auxin transport by flavonoids. However, since the targets of flavonoids in HCT-silenced plants have not been uncovered, indirect effects leading to the inhibition of auxin transport and growth reduction cannot be completely excluded.
Apical-basal flow of auxin has long been suspected to be involved in vascular tissue differentiation (Berleth et al., 2000). Consistent with this assumption, a pin1 mutant that is affected in polar auxin transport showed abnormal xylem proliferation in the inflorescence axis (Galweiler et al., 1998). Moreover, polar auxin transport inhibition provoked by NPA treatment of wild-type plants caused similar alterations in vascular pattern formation (Galweiler et al., 1998; Mattsson et al., 1999). It is noteworthy that PIN1 protein, an auxin efflux carrier, has been shown to localize to cambial and young xylem parenchyma cells of stems (Galweiler et al., 1998), which express lignin synthesis genes and are actively lignifying (Hoffmann et al., 2004). The PINOID protein kinase that regulates PIN localization is also expressed in young vascular tissues (Benjamins et al., 2001), and Pid mutants have been shown to be affected in flower venation (Christensen et al., 2000). Thus, several lines of evidence suggest that auxin transport and phenylpropanoid synthesis are tightly linked and colocalized in vascular tissues. This implies that the rerouting of metabolic flux due to HCT repression is likely to produce flavonols at the right place to rapidly and efficiently inhibit the auxin flow. Moreover, as tracheary elements are distorted in the HCT-silenced lines (Figure 8), the alterations in vascular tissues might contribute to the inhibition of auxin transport.
The inhibition of flavonoid production by repression of CHS in an HCT-deficient background restored normal auxin transport rates and wild-type growth. This result demonstrates that the profound changes in lignin structure that result from HCT repression (Table 2, CHS−/HCT− plants) are not responsible for the reduction of plant growth. This observation opens the way to new strategies for avoiding detrimental effects on the development of plants engineered to produce modified lignin and better pulp.
In plants, flavonoids have been shown to localize to different cell compartments, indicating multiple physiological roles. Subcellular sites of flavonoid accumulation include the nucleus, where some of the biosynthetic enzymes are also found (Saslowsky et al., 2005). These latter findings are consistent with the idea that some flavonoids could exert direct control on the transcription of genes involved in plant growth and development. In animals, flavonoids have many interesting pharmacological activities as antioxidant and anticancer agents, being implicated, for instance, in inhibition of cell growth or apoptosis induction (for a review, see Le Marchand, 2002). Interestingly, MDR/P-glycoproteins are targets of flavonoids in animal cells (Conseil et al., 1998; Chung et al., 2005; Zhang et al., 2005), as is probably also the case for plant MDR/P-glycoproteins involved in polar auxin transport (Noh et al., 2001, 2003; Murphy et al., 2002; Blakeslee et al., 2005). Indeed, auxin efflux activities of Arabidopsis P-glycoproteins PGP1 and PGP19 have been shown to be inhibited by quercetin (Geisler et al., 2005; Bouchard et al., 2006). Among other known targets of flavonoids in animal cells are numerous ATP binding proteins, including several distinct ATPases and protein kinases (Conseil et al., 1998; Vijayababu et al., 2005; Zhang et al., 2005). There is no doubt that knowledge of whether flavonoids regulate such activities in planta would significantly increase our understanding of the mechanisms underlying flavonoid functions in plant development.
METHODS
Plant Material and Culture Conditions
Arabidopsis thaliana Columbia-0 ecotype was used. HCT RNAi and CHS RNAi lines were obtained as previously described (Wesley et al., 2001; Dunoyer et al., 2004; Hoffmann et al., 2004). For germination, seeds were surface-sterilized and placed on Murashige and Skoog medium (Duchefa) supplemented with 10 g/L sucrose and 10 mg/L Basta herbicide (glufosinate) if required. After the seeds had undergone a cold treatment for homogenous germination (2 d at 4°C in the dark), they were exposed to 20°C and 70 μmol·m−2·s−1 light intensity under a light/dark cycle of 16 h/8 h. After 12 d, plants were transferred to a growth chamber under 70 μmol·m−2·s−1 light intensity and a light/dark cycle of 12 h/12 h. The temperature was maintained at 20°C. Inflorescence stems were induced 6 weeks after germination under a light/dark cycle of 16 h/8 h. For experimentation, various light intensities were applied, corresponding to 20 μmol·m−2·s−1 in the case of low lighting and to 190 μmol·m−2·s−1 to induce light stress.
