Abstract
The histone variant H2A.Z has been implicated in numerous chromatin-mediated processes, including transcriptional activation, euchromatin maintenance, and heterochromatin formation. In yeast and humans, H2A.Z is deposited into chromatin by a conserved protein complex known as SWR1 and SRCAP, respectively. Here, we show that mutations in the Arabidopsis thaliana homologs of two components of this complex, ACTIN-RELATED PROTEIN6 (ARP6) and PHOTOPERIOD-INDEPENDENT EARLY FLOWERING1 (PIE1), produce similar developmental phenotypes and result in the misregulation of a common set of genes. Using H2A.Z-specific antibodies, we demonstrate that ARP6 and PIE1 are required for the deposition of H2A.Z at multiple loci, including the FLOWERING LOCUS C (FLC) gene, a central repressor of the transition to flowering. Loss of H2A.Z from chromatin in arp6 and pie1 mutants results in reduced FLC expression and premature flowering, indicating that this histone variant is required for high-level expression of FLC. In addition to defining a novel mechanism for the regulation of FLC expression, these results support the existence of a SWR1-like complex in Arabidopsis and show that H2A.Z can potentiate transcriptional activation in plants. The finding that H2A.Z remains associated with chromatin throughout mitosis suggests that it may serve an epigenetic memory function by marking active genes and poising silenced genes for reactivation.
INTRODUCTION
To ensure reproductive success, plants must align their transition to flowering with favorable environmental conditions. This is achieved through the integration of signals from multiple pathways that sense and respond to various environmental and endogenous cues, such as daylength, temperature, and developmental state (Simpson and Dean, 2002). In Arabidopsis thaliana, several of these signaling pathways converge on the central floral repressor gene FLOWERING LOCUS C (FLC) and either activate or repress its expression to suppress or promote the transition to flowering, respectively (Henderson and Dean, 2004; Simpson, 2004; Sung and Amasino, 2004). The terminal effectors of these pathways controlling FLC expression are now known to represent many types of chromatin-modifying factors, including histone acetyltransferases, histone deacetylases, histone methyltransferases, polycomb-type proteins, and a putative histone demethylase (He and Amasino, 2005; Reyes, 2006). Thus, FLC serves as a model for how chromatin remodeling and modification can regulate a critical developmental switch. In contrast with histone modifications, there has been no evidence to date for the role of histone variants in the control of FLC expression.
The fundamental repeating unit of chromatin, known as the nucleosome, consists of ∼150 bp of DNA wrapped around a protein particle composed of two copies of each of the four core histones: H2A, H2B, H3, and H4. These histone proteins are encoded by multiple gene copies and are produced in large quantities to accommodate the nascent genome during DNA replication. In addition to these bulk histones, eukaryotic genomes also encode variant histones that are deposited independently of DNA replication and serve to functionally specialize or differentiate specific chromatin regions. In Saccharomyces, Drosophila, and human, the histone variant H2A.Z is deposited into chromatin by a conserved protein complex known as SWR1, Tip60, and SRCAP, respectively (Krogan et al., 2003; Kobor et al., 2004; Kusch et al., 2004; Mizuguchi et al., 2004; Ruhl et al., 2006). The yeast H2A.Z variant plays important roles in both activating transcription and antagonizing the spread of heterochromatin into euchromatic regions (Santisteban et al., 2000; Adam et al., 2001; Larochelle and Gaudreau, 2003; Meneghini et al., 2003). Several genome-wide studies of yeast H2A.Z occupancy have shown that this variant localizes to the promoters of thousands of euchromatic genes, and its presence is required for the optimal transcriptional activation of many of these genes (Guillemette et al., 2005; Li et al., 2005; Raisner et al., 2005; Zhang et al., 2005). In contrast with the situation in yeast, cytological and genetic studies suggest that in metazoans H2A.Z may also play a role in heterochromatin formation and/or maintenance (Rangasamy et al., 2003; Swaminathan et al., 2005), although this variant has been observed at the 5′ ends of several active genes in human and chicken cell lines (Bruce et al., 2005; Farris et al., 2005). However, null mutations in mouse and Drosophila H2A.Z result in embryonic lethality (van Daal and Elgin, 1992; Faast et al., 2001), making it difficult to study the developmental functions of H2A.Z in these organisms.
Previous work in Arabidopsis showed that loss-of-function mutations in two genes encoding putative homologs of components of the SWR1/SRCAP complex, ACTIN-RELATED PROTEIN6 (ARP6) and the Snf2 protein PHOTOPERIOD-INDEPENDENT EARLY FLOWERING1 (PIE1), led to premature flowering as a result of reduced FLC expression (Noh and Amasino, 2003; Choi et al., 2005; Deal et al., 2005; Martin-Trillo et al., 2006). However, the mechanism by which these proteins regulate FLC expression has not been addressed. Here, we provide evidence for the existence of a SWR1-like complex in plants and show that ARP6 and PIE1 are both required for the deposition of H2A.Z into chromatin at FLC and the FLC homologous genes MADS AFFECTING FLOWERING4 (MAF4) and MAF5. Loss of H2A.Z from chromatin in arp6 and pie1 mutants results in reduced expression of FLC, MAF4, and MAF5, indicating that H2A.Z acts to potentiate the transcriptional activation of these genes. Thus, in addition to the host of chromatin-modifying factors identified previously, the histone variant H2A.Z and its deposition machinery define a novel mechanism for promoting FLC expression, thereby ensuring the proper timing of the transition from vegetative growth to flowering.
RESULTS
arp6 and pie1 Mutants Have Similar Developmental and Molecular Phenotypes
To investigate the similarities in phenotype caused by disruption of the ARP6 and PIE1 genes, we compared the null mutants arp6-1 (Deal et al., 2005) and pie1-5 (Figure 1). Our observations indicated that arp6 and pie1 showed a strikingly similar array of developmental phenotypes, including reduced leaf size and early flowering (Figure 2A). In addition to these phenotypes, real-time RT-PCR analysis showed that a common set of seven genes was downregulated in both arp6-1 and pie1-5 mutants (Figure 2B). Among these genes was the MADS box floral repressor FLC (Michaels and Amasino, 1999), which was reduced by ∼10-fold in each mutant compared with the wild type. Expression of the FLC homologs MAF4 and MAF5 (Ratcliffe et al., 2003) was also decreased in arp6-1 and pie1-5, but the reduction of both transcripts was more severe in the pie1-5 mutant, suggesting that PIE1 has additional effects on these genes that are independent of ARP6. Preliminary microarray experiments comparing wild-type and arp6-1 plants (R.B. Deal and R.B. Meagher, unpublished data) revealed a large number of genes whose expression was disrupted by the loss of ARP6. As such, we chose to examine the expression of several of these candidate genes in both arp6-1 and pie1-5 by real-time RT-PCR. We found that both mutants were defective in the expression of a CONSTANS-LIKE (COL) transcription factor, the WRK70 transcription factor, and the putative disease resistance genes PRB1 and TIR (Figure 2B). We also found several transcripts that were unchanged in the mutants and one whose expression was increased in both arp6-1 and pie1-5 (data not shown). The common developmental phenotypes and gene expression defects observed in these mutants suggested that ARP6 and PIE1 act in a common pathway and perhaps in the same protein complex.
