Abstract
Arabidopsis thaliana GAMT1 and GAMT2 encode enzymes that catalyze formation of the methyl esters of gibberellins (GAs). Ectopic expression of GAMT1 or GAMT2 in Arabidopsis, tobacco (Nicotiana tabacum), and petunia (Petunia hybrida) resulted in plants with GA deficiency and typical GA deficiency phenotypes, such as dwarfism and reduced fertility. GAMT1 and GAMT2 are both expressed mainly in whole siliques (including seeds), with peak transcript levels from the middle until the end of silique development. Within whole siliques, GAMT2 was previously shown to be expressed mostly in developing seeds, and we show here that GAMT1 expression is also localized mostly to seed, suggesting a role in seed development. Siliques of null single GAMT1 and GAMT2 mutants accumulated high levels of various GAs, with particularly high levels of GA1 in the double mutant. Methylated GAs were not detected in wild-type siliques, suggesting that methylation of GAs by GAMT1 and GAMT2 serves to deactivate GAs and initiate their degradation as the seeds mature. Seeds of homozygous GAMT1 and GAMT2 null mutants showed reduced inhibition of germination, compared with the wild type, when placed on plates containing the GA biosynthesis inhibitor ancymidol, with the double mutant showing the least inhibition. These results suggest that the mature mutant seeds contained higher levels of active GAs than wild-type seeds.
INTRODUCTION
Gibberellins (GAs) form a large group of tetracyclic diterpenoid carboxylic acids, certain members of which function as natural regulators of a variety of growth and development processes in plants, such as germination, leaf expansion, stem elongation, bolting, flower induction, flower development, seed set, and fruit development (Davies, 1995). To date, >135 structurally distinct GAs have been identified from natural sources (http://www.plant-hormones.info/gibberellins.htm), most of which are not active in plants and may represent intermediates in the biosynthesis or degradation of bioactive GAs (Hedden and Phillips, 2000; Yamaguchi and Kamiya, 2000). In general, concentrations of active GAs in plants are highest in areas of rapidly elongating cells, such as stems, shoots, and developing seeds.
GA-deficient plants are extremely dwarfed in size and are often sterile or exhibit high rates of seed abortion, while plants that synthesize excess amounts of GAs are taller, with long, light-green leaves and decreased seed dormancy compared with wild-type seeds (Jacobsen and Olszewski, 1993; Huang et al., 1998). In Arabidopsis thaliana, the main bioactive GAs, GA4 and GA1 (Figure 1), have been shown to be involved in important aspects of plant development. Mutants with mildly reduced levels of GA4 and GA1 showed a semidwarf growth habit (Talon et al., 1990; Schomburg et al., 2003). GA4 and GA1 in Arabidopsis have been previously detected in seedlings, shoots, developing seeds, germinating seeds, and mature seeds.
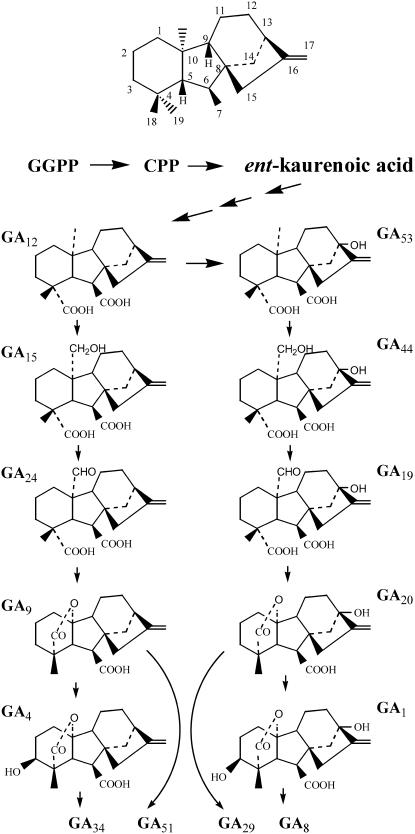
Figure 1.
The GA Biosynthetic Pathway.
The pathway for biosynthesis of some active GAs, and their catabolic fates, are shown. GA34, GA51, GA29, and GA8 are catabolic products of GA degradation. The basic structure of ent-gibberellane with the numbering system of the carbons is shown at the top. GGPP, geranylgeranyl diphosphate; CPP, ent-copalyl diphosphate.
The roles of GAs in developing and mature seeds are particularly intriguing. For example, it has been reported that during early seed development in many plant species, GA biosynthesis increases until the seeds mature, at which point the levels of active GAs begin to decrease in concert with the onset of dormancy (Kim et al., 2005; Swain et al., 1995). De novo biosynthesis of GAs is required during imbibition in some plant species, as was concluded from the observation that the presence of inhibitors of GA biosynthesis, such as paclobutrazol and tetcyclacis, prevented germination (Karssen et al., 1989; Schomburg et al., 2003). It is not clear whether active GAs synthesized during embryogenesis are degraded when the seed mature or whether they are stored for later use, and if so, what the storage form is and how they are converted back to active GAs.
Deactivation mechanisms are necessary for effective regulation of bioactive hormone levels. So far, GA 2β-hydroxylation catalyzed by GA 2-oxidase has been the best-characterized GA deactivation reaction, which includes the conversions of GA4 and GA1 to GA34 and GA8, respectively (Figure 1). GA 2-oxidases belong to the 2-oxoglutarate–dependent dioxygenase family (Thomas et al., 1999; Schomburg et al., 2003). Recently, another deactivation mechanism, involving epoxidation of the 16,17-double bond of GAs by a cytochrome P450 monooxygenase, has been found in rice (Oryza sativa) internodes (Zhu et al., 2006). Deactivation may also result from GA conjugation, which involves the formation of glucosyl ethers and esters (Schneider et al., 1992), although genes encoding these enzymes have not been identified yet.
Here, we report that two members of the SABATH methyltransferase gene family in the Arabidopsis genome, GAMT1 and GAMT2, encode enzymes that use S-adenosine-l-methionine (SAM) as a methyl donor to methylate the carboxyl group of GAs, resulting in the methyl esters of GAs (MeGAs). Both genes are expressed most highly in the siliques during seed development. Siliques of GAMT1 and GAMT2 mutants show an increase in concentrations of GAs, and mature seeds of mutants appear to have higher concentrations of active GAs. Thus, methylation of GAs may constitute an additional mechanism for modulating cellular GA concentrations in addition to the oxidative pathways previously described (Thomas et al., 1999; Schomburg et al., 2003; Zhu et al., 2006).
RESULTS
Two Members of the Arabidopsis SABATH Family Methylate GAs
As part of a project aimed at identifying the substrates of each of the 24 carboxyl methyltransferases encoded by the SABATH gene family in Arabidopsis (Chen et al., 2003; D'Auria et al., 2003), we expressed full-length cDNAs of genes At4g26420 and At5g56300 in Escherichia coli, purified the proteins, and tested their activity with radioactive 14C-SAM and a battery of potential substrates, using the methodology and array of substrates previously developed (Yang et al., 2006). Since the proteins belonging to these methyltransferases can methylate a deprotonated oxygen that is part of a carboxyl group, but cannot methylate the protonated oxygen on a hydroxyl group (Zubieta et al., 2003), the tested substrates included plant compounds with one or more carboxyl groups (Yang et al., 2006). In these high-throughput assays, GAMT1 (encoded by At4g26420) and GAMT2 (encoded by At5g56300) showed activity with GA3, the one GA included in the high-throughput assay. No activity was observed with any other substrates included in this test.
