Abstract
Plants make full use of light signals to determine the timing of flowering. In Arabidopsis thaliana, a blue/UV-A photoreceptor, CRYPTOCHROME 2 (cry2), and a red/far-red photoreceptor, PHYTOCHROME B (phyB), are two major photoreceptors that control flowering. The light stimuli for the regulation of flowering are perceived by leaves. We have recently shown that phyB expression in mesophyll but not in vascular bundles suppresses the expression of a key flowering regulator, FLOWERING LOCUS T (FT), in vascular bundles. In this study, we asked where in the leaf cry2 perceives light stimuli to regulate flowering. To answer this question, we established transgenic Arabidopsis lines in which the cry2–green fluorescent protein (GFP) fusion was expressed under the control of organ/tissue-specific promoters in a cry2-deficient mutant background. Analysis of these lines revealed that expression of cry2-GFP in vascular bundles, but not in epidermis or mesophyll, rescued the late flowering phenotype. We further confirmed that cry2-GFP expressed in vascular bundles increased FT expression only in vascular bundles. Hence, in striking contrast with phyB, cry2 most likely regulates FT expression in a cell-autonomous manner.
INTRODUCTION
Environmental factors, such as temperature, nutrition, and light, strongly affect the growth and development of plants. Transition from vegetative to reproductive growth, namely flowering, is an important developmental step for plants. The timing of flowering is under strict control of the light environment (Simpson and Dean, 2002). Importantly, plants monitor daylength, which determines when to flower, ensuring that flowering occurs at the proper time of the year. In addition, the light quality affects the flowering time. In the shade of neighboring vegetation, flowering is advanced due to the shade avoidance response.
To perceive light signals, plants use a set of photoreceptors, including blue/UV-A photoreceptors, cryptochromes (cry) (Cashmore et al., 1999), and phototropins (Briggs and Christie, 2002), and red/far-red light photoreceptors, phytochromes (phy) (Quail, 2002). In Arabidopsis thaliana, cry2, phyB, and phyA are the major photoreceptors that regulate flowering. cry2 and phyA mediate the response to daylength (Koornneef et al., 1991; Guo et al., 1998; Yanovsky and Kay, 2002; Mockler et al., 2003), whereas phyB acts as the major photoreceptor for the shade avoidance responses (Smith and Whitelam, 1997).
cry2 is a nuclear protein, which binds the flavin chromophore and shares sequence similarity with prokaryotic DNA photolyases (Lin et al., 1998; Guo et al., 1999; Kleiner et al., 1999). cry2 is thought to transduce the light signal through the interaction with CONSTITUTIVE PHOTOMORPHOGENIC1 (COP1), a negative regulator of photomorphogenesis, in the nucleus (Deng et al., 1992). phyA and phyB are large, soluble proteins carrying a single covalently linked linear tetrapyrrole chromophore (Lagarias and Rapoport, 1980). Upon light activation, both phyA and phyB are accumulated in the nucleus (Kircher et al., 1999; Yamaguchi et al., 1999) and regulate gene expression (Martinez-Garcia et al., 2000). Since cry2 fails to affect flowering in the absence of phyB, cyr2 is proposed to regulate flowering by modifying the function of phyB (Guo et al., 1998; Mockler et al., 1999).
The molecular mechanism of the regulation of flowering by light has been intensively studied in Arabidopsis. An external coincidence model is now widely accepted to explain how Arabidopsis recognizes the daylength and regulates flowering (Davis, 2002). The CONSTANS (CO) protein plays a critical role in this process. Expression of the CO mRNA is regulated by the circadian clock to peak at dusk. In the presence of sufficient light, cry2 and phyA stabilize the CO protein (Valverde et al., 2004). In insufficient light, CO protein is destabilized. Consequently, CO cannot accumulate to the level required to induce flowering. In this way, plants perceive the daylength.
In case of the shade avoidance response, phyB destabilizes the CO protein (Valverde et al., 2004). PHYTOCHROME AND FLOWERING TIME1 has been reported to be involved in this process (Cerdan and Chory, 2003). In both cases, the stabilized CO upregulates the expression of another flowering regulator, FLOWERING LOCUS T (FT), which is the proximal inducer of flowering (Kardailsky et al., 1999; Kobayashi et al., 1999).
Spot light irradiation and grafting experiments have demonstrated that leaves are the major organs to sense both daylength and shade for the regulation of flowering (Knott, 1934; Chailakhyan, 1936; Zeevaart, 1976; Bernier et al., 1993). The leaf includes three major tissues: mesophyll, vascular bundle, and epidermis. Spatial expression patterns of the CO and FT proteins have been recently revealed. These proteins are expressed specifically in vascular bundles of leaves (Takada and Goto, 2003). In addition, the late flowering phenotype of the co mutation is complemented when CO or FT is ectopically expressed in vascular bundles (An et al., 2004).
In contrast with CO and FT, photoreceptors are expressed in almost all tissues. phyB, for example, is expressed in the epidermis, mesophyll, and vascular bundles in cotyledons (Somers and Quail, 1995; Goosey et al., 1997). We have examined the functional site of phyB for the regulation of flowering using the enhancer-trap system with modifications (Endo et al., 2005). phyB–green fluorescent protein (GFP) fusion protein has been expressed in the phyB mutant background under the control of the cauliflower mosaic virus 35S minimum promoter. Depending on the insertion site, the introduced gene is expressed in distinct tissue-specific patterns. The analysis of these lines has demonstrated that phyB-GFP expressed in mesophyll cells affects the flowering, whereas phyB-GFP expressed in vascular bundles does not. Furthermore, phyB-GFP expressed in mesophyll cells suppresses FT expression in vascular bundles (Endo et al., 2005). Hence, intertissue signaling between the mesophyll and the vascular bundles appears to exist.
It is essential to know the functional site of cry2 at the tissue level to elucidate the molecular mechanism of its action. As is the case with phyB, the CRY2 gene is expressed in almost all organs/tissues (Toth et al., 2001). However, it is unlikely that cry2 expression in every organ/tissue contributes equally to the response. Here, we expressed a cry2-GFP fusion protein in different tissues of leaves with the aid of tissue-specific promoters. The resultant transgenic lines were compared with respect to their flowering phenotypes. The results showed that cry2-GFP expression in vascular bundles, but not cry2-GFP expression in other tissues, advanced flowering by promoting the expression of FT. Hence, the two photoreceptors cry2 and phyB, acting in different tissues within the leaf, coordinately regulate flowering in response to light stimuli.
RESULTS
Preparation of Transgenic Arabidopsis Lines That Express cry2-GFP in an Organ/Tissue-Specific Pattern
To determine the functional site of cry2 in leaves, we established transgenic lines that expressed cry2-GFP under the control of tissue-specific promoters on the cry2 mutant background. For this purpose, the CAB3 (for mesophyll) (Susek et al., 1993), SUC2 (for vascular bundles) (Truernit and Sauer, 1995), Sultr1;3 (for vascular bundles) (Yoshimoto et al., 2003), At ML1 (for epidermis) (Lu et al., 1996), and CER6 (for epidermis) (Hooker et al., 2002) promoters were employed. As negative controls, we expressed cry2-GFP under the control of the UNUSUAL FLORAL ORGAN (UFO) (for the shoot apex) (Ingram et al., 1995; Lee et al., 1997) and the At3g25820/25830 (for roots) (Chen et al., 2004) promoters.