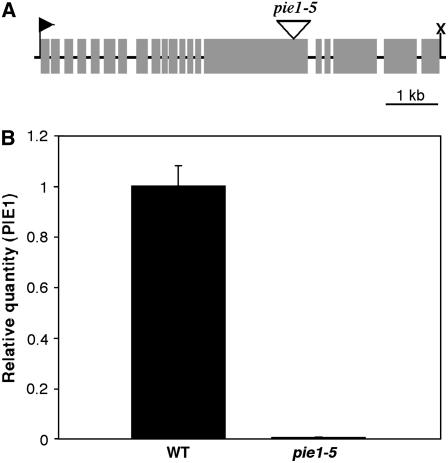
Figure 1.
The pie1-5 Mutant Allele Is Null.
(A) Diagram of the PIE1 gene. The transcription start site is shown as a right-facing arrowhead, and the termination site is indicated by an X. Exons are depicted as gray boxes. The location of the T-DNA insertion in pie1-5 is shown as a triangle above exon 15.
(B) Relative quantity of the PIE1 transcript in wild-type and pie1-5 plants as detected by real-time RT-PCR. The transcript is essentially undetectable in the pie1-5 mutant, indicating that the allele is null. Error bars represent se (n = 3).
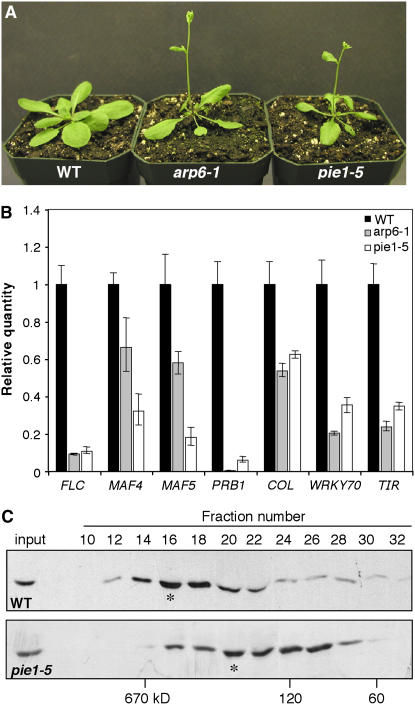
Figure 2.
arp6 and pie1 Mutants Exhibit Similar Phenotypes.
(A) Twenty-day-old wild-type, arp6-1, and pie1-5 plants grown under long-day conditions.
(B) Real-time RT-PCR data showing relative expression of the indicated genes in wild-type, arp6-1, and pie1-5 plants. Average relative quantities ± se (n = 3) are shown for each sample.
(C) Protein gel blots of even-numbered gel filtration fractions from a Sephacryl S-300 column run with extract from wild-type (top panel) or pie1-5 (bottom panel) plants. Blots were probed with an ARP6 monoclonal antibody. The input lanes in each blot were loaded with ∼25 μg of the unfractionated protein extract from each genotype. Asterisks indicate the ARP6 peak fractions, and calibrated molecular masses are given below the blots.
We next sought to determine whether the ARP6 and PIE1 proteins were actually part of the same protein complex, as predicted based on their homology with yeast SWR1 complex components. Using gel-filtration chromatography and an ARP6 monoclonal antibody (Deal et al., 2005), we found that the major native form of ARP6 migrated with a molecular mass of ∼600 kD. In these experiments, the 50-kD monomeric ARP6 was not detectable as a discrete peak, suggesting that ARP6 exists mainly as part of a protein complex in vivo (Figure 2C). Based on a previous genomic survey, the observed mass of 600 kD is consistent with the expected size of an Arabidopsis SWR1 complex (Meagher et al., 2005). When the same experiment was performed with pie1-5 mutant extracts, the peak of native ARP6 shifted down to ∼250 kD (Figure 2C), as would be expected if the 230-kD PIE1 protein, and perhaps other subunits, were lost from the complex. These results strongly suggested that PIE1 and ARP6 are normally part of the same protein complex and that at least some of the components of the complex remain associated with ARP6 in the absence of PIE1. This result is consistent with the previous finding that yeast ARP6 forms a subcomplex with the Swc2, Swc3, and Swc6 components of the SWR1 complex (Wu et al., 2005).
ARP6 Interacts with H2A.Z
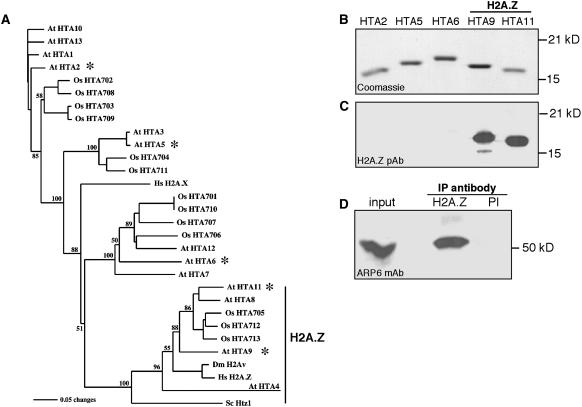
The Arabidopsis genome encodes 13 H2A histones that fall into four distinct phylogenetic classes, each of which is also conserved in the monocot Oryza sativa. Of these 13 H2As, 4 are clearly H2A.Z variants, being more closely related to yeast and metazoan H2A.Z proteins than to other plant H2As (Figure 3A). Using synthetic peptides as antigens, we raised polyclonal antibodies that reacted with the relatively conserved N termini of the Arabidopsis H2A.Z subclass proteins HTA9 and HTA11 but not with representatives of any of the other three H2A subclasses (Figures 3B and 3C). Immunoprecipitations performed on plant protein extracts with the H2A.Z-specific antibodies efficiently precipitated ARP6, whereas parallel experiments using preimmune rabbit serum as primary antibody failed to precipitate ARP6 (Figure 3D). These results confirmed that ARP6 interacts with H2A.Z, either directly or indirectly, and further support the notion that ARP6 is involved in the deposition of H2A.Z into chromatin.
Figure 3.
H2A Phylogeny and H2A.Z-Specific Antibodies.
(A) Neighbor-joining protein sequence phylogeny showing H2A proteins from Arabidopsis (At), rice (Os), Drosophila (Dm), human (Hs), and Saccharomyces (Sc). Bootstrap values are shown on the branch points of the tree. The H2A.Z clade is indicated by a vertical bar at right, and asterisks indicate proteins used in (B) and (C).