Based on these initial results, we tested additional GAs, including four that are hydroxylated at position 13 (GA1, GA3, GA19, and GA20) and five that are not hydroxylated at this position (GA4, GA9, GA12, GA34, and GA51) (Figure 1). These GAs have been previously detected in Arabidopsis (Talon et al., 1990; Huang et al., 1998; Eriksson et al., 2006). GAMT1 showed highest activity with GA9 and GA20, followed by GA3, GA4, GA34, GA51, and GA1 (Table 1). GAMT2 methylated GA4 with the highest efficiency, followed by GA34, GA9, GA3, GA1, GA51, and GA20. GAMT1 was able to methylate GA19 and GA12, which do not contain a γ-lactone, albeit at a low rate, while GAMT2 did not act on these two GAs. Neither GAMT1 nor GAMT2 methylated additionally tested compounds with structural relatedness to GAs, such as the diterpenes abietic acid and ent-kaurenoic acid. Products from the enzyme assays were verified to be methylated by comparing the retention time of the MeGA products with authentic standards on thin layer chromatography (TLC) and by gas chromatography–mass spectrometry after derivatization (see Supplemental Figure 1 online).
Table 1.
Relative Activity of GAMT1 and GAMT2 with Several GAs
| Relative Activity (%)
|
||
|---|---|---|
| Substrate | GAMT1 | GAMT2 |
| GA1 | 25 | 30 |
| GA3 | 80 | 45 |
| GA4 | 69 | 100 |
| GA9 | 100 | 60 |
| GA12 | 9 | 0 |
| GA19 | 19 | 0 |
| GA20 | 95 | 15 |
| GA34 | 46 | 80 |
| GA51 | 32 | 16 |
Enzymatic Properties of GAMT1 and GAMT2
The steady state kinetic parameters of each of the proteins were determined (Table 2). The Km values of both enzymes with several GAs were in the range of 1.9 to 15.8 μM. GAMT1 had the lowest Km value, 5.4 μM, with GA4, and GAMT2 had the lowest Km value, 1.9 μM, with GA9. The Km values for these enzymes with SAM in combination with various GAs were in the range of 30 to 50 μM. All these Km values and the observed Kcat values (Table 2) are similar to those obtained for other SABATH methyltransferases with their in vivo substrates (Ross et al., 1999; Zubieta et al., 2003). The pH optimum of GAMT2 was determined to be 8.0, whereas the pH optimum for GAMT1 was 7.5. The cations K+ and NH4+ in concentration of 5 mM were found to increase the activity of GAMT1 by threefold and fourfold, respectively, while Zn2+, Cu2+, Fe2+, and Fe3+ at this concentration inhibited >90% GAMT1 activity. GAMT2 activity was not enhanced nor inhibited by K+, NH4+, Na+, Ca2+, Fe2+, Mg2+, and Mn2+ but was inhibited similarly to GAMT1 by Fe3+, Zn2+, and Cu2+.
Table 2.
Kinetic Parameters of GAMT1 and GAMT2
| Km (μM) | Kcat (s−1) | Kcat/Km (nM−1 s−1) | |
|---|---|---|---|
| GAMT1 | |||
| GA4 | 5.4 | 1.0 × 10−2 | 1.8 |
| GA9 | 15.8 | 2.6 × 10−2 | 1.6 |
| GAMT2 | |||
| GA4 | 5.9 | 1.5 × 10−3 | 0.2 |
| GA9 | 1.9 | 1.8 × 10−3 | 0.9 |
Protein Sequences of GAMT1 and GAMT2
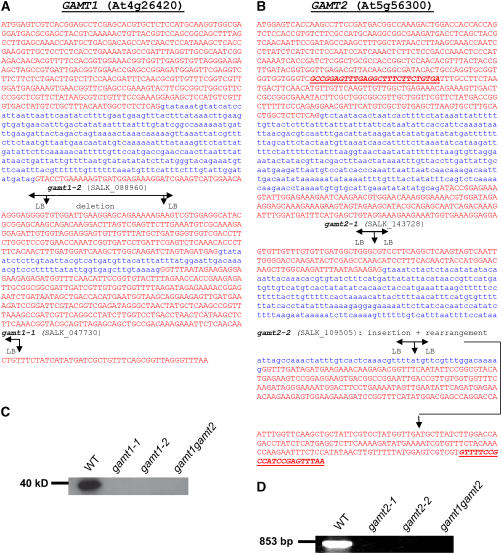
GAMT1 and GAMT2 are part of the SABATH methyltransferase family with 24 members in Arabidopsis (Chen et al., 2003; D'Auria et al., 2003). GAMT1 encodes a protein of 376 amino acids, with a calculated molecular mass of 41.5 kD, and GAMT2 encodes a protein of 388 amino acids, with a calculated molecular mass of 43.3 kD. The Arabidopsis enzyme with the most similar sequence to GAMT1 within this family is GAMT2, and vice versa. An alignment of the sequences indicates that they are 58% identical (Figure 2). The next most similar Arabidopsis protein, with 34% identity to both GAMT1 and GAMT2, is IAMT1, an enzyme that is capable of methylating auxin (indole-3-acetic acid [IAA]) to produce methyl IAA (Zubieta et al., 2003; Qin et al., 2005). It should be noted that the current annotation of GAMT1 (At4g26420) on The Arabidopsis Information Resouce (TAIR) website (http://www.arabidopsis.org/) shows a third intron occurring toward the end of the third exon shown in Figure 3A, then a fourth exon, a fourth intron, and a fifth exon. Such a gene would encode a protein of 620 amino acids, with a calculated molecular mass of 68.2 kD. There are multiple lines of evidence indicating that this annotation is erroneous. First, all the full-length clones that we have isolated, as well as the full-length clones shown on the TAIR website, support the three-exon, two-intron structure shown in Figure 3A, encoding the protein whose sequence is shown in Figure 2 (this is the protein whose biochemical characterization is presented here). Furthermore, all SABATH methyltransferases, including the GAMT1 protein encoded by the three-exon, two-intron gene shown in Figure 3A, are ∼350 to 400 amino acids long. Finally, the GAMT1 protein in the plant, as detected by antibodies specific to GAMT1, migrates on SDS-PAGE gel as an ∼40-kD protein (Figure 3C), a mass consistent with the calculated molecular mass (41.5 kD) of the GAMT1 protein encoded by the gene whose structure is shown in Figure 3A but inconsistent with the present annotation in TAIR. The updated annotation of At4g26420 will be available from TAIR and the National Center for Biotechnology Information (NCBI) following the next genome release (TAIR7) scheduled for January, 2007.
Figure 2.
Alignment of the Protein Sequences of Arabidopsis GAMT1, GAMT2, and IAMT1.
Residues at a given position that are identical in at least two of the three proteins are shown in white letters on black background. IAMT1 is encoded by gene At5g55250. The sequences of the proteins and the genes encoding them can be accessed at http://www.biology.lsa.umich.edu/research/labs/pichersky/2010%20NSF/database/database1.htm.
Figure 3.
Analysis of T-DNA Insertional Mutants in GAMT1 and GAMT2.
(A) The correct gene structure of GAMT1 (At4g26420) based on comparisons of full-length cDNAs with the genomic sequence. Exon sequences are shown in red, and intron sequences are shown in blue. The position of the T-DNA insertions in gamt1-1 and gamt1-2 are shown based on sequencing of the amplified border segments. In gamt1-2, a left border (LB) is present at both ends of the T-DNA insertion, and the genomic sequence in between has been deleted.
(B) The gene structure of GAMT2 (At5g56300) based on the annotation in TAIR and the comparisons of full-length cDNAs with the genomic sequence. The position of the T-DNA insertions in gamt2-1 and gamt2-2 are shown based on sequencing of the amplified border segments. In gamt2-2, the region downstream from the insertion is rearranged so that the outlined part of the third exon is missing and a segment of intron 2 plus additional unknown segments are present. The bold and underlined sequences indicate the position of the primers used in the RT-PCR experiment whose results are shown in (D).
(C) A protein gel blot using anti-GAMT1 antibodies with crude protein extracts from siliques of wild-type plants, gamt1-1, gamt1-2, and the double mutant gamt1-2 gamt2-2. The GAMT1 protein, present in the wild-type plants but not in the mutants, migrates on SDS-PAGE gels as an ∼40-kD protein.