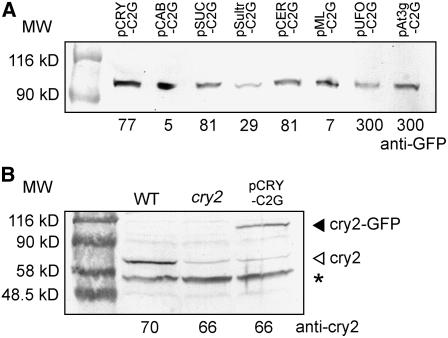
The CRY2-GFP gene was fused to these promoters and introduced into the cry2 mutant of Arabidopsis by the Agrobacterium tumefaciens–mediated method. As a control, the CRY2-GFP gene was expressed under the control of the authentic CRY2 promoter (Toth et al., 2001). We established several homozygous lines for each construct. The lines with the CAB3, SUC2, Sultr1;3, At ML1, CER6, UFO, At3g25820/25830, and CRY2 promoters are referred to as pCAB-C2G, pSUC-C2G, pSultr-C2G, pML-C2G, pCER-C2G, pUFO-C2G, pAt3g-C2G, and pCRY-C2G lines, respectively. Immunoblotting analysis with an anti-GFP antibody revealed that a protein of the predicted size (96 kD) was expressed in all of the lines (Figure 1A).
Figure 1.
Immunoblot Detection of cry2-GFP and Endogenous cry2.
Proteins were extracted from 10-d-old seedlings grown under LD. The numbers below the panel indicate loading amounts of the proteins (μg). MW, molecular mass (kD). The lines shown are pCRY-C2G-16, pCAB-C2G-6, pSUC-C2G-2, pSultr-C2G-10, pML-C2G-6, pCER-C2G-8, pUFO-C2G-13, and pAt3g-C2G-7.
(A) Immunoblotting analysis with an anti-GFP antibody.
(B) Immunoblotting analysis with an anti-cry2 antibody. The asterisk indicates nonspecific bands.
Expression Patterns of cry2-GFP
We first examined the expression levels of cry2-GFP protein in independent pCRY-C2G lines with an anti-cry2 antibody to choose a standard line in which cry2-GFP was expressed at the endogenous level (Figure 1B). Seedlings of this line were then observed under a laser scanning confocal microscope (Figures 2A, 2I, 3A, 3I, and 3Q). GFP fluorescence was observed exclusively in the nucleus, as has been reported previously (Guo et al., 1999). cry2-GFP expression was detected in all three major organs, namely, cotyledons, the hypocotyl, and the root in pCRY-C2G. In these organs, cry2-GFP was expressed in every tissue, including epidermis, mesophyll, cortex, and vascular bundles, except the root tip. In addition, expression in the shoot apex was detected. These results were consistent with the previously reported patterns of authentic cry2 expression (Toth et al., 2001).
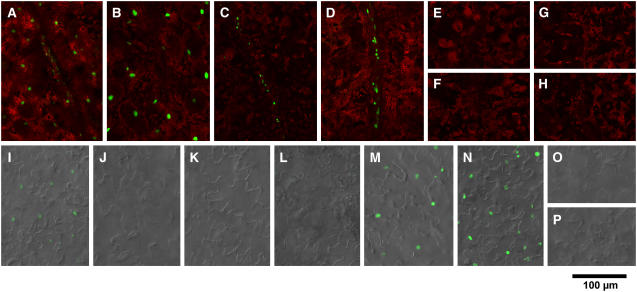
Figure 2.
Confocal Microscopic Observation of cry2-GFP Nuclear Accumulation (Guo et al., 1999) in Mesophyll, Vascular Bundles, and Epidermis of Cotyledons.
Seedlings were grown for 10 d under LD. Green fluorescence from GFP and red fluorescence from chlorophyll were overlaid electronically. In addition, differential interference contrast images were overlaid for epidermis. Seedlings of pCRY-C2G-16 ([A] and [I]), pCAB-C2G-6 ([B] and [J]), pSUC-C2G-2 ([C] and [K]), pSultr-C2G-10 ([D] and [L]), pML-C2G-6 ([E] and [M]), pCER-C2G-8 ([F] and [N]), pUFO-C2G-13 ([G] and [O]), and pAt3g-C2G-7 ([H] and [P]) are shown. Bar = 100 μm.
(A) to (H) cry2-GFP fluorescence in mesophyll/vascular bundles.
(I) to (P) cry2-GFP fluorescence in epidermis.
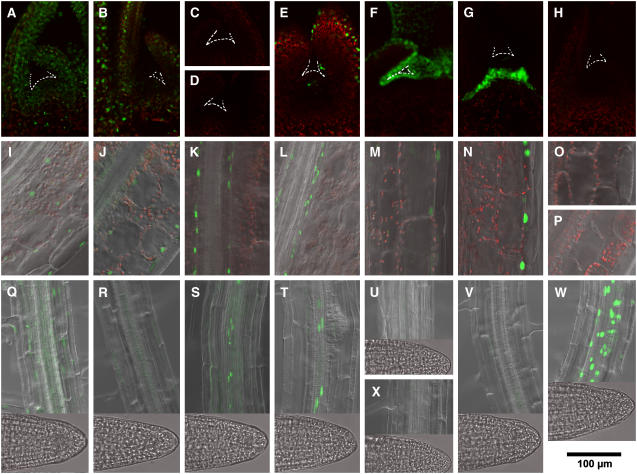
Figure 3.
Confocal Microscopic Observation of cry2-GFP Nuclear Accumulation (Guo et al., 1999) in Shoot Apex, the Hypoctyl, and the Root.
Seedlings were grown for 10 d under LD. Green fluorescence from GFP and red fluorescence from chlorophyll were overlaid electronically. In addition, differential interference contrast images were overlaid for hypocotyl and root. Images of pCRY-C2G-16 ([A], [I], and [Q]), pCAB-C2G-6 ([B], [J], and [R]), pSUC-C2G-2 ([C], [K], and [S]), pSultr-C2G-10 ([D], [L], and [T]), pML-C2G-6 ([E], [M], and [U]), pCER-C2G-8 ([F], [N], and [X]), pUFO-C2G-13 ([G], [O], and [V]), and pAt3g-C2G-7 ([H], [P], and [W]) seedlings are shown. Bar = 100 μm.
(A) to (H) cry2-GFP fluorescence in the shoot apex. Dotted lines indicate the edges of shoot apex and leaf primordia.
(I) to (P) cry2-GFP fluorescence in the hypocotyl.
(Q) to (X) cry2-GFP fluorescence in the root and root tip.