(B) Coomassie blue–stained SDS-PAGE gel showing purified recombinant Arabidopsis H2A histones. The H2A.Z variants are indicated by a horizontal bar above the protein names. Molecular masses are shown at right.
(C) Protein gel blot of a gel loaded as in (B), probed with the H2A.Z-specific antibody. Molecular masses are shown at right. The polyclonal antibody (pAb) recognized HTA9 and HTA11 and was predicted to react with HTA8, because it is nearly identical to HTA11 in the N-terminal region. The HTA4 protein is highly divergent from the other H2A.Zs at the N terminus and is not expected to be recognized by the antibody.
(D) Protein gel blot of immunoprecipitates probed with an ARP6 monoclonal antibody (mAb). The input sample was 5% of the total protein used in each immunoprecipitation. The antibody used for immunoprecipitation is indicated above the blot: either the H2A.Z antibody or preimmune serum (PI) from the same rabbit.
H2A.Z Is Excluded from Chromocenters and Retained in Mitotic Chromosomes
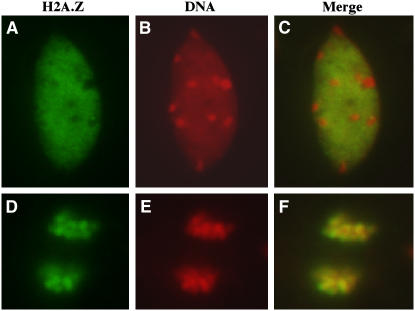
To examine the distribution of H2A.Z in chromatin, we performed immunolocalization on isolated leaf and root cells with the H2A.Z antibody. We found that H2A.Z was dispersed widely throughout the nucleus but was conspicuously absent from the chromocenters (Figures 4A to 4C), which are composed of highly condensed heterochromatin containing centromeric and pericentromeric repeats (Fransz et al., 2002). This finding indicated that H2A.Z is found mainly in euchromatic regions and is likely responsible for regulating the expression of many genes. The incorporation pattern observed is in contrast with that of mammalian and Drosophila H2A.Zs, which accumulate in both heterochromatic and euchromatic chromatin (Leach et al., 2000; Rangasamy et al., 2003). In dividing cells, we observed H2A.Z throughout the condensed chromosomes (Figures 4D to 4F), indicating that this variant remains in the chromatin through mitosis and therefore may serve as a stable epigenetic mark. From these data, we cannot yet discern whether H2A.Z is simply inherited through DNA replication or is reincorporated after replication, before mitosis.
Figure 4.
H2A.Z Localizes to Euchromatic Regions but Not to Heterochromatic Chromocenters.
(A) Isolated leaf cell nucleus probed with the H2A.Z antibody.
(B) The 4′,6-diamidino-2-phenylindole (DAPI) channel image of the nucleus shown in (A). Chromocenters appear as densely stained spots throughout the nucleus.
(C) Merge of images shown in (A) and (B).
(D) Anaphase-stage root cell probed with the H2A.Z antibody.
(E) DAPI channel image of the cell shown in (D).
(F) Merge of images shown in (D) and (E).
ARP6 and PIE1 Are Required for H2A.Z Deposition
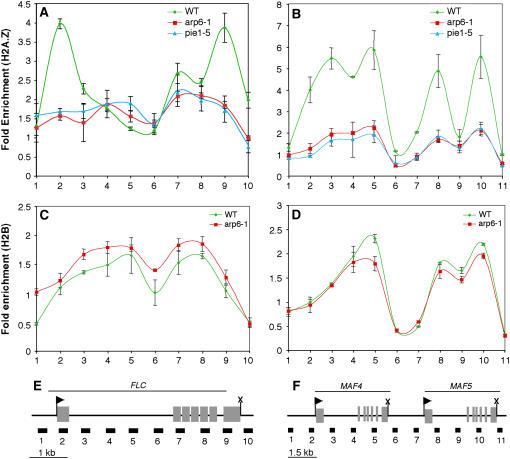
We next sought to examine the possibility that ARP6 and PIE1 were involved in the deposition of H2A.Z at the FLC gene, because FLC expression was greatly reduced in both arp6-1 and pie1-5 (Figure 2B). Chromatin immunoprecipitation (ChIP) using the Arabidopsis H2A.Z antibody was used to examine H2A.Z abundance across the FLC gene in wild-type and mutant plants. In wild-type plants, H2A.Z was predominantly enriched in two discrete domains, one at each end of the transcribed region, with relatively low H2A.Z levels between these two domains (Figures 5A and 5E). By contrast, the arp6-1 and pie1-5 mutants showed very little H2A.Z enrichment across the FLC gene, and the pattern of enrichment was nearly indistinguishable between the two mutants (Figure 5A).
Figure 5.
ARP6 and PIE1 Are Required for the Deposition of H2A.Z at FLC, MAF4, and MAF5.
(A) Enrichment of H2A.Z on the FLC gene in the wild type and mutants as measured by ChIP with the H2A.Z antibody. The graph shows average fold enrichment ± se as measured by real-time PCR.
(B) Enrichment of H2A.Z on the MAF4 and MAF5 genes in the wild type and mutants as measured by ChIP and real-time PCR.
(C) Enrichment of H2B on the FLC gene in the wild type and arp6-1.
(D) Enrichment of H2B on the MAF4 and MAF5 genes in the wild type and arp6-1. For each ChIP experiment ([A] to [D]), n = 3.
(E) Diagram of the FLC gene with exons indicated as gray boxes. The transcription start site is shown as an arrowhead, and the termination site is shown as an X. PCR primer sets are shown as black boxes below the diagram. Primer set numbers correspond to the numbers on the x axis of the graphs in (A) and (C).
(F) Diagram of the MAF4 and MAF5 genes and locations of PCR primer sets, depicted as in (E). Primer set numbers correspond to the numbers on the x axis of the graphs in (B) and (D).
ChIP experiments examining the MAF4 and MAF5 genes in wild-type plants revealed a pattern of H2A.Z distribution similar to that found at FLC, with the highest levels near the beginning and end of each transcribed region (Figures 5B and 5F). In the case of MAF4, we observed only a small decrease in H2A.Z over the middle of the transcribed region, whereas H2A.Z levels were quite low in the middle of the MAF5 gene (Figures 5B and 5F). Again, in contrast with the wild type, there was little or no H2A.Z accumulation across the MAF4 and MAF5 genes in arp6-1 and pie1-5. Control ChIP experiments using a histone H2B antibody, which should react with all nucleosomes, showed no significant difference in overall H2B distribution between the wild type and arp6-1 on FLC, MAF4, or MAF5 (Figures 5C and 5D). Thus, the observed differences in H2A.Z enrichment on these genes were not simply attributable to gross differences in nucleosome occupancy among genotypes. These results indicated that ARP6 and PIE1 were both required for the deposition of H2A.Z at the three genes examined, supporting the existence of a SWR1/SRCAP-like complex in Arabidopsis. In addition, H2A.Z is normally distributed across the FLC and MAF genes with the highest levels at the beginning and end of the transcribed regions, and loss of H2A.Z from chromatin in the mutants is correlated with decreased gene expression and early flowering. Thus, Arabidopsis H2A.Z is required for high-level expression of the FLC, MAF4, and MAF5 genes, suggesting that it plays a role similar to yeast H2A.Z in promoting transcription.