(D) Results of RT-PCR using the oligonucleotide primers indicated in (B) and RNA extracted from siliques of wild-type plants and gamt2-1, gamt2-2, and gamt1-2 gamt2-1. The predicted product of 853 nucleotides was obtained from wild-type plants but not from mutants.
Overexpression of Arabidopsis GAMT1 and GAMT2 in Arabidopsis, Tobacco, and Petunia Plants Results in Dwarf Phenotypes
We obtained Arabidopsis plants that expressed GAMT1 and GAMT2 under the control of the 35S cauliflower mosaic virus promoter. We also obtained an Arabidopsis line in which an enhancer tag was inserted upstream of the GAMT1 gene (Weigel et al., 2000; Yu et al., 2004). The majority of independently obtained transgenic Arabidopsis lines (>10) expressing GAMT1 either under the control of the 35S promoter or the enhancer tag (verified by RT-PCR and RNA gel blot analysis; data not shown) exhibited a similar phenotype that included small, dark-green rosette leaves (Figure 4A) that grew exceedingly slow, and they developed into dwarfed, bushy plants (Figure 4B). Some never bloomed, and those that did bloom started doing so after 3 months (whereas wild-type plants under these conditions began blooming in 6 weeks). Their flowers were small and mostly sterile. Spraying plants with GA4 recovered some fertility and caused partial amelioration of the dwarf phenotype of the whole plant. Spraying with MeGA4 did not ameliorate the dwarf phenotype.
Figure 4.
Phenotypes of Arabidopsis Col-0 Overexpressing GAMT1 and GAMT2.
(A) A 4-week-old Arabidopsis wild-type plant (left) next to a same-age plant overexpressing GAMT1 due to enhancer tagging.
(B) A 3-month-old plant overexpressing GAMT1 due to enhancer tagging, showing the bushy, dwarf phenotype.
(C) A 2-month-old flowering Arabidopsis plant overexpressing GAMT2 under the control of the 35S promoter, showing the bushy, semidwarf phenotype with very small flowers.
All plants are shown to roughly equal scale.
A similar phenotype was observed in transgenic tobacco (Nicotiana tabacum) and petunia (Petunia hybrida) plants expressing GAMT1 under the control of the 35S promoter (Figure 5). The transgenic tobacco plants were slow growers and had small flowers with reduced seed set. The petunia plants bloomed only under long-day conditions, and the flowers were small and sterile.
Figure 5.
Phenotypes of Petunia and Tobacco Plants Overexpressing GAMT1 under the Control of the 35S Promoter.
(A) Tobacco wild-type plant.
(B) A tobacco plant of the same age as in (A) expressing GAMT1 under the control of the 35S promoter.
(C) Petunia wild-type.
(D) A petunia plant of the same age as in (C) expressing GAMT1 under the control of the 35S promoter.
Expression of GAMT2 in transgenic Arabidopsis plants under the control of the 35S promoter did not produce a dwarf phenotype at the early stage of plant development when grown on Murashige and Skoog (MS) medium, and seedlings were indistinguishable from wild-type seedlings. However, after transfer to soil on the third week after germination, the majority of transgenic plants (>10 independent lines) showed a different growth pattern from the wild type, growing slowly and exhibiting a higher number of branches compared with wild-type plants, giving them a semidwarf, bushy look (Figure 4C). While some of these plants had reduced fertility, the majority had normal seed set. The differences in the phenotypes of GAMT2-overexpressing lines and GAMT1-overexpresing lines persisted under different light regimes.
GAMT1- and GAMT2-Overexpressing Lines Are Deficient in GAs
To verify whether overexpression of GAMT genes caused GA deficiency in Arabidopsis plants, we measured endogenous levels of precursor and bioactive GAs in aerial parts of GAMT1- and GAMT2-overexpressing plants just after the start of flowering. As shown in Table 3, the levels of GA4, the major bioactive form at this stage in Arabidopsis, were much lower in plants overexpressing GAMT1 (<10%) than those in wild-type plants. In addition, all precursor GAs that we measured were less abundant in GAMT1 overexpression lines than in wild-type controls. Plants overexpressing GAMT2 contained lower levels of bioactive GA4 than did wild-type plants, but in general, the depletions of endogenous GAs in GAMT2-overexpressing lines was not as severe as those in GAMT1-overexpressing lines, and some GAs showed no difference in levels with control or even higher levels (e.g., GA15). These results are consistent with the interpretation that the dwarfed and semidwarfed phenotypes of plants overexpressing GAMT1 and GAMT2, respectively, are attributable to reductions in bioactive GA levels.
Table 3.
Endogenous GA Levels in Arabidopsis Plants Overexpressing GAMT1 or GAMT2
| Wild-Type Control for GAMT1-OEa | GAMT1-OE Line 13-5a | GAMT1-OE Line 13-6a | Wild-Type Control for GAMT2-OEb | GAMT2-OE Line 12-3b | GAMT2-OE Line 12-5b | ||
|---|---|---|---|---|---|---|---|
| 13-H | GA12 | 15.4/13.2 | 0.7/4.1 | 4.3/1.2 | 21.5/14.0 | 16.9/10.2 | 10.7/15.0 |
| GAs | GA15 | 1.5/3.2 | 0.2/1.3 | 0.4/1.2 | 4.4/1.7 | 10.8/7.8 | 11.8/10.6 |
| GA24 | –/21.7 | –/2.9 | –/2.9 | 24.6/16.9 | 14.6/10.8 | 10.7/15.2 | |
| GA9 | 0.4/ND | 0.1/ND | 0.1/ND | ND/ND | ND/ND | ND/ND | |
| GA4 | 0.8/2.3 | 0.1/0.1 | ND/0.1 | 0.9/0.4 | 0.1/0.1 | 0.2/0.2 | |
| 13-OH | GA53 | 2.4/2.4 | 0.6/0.4 | 0.1/0.3 | 2.8/1.6 | 1.4/1.1 | 0.6/1.0 |
| GAs | GA44 | ND/ND | ND/ND | ND/ND | ND/ND | ND/ND | ND/ND |
| GA19 | –/16 | –/3.6 | –/2.9 | 8.2/3.0 | 9.5/6.3 | 8.2/10.5 | |
| GA20 | 0.4/0.4 | 0.1/ND | 0.1/ND | 0.1/ND | 0.1/ND | 0.1/0.1 | |
| GA1 | 0.2/0.5 | ND/ND | ND/ND | ND/ND | ND/ND | ND/ND |
GA measurements were performed twice using independently prepared plant materials. Values (ng/g dry weight) from both the first (left) and second (right) measurements are shown. –, Detectable, but could not be quantified reliably due to comigration of impurities. ND, not detected or could not be quantified reliably due to low abundance.
These two sets of plant materials were grown on different occasions. OE, overexpressing.
GAMT1 and GAMT2 Are Expressed in Developing Seeds in Arabidopsis
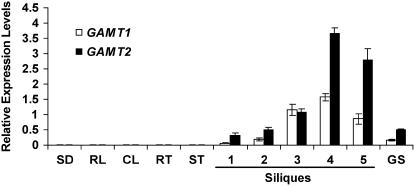
The results described above indicated that Arabidopsis possesses enzymes capable of methylating GAs. To determine the role of these enzymes in the plant, we next examined the expression profile of the two genes encoding these enzymes. Quantitative RT-PCR (qRT-PCR) was performed on RNA isolated from cauline and rosette leaves, stem, flowers, roots, siliques (including seeds), germinating seeds, and 2-week-old seedlings. Little expression of either of these two genes was observed in tissues other than siliques (Figure 6), with the exception of weak but detectable expression in germinating seeds. Examination of the levels of GAMT1 and GAMT2 transcripts from siliques at different stages of development (Bowman, 1994) showed that expression of both genes began at early stages of silique development, peaked in the second half of this process, and then began to decrease after the start of desiccation (Figure 6). Germinating seeds contained detectable levels of GAMT1 and GAMT2 transcripts, but they were much lower than the maximum levels in developing siliques.