In contrast with pCRY-C2G, other lines exhibited organ/tissue-specific expression patterns (Figures 2 and 3). In pCAB-C2G, expression was observed exclusively in the mesophyll of both cotyledons and young true leaves (Figures 2B, 2J, and 3B). Semiquantitative analysis of GFP fluorescence indicated that the expression level in pCAB-C2G mesophyll was ∼1.8 times higher than that in pCRY-C2G mesophyll. In addition, cry2-GFP was expressed in the cortex of hypocotyl at lower levels (Figure 3J). In pSUC-C2G and pSultr-C2G, GFP expression was observed only in vascular bundles in all organs (Figures 2C, 2D, 3K, 3L, 3S, and 3T). No expression was observed in the shoot apexes. Semiquantitative analysis indicated that GFP expression levels in pSUC-C2G and pSultr-C2G in cotyledonous vascular bundles were ∼1.5 and 2.7 times higher than that in pCRY-C2G, respectively. Expression was restricted to epidermis of the aerial part of the seedlings in pML-C2G and pCER-C2G (Figures 2M, 2N, 3M, and 3N). Expression levels in pML-C2G and pCER-C2G in cotyledons were ∼3.8 and 4.7 times higher, respectively, than in pCRY-C2G. We also confirmed that expression was restricted to the shoot apex in pUFO-C2G and to the root in pAt3g-C2G (Figures 2G and 3W).
cry2-GFP in Vascular Bundles Regulates the Flowering Time
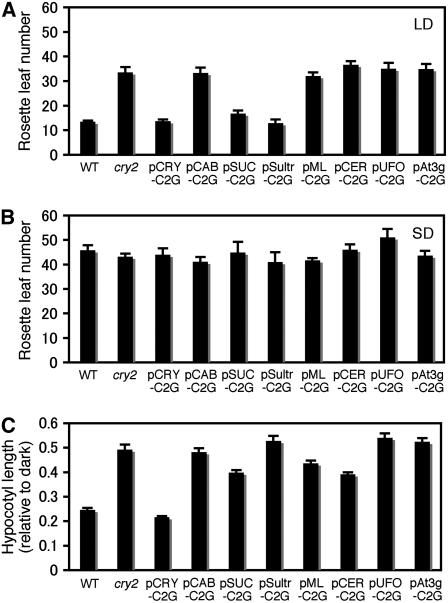
A late flowering phenotype is exhibited by cry2 mutants under long-day (LD) conditions in the presence of phyB (Guo et al., 1998). We examined whether this phenotype was complemented in our transgenic lines (Figure 4A). As expected, full complementation was observed in pCRY-C2G. The flowering time was neither advanced nor delayed compared with the wild type. In the pSUC-C2G and pSultr-C2G lines, which expressed cry2-GFP only in vascular bundles, the late flowering phenotype was also complemented. The other lines, pCAB-C2G (mesophyll), pML-C2G (epidermis), pCER-C2G (epidermis), pUFO-C2G (shoot apex), and pAt3g-C2G (root), did not complement and flowered as late as the parental cry2 mutant. Hence, cry2-GFP expression in vascular bundles, but not in other tissues, effectively accelerated the flowering in LD.
Figure 4.
Flowering Times and Hypocotyl Lengths in the Lines Expressing cry2-GFP in Specific Organs/Tissues.
(A) and (B) Flowering times were scored by determining the number of rosette leaves when the first flower opened. The number is inversely correlated with the flowering time. Plants were grown under LD (A) or SD (B). Mean ± se (n ≥ 15).
(C) Hypocotyl lengths of seedlings grown under continuous blue light for 7 d. Mean ± se (n = 25).
The lines shown are pCRY-C2G-16, pCAB-C2G-6, pSUC-C2G-2, pSultr-C2G-10, pML-C2G-6, pCER-C2G-8, pUFO-C2G-13, and pAt3g-C2G-7.
We then examined whether or not pSUC-C2G and pSultr-C2G responded normally to changes in daylength. In the short-day (SD) condition, flowering is not delayed in the cry2 mutant compared with the wild type (Guo et al., 1998). As expected, pSUC-C2G and pSultr-C2G flowered as late as pCRY-C2G and the wild type (Figure 4B). Hence, cry2-GFP in pSUC-C2G and pSultr-C2G did not accelerate flowering in SD. Flowering times in other lines were not affected by the transgene and were the same as the control plants.
The cry2 mutant exhibits long hypocotyl phenotype under blue light (Lin et al., 1998). Hence, we checked whether this phenotype was complemented in the transgenic lines (Figure 4C). As expected, full complementation was observed in pCRY-C2G. However, in contrast with the flowering phenotype, none of the lines showed full complementation, although partial complementation was observed in pSUC-C2G, pML-C2G, and pCER-C2G.
To confirm the tissue-specific expression of cry2-GFP in pSUC-C2G and pSultr-C2G, the CRY2-GFP mRNA levels in isolated vascular bundles and mesophyll protoplasts were quantified by RT-PCR (Figure 5). Mesophyll cells and vascular bundles were isolated from cotyledons as described (Endo et al., 2005). We confirmed the purity of the samples by checking the expression levels of marker genes, Sultr and RbcS (data not shown) (Endo et al., 2005).
Figure 5.
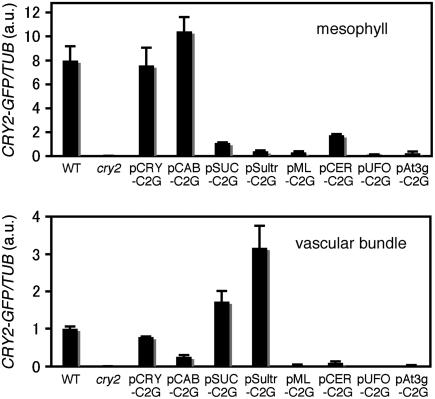
CRY2-GFP mRNA Expression in Mesophyll and Vascular Bundles.
Mesophyll protoplasts (top panel) and vascular bundles (bottom panel) were isolated from cotyledons in the seedlings grown for 10 d under LD. The samples were analyzed by quantitative RT-PCR. The averages of three biological replicates are shown. TUB2/TUB3 was used as an internal control for calculating relative levels of CRY2-GFP mRNA in C2G lines and CRY2 mRNA in the wild type. Data were normalized to the level of CRY2 mRNA in vascular bundles of the wild type, which was set to 1 arbitrary unit (a.u.). Mean ± se (n = 3). The lines shown are pCRY-C2G-16, pCAB-C2G-6, pSUC-C2G-2, pSultr-C2G-10, pML-C2G-6, pCER-C2G-8, pUFO-C2G-13, and pAt3g-C2G-7.
Consistent with microscopic observation (Figures 2 and 3), the CRY2-GFP mRNA levels in mesophyll cells were much higher in pCRY-C2G and pCAB-C2G than in other lines (Figure 5). Consistent with the semiquantification of the GFP fluorescence (see above), the level was higher in pCAB-C2G than in pCRY-C2G. Conversely, the CRY2-GFP mRNA was detected in vascular bundles in pSUC-C2G, pSultr-C2G, and pCRY-C2G (Figure 5). The levels were much lower in other lines, except pCAB-C2G. This might be due to the contamination of the vascular bundle samples with mesophyll cells. It should be noted that cry2-GFP in pCAB-C2G failed to affect flowering regardless of the high CRY2-GFP mRNA expression in the mesophyll (∼10 times higher than that in the vascular bundles of pCRY-C2G).
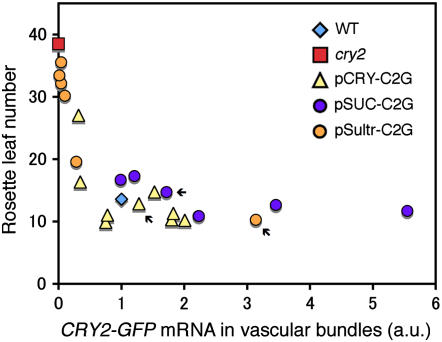
To examine quantitative relationships between cry2-GFP expression in vascular bundles and flowering times, we examined several independent pSUC-C2G, pSultr-C2G, and pCRY-C2G lines under LD. The expression levels of CRY2-GFP mRNA in vascular bundles were determined by RT-PCR and plotted against flowering times (Figure 6). The expression level and the flowering time correlated well. All points were laid on a single curve containing the point for the wild-type plant. Flowering was advanced depending on the expression levels of CRY2-GFP up to about the endogenous level. The response was then more or less saturated. These results further supported that the flowering time was dependent on the expression of cry2-GFP in vascular bundles but not in other tissues.