Production and Use of Polyclonal Antibodies
The Arabidopsis HCT gene (At5g48930) was cloned in the pGEX-KG plasmid, and heterologous expression in Escherichia coli cells and purification of the HCT protein were performed as described previously (Hoffmann et al., 2003). Polyclonal antibodies were raised in rabbit by four successive injections of 100 μg of the purified enzyme. Protein was emulsified with Freund's complete adjuvant for the first injection and with incomplete adjuvant every 2 weeks for the three following injections. The first injection was intramuscular, whereas the next three were subcutaneous. Anti-HCT antibodies were used at 1/10,000 dilution in protein blot experiments after overnight preincubation with an acetonic powder of E. coli bacteria to eliminate any aspecific signal.
Protein Gel Blot Analysis
The basic procedures for the electrophoresis of proteins under denaturing conditions and immunoblotting were as described previously (Geoffroy et al., 1990), except that phosphatase activity was detected with a chemiluminescent substrate (CDP-Star; Bio-Rad ).
RNA Gel Blot Analysis
Total RNA extraction was performed with Tri-Reagent (Sigma-Aldrich) according to the manufacturer's instructions. RNA samples (5 μg) prepared in 1× HEPES/50% formamide/15% formaldehyde was denatured and separated by electrophoresis in a 1% agarose/20% formaldehyde/1× HEPES gel. The gel was transferred by capillary elution to a Hybond N+ membrane (Amersham Biosciences). The [α-32P]dCTP-labeled probe was synthesized from a 256-bp fragment of Arabidopsis CHS gene (Dunoyer et al., 2004) by random priming (Promega). RNA gel blot prehybridization and hybridization were performed with Perfect-Hyb buffer (Sigma-Aldrich) at 65°C.
HCT Activity Assays
HCT activity measurements were performed on plant extracts as previously described (Hoffmann et al., 2004) using 5 μg of protein and an incubation time of 15 min at 30°C. At the end of incubation, the enzymatic reaction was stopped by the addition of 1 volume of HPLC solvent A containing 2 n HCl, and reaction products were analyzed and quantified by HPLC.
Histochemical Analysis
Histochemical analysis was performed on 2-month-old stems. Mäule staining was performed as previously described (Atanassova et al., 1995) on hand-cut sections. The observations were performed with a Leica MZ12 stereomicroscope. For toluidine blue staining, stem segments were fixed with a solution containing 3.7% formaldehyde, 50% ethanol, and 5% acetic acid in water, dehydrated in an ethanol series, and gradually embedded in paraplast. Transversal sections (10-μm thick) were made with the Leica RM2155 microtome. Stem sections were hydrated and stained with 0.4% toluidine blue for 5 min, rinsed, dehydrated in ethanol series, and mounted on slides with Eukitt. The observations were made with a Nikon E800 microscope.
Lignin Analysis
Lignin content analysis was performed on air-dried stems of 2-month-old plants by thioacidolysis as described (Lapierre, 1986; Hoffmann et al., 2004). Quantification of the main p-hydroxyphenyl (H), guaiacyl (G), and syringyl (S) lignin-derived monomers, analyzed as their trimethylsilylated derivatives, was performed from specific ion chromatograms reconstructed at m/z of 239 for the H monomers, m/z 269 for G monomers, and m/z 299 for S monomers after an appropriate calibration relative to the docosane internal standard.
Extraction of Soluble Phenolic Compounds
Samples (100 mg) of leaves or stems were frozen in liquid nitrogen and quickly ground in 500 μL methanol. After centrifugation at 500 rpm, the supernatant was collected and the residual pellet was reextracted with 200 μL of 70% methanol. The supernatants were combined, clarified at 2000 rpm for 30 min, and analyzed by HPLC.