Figure 6.
Detection of GAMT1 and GAMT2 Transcripts in Different Arabidopsis Organs and Stages of Seed Development by qRT-PCR.
Relative levels of GAMT1 and GAMT2 transcripts (compared with levels of transcripts of the control, the ubi10 gene) are presented as mean values ± sd, calculated from three sets of independent experiments. Silique samples are as follows: 1, flower buds and stage 1 siliques; 2, stages 2 and 3; 3, stages 4 and 5; 4, stages 6, 7, and 8; 5, stages 9 and 10. The 10 stages of the siliques were defined according to Bowman (1994), and they span the development process from the smallest silique (stage 1) to fully mature and dry siliques (stage 10). SD, 2-week-old seedlings; RL, rosette leaf; CL, cauline leaf; RT, root; ST, stem; GS, germinating seeds.
We also performed immunoblot measurements of the levels of GAMT1 protein in different Arabidopsis organs using polyclonal antibodies specific against GAMT1 (these antibodies did not react with GAMT2 protein extracted from the plant [Figure 3C] or with GAMT2 protein produced in E. coli [data not shown]; raising antibodies against GAMT2 was not successful). The results of these immunoblots (Figure 7A) were consistent with the qRT-PCR results, showing the presence of the protein in siliques only. A more detailed examination of the levels of GAMT1 protein in developing siliques at different stages of development showed that protein levels closely correlated with GAMT1 transcript levels, peaking toward the end of the developmental process of the silique (Figure 7B).
Figure 7.
Levels of GAMT1 Protein in Different Tissues and Developmental Stages of Arabidopsis Plants.
(A) Immunoblot analysis of the distribution of GAMT1 protein in different plant organs using anti-GAMT1 antibodies. CL, cauline leaf; RL, rosette leaf; FL, flower; ST, stem; SD, 2-week-old seedlings; SI, siliques (stages 6, 7, and 8); RT, root.
(B) Immunoblot analysis of GAMT1 in different stages of silique development and in germinating seeds. Lanes 1 to 5 are samples from siliques of same stages denoted by samples 1 to 5 in Figure 6. GS, germinating seeds.
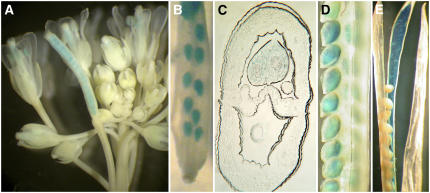
Plants expressing a β-glucuronidase (GUS) reporter gene under the control of the GAMT1 promoter (GAMT1-GUS) and stained for GUS activity showed staining in the developing seeds and in the anthers (Figures 8A to 8D). In later stages of seed development, some GUS staining was also observed in the septum and in the funiculus as well (Figure 8E). GUS staining of transgenic plants with a GAMT2 promoter–GUS fusion gene indicated expression in the anthers but, in contrast with the qRT-PCR results, no staining in the developing seeds (data not shown).
Figure 8.
Staining for GUS Activity with Plants Transgenic for the GAMT1 Promoter–GUS Transgene.
(A) Inflorescence with flowers and siliques at different stages of development.
(B) A young silique showing staining in the developing seeds.
(C) Cross section of a developing seed.
(D) Maturing seeds.
(E) A mature silique showing staining in the septum and funiculus.
Siliques of Mutant Plants Have Higher Levels of GAs Than Wild-Type Plants
We obtained Arabidopsis Columbia (Col) plants that contained T-DNA insertions in GAMT1 and GAMT2 (Figure 3). gamt1-1 had an insertion in exon 3, and gamt1-2 had an insertion in exon 2 that included a small deletion of the coding region (Figure 3A). Lines homozygous for either gamt1-1 or gamt1-2 produced no GAMT1 protein (Figure 3C). gamt2-1 had an insertion in exon 2 (Figure 3B), and gamt2-2 had an insertion intron 2 that caused a partial deletion and rearrangement of exon 3 (Figure 3B). Lines homozygous for either mutation contained no functional transcript (Figure 3D). In addition, we obtained the homozygous double mutant line by crossing gamt1-2 and gamt2-2 mutants. None of the single mutant or double mutant plants showed visible phenotypes, such as altered germination rate, growth habit, or seed set, under normal growth conditions. These verified null mutants (Figures 3C and 3D) grew normally and had no visible morphological differences with wild-type plants.
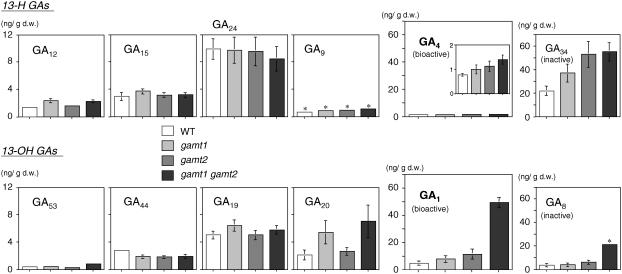
We measured the content of some GAs in the siliques of homozygous gamt1-2, gamt2-2, and gamt1-2 × gamt2-2 plants using siliques of developmental stages 5, 6, and 7, where GAMT1 and GAMT2 expression levels are close to or at their highest. The levels of bioactive GA4 and GA1 in siliques were significantly higher in the gamt1 gamt2 double mutant than those in wild-type plants, with the level of GA1 especially elevated in the double mutant (∼10-fold) compared with that in the wild type (Figure 9). GA4 and GA1 levels were slightly more abundant in gamt1 and gamt2 single mutants than in wild-type plants as well (Figure 9). The effects of gamt single and/or double mutations were observed on the levels of the deactivated forms GA34 and GA8 and on some precursor GAs in the gamt1 plants. These results support the conclusion that GAs are endogenous substrates for GAMT1 and GAMT2 in wild-type siliques.
Figure 9.
GA Levels in Developing Siliques of gamt Single and Double Mutants.
Shown are endogenous levels of GAs in developing siliques of the wild type, gamt1-1 and gamt2-2 single mutants, and the gamt1-2 gamt2-2 double mutant plants. Results of respective GAs are displayed according to their order in the biosynthesis pathway (see Figure 1). Results of GA4 are also indicated in the inset with a different y axis scale to clarify the differences among the genotypes. Note that the y axis scale for precursor GAs (GA12, GA15, GA24, GA9, GA53, GA44, GA19, and GA20) is different from that for bioactive (GA4 and GA1) and deactivated (GA34 and GA8) forms. GA measurements were conducted three times using independently prepared plant extracts. Means with se (which are displayed only when they are >0.2) from triplicates are shown. Asterisks indicate that these GAs could be measured reliably only once due to comigration of impurities on the liquid chromatograph. d.w., dry weight.
Analysis of Endogenous MeGAs
To our knowledge, MeGAs have not been identified as endogenous compounds in angiosperms. Elevated GA levels in siliques of gamt mutants suggested that MeGAs were produced by GAMTs and that they might accumulate in wild-type siliques. To address this question, we analyzed endogenous MeGA1, MeGA4, and MeGA20 using isotope-labeled MeGAs as internal standards by gas chromatography–selected ion monitoring (GC-SIM). We chose these MeGAs to be analyzed because the levels of the corresponding free GAs increased in the gamt1 gamt2 mutant (Figure 9) and/or because they were preferred substrates of at least one of the GAMTs in vitro (Table 1). Using the procedure detailed in the Supplemental Methods online, we were able to purify and detect all isotope-labeled MeGAs added to the extracts. However, none of the endogenous ones was detected in wild-type and gamt1 gamt2 mutant (included as a control) siliques (see Supplemental Figure 2 online). We also analyzed these MeGAs in plants overexpressing GAMT1 (lines 13-5 and 13-6; Table 3) and wild-type plants (as a control) just after flowering. Again, endogenous MeGAs were undetectable, while all internal standards were detected.