Figure 6.
Quantitative Analysis of the Effects of CRY2-GFP Expression in Vascular Bundles.
The flowering times (ordinate) versus the levels of CRY2-GFP mRNA in vascular bundles (abscissa) in pCRY-C2G, pSUC-C2G, and pSultr-C2G lines under LD are shown. Each point represents an individual line. Arrows indicate pCRY-C2G-16, pSUC-C2G-2, and pSultr-C2G-10. The levels of mRNA in vascular bundles were determined by quantitative RT-PCR in 10-d-old seedlings. Relative levels of CRY2-GFP and CRY2 mRNA were calculated as for Figure 5.
cry2-GFP in Vascular Bundles Regulates FT Expression in Vascular Bundles
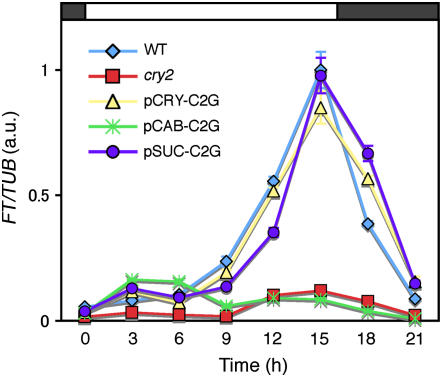
cry2 advances flowering by inducing the expression of a key flowering regulator, FT (Yanovsky and Kay, 2002). FT expression in the wild type exhibits diurnal oscillation in LD (Suárez-López et al., 2001; Yanovsky and Kay 2002). Therefore, to confirm that cry2-GFP expressed in vascular bundles regulated flowering by increasing FT expression, we monitored the levels of FT expression during a 24-h period on day 10 in pCRY-C2G, pCAB-C2G, and pSUC-C2G.
Total mRNA was extracted from cotyledons every 3 h and subjected to RT-PCR analysis. The FT expression patterns in pCRY-C2G and pSUC-C2G were almost identical to that in the wild type (Figure 7). The expression peaked at the end of the light period in these lines. Hence, the effects of cry2-GFP expressed in vascular bundles were indistinguishable from those of the endogenous cry2. By contrast, the levels of FT expression remained low in pCAB-C2G and the cry2 mutant. These results were fully consistent with the flowering phenotype (Figure 4A).
Figure 7.
Diurnal Expression of FT.
Seedlings were grown under LD for 9 d, and then RNA was extracted from cotyledons every 3 h over a 24-h period in LD. The samples were analyzed by quantitative RT-PCR. The averages of four biological replicates are shown. TUB2/TUB3 was used as an internal control to calculate relative FT mRNA levels. Relative amounts of FT mRNA in arbitrary units (a.u.) are shown. The peak level of FT mRNA in cotyledons of the wild type was set to 1 arbitrary unit. Mean ± se (n = 4). The lines shown are pCRY-C2G-16, pCAB-C2G-6, and pSUC-C2G-2.
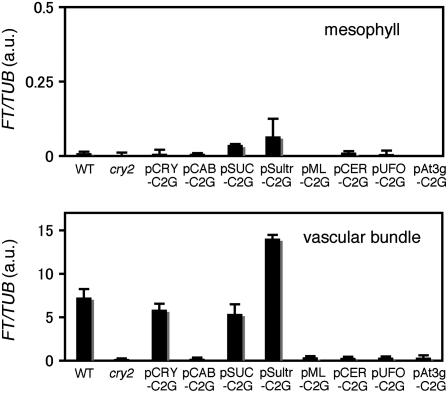
It has been shown that FT expression is restricted to vascular bundles in either the presence or absence of phyB (Takada and Goto, 2003; Endo et al., 2005). We examined whether this was true in pSUC-C2G and pSultr-C2G. Mesophyll cells and vascular bundles were isolated from cotyledons of 10-d-old seedlings and examined for FT mRNA expression by RT-PCR (Figure 8). As expected, induction of FT mRNA expression was observed only in vascular bundles. Although cyr2-GFP was expressed at a high level in mesophyll in pCAB-C2G (Figure 5), FT induction was not observed in either mesophyll or in vascular bundles in this line.
Figure 8.
FT Expression in Mesophyll and Vascular Bundles.
Mesophyll protoplasts (top panel) and vascular bundles (bottom panel) were isolated from cotyledons of 10-d-old seedlings grown under LD. The samples were analyzed by quantitative RT-PCR. The averages of four biological replicates are shown. Relative FT mRNA levels were calculated as for Figure 7. Note that 1 arbitrary unit (a.u.) was defined as the peak level in wild-type cotyledons (Figure 7). Mean ± se (n = 4). The lines shown are pCRY-C2G-16, pCAB-C2G-6, pSUC-C2G-2, pSultr-C2G-10, pML-C2G-6, pCER-C2G-8, pUFO-C2G-13, and pAt3g-C2G-7.
DISCUSSION
cry2-GFP Expression Using Organ/Tissue- Specific Promoters
In this study, we employed tissue-specific promoters to express cry2-GFP in different tissues. Our main focus was on three major tissues in cotyledons: namely, mesophyll, vascular bundles, and epidermis. As negative controls, we expressed cry2-GFP in other organs/tissues, such as the root and the shoot apex.
For mesophyllic expression, the CRY2-GFP gene was driven by the CAB promoter. The CAB gene encodes chlorophyll a/b binding proteins that are highly expressed in mesophyll. As was expected from previous results using a reporter gene analysis (Susek et al., 1993), cry2-GFP was expressed in mesophyll cells at a high level and in hypocotyl cortex cells at a lower level (Figures 2, 3, and 5). For vascular expression, the promoters for sucrose (SUC2) and sulfate (Sultr1;3) transporter genes were chosen. These genes are expressed specifically in vascular bundles (Truernit and Sauer, 1995). More recently, they have been shown to be expressed preferentially in the companion cells (Stadler and Sauer, 1996; Yoshimoto et al., 2003). The SUC2 promoter has been successfully used to examine functions of the CO and FT genes in vascular bundles (An et al., 2004). As expected, the cry2-GFP was expressed only in the vascular bundle region in pSUC-C2G and pSultr-C2G at relatively high levels (Figures 2, 3, and 5).
Epidermal expression of cry2-GFP was examined using the At ML1 and CER6 promoters. The At ML1 and CER6 genes, which encode a homeobox protein and a very-long-chain fatty acid condensing enzyme, respectively (Lu et al., 1996; Millar et al., 1999), are expressed specifically in epidermis (Sessions et al., 1999; Hooker et al., 2002). The At ML1 promoter has been successfully used to examine functions of the FT genes in epidermis (An et al., 2004). As expected, the expression driven by these promoters was highly specific (Figures 2 and 3). In addition, we employed the UFO and At3g25820/25830 promoters for the shoot apex and root-specific expression (Ingram et al., 1995; Lee et al., 1997; Chen et al., 2004). Microscopic observation of pUFO-C2G and pAT3g-C2G indicated that cry2-GFP was expressed specifically in the expected locations (Figure 3).
Tissue-specific promoters can be a powerful tool to examine tissue-specific functions of photoreceptors and other factors. Although such possibilities in plants have not been explored fully yet, they would be applicable to various aspects of the light signal transduction. It is now possible to suppress the function of endogenous genes by RNA interference technology (Fire et al., 1998). Hence, not only the gain-of-function but also the loss-of-function types of analyses are feasible. However, not all the promoters work properly. In addition to the above promoters, we examined several others, but the specificity was quite low for unknown reasons. It would be necessary to try several promoters to obtain satisfactory results.