HPLC Analysis of Plant Phenolic Compounds and Liquid Chromatography–Mass Spectrometry Characterization
Phenolic compounds were resolved on a Novapak RP C18 column (4 μm, 4.6 × 250 mm; Waters) using an increasing gradient of acetonitrile in water containing 0.1% formic acid. Gradient conditions at 1 mL/min flow rate were as follows: 100% solvent A to 50% solvent B for 50 min, 50% solvent B to 100% solvent B for 5 min, 100% solvent B to 100% solvent A for 5 min, and then 10 min reequilibration in 100% solvent A. Solvent A contained acetonitrile:water:formic acid (10:89.9:0.1) and solvent B acetonitrile:water:formic acid (80:19.9:0.1). Compounds were first characterized by their elution time and their UV absorption spectrum recorded with a photodiode array detector (Waters). The quantitative evaluation was performed on 345-nm absorbance profiles for flavonol glycosides and on 530-nm profiles for anthocyanin derivatives. Sinapoylmalate was quantified by its fluorescence. Kaempferol, cyanidin, and sinapic acid were used as standards for quantification of flavonol and anthocyanin derivatives and sinapoylmalate, respectively.
For liquid chromatography–mass spectrometry, an HPLC-PDA/electrospray ionization-MS system was used comprising a Finnigan LCQ-DECA mass spectrometer (Thermo Electron). The mass spectral data were collected in negative or positive mode with the ion trap mass spectrometer equipped with a heated capillary electrospray interface. The sprayer needle voltage was set to 4 kV, and the capillary was heated to 350°C. Product peaks were identified by their mass spectrometric fragmentation pattern and relative to data reported in the literature (Veit and Pauli, 1999; Bloor and Abrahams, 2002; Jones et al., 2003; Tohge et al., 2005). Cyanidin derivatives could be identified by electrospray mass spectrometry in the positive mode, whereas sinapoyl malate could be identified only in the negative mode. The flavonol glycosides were preferably analyzed in the negative mode due to a higher detection sensitivity.
Auxin Transport Assays
Auxin transport was measured using a modification of previously published procedures (Okada et al., 1991; Brown et al., 2001). A 2.5-cm stem segment of primary inflorescence was used that spanned a zone starting from 5 cm above the base of the stem for wild-type, CHS−, and HCT−i plants and from 2.5 cm above the base for HCT−s plants. Segments were placed in a 1.5-mL microcentrifuge tube with the apical end submerged in 30 μL of MES buffer (5 mM MES and 1% sucrose, pH 5.5) containing 1 μM total indole-3-acetic acid with 66 nM of tritiated indole-3-acetic acid, in the presence or absence of 1 μM NPA. After 4 h of incubation in the dark, the segment was removed and the last 5 mm of the nonsubmerged end was excised and placed in 2.5 mL of liquid scintillation cocktail (MP solution; Beckman). The samples were slowly shaken overnight before counting radioactivity in a scintillation counter (Beckman LS6500).
Accession Numbers
Arabidopsis Genome Initiative codes for the sequence data from this article are At5g48930 (Arabidopsis HCT) and At5g13930 (Arabidopsis CHS).
Acknowledgments
We thank Patrice Dunoyer and Olivier Voinnet for CHS RNAi seeds and stimulating discussions. We also thank Kenneth Richards for critical reading of the manuscript and Danielle Werck-Reichhart for fruitful discussions. The assistance of Denise Meyer for histochemical analysis and Laurent Cézard for thioacidolysis is gratefully acknowledged. This work was supported by doctoral fellowships of the Ministère de l'Education Nationale, de l'Enseignement Supérieur et de la Recherche to S.B. and L.H.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Michel Legrand (michel.legrand@ibmp-ulp.u-strasbg.fr).