GAMT1 and GAMT2 Mutants Display an Excess GA Phenotype in the Seeds
The expression profiles of GAMT1 and GAMT2 suggest that the enzymes they encode play a specific role during seed development. To examine the effect of the methylation of GAs in Arabidopsis seeds, the homozygous single and double mutant seeds were placed on plates containing MS basal medium that was supplemented with ancymidol, an inhibitor of GA biosynthesis (Rademacher, 2000). Differences in germination rates were observed between mutants and wild-type plants. Germination (defined as the appearance of the radicle) of wild-type seeds was not inhibited by 10 μM concentration of ancymidol but was ∼90% inhibited by the presence of 20 μM concentration of ancymidol and completely inhibited at 30 μM (Figure 10). By contrast, at 30 μM ancymidol, 25 to 60% of the single and the double mutant seeds still germinated, with the double mutant seeds showing the highest germination rate (Figure 10), and their cotyledons were green, although they eventually stopped growing. Some germination of the single and double mutants was still observed at 40 μM concentration of ancymidol in the medium. Consistent with these results, germination of seeds from plants overexpressing GAMT1and GAMT2 with the 35S promoter was 85% inhibited by 10 μM ancimydol, and germination of seeds from plants expressing GAMT1 via an enhancer tag was inhibited 50% (Figure 10).
Figure 10.
Inhibition of Seed Germination by Ancymidol.
Seeds were germinated on MS plates containing the indicated concentration of ancymidol. GAMT1-1 is the enhancer-tagged line overexpressing GAMT1, GAMT1-2 is the GAMT1-overexpressing line 13-5 (Table 1), and GAMT2-1 and GAMT2-2 are two independent GAMT2-overexpressing lines 12-3 and 12-5, respectively. Values are means ± sd calculated from three sets of independent experiments.
DISCUSSION
Arabidopsis At4g26420 and At5g56300 Encode GA Methyltransferases That Are Expressed Predominantly in Siliques
GAMT1 and GAMT2, which we have shown here to methylate the carboxyl group of various GAs to form the corresponding MeGA esters, belong to the plant SABATH family of methyltransferases. This family also includes enzymes that methylate the carboxyl group of other growth substances, such as salicylic acid (SA), jasmonic acid (JA), and auxin (IAA), to form the respective methyl esters MeSA, MeJA, and MeIAA (Ross et al., 1999; Seo et al., 2001; Zubieta et al., 2003). The Arabidopsis genome has 24 genes belonging to this family (Chen et al., 2003; D'Auria et al., 2003), and among the proteins encoded by these 24 genes, GAMT1 and GAMT2 are the most similar proteins to each other. The level of divergence between GAMT1 and GAMT2 proteins (58% identity) indicates that the duplication that gave rise to the genes encoding them (At4g26420 and At5g56300) is not very recent.
We show here that GAMT1 and GAMT2 both are expressed primarily in developing siliques, with peak transcript levels toward the end of this process. Our qRT-PCR results concerning the expression of GAMT2 are consistent with data obtained from microarray experiments and available online (Zimmermann et al., 2004; https://www.genevestigator.ethz.ch; Schmid et al., 2005; http://www.weigelworld.org/resources/microarray/AtGenExpress/). Because of the mistake in the annotation of GAMT1 in TAIR, and the fact that based on this erroneous annotation all 11 oligonucleotides chosen by Affimatrix to represent GAMT1 on the ATH1 microarray are derived from sequences downstream of GAMT1 that are not part of the GAMT1 gene, there is no relevant information on the expression of GAMT1 on these websites. Nonetheless, our qRT-PCR measurements of GAMT1 transcripts showed that this gene has a very similar pattern of expression to GAMT2 throughout the developmental process of the siliques, with little expression seen elsewhere. Measurements of GAMT1 protein levels by protein blotting experiments with GAMT1-specific antibodies were consistent with the qRT-PCR results.
Staining of siliques of transgenic plants expressing GUS under the control of the GAMT1 promoter indicated expression mostly in the developing seeds. We did not observe any GUS staining in the developing seeds of the transgenic plants carrying the GAMT2 promoter–GUS fusion gene, but this might have been the result of an incomplete promoter segment being used, as data provided on both https://www.genevestigator.ethz.ch and http://www.weigelworld.org/resources/microarray/AtGenExpress/ indicate expression specifically in seeds, in addition to expression in whole siliques.
In in vitro assays, GAMT1 and GAMT2 methylated the carboxyl group (position 7) of a variety of GAs to produce the corresponding methyl esters. While we did not test all possible GAs, the results obtained with the substrates we used indicated that both enzymes have a broad specificity, although GAMT1 appears to have somewhat broader specificity (Table 1). Transgenic Arabidopsis plants overexpressing either GAMT1 or GAMT2 show depletion in various GAs (Table 3), as would be expected if GAs were the substrates of these enzymes. While the specific level of depletion of each GA depends on a complex set of parameters, including the internal concentrations of each GA, the internal concentration of the methyltransferase, the Km values of each enzyme for each substrate, and the flux of the biosynthetic and catabolic pathways that interconvert all these GAs, some correlations between depletion levels and preference of the enzymes for specific substrates could be discerned. For example, GAMT1 appears to have a higher activity with GA20 than GAMT2 does, and the depletion of GA20 in the GAMT1-overexpressing lines is more noticeable than in GAMT2-overexpressing lines. However, this correlation is not perfect and with our present limited knowledge may be too difficult to predict. In addition, several genes encoding enzymes in the late stage of GA biosynthesis have been shown to be under negative feedback regulation via bioactive GAs. Therefore, the observed decrease in the levels of some precursor GAs in GAMT-overexpressing plants might be due to increased GA 20-oxidase activity as a consequence of reduced GA4 levels. So, while the clear phenotypic differences between Arabidopsis plants overexpressing GAMT1 and GAMT2, which are correlated with less severe reduction in active and nonactive GAs in GAMT2 overexpressors compared with GAMT1 overexpressors, are most likely due to the differences in enzymatic activities of the two enzymes with various GAs, the direct causes of these differences are difficult to identify at this point.
An even stronger indication that GAs are the endogenous substrates of GAMT1 and GAMT2 is the observation that in gamt1, gamt2, and gamt1 gamt2 mutant lines, the levels of several GAs in the siliques, and in particular the levels of the active GA1 and GA4, are higher than in the siliques of wild-type plants. Here, too, a general but imperfect correlation is seen between increases in levels of individual GAs and the relative activity levels of GAMT1 and GAMT2 with these compounds. For example, GAMT1 has strong in vitro activity with GA20, and gamt1 shows increased levels of GA20 in silqiues, while GAMT2 has weak activity with GA20 in vitro, and the gamt2 mutant shows little or no differences in GA20 levels in siliques. In some cases, when both enzymes appear to have similar activity with a given GA, for example GA1, the level of this GA increases only a little in either gamt1 or gamt2, while the level of this GA is much increased in the gamt1 gamt2 double mutant.
Curaba et al. (2004) reported that the level of GA1 was similar to that of GA4 in the developing Arabidopsis seeds, unlike in other Arabidopsis tissues that were found to contain higher levels of GA4 than those of GA1, including vegetative organs, flowers, and germinating seeds (Talon et al., 1990; Huang et al., 1998; Ogawa et al., 2003; Schomburg et al., 2003; Fei et al., 2004). Our results here show that GA1 was more abundant than GA4 in developing siliques (including seeds) of wild-type plants (Figure 9). Interestingly, the level of GA1 was drastically elevated, whereas that of GA4 increased only slightly in siliques of gamt1 gamt2 mutant plants compared with wild-type siliques (Figure 9). These differences in the levels of GA4 and GA1 among the genotypes cannot be readily explained by the substrate specificity of GAMTs because their recognition of the 13-hydroxyl group does not appear to be strict in vitro (Table 1). These results suggest an unknown mechanism that allows accumulation of GA1 specifically in developing siliques and/or seeds; possibly, GA 13-oxidase (for which the gene has not yet been identified) and GA catabolic enzymes that strictly distinguish the 13-hydroxyl group (such as GA 16,17-epoxidase of rice; Zhu et al., 2006) might be involved in this mechanism.