Vascular Bundle as a Functional Site of cry2
It has been known for many years that photoperiodic stimuli are perceived by leaves (Knott, 1934; Chailakhyan, 1936; Zeevaart, 1976). A leaf consists of three major tissues, including mesophyll, vascular bundles, and epidermis. Our results indicate that the vascular bundles are the major functional site of cry2-mediated regulation of flowering. Flowering times correlated well with the expression levels of cry2-GFP in vascular bundles (Figure 6). By contrast, flowering was unaffected by cry2-GFP expression in mesophyll or epidermis, even though cry2-GFP expression was at a high level (Figures 4 and 5).
Although cry2-GFP expression in vascular bundles was required for regulation of flowering, cry2 is expressed in other tissues as well (Toth et al., 2001) (Figures 2, 3, and 5). Hence, the question arises as to what are the functions of cry2 in other tissues. In addition to regulating flowering, cry2 regulates a number of photomorphogenic responses, including hypocotyl elongation, cotyledon expansion, and chlorophyll synthesis (Lin et al., 1998; Usami et al., 2004). The responses related to the development of chloroplasts, for example, might be controlled by cry2 in photosynthetic tissues in a cell-autonomous manner.
With respect to the hypocotyl elongation, we did not observe dramatic effects of cry2-GFP in any lines except pCRY-C2G (Figure 4C). Hence, our set of lines may not be covering all of the functional sites of cry2. Alternatively, cry2-GFP expression in multiple tissues may be required for these photomorphogenic responses. This was in striking contrast with phyB. Our previous work has demonstrated that phyB in mesophyll cells principally regulates hypocotyl elongation (Endo et al., 2005). Consistent with these observations, cry2 and phyB independently regulate hypocotyl elongation (Mockler et al., 1999). Furthermore, the state of phytochrome in leaves determines the stem elongation rate (Casal and Smith, 1988). By contrast, blue light is perceived by the stem to regulate its elongation (Black and Shuttleworth, 1974). Such a discrepancy may be explained by the difference in the functional sites of cry2 and phyB for the regulation of stem elongation.
Possible Mechanism of cry2 Function in Vascular Bundles
It is intriguing that cry2 functions in vascular bundles because the key flowering regulators acting downstream of cry2, namely, CO and FT, are expressed specifically in leaf vascular bundles (Takada and Goto, 2003; An et al., 2004). cry2 is proposed to increase CO protein stability (Valverde et al., 2004). Hence, this study suggests that cry2 regulates CO protein levels in a cell-autonomous manner.
One possible mechanism by which cry2 stabilizes CO is through a physical interaction with COP1, which has E3 ubiquitin ligase activity (Wang et al., 2001). The observation that the cop1 mutation suppresses the late flowering phenotype of the cry2 mutant (Nakagawa and Komeda, 2004) suggests that COP1 may trigger the degradation of CO. Another factor that may be involved in the CO degradation is SPA proteins. SPA proteins contain a COP1-like WD repeat domain, a coiled-coil domain, and a kinase-like domain (Hoecker et al., 1999). COP1 and SPA1 physically interact (Hoecker and Quail, 2001). The CO protein is more stable in the spa1 spa3 spa4 mutant than in the wild type (Laubinger et al., 2006). Hence, cry2 might regulate CO protein stability in vascular bundle cells through the physical interaction with COP1 and SPA proteins.
Flowering Regulation by phyB and cry2
Genetic and physiological analyses have indicated that cry2 advances flowering by suppressing the inhibitory effect of phyB on flowering (Mockler et al., 1999). Our previous work has demonstrated that phyB expression in mesophyll regulates flowering (Endo et al., 2005). This work, however, indicated that cry2 was functioning in vascular bundles. Therefore, the genetic interaction between cry2 and phyB does not necessarily mean a direct interaction between these photoreceptors. Although physical interaction between cry2 and phyB has been reported (Mas et al., 2000), its biological relevance remains obscure.
It is not clear if there is any advantage for plants to regulate flowering using photoreceptors in different tissues. A possible explanation is as follows: The timing is a key issue in the daylength perception by cry2. In LD plants such as Arabidopsis, daylength is thought to be recognized in the following way. CO mRNA accumulation is controlled by the circadian clock (Yanovsky and Kay, 2002). If, at dusk, it is still light, namely under LD, cry2 is activated (by light) and functionally stabilizes CO (Valverde et al., 2004). Under SD, low light at dusk fails to activate cry2, which, consequently, is unavailable to stabilize CO. For this mechanism to work properly, the activation state of cry2 is intimately related to the stability of CO. Hence, cry2 might have to function in the same cell as CO.
It remains unclear why phyB regulates the flowering in mesophyll. Timing may be less important for the phyB responses because it mainly mediates the shade avoidance responses. However, it should be mentioned here that phyB is also involved in photoperiodic flowering in other species (Hanumappa et al., 1999; Weller et al., 2001). phyB perceives the reduction in the ratio of red to far-red light. This mechanism is unlikely to occur in vascular bundles because vascular bundles are embedded in the mesophyll where the red:far-red ratio would be substantially reduced due to surrounding cells. In addition, phyB might require a greater number of cells than are available in vascular bundles to regulate the developmental processes in whole plants. Hence, there might be good reasons for plants to use phyB in mesophyll rather than vascular bundles.
cry2 and phyB are not the only photoreceptors involved in the flowering regulation. phyA mediates photoperiodic responses by far-red light (Yanovsky and Kay, 2002; Mockler et al., 2003), which resembles the cry2 function. Hence, phyA may function in vascular bundles to regulate flowering. By contrast, phyC delays flowering in SD (Monte et al., 2003). Hence, it may function in the mesophyll, as is the case with phyB.
METHODS
Plant Materials and Growth Conditions
The Arabidopsis thaliana cry2 mutant used for this report was cry2-2 (Guo et al., 1998) on the Columbia (Col) ecotype background, in which the CRY2 gene is completely deleted. Seeds were surface-sterilized and sown on 0.6% agar plates containing Murashige and Skoog medium without sucrose. The plates were kept in the dark at 4°C for 24 h and then placed under LD for 10 d to obtain seedlings. Seeds were sown directly on rockwool for measurement of flowering times. Flowering times were scored by determining the number of rosette leaves when the first flower opened (Koornneef et al., 1991). Plants grown in LD (16 h of white light at ∼35 μmol m−2 s−1 from white fluorescent light tubes [FLR40SW/M/36-B; Hitachi] and 8 h of darkness) and SD (8 h of white light at ∼70 μmol m−2 s−1 from white fluorescent light tubes and 16 h of darkness) received the same total fluence of light. For hypocotyl length measurements, plants were grown in continuous blue light (3 μmol m−2 s−1 from fluorescent light tubes [FL20S·B; Toshiba]) for 7 d. Hypocotyl lengths were measured by ImageJ software (National Institutes of Health).
Plasmid Construction and Plant Transformation
The binary vector pPZP211/NP was derived from the binary plant transformation vector pPZP211 (Hajdukiewicz et al., 1994). pPZP211/NP has a neomycin phosphotransferase II gene driven by the nopaline synthase promoter for selection of plants on kanamycin.