H2A.Z Occupancy Is Inversely Correlated with FLC Transcript Levels
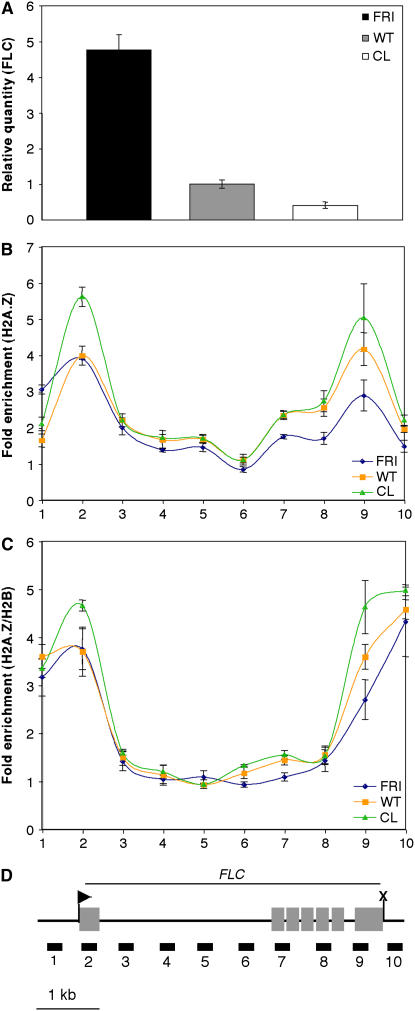
To gain insight into the role of H2A.Z in promoting the high-level expression of FLC, we examined H2A.Z distribution in plant samples with widely different FLC transcript levels. H2A.Z accumulation on FLC was measured in wild-type cauline leaves, 10-d-old wild-type shoots, and 10-d-old shoots carrying the strong FLC activator FRIGIDA (FRI) (Lee and Amasino, 1995). Among these samples, the FRI-expressing line showed the highest levels of FLC expression, the wild type was intermediate, and cauline leaves had a 10-fold lower level of FLC than did the FRI line (Figure 6A). Interestingly, the spatial distribution of H2A.Z on FLC was the same in each sample, and the overall levels of H2A.Z in each were also surprisingly similar, showing only minor differences except at the 5′ and 3′ ends of the gene, where an inverse correlation between transcript level and H2A.Z occupancy was observed (Figures 6B and 6D). This correlation was most clear at the 3′ end of the gene.
Figure 6.
Relationship between FLC Expression Levels and H2A.Z Abundance on the Gene.
(A) Real-time RT-PCR results showing the relative FLC level in each genotype or tissue. FRI indicates a Columbia line carrying the strong FRI allele from the Sf-2 ecotype (Lee and Amasino, 1995), WT indicates Columbia wild type, and CL indicates cauline leaf. The graph shows average relative quantities ± se (n = 3).
(B) Enrichment of H2A.Z on the FLC gene in the indicated samples as measured by ChIP and real-time PCR. The graph shows average fold enrichment ± se.
(C) ChIP was performed on the indicated samples using either an H2A.Z antibody or an H2B antibody, and data are reported as H2A.Z enrichment/H2B enrichment to correct the H2A.Z levels for total nucleosome occupancy. The graph shows average fold enrichment ± se. For each ChIP experiment ([B] and [C]), n = 3.
(D) Diagram of the FLC gene with exons indicated as gray boxes. The transcription start site is shown as an arrowhead, and the termination site is shown as an X. PCR primer sets are shown as black boxes below the diagram. Primer set numbers correspond to the numbers on the x axis of the graphs in (B) and (C).
Because nucleosome occupancy on FLC might have differed among these samples, we performed parallel ChIP experiments with an H2B antibody to quantify total nucleosome distribution, allowing us to measure H2A.Z enrichment relative to that of H2B. We found that the H2A.Z:H2B ratio across the FLC gene was also nearly indistinguishable among the three samples at most sites tested, but the H2A.Z:H2B ratio at the 5′ and 3′ ends of the gene still showed an inverse correlation with FLC transcript levels (Figures 6C and 6D). Thus, the higher the transcript level, the less H2A.Z was present at the 5′ and 3′ ends of the gene, similar to the trend observed previously for yeast H2A.Z at the 5′ ends of genes (Guillemette et al., 2005; Zhang et al., 2005). Collectively, our results indicate that H2A.Z is required for the high-level expression of FLC but that its presence does not directly induce transcriptional activation, suggesting that this variant poises the gene in a state competent for activation by other factors.
DISCUSSION
Previous studies of Arabidopsis ARP6 and PIE1 revealed that these proteins were both involved in regulating multiple developmental processes, including leaf development, inflorescence and flower development, and repression of the transition to flowering. In the case of flowering time control, both ARP6 and PIE1 were shown to be required for high-level expression of the floral repressor gene FLC (Noh and Amasino, 2003; Choi et al., 2005; Deal et al., 2005; Martin-Trillo et al., 2006), indicating that these proteins were likely involved in transcriptional regulation. Concurrent with the elucidation of ARP6 and PIE1 function in Arabidopsis, several groups discovered that in Saccharomyces cerevisiae, ARP6 was a component of the SWR1 chromatin-remodeling complex (Krogan et al., 2003; Kobor et al., 2004; Mizuguchi et al., 2004). This complex was shown to have the novel activity of replacing histone H2A with the variant H2A.Z in particular nucleosomes, thus functionally specializing the surrounding chromatin and in many cases potentiating the expression of nearby genes. Given this information, we considered the possibility that such a complex might also exist in Arabidopsis. A comparison of the yeast SWR1 ATPase subunit with all Arabidopsis proteins indicated that of the 42 Swi2/Snf2 family proteins encoded by the Arabidopsis genome, PIE1 was the most closely related to SWR1. In addition to ARP6 and PIE1, the Arabidopsis genome also encodes clear orthologs of most other SWR1 complex components (Meagher et al., 2005) as well as multiple H2A.Z isovariants (Figure 3). Could the developmental functions of Arabidopsis ARP6 and PIE1 be attributable to their activity within a plant SWR1-like complex? In yeast, the SWR1 complex deposits the histone H2A.Z variant into euchromatic regions near telomeres (Krogan et al., 2003; Kobor et al., 2004; Mizuguchi et al., 2004) and in the promoters of many euchromatic genes (Guillemette et al., 2005; Li et al., 2005; Raisner et al., 2005; Zhang et al., 2005), thereby preventing the spread of silent heterochromatin into euchromatic regions (Meneghini et al., 2003) and promoting transcriptional activation, respectively. Perhaps a plant SWR1-like complex could have an analogous function of depositing H2A.Z into FLC chromatin, ensuring the competence of the gene for high-level expression and thus allowing the flowering program to be repressed and vegetative growth to continue. In this investigation, we explored the hypothesis that the contribution of ARP6 and PIE1 to the developmental program in Arabidopsis was a manifestation of their role in depositing H2A.Z into chromatin and thus regulating the expression of multiple developmentally important genes.