We chose to measure GAs in siliques at the developmental stage in which GAMT1 and GAMT2 show peak expression to maximize differences between wild-type and mutant plants. We therefore do not know the exact concentrations of GAs in the mature seeds. However, the observation that the germination of gamt1 and gamt2 mutant seeds is inhibited less by the GA biosynthesis inhibitor ancymidol (a pyrimidine that acts as an inhibitor of monoxygenases catalyzing the oxidative step from ent-kaurene to ent-kaurenoic acid; Rademacher, 2000) (Figure 10) suggests that the mature seeds of the mutants also contain higher levels of active GAs than in the wild-type seeds and are thus capable of initiating germination without de novo GA biosynthesis. Alternatively, weak but detectable expression of GAMT1 and GAMT2 genes in germinating seeds revealed by qRT-PCR (Figure 6) suggests that these methyltransferases might be involved in GA deactivation in imbibed seeds as well. If this was the case, the observed differences in germination rates among the genotypes in the presence of ancymidol (Figure 10) might be due to the effect of gamt mutations on GA levels after the start of imbibition.
Putative Role of GA Methylation in Seed Development and Germination
It is generally known that GAs are synthesized during seed development and during germination (Ogawa et al., 2003), but little is known about the distribution and specific roles of GAs in Arabidopsis developing siliques and seeds (Curaba et al., 2004; Kim et al., 2005). In pea (Pisum sativum), an active GA, GA1, is found in high levels in the seeds (4 to 7 d after anthesis) and at lower levels in the pod (MacKenzie-Hose et al., 1998). It has also been reported that exchange of hormones in the fruit between the pod and the seeds is necessary for proper seed development in P. sativum and that lack of sufficient levels of GAs in either the pod or in the embryo leads to seed abortion (Eeuwens and Schwabe, 1975; MacKenzie-Hose et al., 1998).
Although most enzymes of the GA biosynthetic pathway and the enzymes for the catabolic pathway have been identified (Hedden and Kamiya, 1997; Olszewski et al., 2002), roles of many of these enzymes in plant development remain to be fully characterized. In particular, while several homologous 2β-oxidases that can deactivate GAs have been identified in Arabidopsis (Thomas et al., 1999; Schomburg et al., 2003; Wang et al., 2004) and some of these show expression in developing seeds, a physiological role in GA deactivation in the seeds for these oxidases has not yet been fully established. In pea, the importance of GA deactivation has been shown by the phenotypic and biochemical studies of the slender (sln) mutant, which lacks a GA 2-oxidase that is involved in the conversion of GA20 to GA29 during seed development (Lester et al., 1999; Martin et al., 1999). In the sln mutant, GA20 accumulates to a high level in mature seeds and then is converted to GA1 upon germination, resulting in a GA overproduction phenotype during seedling growth.
Methyl esters of GAs have been shown to be inactive in several angiospermous species (Weiss et al., 1995; Cowling et al., 1998), and MeGA4 did not bind significantly to the soluble receptor GID1 from rice (Ueguchi-Tanaka et al., 2005) and its Arabidopsis homologs (Nakajima et al., 2006), although some methylated GAs, such as MeGA9 and MeGA73, appear to serve as bioactive molecules in ferns during the formation of antheridium (Yamauchi et al., 1996). The observation that transgenic Arabidopsis plants overexpressing GAMT1 or GAMT2 have lower levels of GAs, show varying levels of dwarfism, delayed flowering, and sterility and that these phenotypes can be partially reversed by the external application of GAs suggest that MeGAs are indeed inactive in Arabidopsis as well. We also directly tested the activity of MeGA4 by applying it to the GAMT1-overexpressing lines and saw no amelioration in the dwarf phenotype.
Thus, methylation of GAs might be part of the mechanism affecting the levels of active GAs in developing siliques and/or seeds. It is possible that methylation of GAs is a step in the irreversible deactivation of GAs, perhaps tagging it for further catabolism by epoxidation, oxidation, or glycosylation, reactions known to occur in other parts of the plant or other plant species (Christmann and Doumas, 1998; Thomas et al., 1999; Zhu et al., 2006). Methylated GAs have a more hydrophobic character, and methylation thus may be necessary in part to allow the modified GAs to diffuse through membranes to reach their site of degradation. The lack of detection of MeGAs in both wild-type siliques and in leaves of plants overexpressing GAMT1 also suggests that MeGAs are quickly converted to unknown metabolites, although a more extensive analysis of possible metabolites of the methylation reaction is required before a firm conclusion about the fate of MeGAs can be reached. The expression of GAMT1 and GAMT2 in the siliques peaks at the later stages of silique development, when the embryos in the siliques are nearing the end of their fast cell division and elongation, a process that is controlled by GAs, further suggesting a role for GAMT1 and GAMT2 in inactivation of GAs.
With some plant signaling molecules, methylation is known to be a reversible process. For example, an esterase that converts MeJA back to JA has been reported from tomato (Solanum lycopersicum) (Stuhlfelder et al., 2004) and potato (Solanum tuberosum) (accession number AY684102), and a related enzyme that hydrolyzes MeSA to SA has been reported from tobacco (Forouhar et al., 2005). Interestingly, the GA receptor recently reported from rice (Ueguchi-Tanaka et al., 2005) is a member of the same α/β-hydrolase superfamily to which the MeSA and MeJA esterases belong (Marshall et al., 2003), although it was reported that the angiosperm GA receptor is unlikely to be an active esterase. While it is possible that methylation of GAs, like GA glycosylation (Schneider et al., 1992), is in some cases a reversible process in plants, the results of the ancymidol inhibition experiments indicated that the wild-type Arabidopsis seeds were unable to recover the GAs that had been lost due to the activities of GAMT1 and GAMT2. Thus, the overall data we obtained in this study tentatively suggest that the function of methylating GAs in the developing seeds is in the irreversible inactivation of GAs. Since both GAMT1 and GAMT2 are expressed mainly in the developing siliques (including seeds), the enzymes they encode have partially overlapping substrate specificities, and double mutants show an additive effect of GAMT1 and GAMT2 expression on levels of some GAs, it can be concluded that the two genes have partially overlapping functions in silique and seed development. However, the specific physiological role of GA methylation in siliques, seeds, or in other parts of the plant remains to be determined.
METHODS
Plant Material and Growth Conditions
Wild-type Arabidopsis thaliana ecotype Col-0 was used in this study for all experiments. Plants were grown in soil or on MS basal salt medium containing 0.8 g/L agar with 12 h of light at 22°C and 12 h of dark at 18°C. cDNA clones and four SALK knockout mutant lines of GAMT1 (SALK_088960 and SALK_047730) and GAMT2 (SALK_109505 and SALK_143728) with Col-0 background were obtained from the ABRC (Alonso et al., 2003). Transgenic plants were obtained by Agrobacterium tumefaciens–mediated floral dip transformation (Clough and Bent, 1998) of wild-type plants (Col-0) with cDNA of GAMT1 or cDNA of GAMT2 in sense orientation, driven by the cauliflower mosaic virus 35S promoter. Promoter-GUS transgenic plants with Col-0 background were used for histochemical analysis of GUS activity to monitor the expression pattern under GAMT1- and GAMT2-specific promoters.