The full-length CRY2 cDNA was PCR-amplified from a cDNA library using specific primers with XbaI-SpeI and ClaI tails. The GFP was PCR-amplified from the pPZP211/Bpro (Endo et al., 2005) plasmid using specific primer sequences with ClaI and SacII tails. CRY2 and GFP were fused in the binary vector pPZP211/NP (pPZP211/NP/CRY2-GFP).
Organ/tissue-specific promoters were inserted into pPZP211/NP/CRY2-GFP using the SalI, XbaI, or SpeI site. The CRY2 (∼1200 bp), CAB3 (∼1550 bp), SUC2 (∼3500 bp), Sultr1;3 (∼2700 bp), At ML1 (∼3700 bp), CER6 (∼3500 bp), UFO (∼3800 bp), and At3g25820/25830 (∼2450 bp) promoters were PCR-amplified from Col genomic DNA using specific primers with SalI, XbaI, or SpeI tails.
Specific sequences for each primer pair were as follows: CRY2-cDNA-F, 5′-GCTCTAGAACTAGTATGAAGATGGACAAAAAGAC-3′; CRY2-cDNA-R, 5′-CCATCGATTTTGCAACCATTTTTTCCCAAACTTG-3′; CRY2-pro-F, 5′-ACTTGTCGACAATTGCAAAAAGAAATGCTACTC-3′; CRY2-pro-R, 5′-GCTCTAGAGTTATTATGATCACAGATGAATCAAAGAT-3′; CAB3-pro-F, 5′-CGTCTAGAAATCAAGAGAAAATGTGATTCTCGG-3′; CAB3-pro-R, 5′-GCACTAGTGAAACTTTTTGTGTTTTTTTTTTTTTTTG-3′; SUC2-pro-F, 5′-ACTTGTCGACTTTGTCATACATTTATTTGCCACAAG-3′; SUC2-pro-R, 5′-GCTCTAGATTTGACAAACCAAGAAAGTAAGAAAAAAA-3′; Sultr1;3-pro-F, 5′-CGTCTAGAGTTTTTCTTCATAGGTTTCGTGATAAT-3′; Sultr1;3-pro-R, 5′-GCTCTAGATGCTATGTGTGTTTTGTAGCAAAC-3′; At ML1-pro-F, 5′-ACTTGTCGACCAAGAGATATTGGGTTGCTACACAA-3′; At ML1-pro-R, 5′-GGACTAGTGATGATGATGGATGCCTATCAATTTTT-3′; CER6-pro-F, 5′-ACTTGTCGACAATGATGAGCAAAAGTGTTTGC-3′; CER6-pro-R, 5′-TGCTCTAGACGTCGGAGAGTTTTAATGTATAATTG-3′; UFO-pro-F, 5′-ACTTGTCGACGAATTCTCTGTTTTAATTGCCCCA-3′; UFO-pro-R, 5′-GCTCTAGATTTAGCTGAAAAATGAAAAGATTTGG-3′; At3g25820/25830-pro-F, 5′-CGTCTAGATGTCAATTTTCATGGCACATCGAG-3′; and At3g25820/25830-pro-R, 5′-GGACTAGTGATTTAGTAGACTATTCTCTTATTCGTGGC-3′.
The Arabidopsis cry2 mutant was transformed with vectors described above by the Agrobacterium tumefaciens–mediated floral dip method (Clough and Bent, 1998). Transformed plants were selected on agar medium containing 25 mg/L of kanamycin.
Immunochemical and Microscopic Detection of cry2-GFP
cry2 and cry2-GFP proteins were detected by immunoblotting of protein extracts from seedlings. Protein extraction, SDS-PAGE, protein blotting, and immunodetection were performed as described by Yamaguchi et al. (1999). Proteins were extracted from the seedlings at the end of the light period on day 10 and subjected to immunoblotting analysis with anti-Arabidopsis cry2 antibody (Lin et al., 1998) and anti-GFP antibody (Nacalai Tesque).
Ten-day-old seedlings grown under LD were observed at around the end of the light period with a confocal laser scanning microscope (Zeiss LSM510). Seedlings were vacuum-infiltrated in water before observation. For shoot apex and hypocotyl observations, seedlings were embedded in 5% low melting point agarose to prepare longitudinal sections without fixation. Green fluorescence from GFP (observation, 500 to 530 nm; excitation, 488 nm) and red fluorescence from chlorophyll (observation, >560 nm; excitation, 543 nm) were overlaid electronically. To semiquantify the GFP fluorescence, the gain was set to 720 and the fluorescence intensity within the nuclear region was integrated for each nucleus. For each line, >20 nuclei from several samples were quantified and averaged.
Isolation of Mesophyll Cells and Vascular Bundles
Mesophyll protoplasts and vascular bundles were isolated from the 10-d-old seedlings at around the end of the light period as described (Endo et al., 2005).
RNA Extraction, cDNA Synthesis, and Real-Time PCR
Total RNA was extracted from the whole seedlings using Sepazol super (Nacalai Tesque) following the manufacturer's instructions. For tissue specificity analysis, total RNA was extracted from 1 × 104 mesophyll protoplasts or from vascular bundles prepared from 20 cotyledons using the RNAqueous Micro isolation kit (Ambion) following the manufacturer's instructions (Endo et al., 2005). The samples were treated with RQ1 RNase-free DNase (Promega) following the manufacturer's instructions. Reverse transcription was performed with the oligo(dT) primer using the SuperScript first-strand synthesis system for RT-PCR (Invitrogen) according to the manufacturer's instructions.
The TUB2/TUB3 genes, which are known to be expressed at similar levels in different tissues, were used as an internal control for normalization of the PCR reaction. Note that no CRY2 mRNA is expressed in the parental cry2-2 mutant because the CRY2 gene is deleted in this allele.
Specific sequences for each primer pair were as follows: CRY2-RT-F, 5′-AATCCCGCGTTACAAGGC-3′; CRY2-RT-R, 5′-TTCCGAGTTCCACACCAG-3′; FT-RT-F, 5′-TATCTCCATTGGTTGGTGACTG-3′; FT-RT-R, 5′-GGGACTTGGATTTTCGTAACAC-3′; TUB2/3-RT-F, 5′-CCAGCTTTGGTGATTTGAAC-3′; and TUB2/3-RT-R, 5′-CAAGCTTTCGGAGGTCAGAG-3′.
Quantitative RT-PCR was performed in 200-μL tubes with a Rotor-Gene RG-3000A (Corbett Research) using SYBR green to monitor double-strand DNA synthesis. The reaction mixture contained 7.5 μL of Platinum SYBR Green qPCR SuperMix UDG (Invitrogen), 1 μL of cDNA, and 200 nM gene-specific primers in a final volume of 15 μL. The following thermal cycling profile was used for all PCRs: 95°C for 20 s, ∼55 cycles of 95°C for 5 s, and 60°C for 20 s. Data were analyzed using Rotor-Gene 6.0.16 software (Corbett Research). Negative template controls were run in these experiments, and no signal was observed (data not shown).
Accession Numbers
Arabidopsis Genome Initiative locus identifiers for the genes mentioned in this article are as follows: CRY2 (At1g04400), CAB3 (At1g29910), SUC2 (At1g22710), Sultr1;3 (At1g22150), At ML1 (At4g21750), CER6 (At1g68530), UFO (At1g30950) TUB2 (At5g62690), TUB3 (At5g62700), FT (At1g65480), and CO (At5g15840).