References
- Abdulrazzak, N., et al. (2005). A coumaroyl-ester-3-hydroxylase insertion mutant reveals the existence of nonredundant meta-hydroxylation pathways and essential roles for phenolic precursors in cell expansion and plant growth. Plant Physiol. 140 30–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atanassova, R., Favet, N., Martz, F., Chabbert, B., Thollier, M.T., Monties, B., Fritig, B., and Legrand, M. (1995). Altered lignin composition in transgenic tobacco expressing O-methyltransferase sequences in sense and antisense orientation. Plant J. 8 465–477. [Google Scholar]
- Benjamins, R., Quint, A., Weijers, D., Hooykaas, P., and Offringa, R. (2001). The PINOID protein kinase regulates organ development in Arabidopsis by enhancing polar auxin transport. Development 128 4057–4067. [DOI] [PubMed] [Google Scholar]
- Berleth, T., Mattsson, J., and Hardtke, C.S. (2000). Vascular continuity and auxin signals. Trends Plant Sci. 5 387–393. [DOI] [PubMed] [Google Scholar]
- Blakeslee, J.J., Peer, W.A., and Murphy, A.S. (2005). Auxin transport. Curr. Opin. Plant Biol. 8 494–500. [DOI] [PubMed] [Google Scholar]
- Bloor, S.J., and Abrahams, S. (2002). The structure of the major anthocyanin in Arabidopsis thaliana. Phytochemistry 59 343–346. [DOI] [PubMed] [Google Scholar]
- Boerjan, W., Ralph, J., and Baucher, M. (2003). Lignin biosynthesis. Annu. Rev. Plant Biol. 54 519–546. [DOI] [PubMed] [Google Scholar]
- Bouchard, R., Bailly, A., Blakeslee, J.J., Oehring, S.C., Vincenzetti, V., Lee, O.R., Paponov, I., Palme, K., Mancuso, S., Murphy, A.S., Schulz, B., and Geisler, M. (2006). Immunophilin-like TWISTED DWARF1 modulates auxin efflux activities of Arabidopsis P-glycoproteins. J. Biol. Chem. 281 30603–30612. [DOI] [PubMed] [Google Scholar]
- Brown, D.E., Rashotte, A.M., Murphy, A.S., Normanly, J., Tague, B.W., Peer, W.A., Taiz, L., and Muday, G.K. (2001). Flavonoids act as negative regulators of auxin transport in vivo in Arabidopsis. Plant Physiol. 126 524–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buer, C.S., and Muday, G.K. (2004). The transparent testa4 mutation prevents flavonoid synthesis and alters auxin transport and the response of Arabidopsis roots to gravity and light. Plant Cell 16 1191–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen, S.K., Dagenais, N., Chory, J., and Weigel, D. (2000). Regulation of auxin response by the protein kinase PINOID. Cell 100 469–478. [DOI] [PubMed] [Google Scholar]
- Chung, S.Y., Sung, M.K., Kim, N.H., Jang, J.O., Go, E.J., and Lee, H.J. (2005). Inhibition of P-glycoprotein by natural products in human breast cancer cells. Arch. Pharm. Res. 28 823–828. [DOI] [PubMed] [Google Scholar]
- Conseil, G., Baubichon-Cortay, H., Dayan, G., Jault, J.M., Barron, D., and Di Pietro, A. (1998). Flavonoids: A class of modulators with bifunctional interactions at vicinal ATP- and steroid-binding sites on mouse P-glycoprotein. Proc. Natl. Acad. Sci. USA 95 9831–9836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Auria, J.C. (2006). Acyltransferases in plants: A good time to be BAHD. Curr. Opin. Plant Biol. 9 331–340. [DOI] [PubMed] [Google Scholar]
- Delbarre, A., Muller, P., and Guern, J. (1998). Short-lived and phosphorylated proteins contribute to carrier-mediated efflux, but not to influx, of auxin in suspension-cultured tobacco cells. Plant Physiol. 116 833–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunoyer, P., Lecellier, C.H., Parizotto, E.A., Himber, C., and Voinnet, O. (2004). Probing the microRNA and small interfering RNA pathways with virus-encoded suppressors of RNA silencing. Plant Cell 16 1235–1250. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Feinbaum, R.L., and Ausubel, F.M. (1988). Transcriptional regulation of the Arabidopsis thaliana chalcone synthase gene. Mol. Cell. Biol. 8 1985–1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke, R., Humphreys, J.M., Hemm, M.R., Denault, J.W., Ruegger, M.O., Cusumano, J.C., and Chapple, C. (2002). The Arabidopsis REF8 gene encodes the 3-hydroxylase of phenylpropanoid metabolism. Plant J. 30 33–45. [DOI] [PubMed] [Google Scholar]
- Friml, J. (2003). Auxin transport - Shaping the plant. Curr. Opin. Plant Biol. 6 7–12. [DOI] [PubMed] [Google Scholar]