For the ancymidol treatment experiment, dry Arabidopsis seeds stored for at least 1 month at room temperature were used. After sterilization, 50 to 100 seeds were placed on plates containing MS basal medium containing various concentrations of ancymidol (Sigma-Aldrich). Plates were placed at 4°C for 3 d and then transferred to a growth chamber at 22°C and a light/dark regime of 14/10 h, respectively, for 2 weeks.
For the qRT-PCR experiments, germinating seeds were obtained by placing seeds on soaked filter paper in the dark at 4°C for 3 d and then transferring the plate to 22°C in constant light for 48 h. At this point, the radicle began to protrude from the seeds and the material was harvested for RNA extraction.
Screening of T-DNA Insertion Mutants
The T-DNA insertion sites in the GAMT1 and GAMT2 genes were verified first by PCR (Siebert et al., 1995). For GAMT1, we used the genomic primers GAMT1 forward (5′-GGGAAGAAGCTAGCCGTGATTGACG-3′) and GAMT1 reverse (5′-CCACCGAAGAGATTGCGGCGGCGATTG-3′) to characterize the insertion lines SALK_047730 (designated here as gamt1-1) and SALK_088960 (designated as gamt1-2). For GAMT2, we used the primers GAMT2 forward (5′-GCCGGAGTTTGAGGCTTTCTTCTGTG-3′) and GAMT2 reverse (5′-GTTTTCCGCCATCCGAGTTTAA-3′) to characterize insertion lines SALK _143728 (designated here as gamt2-1) and SALK_109505 (designated as gamt2-2). In these experiments, we also used the T-DNA–specific primer LBB1 (5′-GCGTGGACCGCTTGCTGCAACT-3′). All PCR products were further verified by sequencing. Homozygous lines for the four mutants were confirmed by PCR with specific primers and subsequent DNA gel blot analysis.
Plasmid Construction
cDNA of GAMT1 and GAMT2 (ABRC) were cloned into the GATEWAY donor vector pDONR207 (Invitrogen). cDNA clones were transferred from pDONR207 to a destination vector pCHF3 (gift from Jianming Li, University of Michigan) modified with a GATEWAY cassette (Invitrogen). For expression of recombinant methyltransferases in Escherichia coli, cDNA clones GAMT1 and GAMT2 were transferred to pET28A vector (Novagen, EMD Biosciences), modified for GATEWAY.
RT-PCR and qRT-PCR Analyses
For RT-PCR analysis of the mutants, siliques half-way through maturation were used for extraction of total RNA (Maes and Messens, 1992). First-strand cDNA was synthesized by AMV reverse transcriptase (Promega) with poly(T)18 primer. PCR was performed with 1 μL of the first-strand cDNA reaction. Specific forward and reverse primers for GAMT1 were 5′-GGGAAGAAGCTAGCCGTGATTGACG-3′ and 5′-TACTACGCATGTTATGTACATATA-3′, respectively, and for GAMT2 5′-GCCGGAGTTTGAGGCTTTCTTCTGTG-3′ and 5′-TTAAACTCGGATGGCGGAAAAC-3′, respectively.
For qRT-PCR analysis, total RNA was isolated from different organs of Arabidopsis and from siliques at different stages of development as defined by Bowman (1994) using the RNA procedure described by Maes and Messens (1992). The RNA was subjected to DNase treatment using the DNA-free kit (Ambion), and first-strand cDNA was synthesized by AMV reverse transcriptase with poly(T)18 primer in parallel with a negative control reaction in which no AMV reverse transcriptase was added. All samples were brought to 100 μL in volume, after which 1 μL was used in a 20 μL of qPCR reaction containing 20 mM Tris, pH 8.4, 50 mM KCl, 3 mM MgCl2, 5% DMSO, 250 μM deoxynucleotide triphosphate, 200 nM primer, 0.5 units of Taq polymerase B (Promega), 1× CYBR-Green I dye (Molecular Probes),and 10 nM fluorescein (Bio-Rad). GAMT1 and GAMT2 gene-specific primers were designed as follows: GAMT1 forward 5′-TGTTGTTTATGCTGATGGGTGGTC-3′ and GAMT1 reverse 5′-CGCAATCTCTTCGGTGGTTCTAA-3′; GAMT2 forward 5′-CGTCCTTCAGGCTCAAGTAGTC-3′ and GAMT2 reverse 5′-CCCTATCTTGAAACCACCACAACGGTC-3′. Amplification of the ubiquitin gene ubq10 (At4g05320) using the forward primer 5′-AGGAGTCCACACTTCACTTGGTC-3′ and the reverse primer 5′-GGTGTCAGAGCTCTCTACCTCCA-3′ was used as internal control. The qPCR was performed on an iCycler thermocycler (Bio-Rad), with the following conditions: 95°C for 3 min, 50 cycles of 95°C for 15 s, 60°C for 30 s, and 72°C for 30 s, followed by a melting cycle of 55 to 95°C with an increasing gradient of 0.5°C, and a 10-s pause at each temperature. All reactions were performed in duplicate. Data processing was done by iCycler real-time detection system software (version 3.0). cDNA of RNA from siliques at half-way through development was serially diluted to generate the standard curve. Analyses of the melting curves were performed to ensure amplification of one specific gene product. At least three sets of independent experiments were performed to calculate a mean value and standard deviation. The cycle threshold (Ct) values generated by the iCycler software for each cDNA sample were used only when they met the following parameters: significant difference in Ct numbers between cDNA sample and the negative control was observed and the melting curve of the PCR product generated by the melting cycles was indicative of specific amplification.
Construction of Promoter-GUS Reporter Gene Fusion and Histochemical Localization of GUS Activity
The upstream regions of GAMT1 and GAMT2 were amplified from genomic DNA of Arabidopsis, cloned into pTOPO-ENTR, and subsequently transferred to pDW137 (Blazquez et al., 1997), modified for GATEWAY cloning by LR clonase reaction (Invitrogene). For GAMT1, the forward primer 5′-CACTTTGAAGAAGCGTGAGTGTAGGTAGTGC-3′ and the reverse primer 5′-GTTGTAAATGTGTAAGAAAAGGTGTGAACG-3′ were used to amplify a 1543-bp fragment upstream of the initiating ATG. For GAMT2, the forward primer 5′-GGATGGATAAATAGAGACATCGATCTG-3′ and the reverse primer 5′-GCTCTGTCTCTTCTCTCTTATTGGAAGTTGTAG-3′ were used to amplify a 1300-bp fragment. Five independent transgenic lines were obtained and tested for β-glucuronidase activity (Jefferson, 1987). Different plant organs were placed in staining solution containing 100 mM phosphate buffer, pH 7.0, 1 mM X-Gluc, 10 mM EDTA, 0.1% Triton-X and incubated at 37°C overnight. Plant material was destained with 70% ethanol. Cross-sectioning of GUS-stained siliques was performed with the JB-4 Plus embedding kit (Polysciences) using a protocol developed by Lee and Schiefelbein (2002).
Expression of GAMT1 and GAMT2 in E. coli and Protein Purification
Purification of GAMT1 and GAMT2 was performed by nickel–nitrilotriacetic acid agarose purchased from Qiagen. The expression and the purification procedure were performed by a modified procedure as described by Zubieta et al. (2003), but the elution step was the final step. The N-terminal His-tag was not cleaved for the subsequent methyltransferase assay. Purified protein of GAMT1 was used for production of rabbit polyclonal antibodies (Cocalico Biologicals).
Immunoblotting
Crude extracts were prepared from different stages of silique development or different parts from Arabidopsis. Immunodetection was performed using rabbit anti-GAMT1 polyclonal antibodies (1:2000 dilution). Goat anti-rabbit IgG horseradish peroxidase conjugate (1:20,000 dilution) was used as a secondary antibody. Subsequent chemiluminescence was detected and exposed on Kodak BioMax XAR film using the Western Lightning Chemiluminiscence Reagent Plus kit (Perkin-Elmer Life Sciences).