A direct comparison of arp6 and pie1 indicated that these mutants shared many developmental and molecular phenotypes. Both mutations caused aberrations in leaf development and early flowering and resulted in the misregulation of a common set of genes, including the flowering regulators FLC, MAF4, and MAF5. In addition, we found that ARP6 was a component of a high-molecular-mass protein complex and that the size of this complex was reduced dramatically in the absence of PIE1, suggesting that ARP6 and PIE1 were indeed part of the same protein complex (Figure 2). Furthermore, polyclonal antibodies that reacted with at least two of the four Arabidopsis H2A.Z proteins efficiently immunoprecipitated ARP6 from plant extracts, confirming an interaction between ARP6 and H2A.Z (Figure 3). Collectively, these results suggested that ARP6 and PIE1 were part of a plant SWR1-like complex.
ChIP was used to determine whether ARP6 and PIE1 were involved in the deposition of H2A.Z into chromatin at the FLC and MAF genes, because the expression of these genes was reduced in both arp6 and pie1 mutants. We found that H2A.Z was normally enriched in two discrete domains on each of these genes, one near the transcription start site and another near the end of the gene, and that very little H2A.Z accumulated in the chromatin of arp6 or pie1 mutants (Figure 5). These results indicated that ARP6 and PIE1 were indeed necessary for the deposition of H2A.Z into chromatin at the three loci examined. However, H2A.Z was still detectable at several sites on the FLC and MAF genes in arp6-1and pie1-5, indicating that the variant can be incorporated into chromatin at very low levels independently of ARP6 and PIE1. Loss of H2A.Z from chromatin in arp6-1 and pie1-5 was correlated with a reduced expression of FLC, MAF4, and MAF5, indicating that H2A.Z is normally required to promote the expression of each of these genes. This observation suggested that, like yeast H2A.Z, the Arabidopsis variant can also potentiate transcriptional activation. ChIP analysis of the PRB1, COL, WRKY70, and TIR genes, whose transcript levels were also reduced in the mutants (Figure 2B), indicated that even in wild-type plants these loci did not accumulate significant amounts of H2A.Z (data not shown); thus, these genes were likely to be indirectly regulated by ARP6 and PIE1. This finding indicated that not all Arabidopsis genes require H2A.Z for high-level expression and that many of the transcriptional defects in arp6 and pie1 mutants were likely secondary effects resulting from the misregulation of a smaller number of primary target genes that do require H2A.Z for proper expression.
In contrast with the situation in yeast, in which H2A.Z is normally found mainly in nucleosomes around the transcription start site (Guillemette et al., 2005; Li et al., 2005; Raisner et al., 2005; Zhang et al., 2005), Arabidopsis H2A.Z occupies regions near both the transcription start and termination sites on the three genes examined. Previous studies have shown a role for H2A.Z in recruiting RNA polymerase (Adam et al., 2001) and acting in concert with nucleosome-remodeling complexes (Santisteban et al., 2000) to initiate transcription. However, the data presented here suggest that in Arabidopsis this histone variant also has a function beyond the 5′ end of genes, perhaps in facilitating transcript elongation and/or termination. The tendency of H2A.Z to destabilize nucleosomes that contain it (Suto et al., 2000; Abbott et al., 2001; Zhang et al., 2005) may facilitate nucleosome remodeling and thus polymerase passage and transcript elongation. In addition, multiple lines of evidence now indicate that chromatin-remodeling enzymes regulate all phases of the transcription cycle, including termination (Alen et al., 2002; Morillon et al., 2003). Thus, Arabidopsis H2A.Z may facilitate remodeling at both ends of the gene to effect the proper initiation and termination of transcription. The occupancy of H2A.Z at sites beyond the transcription initiation region has also been observed for several other Arabidopsis genes (A.P. Smith, R.B. Deal, and R.B. Meagher, unpublished data), suggesting that this phenomenon is not specific to the MADS box transcription factor genes.
As a means of gaining insight into the role of H2A.Z in promoting transcriptional activation, we examined the relationship between FLC transcript levels and H2A.Z abundance on the FLC gene. H2A.Z occupancy on FLC was measured in three different tissues with FLC expression levels spanning a 10-fold range, from very high to very low. In each sample, we observed essentially the same spatial distribution of H2A.Z across the gene, with a peak at the beginning and end of the transcribed region. However, there was an inverse correlation between FLC expression levels and H2A.Z abundance on the gene, such that the higher the transcript level, the less H2A.Z was present on the gene (Figure 6). This inverse relationship between H2A.Z occupancy and gene expression may result from a shift in the balance between ARP6/PIE1-mediated deposition of H2A.Z and loss of H2A.Z as a result of nucleosome disruption by RNA polymerase. During high-level transcription, the rate of transcription-induced depletion of H2A.Z might exceed the rate of replacement by ARP6/PIE1, resulting in an inverse correlation between transcript level and H2A.Z occupancy. In any case, these results clearly indicated that there was no positive correlation between FLC expression level and H2A.Z abundance on the gene, even over a 10-fold range of FLC transcript levels. This finding suggests that the H2A.Z variant serves to poise the gene in a state competent for activation by other factors, rather than activating transcription directly. This may reflect the ability of H2A.Z to facilitate nucleosome remodeling (Santisteban et al., 2000) and/or to recruit the transcription machinery (Adam et al., 2001) or other activators to allow high-level transcription under the appropriate conditions. Thus, in the absence of H2A.Z in arp6 and pie1 mutants, FLC levels remain low even in the presence of strong activators such as FRI (Noh and Amasino, 2003; Choi et al., 2005; Deal et al., 2005; Martin-Trillo et al., 2006), resulting in early flowering.
In addition to ChIP studies on the distribution of H2A.Z across single genes, we also used our H2A.Z antibodies to examine the nuclear localization of this histone variant during interphase and mitosis. In contrast with mammalian and Drosophila H2A.Zs, which accumulate in both heterochromatic and euchromatic chromatin (Leach et al., 2000; Rangasamy et al., 2003), we found that Arabidopsis H2A.Z was widely distributed throughout euchromatin but was excluded from the heterochromatic chromocenters (Figure 4). This finding indicated that H2A.Z is likely responsible for regulating the expression of many genes in euchromatin but is not likely involved in constitutive heterochromatin formation or maintenance in Arabidopsis. The observation that H2A.Z is incorporated into euchromatin during interphase and remains in chromatin through mitosis suggests that it may serve an epigenetic memory function by marking actively transcribed genes and providing competence for the reactivation of silenced genes. Such a function, coupled with the ability of H2A.Z to promote transcriptional activation, is likely to be important in the establishment and maintenance of cell fate during development.