- Friml, J., et al. (2004). A PINOID-dependent binary switch in apical-basal PIN polar targeting directs auxin efflux. Science 306 862–865. [DOI] [PubMed] [Google Scholar]
- Galweiler, L., Guan, C., Muller, A., Wisman, E., Mendgen, K., Yephremov, A., and Palme, K. (1998). Regulation of polar auxin transport by AtPIN1 in Arabidopsis vascular tissue. Science 282 2226–2230. [DOI] [PubMed] [Google Scholar]
- Geisler, M., et al. (2005). Cellular efflux of auxin catalyzed by the Arabidopsis MDR/PGP transporter AtPGP1. Plant J. 44 179–194. [DOI] [PubMed] [Google Scholar]
- Geisler, M., et al. (2003). TWISTED DWARF1, a unique plasma membrane-anchored immunophilin-like protein, interacts with Arabidopsis multidrug resistance-like transporters AtPGP1 and AtPGP19. Mol. Biol. Cell 14 4238–4249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geldner, N., Anders, N., Wolters, H., Keicher, J., Kornberger, W., Muller, P., Delbarre, A., Ueda, T., Nakano, A., and Jurgens, G. (2003). The Arabidopsis GNOM ARF-GEF mediates endosomal recycling, auxin transport, and auxin-dependent plant growth. Cell 112 219–230. [DOI] [PubMed] [Google Scholar]
- Geldner, N., Friml, J., Stierhof, Y.D., Jurgens, G., and Palme, K. (2001). Auxin transport inhibitors block PIN1 cycling and vesicle trafficking. Nature 413 425–428. [DOI] [PubMed] [Google Scholar]
- Geoffroy, P., Legrand, M., and Fritig, B. (1990). Isolation and characterization of a proteinaceous inhibitor of microbial proteinases induced during the hypersensitive reaction of tobacco to tobacco mosaic virus. Mol. Plant Microbe Interact. 3 327–333. [DOI] [PubMed] [Google Scholar]
- Gil, P., Dewey, E., Friml, J., Zhao, Y., Snowden, K.C., Putterill, J., Palme, K., Estelle, M., and Chory, J. (2001). BIG: A calossin-like protein required for polar auxin transport in Arabidopsis. Genes Dev. 15 1985–1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goujon, T., Ferret, V., Mila, I., Pollet, B., Ruel, K., Burlat, V., Joseleau, J.P., Barriere, Y., Lapierre, C., and Jouanin, L. (2003). Down-regulation of the AtCCR1 gene in Arabidopsis thaliana: Effects on phenotype, lignins and cell wall degradability. Planta 217 218–228. [DOI] [PubMed] [Google Scholar]
- Graham, T.L. (1998). Flavonoid and flavonol glycoside metabolism in Arabidopsis. Plant Physiol. Biochem. 36 135–144. [Google Scholar]
- Hoffmann, L., Besseau, S., Geoffroy, P., Ritzenthaler, C., Meyer, D., Lapierre, C., Pollet, B., and Legrand, M. (2004). Silencing of hydroxycinnamoyl-coenzyme A shikimate/quinate hydroxycinnamoyltransferase affects phenylpropanoid biosynthesis. Plant Cell 16 1446–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann, L., Maury, S., Martz, F., Geoffroy, P., and Legrand, M. (2003). Purification, cloning and properties of an acyltransferase controlling shikimate and quinate ester intermediates in phenylpropanoid metabolism. J. Biol. Chem. 278 95–103. [DOI] [PubMed] [Google Scholar]
- Jacobs, M., and Rubery, P.H. (1988). Naturally occurring auxin transport regulators. Science 241 346–349. [DOI] [PubMed] [Google Scholar]
- Jones, L., Ennos, A.R., and Turner, S.R. (2001). Cloning and characterization of irregular xylem4 (irx4): A severely lignin-deficient mutant of Arabidopsis. Plant J. 26 205–216. [DOI] [PubMed] [Google Scholar]
- Jones, P., Messner, B., Nakajima, J., Schaffner, A.R., and Saito, K. (2003). UGT73C6 and UGT78D1, glycosyltransferases involved in flavonol glycoside biosynthesis in Arabidopsis thaliana. J. Biol. Chem. 278 43910–43918. [DOI] [PubMed] [Google Scholar]
- Kubasek, W.L., Shirley, B.W., McKillop, A., Goodman, H.M., Briggs, W., and Ausubel, F.M. (1992). Regulation of flavonoid biosynthetic genes in germinating Arabidopsis seedlings. Plant Cell 4 1229–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapierre, C. (1986). Thioacidolysis of poplar lignins: Identification of monomeric syringyl products and characterization of guaiacyl-syringyl rich fractions. Holzforschung 40 113–118. [Google Scholar]
- Le Marchand, L. (2002). Cancer preventive effects of flavonoids - A review. Biomed. Pharmacother. 56 296–301. [DOI] [PubMed] [Google Scholar]
- Lomax, T.L., Muday, G.K., and Rubery, P.H. (1995). Auxin transport. In Plant Hormones: Physiology, Biochemistry and Molecular Biology, P.J. Davies, ed (Dordrech, The Netherlands: Kluwer Academic Publishers), pp. 509–530.