Acknowledgments
We thank T. Nishimura for providing the pPZP211/NP vector. We thank C. Lin for providing the anti-cry2 antibody. We thank BioMed Proofreading for English proofreading. This work was partially supported by a Grant-in-Aid for Scientific Research (B) 17370018 (to A.N.), a Grant-in-Aid for Scientific Research on Priority Areas 17084002 (to A.N.), a Grant-in-Aid for Japan Society for the Promotion of Science Fellows 02709 (to M.E.), and a Grant-in-Aid for 21st Century Circle of Excellence Research, Kyoto University (A14).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Akira Nagatani (nagatani@physiol.bot.kyoto-u.ac.jp).
References
- An, H., Roussot, C., Suarez-Lopez, P., Corbesier, L., Vincent, C., Pineiro, M., Hepworth, S., Mouradov, A., Justin, S., Turnbull, C., and Coupland, G. (2004). CONSTANS acts in the phloem to regulate a systemic signal that induces photoperiodic flowering of Arabidopsis. Development 131 3615–3626. [DOI] [PubMed] [Google Scholar]
- Bernier, G., Havelange, A., Houssa, C., Petitjean, A., and Lejeune, P. (1993). Physiological signals that induce flowering. Plant Cell 5 1147–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black, M., and Shuttleworth, J.E. (1974). The role of the cotyledons in the photocontrol of hypocotyl extension in Cucumis sativus L. Planta 117 57–66. [DOI] [PubMed] [Google Scholar]
- Briggs, W.R., and Christie, J.M. (2002). Phototropins 1 and 2: Versatile plant blue-light receptors. Trends Plant Sci. 7 204–210. [DOI] [PubMed] [Google Scholar]
- Casal, J.J., and Smith, H. (1988). Persistent effects of changes in phytochrome status on internode growth in light-grown mustard: Occurrence, kinetics and locus of perception. Planta 175 214–220. [DOI] [PubMed] [Google Scholar]
- Cashmore, A.R., Jarillo, J.A., Wu, Y.J., and Liu, D. (1999). Cryptochromes: Blue light receptors for plants and animals. Science 284 760–765. [DOI] [PubMed] [Google Scholar]
- Cerdan, P.D., and Chory, J. (2003). Regulation of flowering time by light quality. Nature 423 881–885. [DOI] [PubMed] [Google Scholar]
- Chailakhyan, M.K. (1936). On the hormonal theory of plant development. Dokl. Akad. Nauk SSSR 12 443–447. [Google Scholar]
- Chen, F., Ro, D.K., Petri, J., Gershenzon, J., Bohlmann, J., Pichersky, E., and Tholl, D. (2004). Characterization of a root-specific Arabidopsis terpene synthase responsible for the formation of the volatile monoterpene 1,8-cineole. Plant Physiol. 135 1956–1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Davis, S.J. (2002). Photoperiodism: The coincidental perception of the season. Curr. Biol. 12 841–843. [DOI] [PubMed] [Google Scholar]
- Deng, X.W., Matsui, M., Wei, N., Wagner, D., Chu, A.M., Feldmann, K.A., and Quail, P.H. (1992). COP1, an Arabidopsis regulatory gene, encodes a protein with both a zinc-binding motif and a G beta homologous domain. Cell 71 791–801. [DOI] [PubMed] [Google Scholar]
- Endo, M., Nakamura, S., Araki, T., Mochizuki, N., and Nagatani, A. (2005). Phytochrome B in the mesophyll delays flowering by suppressing FLOWERING LOCUS T expression in Arabidopsis vascular bundles. Plant Cell 17 1941–1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire, A., Xu, S., Montgomery, M.K., Kostas, S.A., Driver, S.E., and Mello, C.C. (1998). Potent and specific genetic interference by double stranded RNA in Caenorhabditis elegans. Nature 391 806–811. [DOI] [PubMed] [Google Scholar]
- Goosey, L., Palecanda, L., and Sharrock, R.A. (1997). Differential patterns of expression of the Arabidopsis PHYB, PHYD, and PHYE phytochrome genes. Plant Physiol. 115 959–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, H., Duong, H., Ma, N., and Lin, C. (1999). The Arabidopsis blue light receptor cryptochrome 2 is a nuclear protein regulated by a blue light-dependent post-transcriptional mechanism. Plant J. 19 279–287. [DOI] [PubMed] [Google Scholar]
- Guo, H., Yang, H., Mockler, T.C., and Lin, C. (1998). Regulation of flowering time by Arabidopsis photoreceptors. Science 27 1360–1363. [DOI] [PubMed] [Google Scholar]
- Hajdukiewicz, P., Svab, Z., and Maliga, P. (1994). The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol. Biol. 25 989–994. [DOI] [PubMed] [Google Scholar]
- Hanumappa, M., Pratt, L.H., Cordonnier-Pratt, M.M., and Deitzer, G.F. (1999). A photoperiod-insensitive barley line contains a light-labile phytochrome B. Plant Physiol. 119 1033–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoecker, U., and Quail, P.H. (2001). The phytochrome A-specific signaling intermediate SPA1 interacts directly with COP1, a constitutive repressor of light signaling in Arabidopsis. J. Biol. Chem. 276 38173–38178. [DOI] [PubMed] [Google Scholar]
- Hoecker, U., Tepperman, J.M., and Quail, P.H. (1999). SPA1, a WD-repeat protein specific to phytochrome A signal transduction. Science 284 496–499. [DOI] [PubMed] [Google Scholar]
- Hooker, T.S., Millar, A.A., and Kunst, L. (2002). Significance of the expression of the CER6 condensing enzyme for cuticular wax production in Arabidopsis. Plant Physiol. 129 1568–1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingram, G.C., Goodrich, J., Wilkinson, M.D., Simon, R., Haughn, G.W., and Coen, E.S. (1995). Parallels between UNUSUAL FLORAL ORGANS and FIMBRIATA, genes controlling flower development in Arabidopsis and Antirrhinum. Plant Cell 7 1501–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kardailsky, I., Shukla, V.K., Ahn, J.H., Dagenais, N., Christensen, S.K., Nguyen, J.T., Chory, J., Harrison, M.J., and Weigel, D. (1999). Activation tagging of the floral inducer FT. Science 286 1962–1965. [DOI] [PubMed] [Google Scholar]
- Kircher, S., Kozma-Bognar, L., Kim, L., Adam, E., Harter, K., Schafer, E., and Nagy, F. (1999). Light quality-dependent nuclear import of the plant photoreceptors phytochrome A and B. Plant Cell 11 1445–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleiner, O., Kircher, S., Harter, K., and Batschauer, A. (1999). Nuclear localization of the Arabidopsis blue light receptor cryptochrome 2. Plant J. 19 289–296. [DOI] [PubMed] [Google Scholar]
- Knott, J.E. (1934). Effect of localized photoperiod on spinach. Proc. Soc. Hort. Sci. 31 152–154. [Google Scholar]
- Kobayashi, Y., Kaya, H., Goto, K., Iwabuchi, M., and Araki, T. (1999). A pair of related genes with antagonistic roles in mediating flowering signals. Science 286 1960–1962. [DOI] [PubMed] [Google Scholar]
- Koornneef, M., Hanhart, C.J., and van der Veen, J.H. (1991). A genetic and physiological analysis of late flowering mutants in Arabidopsis thaliana. Mol. Gen. Genet. 229 57–66. [DOI] [PubMed] [Google Scholar]