In conclusion, it appears that ARP6 and PIE1 act together to control multiple developmental processes, most likely by regulating the expression of a large number of genes through the incorporation of H2A.Z into chromatin. Our results support the notion that the H2A.Z deposition machinery is conserved in plants as it is in yeast and metazoans, requiring both ARP6 and a Snf2 protein of the SWR1/SRCAP class. Furthermore, the loss of H2A.Z from chromatin in arp6-1 and pie1-5 mutants correlates with decreased expression of FLC, MAF4, and MAF5, indicating that H2A.Z deposition is essential for the full transcriptional activation of these genes. Thus, H2A.Z can act to potentiate the transcriptional activation of Arabidopsis genes, similar to its role in yeast. In terms of flowering time control, H2A.Z and its deposition machinery now define a novel mechanism for promoting FLC expression and thus repressing the transition from vegetative to reproductive development in Arabidopsis. During vegetative growth, H2A.Z allows high-level FLC expression and floral repression, yet it remains associated with the inactive gene after flowering, perhaps to poise the gene for reactivation and reestablishment of the vegetative growth program in the next generation.
Future work in this area will be focused on large-scale approaches to identifying all of the genes that require H2A.Z for proper expression and elucidating the DNA sequence determinants and other factors that promote the deposition of H2A.Z into chromatin at particular loci. Insight into the mechanism by which H2A.Z promotes transcriptional activation could be gleaned from protein–protein interaction studies and the identification of second-site mutations that suppress the early flowering of arp6 and pie1 mutants. Because ARP6 and PIE1 act as a hub through which the expression of many genes is controlled, identification of the full set of genes misregulated in arp6 and pie1 mutants should allow the assignment of many currently anonymous genes to particular developmental pathways.
METHODS
Plants and Growth Conditions
Arabidopsis thaliana plants were of the Columbia ecotype and were germinated by sowing on wet soil and storing at 4°C for 2 d before moving to the growth chamber. Plants were grown at 22°C under 16 h of light per day. The pie1-5 mutation is a T-DNA insertion allele from the Salk Institute (SALK_011204). The T-DNA insertion is in exon 15 and is null based on RNA levels (Figure 1). The arp6-1 allele was described previously (Deal et al., 2005).
Gel Filtration Chromatography
Gel filtration was performed on a Sephacryl S-300HR column (Amersham Biosciences) in a buffer consisting of 20 mM Tris, pH 7.5, 200 mM NaCl, 10 mM MgCl2, and 10% (v/v) glycerol. The column was calibrated with a mixture of standard proteins ranging in size from 670 to 60 kD. Plant extracts were prepared by grinding a mixture of leaf and flower tissue in 2 volumes of the gel filtration buffer supplemented with 1 mM β-mercaptoethanol and Complete protease inhibitors (Roche). After grinding, samples were cleared by centrifugation and filtration. The column was run at room temperature with a flow rate of 0.25 mL/min, and 0.5-mL fractions were collected. Two independent biological replicates of the gel filtration experiment were done, and both gave very similar results.
Phylogenetic Analysis
The following Arabidopsis histone H2A protein sequences were used for phylogenetic analysis: HTA1 (At5g54640), HTA2 (At4g27230), HTA3 (At1g54690), HTA4 (At4g13570), HTA5 (At1g08880), HTA6 (At5g59870), HTA7 (At5g27670), HTA8 (At2g38810), HTA9 (At1g52740), HTA10 (At1g51060), HTA11 (At3g54560), HTA12 (At5g02560), and HTA13 (At3g20670). Other H2A proteins used for the analysis were from Oryza sativa: HTA701 (Os01g31800), HTA702 (Os08g33100), HTA703 (Os12g25120), HTA704 (Os03g51200), HTA705 (Os10g28230), HTA706 (Os05g38640), HTA707 (Os05g02300), HTA708 (Os07g36140), HTA709 (Os07g36130), HTA710 (Os03g17100), HTA711 (Os12g34510), HTA712 (Os03g06670), and HTA713 (Os03g53190); Homo sapiens: Hs H2A.X (NP_002096) and Hs H2A.Z (NP_002097); Drosophila melanogaster: Dm H2Av (NP_524519); and Saccharomyces cerevisiae: Sc Htz1 (NP_014631). Sequences were aligned with ClustalW 1.82 (Higgins and Sharp, 1988) using default settings (protein gap open penalty = 10, protein gap extension penalty = 0.2, protein matrix = gonnet, protein ENDGAP = −1, protein GAPDIST = 4), and phylogenies were constructed with PAUP 4.0 (Swofford, 2003) using the neighbor-joining method. Ties were broken randomly using the initial seed function. Bootstrap support values were based on 1000 replicates using a full heuristic search.
Antibody Preparation
Polyclonal antibodies specific to Arabidopsis H2A.Zs were produced in rabbits by injecting peptides representing the N termini of HTA9 and HTA11. The peptide sequences N-SGKGAKGLIMGKPSGSDKDKDKKKPIT-C (HTA9) and N-AGKGGKGLVAAKTMAANKDKDKDKKKPIS-C (HTA11) were synthesized as fourfold multiple antigenic peptides. The primary injection and three subsequent boosts were done with 250 μg of each peptide. Antibodies were affinity-purified on resin to which the H2A.Z peptides had been coupled.
Immunoprecipitation
Plant extracts were prepared by grinding a mixture of leaf and flower tissue in 2 volumes of immunoprecipitation buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 10 mM MgCl2, 0.1% Nonidet P-40, 1 mM β-mercaptoethanol, and Roche Complete protease inhibitors) followed by centrifugation and filtration to clear the extracts. Extracts were then divided into aliquots in multiple tubes (900 μL each), and 40 μL of protein A–agarose beads (Roche) was added to each tube. Tubes were rocked at 4°C for 1 h to preclear the extracts, and then the beads were removed by centrifugation. Antibodies or preimmune serum were then added, and tubes were kept on ice for 1 h to allow antibody binding. H2A.Z antibodies were used at a 1:100 dilution, and an equivalent amount of preimmune serum was used based on antibody concentration. Antibody–antigen complexes were captured by adding 50 μL of protein A–agarose beads and rocking for 1 h at 4°C. Beads were collected by centrifugation and washed three times for 20 min in immunoprecipitation buffer at 4°C.
Protein Gel Blotting and Immunofluorescence Microscopy
Both of these techniques were performed as described previously (Deal et al., 2005).