- Marigo, G., and Boudet, A.M. (1977). Relations polyphénols-croissance: Mise en évidence d‘un effet inhibiteur des composés phénoliques sur le transport polarisé de l’auxine. Physiol. Plant. 41 197–202. [Google Scholar]
- Mattsson, J., Sung, Z.R., and Berleth, T. (1999). Responses of plant vascular systems to auxin transport inhibition. Development 126 2979–2991. [DOI] [PubMed] [Google Scholar]
- Muday, G.K., and DeLong, A. (2001). Polar auxin transport: Controlling where and how much. Trends Plant Sci. 6 535–542. [DOI] [PubMed] [Google Scholar]
- Muday, G.K., and Murphy, A.S. (2002). An emerging model of auxin transport regulation. Plant Cell 14 293–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy, A., Peer, W.A., and Taiz, L. (2000). Regulation of auxin transport by aminopeptidases and endogenous flavonoids. Planta 211 315–324. [DOI] [PubMed] [Google Scholar]
- Murphy, A.S., Hoogner, K.R., Peer, W.A., and Taiz, L. (2002). Identification, purification, and molecular cloning of N-1-naphthylphthalmic acid-binding plasma membrane-associated aminopeptidases from Arabidopsis. Plant Physiol. 128 935–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noh, B., Bandyopadhyay, A., Peer, W.A., Spalding, E.P., and Murphy, A.S. (2003). Enhanced gravi- and phototropism in plant mdr mutants mislocalizing the auxin efflux protein PIN1. Nature 423 999–1002. [DOI] [PubMed] [Google Scholar]
- Noh, B., Murphy, A.S., and Spalding, E.P. (2001). Multidrug resistance-like genes of Arabidopsis required for auxin transport and auxin-mediated development. Plant Cell 13 2441–2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada, K., Ueda, J., Komaki, M.K., Bell, C.J., and Shimura, Y. (1991). Requirement of the auxin polar transport system in early stages of Arabidopsis floral bud formation. Plant Cell 3 677–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patten, A.M., Cardenas, C.L., Cochrane, F.C., Laskar, D.D., Bedgar, D.L., Davin, L.B., and Lewis, N.G. (2005). Reassessment of effects on lignification and vascular development in the irx4 Arabidopsis mutant. Phytochemistry 66 2092–2107. [DOI] [PubMed] [Google Scholar]
- Peer, W.A., Bandyopadhyay, A., Blakeslee, J.J., Makam, S.N., Chen, R.J., Masson, P.H., and Murphy, A.S. (2004). Variation in expression and protein localization of the PIN family of auxin efflux facilitator proteins in flavonoid mutants with altered auxin transport in Arabidopsis thaliana. Plant Cell 16 1898–1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peer, W.A., Brown, D.E., Tague, B.W., Muday, G.K., Taiz, L., and Murphy, A.S. (2001). Flavonoid accumulation patterns of transparent testa mutants of Arabidopsis. Plant Physiol. 126 536–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen, M., Strack, D., and Matern, U. (1999). Biosynthesis of Phenylpropanoids and Related Compounds. (Sheffield, UK: Sheffield Academic Press).