- Lagarias, J.C., and Rapoport, H. (1980). Chromopeptides from phytochrome: The structure and linkage of the Pr form of the phytochrome chromophore. J. Am. Chem. Soc. 102 4821–4828. [Google Scholar]
- Laubinger, S., Marchal, V., Gentilhomme, J., Wenkel, S., Adrian, J., Jang, S., Kulajta, C., Braun, H., Coupland, G., and Hoecker, U. (2006). Arabidopsis SPA proteins regulate photoperiodic flowering and interact with the floral inducer CONSTANS to regulate its stability. Development 133 3213–3222. [DOI] [PubMed] [Google Scholar]
- Lee, I., Wolfe, D.S., Nilsson, O., and Weigel, D. (1997). A LEAFY co-regulator encoded by UNUSUAL FLORAL ORGANS. Curr. Biol. 7 95–104. [DOI] [PubMed] [Google Scholar]
- Lin, C., Yang, H., Guo, H., Mockler, T., Chen, J., and Cashmore, A.R. (1998). Enhancement of blue-light sensitivity of Arabidopsis seedlings by a blue light receptor cryptochrome 2. Proc. Natl. Acad. Sci. USA 95 2686–2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, P., Porat, R., Nadeau, J.A., and O'Neill, S.D. (1996). Identification of a meristem L1 layer-specific gene in Arabidopsis that is expressed during embryonic pattern formation and defines a new class of homeobox genes. Plant Cell 8 2155–2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Garcia, J.F., Huq, E., and Quail, P.H. (2000). Direct targeting of light signals to a promoter element-bound transcription factor. Science 288 859–863. [DOI] [PubMed] [Google Scholar]
- Mas, P., Devlin, P.F., Panda, S., and Kay, S.A. (2000). Functional interaction of phytochrome B and cryptochrome 2. Nature 408 207–211. [DOI] [PubMed] [Google Scholar]
- Millar, A.A., Clemens, S., Zachgo, S., Giblin, E.M., Taylor, D.C., and Kunst, L. (1999). CUT1, an Arabidopsis gene required for cuticular wax biosynthesis and pollen fertility, encodes a very-long-chain fatty acid condensing enzyme. Plant Cell 11 825–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mockler, T., Yang, H., Yu, X., Parikh, D., Cheng, Y.C., Dolan, S., and Lin, C. (2003). Regulation of photoperiodic flowering by Arabidopsis photoreceptors. Proc. Natl. Acad. Sci. USA 100 2140–2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mockler, T.C., Guo, H., Yang, H., Duong, H., and Lin, C. (1999). Antagonistic actions of Arabidopsis cryptochromes and phytochrome B in the regulation of floral induction. Development 126 2073–2082. [DOI] [PubMed] [Google Scholar]
- Monte, E., Alonso, J.M., Ecker, J.R., Zhang, Y., Li, X., Young, J., Austin-Phillips, S., and Quail, P.H. (2003). Isolation and characterization of phyC mutants in Arabidopsis reveals complex crosstalk between phytochrome signaling pathways. Plant Cell 15 1962–1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa, M., and Komeda, Y. (2004). Flowering of Arabidopsis cop1 mutants in darkness. Plant Cell Physiol. 45 398–406. [DOI] [PubMed] [Google Scholar]
- Quail, P.H. (2002). Phytochrome photosensory signalling networks. Nat. Rev. Mol. Cell Biol. 3 85–93. [DOI] [PubMed] [Google Scholar]
- Sessions, A., Weigel, D., and Yanofsky, M.F. (1999). The Arabidopsis thaliana MERISTEM LAYER 1 promoter specifies epidermal expression in meristems and young primordia. Plant J. 20 259–263. [DOI] [PubMed] [Google Scholar]
- Simpson, G.G., and Dean, C. (2002). Arabidopsis, the Rosetta stone of flowering time? Science 296 285–289. [DOI] [PubMed] [Google Scholar]
- Smith, H., and Whitelam, G.C. (1997). The shade avoidance syndrome: Multiple responses mediated by multiple phytochromes. Plant Cell Environ. 20 840–844. [Google Scholar]
- Somers, D.E., and Quail, P.H. (1995). Temporal and spatial expression patterns of PHYA and PHYB genes in Arabidopsis. Plant J. 7 413–427. [DOI] [PubMed] [Google Scholar]
- Stadler, R., and Sauer, N. (1996). The Arabidopsis thaliana AtSUC2 gene is specifically expressed in companion cells. Bot. Acta 109 299–306. [Google Scholar]
- Suárez-López, P., Wheatley, K., Robson, F., Onouchi, H., Valverde, F., and Coupland, G. (2001). CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature 410 1116–1120. [DOI] [PubMed] [Google Scholar]
- Susek, R.E., Ausubel, F.M., and Chory, J. (1993). Signal transduction mutants of Arabidopsis uncouple nuclear CAB and RBCS gene expression from chloroplast development. Cell 74 787–799. [DOI] [PubMed] [Google Scholar]
- Takada, S., and Goto, K. (2003). TERMINAL FLOWER2, an Arabidopsis homolog of HETEROCHROMATIN PROTEIN1, counteracts the activation of FLOWERING LOCUS T by CONSTANS in the vascular tissues of leaves to regulate flowering time. Plant Cell 15 2856–2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth, R., Kevei, E., Hall, A., Millar, A.J., Nagy, F., and Kozma-Bognar, L. (2001). Circadian clock-regulated expression of phytochrome and cryptochrome genes in Arabidopsis. Plant Physiol. 127 1607–1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truernit, E., and Sauer, N. (1995). The promoter of the Arabidopsis thaliana SUC2 sucrose-H+ symporter gene directs expression of beta-glucuronidase to the phloem: Evidence for phloem loading and unloading by SUC2. Planta 196 564–570. [DOI] [PubMed] [Google Scholar]
- Usami, T., Mochizuki, N., Kondo, M., Nishimura, M., and Nagatani, A. (2004). Cryptochromes and phytochromes synergistically regulate Arabidopsis root greening under blue light. Plant Cell Physiol. 45 1798–1808. [DOI] [PubMed] [Google Scholar]
- Valverde, F., Mouradov, A., Soppe, W., Ravenscroft, D., Samach, A., and Coupland, G. (2004). Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science 303 1003–1006. [DOI] [PubMed] [Google Scholar]
- Wang, H., Ma, L.G., Li, J.M., Zhao, H.Y., and Deng, X.W. (2001). Direct interaction of Arabidopsis cryptochromes with COP1 in light control development. Science 294 154–158. [DOI] [PubMed] [Google Scholar]
- Weller, J.L., Beauchamp, N., Kerckhoffs, L.H.J., Platten, J.D., and Reid, J.B. (2001). Interaction of phytochromes A and B in the control of de-etiolation and flowering in pea. Plant J. 26 283–294. [DOI] [PubMed] [Google Scholar]
- Yamaguchi, R., Nakamura, M., Mochizuki, N., Kay, S.A., and Nagatani, A. (1999). Light-dependent translocation of a phytochrome B-GFP fusion protein to the nucleus in transgenic Arabidopsis. J. Cell Biol. 145 437–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanovsky, M.J., and Kay, S.A. (2002). Molecular basis of seasonal time measurement in Arabidopsis. Nature 419 308–312. [DOI] [PubMed] [Google Scholar]
- Yoshimoto, N., Inoue, E., Saito, K., Yamaya, T., and Takahashi, H. (2003). Phloem-localizing sulfate transporter, Sultr1;3, mediates re-distribution of sulfur from source to sink organs in Arabidopsis. Plant Physiol. 131 1511–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeevaart, J.A.D. (1976). Physiology of flower formation. Annu. Rev. Plant Physiol. 27 321–348. [Google Scholar]