RT-PCR
RNA was isolated from 10-d-old seedlings (minus roots) using the RNeasy plant mini kit (Qiagen). Before reverse transcription, RNA was treated with RQ1 RNase-free DNase I (Promega) according to the manufacturer's instructions. Three micrograms of each RNA was reverse-transcribed with the Superscript III first-strand synthesis kit (Invitrogen). Real-time PCR was used to analyze the cDNA populations using 18S RNA as an endogenous control. The genes assayed by this method were FLC (At5g10140), MAF4 (At5g65070), MAF5 (At5g65080), PRB1 (At2g14580), COL (At5g24930), WRKY70 (At3g56400), and TIR (At1g72930).
ChIP
ChIP was performed as described previously (Gendrel et al., 2005). For each experiment, 1 to 2 g of 10-d-old seedlings (minus roots) was used. For experiments with cauline leaves, 1 to 2 g of leaves from 30-d-old plants was used. The H2A.Z antibody was used at a dilution of 1:100, and the H2B antibody (ab1790; Abcam) was used at a dilution of 1:150. DNA was analyzed by real-time PCR with the ACT2 3′ untranslated region sequence as the endogenous control for all ChIP experiments. The relative quantity value calculated by the 2−ddCt method in ChIP experiments is reported as fold enrichment.
Real-Time PCR
Real-time PCR was performed on an Applied Biosystems 7500 real-time PCR system using SYBR Green detection chemistry (Applied Biosystems). The 2−ddCt method (Livak and Schmittgen, 2001) of relative quantification was used in all experiments. The data presented for both RT-PCR and ChIP experiments are average relative quantities from at least two biological replicates ± se. All primer sequences are available upon request.
Accession Numbers
The accession numbers for the ARP6 and PIE1 genes are At3g33520 and At3g12810, respectively.
Supplemental Data
The following material is available in the online version of this article.
Supplemental Figure 1. Histone H2A Alignments Used for Phylogenetic Analysis.
Supplementary Material
Acknowledgments
We thank Michael Bender and members of the Meagher laboratory for critical reading and discussion of the manuscript. This work was supported by funding from the National Institutes of Health (Grant GM-36397-18) to R.B.M. and a National Institutes of Health training grant (Grant GM-07103-29) to R.B.D. C.N.T. was supported by funding from the National Science Foundation (Grant 0421671).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Richard B. Meagher (meagher@uga.edu).
Online version contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Abbott, D.W., Ivanova, V.S., Wang, X., Bonner, W.M., and Ausio, J. (2001). Characterization of the stability and folding of H2A.Z chromatin particles: Implications for transcriptional activation. J. Biol. Chem. 276 41945–41949. [DOI] [PubMed] [Google Scholar]
- Adam, M., Robert, F., Larochelle, M., and Gaudreau, L. (2001). H2A.Z is required for global chromatin integrity and for recruitment of RNA polymerase II under specific conditions. Mol. Cell. Biol. 21 6270–6279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alen, C., Kent, N.A., Jones, H.S., O'Sullivan, J., Aranda, A., and Proudfoot, N.J. (2002). A role for chromatin remodeling in transcriptional termination by RNA polymerase II. Mol. Cell 10 1441–1452. [DOI] [PubMed] [Google Scholar]
- Bruce, K., Myers, F.A., Mantouvalou, E., Lefevre, P., Greaves, I., Bonifer, C., Tremethick, D.J., Thorne, A.W., and Crane-Robinson, C. (2005). The replacement histone H2A.Z in a hyperacetylated form is a feature of active genes in the chicken. Nucleic Acids Res. 33 5633–5639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, K., Kim, S., Kim, S.Y., Kim, M., Hyun, Y., Lee, H., Choe, S., Kim, S.G., Michaels, S., and Lee, I. (2005). SUPPRESSOR OF FRIGIDA3 encodes a nuclear ACTIN-RELATED PROTEIN6 required for floral repression in Arabidopsis. Plant Cell 17 2647–2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deal, R.B., Kandasamy, M.K., McKinney, E.C., and Meagher, R.B. (2005). The nuclear actin-related protein ARP6 is a pleiotropic developmental regulator required for the maintenance of FLOWERING LOCUS C expression and repression of flowering in Arabidopsis. Plant Cell 17 2633–2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faast, R., Thonglairoam, V., Schulz, T.C., Beall, J., Wells, J.R., Taylor, H., Matthaei, K., Rathjen, P.D., Tremethick, D.J., and Lyons, I. (2001). Histone variant H2A.Z is required for early mammalian development. Curr. Biol. 11 1183–1187. [DOI] [PubMed] [Google Scholar]
- Farris, S.D., Rubio, E.D., Moon, J.J., Gombert, W.M., Nelson, B.H., and Krumm, A. (2005). Transcription-induced chromatin remodeling at the c-myc gene involves the local exchange of histone H2A. J. Biol. Chem. 280 25298–25303. [DOI] [PubMed] [Google Scholar]
- Fransz, P., De Jong, J.H., Lysak, M., Castiglione, M.R., and Schubert, I. (2002). Interphase chromosomes in Arabidopsis are organized as well defined chromocenters from which euchromatin loops emanate. Proc. Natl. Acad. Sci. USA 99 14584–14589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendrel, A.V., Lippman, Z., Martienssen, R., and Colot, V. (2005). Profiling histone modification patterns in plants using genomic tiling microarrays. Nat. Methods 2 213–218. [DOI] [PubMed] [Google Scholar]
- Guillemette, B., Bataille, A.R., Gevry, N., Adam, M., Blanchette, M., Robert, F., and Gaudreau, L. (2005). Variant histone H2A.Z is globally localized to the promoters of inactive yeast genes and regulates nucleosome positioning. PLoS Biol. 3 E384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, Y., and Amasino, R.M. (2005). Role of chromatin modification in flowering-time control. Trends Plant Sci. 10 30–35. [DOI] [PubMed] [Google Scholar]
- Henderson, I.R., and Dean, C. (2004). Control of Arabidopsis flowering: The chill before the bloom. Development 131 3829–3838. [DOI] [PubMed] [Google Scholar]
- Higgins, D.G., and Sharp, P.M. (1988). CLUSTAL: A package for performing multiple sequence alignment on a microcomputer. Gene 73 237–244. [DOI] [PubMed] [Google Scholar]
- Kobor, M.S., Venkatasubrahmanyam, S., Meneghini, M.D., Gin, J.W., Jennings, J.L., Link, A.J., Madhani, H.D., and Rine, J. (2004). A protein complex containing the conserved Swi2/Snf2-related ATPase Swr1p deposits histone variant H2A.Z into euchromatin. PLoS Biol. 2 E131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan, N.J., et al. (2003). A Snf2 family ATPase complex required for recruitment of the histone H2A variant Htz1. Mol. Cell 12 1565–1576. [DOI] [PubMed] [Google Scholar]