- Petrasek, J., et al. (2006). PIN proteins perform a rate-limiting function in cellular auxin efflux. Science 312 914–918. [DOI] [PubMed] [Google Scholar]
- Pinçon, G., Chabannes, M., Lapierre, C., Pollet, B., Ruel, K., Joseleau, J.P., Boudet, A.M., and Legrand, M. (2001. b). Simultaneous down-regulation of caffeic/5-hydroxy ferulic acid-O-methyltransferase I and cinnamoyl-coenzyme A reductase in the progeny from a cross between tobacco lines homozygous for each transgene. Consequences for plant development and lignin synthesis. Plant Physiol. 126 145–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinçon, G., Maury, S., Hoffmann, L., Geoffroy, P., Lapierre, C., Pollet, B., and Legrand, M. (2001. a). Repression of O-methyltransferase genes in transgenic tobacco affects lignin synthesis and plant growth. Phytochemistry 57 1167–1176. [DOI] [PubMed] [Google Scholar]
- Piquemal, J., Lapierre, C., Myton, K., O'Connell, A., Schuch, W., Grima-Pettenati, J., and Boudet, A.M. (1998). Down-regulation of cinnamoyl-CoA reductase induces significant changes of lignin profiles in transgenic tobacco plants. Plant J. 13 71–83. [Google Scholar]
- Rashotte, A.M., DeLong, A., and Muday, G.K. (2001). Genetic and chemical reductions in protein phosphatase activity alter auxin transport, gravity response, and lateral root growth. Plant Cell 13 1683–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruegger, M., Dewey, E., Hobbie, L., Brown, D., Bernasconi, P., Turner, J., Muday, G., and Estelle, M. (1997). Reduced naphthylphthalamic acid binding in the tir3 mutant of Arabidopsis is associated with a reduction in polar auxin transport and diverse morphological defects. Plant Cell 9 745–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saslowsky, D.E., Warek, U., and Winkel, B.S. (2005). Nuclear localization of flavonoid enzymes in Arabidopsis. J. Biol. Chem. 280 23735–23740. [DOI] [PubMed] [Google Scholar]
- Schoch, G., Goepfert, S., Morant, M., Hehn, A., Meyer, D., Ullmann, P., and Werck-Reichhart, D. (2001). CYP98A3 from Arabidopsis thaliana is a 3′-hydroxylase of phenolic esters, a missing link in the phenylpropanoid pathway. J. Biol. Chem. 276 36566–36574. [DOI] [PubMed] [Google Scholar]
- Shirley, B.W., Kubasek, W.L., Storz, G., Bruggemann, E., Koornneef, M., Ausubel, F.M., and Goodman, H.M. (1995). Analysis of Arabidopsis mutants deficient in flavonoid biosynthesis. Plant J. 8 659–671. [DOI] [PubMed] [Google Scholar]
- Taylor, L.P., and Grotewold, E. (2005). Flavonoids as developmental regulators. Curr. Opin. Plant Biol. 8 317–323. [DOI] [PubMed] [Google Scholar]
- Tohge, T., et al. (2005). Functional genomics by integrated analysis of metabolome and transcriptome of Arabidopsis plants over-expressing an MYB transcription factor. Plant J. 42 218–235. [DOI] [PubMed] [Google Scholar]
- Veit, M., and Pauli, G.F. (1999). Major flavonoids from Arabidopsis thaliana leaves. J. Nat. Prod. 62 1301–1303. [DOI] [PubMed] [Google Scholar]
- Vijayababu, M.R., Kanagaraj, P., Arunkumar, A., Ilangovan, R., Aruldhas, M.M., and Arunakaran, J. (2005). Quercetin-induced growth inhibition and cell death in prostatic carcinoma cells (PC-3) are associated with increase in p21 and hypophosphorylated retinoblastoma proteins expression. J. Cancer Res. Clin. Oncol. 131 765–771. [DOI] [PubMed] [Google Scholar]
- Wesley, S.V., et al. (2001). Construct design for efficient, effective and high-throughput gene silencing in plants. Plant J. 27 581–590. [DOI] [PubMed] [Google Scholar]
- Winkel-Shirley, B. (2002). Biosynthesis of flavonoids and effects of stress. Curr. Opin. Plant Biol. 5 218–223. [DOI] [PubMed] [Google Scholar]
- Wisniewska, J., Xu, J., Seifertova, D., Brewer, P.B., Ruzicka, K., Blilou, I., Rouquie, D., Benkova, E., Scheres, B., and Friml, J. (2006). Polar PIN localization directs auxin flow in plants. Science 312 883. [DOI] [PubMed] [Google Scholar]
- Yang, Y., Hammes, U.Z., Taylor, C.G., Schachtman, D.P., and Nielsen, E. (2006). High-affinity auxin transport by the AUX1 influx carrier protein. Curr. Biol. 16 1123–1127. [DOI] [PubMed] [Google Scholar]
- Zhang, S., Yang, X., Coburn, R.A., and Morris, M.E. (2005). Structure activity relationships and quantitative structure activity relationships for the flavonoid-mediated inhibition of breast cancer resistance protein. Biochem. Pharmacol. 70 627–639. [DOI] [PubMed] [Google Scholar]