- Kusch, T., Florens, L., Macdonald, W.H., Swanson, S.K., Glaser, R.L., Yates, J.R., III, Abmayr, S.M., Washburn, M.P., and Workman, J.L. (2004). Acetylation by Tip60 is required for selective histone variant exchange at DNA lesions. Science 306 2084–2087. [DOI] [PubMed] [Google Scholar]
- Larochelle, M., and Gaudreau, L. (2003). H2A.Z has a function reminiscent of an activator required for preferential binding to intergenic DNA. EMBO J. 22 4512–4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leach, T.J., Mazzeo, M., Chotkowski, H.L., Madigan, J.P., Wotring, M.G., and Glaser, R.L. (2000). Histone H2A.Z is widely but nonrandomly distributed in chromosomes of Drosophila melanogaster. J. Biol. Chem. 275 23267–23272. [DOI] [PubMed] [Google Scholar]
- Lee, I., and Amasino, R.M. (1995). Effect of vernalization, photoperiod, and light quality on the flowering phenotype of Arabidopsis plants containing the FRIGIDA gene. Plant Physiol. 108 157–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, B., Pattenden, S.G., Lee, D., Gutierrez, J., Chen, J., Seidel, C., Gerton, J., and Workman, J.L. (2005). Preferential occupancy of histone variant H2AZ at inactive promoters influences local histone modifications and chromatin remodeling. Proc. Natl. Acad. Sci. USA 102 18385–18390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak, K.J., and Schmittgen, T.D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)). Methods 25 402–408. [DOI] [PubMed] [Google Scholar]
- Martin-Trillo, M., Lazaro, A., Poethig, R.S., Gomez-Mena, C., Pineiro, M.A., Martinez-Zapater, J.M., and Jarillo, J.A. (2006). EARLY IN SHORT DAYS 1 (ESD1) encodes ACTIN-RELATED PROTEIN 6 (AtARP6), a putative component of chromatin remodelling complexes that positively regulates FLC accumulation in Arabidopsis. Development 133 1241–1252. [DOI] [PubMed] [Google Scholar]
- Meagher, R.B., Deal, R.B., Kandasamy, M.K., and McKinney, E.C. (2005). Nuclear actin-related proteins as epigenetic regulators of development. Plant Physiol. 139 1576–1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meneghini, M.D., Wu, M., and Madhani, H.D. (2003). Conserved histone variant H2A.Z protects euchromatin from the ectopic spread of silent heterochromatin. Cell 112 725–736. [DOI] [PubMed] [Google Scholar]
- Michaels, S.D., and Amasino, R.M. (1999). FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11 949–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuguchi, G., Shen, X., Landry, J., Wu, W.H., Sen, S., and Wu, C. (2004). ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science 303 343–348. [DOI] [PubMed] [Google Scholar]
- Morillon, A., Karabetsou, N., O'Sullivan, J., Kent, N., Proudfoot, N., and Mellor, J. (2003). Isw1 chromatin remodeling ATPase coordinates transcription elongation and termination by RNA polymerase II. Cell 115 425–435. [DOI] [PubMed] [Google Scholar]
- Noh, Y.S., and Amasino, R.M. (2003). PIE1, an ISWI family gene, is required for FLC activation and floral repression in Arabidopsis. Plant Cell 15 1671–1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raisner, R.M., Hartley, P.D., Meneghini, M.D., Bao, M.Z., Liu, C.L., Schreiber, S.L., Rando, O.J., and Madhani, H.D. (2005). Histone variant H2A.Z marks the 5′ ends of both active and inactive genes in euchromatin. Cell 123 233–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangasamy, D., Berven, L., Ridgway, P., and Tremethick, D.J. (2003). Pericentric heterochromatin becomes enriched with H2A.Z during early mammalian development. EMBO J. 22 1599–1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratcliffe, O.J., Kumimoto, R.W., Wong, B.J., and Riechmann, J.L. (2003). Analysis of the Arabidopsis MADS AFFECTING FLOWERING gene family: MAF2 prevents vernalization by short periods of cold. Plant Cell 15 1159–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes, J.C. (2006). Chromatin modifiers that control plant development. Curr. Opin. Plant Biol. 9 21–27. [DOI] [PubMed] [Google Scholar]
- Ruhl, D.D., Jin, J., Cai, Y., Swanson, S., Florens, L., Washburn, M.P., Conaway, R.C., Conaway, J.W., and Chrivia, J.C. (2006). Purification of a human SRCAP complex that remodels chromatin by incorporating the histone variant H2A.Z into nucleosomes. Biochemistry 45 5671–5677. [DOI] [PubMed] [Google Scholar]
- Santisteban, M.S., Kalashnikova, T., and Smith, M.M. (2000). Histone H2A.Z regulates transcription and is partially redundant with nucleosome remodeling complexes. Cell 103 411–422. [DOI] [PubMed] [Google Scholar]
- Simpson, G.G. (2004). The autonomous pathway: Epigenetic and post-transcriptional gene regulation in the control of Arabidopsis flowering time. Curr. Opin. Plant Biol. 7 570–574. [DOI] [PubMed] [Google Scholar]
- Simpson, G.G., and Dean, C. (2002). Arabidopsis, the Rosetta stone of flowering time? Science 296 285–289. [DOI] [PubMed] [Google Scholar]
- Sung, S., and Amasino, R.M. (2004). Vernalization and epigenetics: How plants remember winter. Curr. Opin. Plant Biol. 7 4–10. [DOI] [PubMed] [Google Scholar]
- Suto, R.K., Clarkson, M.J., Tremethick, D.J., and Luger, K. (2000). Crystal structure of a nucleosome core particle containing the variant histone H2A. Nat. Struct. Biol. 7 1121–1124. [DOI] [PubMed] [Google Scholar]
- Swaminathan, J., Baxter, E.M., and Corces, V.G. (2005). The role of histone H2Av variant replacement and histone H4 acetylation in the establishment of Drosophila heterochromatin. Genes Dev. 19 65–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford, D.L. (2003). PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods), Version 4. (Sunderland, MA: Sinauer Associates).
- van Daal, A., and Elgin, S.C. (1992). A histone variant, H2AvD, is essential in Drosophila melanogaster. Mol. Biol. Cell 3 593–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, W.H., Alami, S., Luk, E., Wu, C.H., Sen, S., Mizuguchi, G., Wei, D., and Wu, C. (2005). Swc2 is a widely conserved H2AZ-binding module essential for ATP-dependent histone exchange. Nat. Struct. Mol. Biol. 12 1064–1071. [DOI] [PubMed] [Google Scholar]
- Zhang, H., Roberts, D.N., and Cairns, B.R. (2005). Genome-wide dynamics of Htz1, a histone H2A variant that poises repressed/basal promoters for activation through histone loss. Cell 123 219–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.