Methyltransferase Enzyme Assay
The enzyme assay contained 1 μL of purified enzyme (∼0.5 pmol), 10 μL of assay buffer (250 mM Bis-Tris-propane-HCl, pH 8.0, and 25 mM KCl), 1 μL of various GA substrates (prepared as a 50 mM solution in ethanol), 0.5 μL of 14C-SAM (58 mCi/mmol; Perkin-Elmer), and water to a final volume of 50 μL. For a negative control, a boiled enzyme was used. The samples were incubated at 30°C for 30 min (over which period the reactions were linear), followed by extraction with 150 μL of ethyl acetate. The tubes were vortexed and briefly centrifuged, and 50 μL of the ethyl acetate phase were transferred to a scintillation vial containing 2 mL of scintillation fluid (Econo-Safe Research Products International) and counted in a scintillation counter (LS-6500 model; Beckman Coulter). For product verification, the same assay was scaled up to 500 μL using nonradioactive SAM for gas chromatography–mass spectrometry analysis or radioactive SAM for radioactive TLC. The nonradioactive products were twice extracted with 0.5 mL of ethyl acetate, dried under N2 flow, and sylilated with 45 μL of pyridine and 60 μL of BSTFA (Sigma-Aldrich) for 30 min at 80°C and analyzed via the Shimadzu 5000 gas chromatograph–mass spectrometer. Separation was performed on an Altech EC-5 column (30 m × 0.32 mm × 1 μm). Helium was the carrier gas (flow rate of 2 mL min−1), a splitless injection (3 μL) was used, and a temperature gradient of 4.5°C min−1 from 70°C (5-min hold) to 310°C was applied. The identities of methylated and sylilated GAs were determined by comparison of retention time and mass spectra to authentic sylilated standards.
Radioactive TLC was performed for all GAs described in the biochemical experiments as putative substrates for GAMT1 and GAMT2. A developing solution of chloroform and methanol in a ratio of 9:1 v/v was used as described by Roberts et al. (1999). The retention time of obtained radioactive products was compared with nonradioactive standards (prepared by L. Mander) or methylated GA standards with (trimethylsilyl)diazomethane (Sigma-Aldrich). For visualization of the nonradioactive GAs and their methylesters, the TLC plate was sprayed with sulfuric acid:ethanol (1:20 [v/v]) and then heated at 110°C for 10 min (Roberts et al., 1999).
Characterization of GAMT Kinetic Parameters
In all enzyme activity measurements, appropriate enzyme concentrations and incubation times were chosen so that the reaction velocity was linear during the incubation time period, and at least three replicates were performed. To determine the Km value for each substrate (GA and SAM), one substrate concentration was fixed at a saturated level (1 mM), and the concentration of the other substrate to be measured was varied. Nine different concentrations were used. Lineweaver-Burk plots were performed to obtain the Km and Kcat values.
To measure the pH optimum for GAMT activity, the reactions were performed in 50 mM Bis-Tris-propane-HCl buffer ranging from pH 6.0 to 9.5. Temperature stability of GAMT1 and GAMT2 was determined by incubating the enzymes at temperatures ranging from 4 to 65°C for 30 min and then chilling the samples on ice, followed by enzyme assays at 28°C.
Enzyme assays were performed with the following cations present in the assay buffer at the final concentration of 1 to 10 mM: Ca2+, Cu2+, Fe2+, Fe3+, K2+, Mg2+, Mn2+, Na+, NH4+, and Zn2+.
GA and MeGA Measurements
Seeds from Arabidopsis Col-0 of the wild type, gamt1, gamt2, and gamt1 gamt2 mutants, and overexpressing lines were sown in a Petri dish with MS medium supplemented with 10% sugar. After 2 weeks at 23°C, the seedlings were transplanted to soil and the plants were grown in a growth chamber at 23°C. For GA measurements for plants overexpressing GAMT1 and GAMT2, the aerial parts of plants that have just begun to flower were collected. For GA measurements of siliques, siliques of developmental stages 5, 6, and 7 were collected.
Quantitative analysis of GAs for the first set of GAMT1-overexpressing lines and their wild-type control (Table 3) was performed by GC-SIM using 2H-labeled GAs as internal standards as described previously (Gawronska et al., 1995). Briefly, an ethylacetate-soluble fraction containing GAs was subjected to HPLC purification using a reverse-phase column (Capcell Pak C18 SG120; Shiseido Fine Chemicals). When necessary, GA-containing fractions were then purified through another round of HPLC using an ion exchange column (Senshu Pak N[CH3]2, 1151-N; Senshu Scientific). The purified fractions were subjected to GC-SIM analysis using a mass spectrometer (Automass Sun; JEOL) equipped with a gas chromatograph (6890N; Agilent Technologies) and a capillary column (DB-1; Agilent Technologies) after derivatization.
GA measurements for other samples (Table 3, Figure 9) were conducted by liquid chromatography–selected reaction monitoring using 2H-labeled GAs as internal standards. We used a liquid chromatography tandem mass spectrometry system consisting of a quadrupole/time-of-flight tandem mass spectrometer (Q-Tof Premier; Waters) and an Acquity Ultra Performance liquid chromatograph (Waters) equipped with a reverse-phase column (Acquity UPLC BEH-C18; Waters). Approximately 200 mg (dry weight) of lyophilized plant materials were used for each measurement. Details about the purification procedures and LC-MS/MS analysis conditions are described in Supplemental Methods and Supplemental Table 1 online.
Endogenous MeGAs were analyzed using 2H-labeled MeGAs as internal standards by GC-SIM after derivatized to TMSi ester. Purification procedures of MeGAs from plant extracts are described in detail in the Supplemental Methods online.
Accession Numbers
The Arabidopsis Genome Initiative locus identifiers for genes mentioned in this article are as follows: GAMT1 (At4g26420), GAMT2 (At5g56300), and IAMT1 (At5g55250). The updated annotation of At4g26420 will be available from TAIR and NCBI following the next genome release (TAIR7) scheduled for January, 2007.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Gas Chromatography–Mass Spectrometry of Authentic MeGA20 and the Product of the Methylation Reaction Catalyzed by GAMT1 and Using GA20 as the Substrate.
Supplemental Figure 2. Analysis of MeGAs in Wild-Type Siliques by GC-Selected Ion Monitoring Using [2H2]MeGAs as Internal Standards.
Supplemental Figure 3. Quantification of GAs by Selected Reaction Monitoring Using LC-MS/MS.
Supplemental Table 1. Parameters for GA Measurements by LC-MS/MS Analysis.
Supplemental Methods.
Supplementary Material
Acknowledgments
We thank S.H. Kwak (University of Michigan) for help with the cross-sectioning of GUS-stained siliques and Jan Zeevaart (Michigan State University) for help with the early stages of the research. This work was supported by National Science Foundation Grants 0312466 to E.P., 0312449 to J.P.N., and 0312857 to V.S., by a Grant-in-Aid for Scientific Research (17770048) to S.Y. from the Ministry of Education, Culture, Sports, Science, and Technology of Japan, and by funds from the Pearlstein Fund for Research in Floriculture from the Hebrew University to R.B. and D.W.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Eran Pichersky (lelx@umich.edu)
Online version contains Web-only data.
References
- Alonso, J.M., Stepanova, A.N., Leisse, T.J., Kim, C.J., Chen, H., Shinn, P., Stevenson, D.K., Zimmerman, J., Barajas, P., and Cheuk, R. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657. [DOI] [PubMed] [Google Scholar]
- Blazquez, M.A., Soowal, L.N., Lee, I., and Weigel, D. (1997). LEAFY expression and flower initiation in Arasbidopsis. Developopment 124 3835–3844. [DOI] [PubMed] [Google Scholar]
- Bowman, J. (1994). Arabidopsis: An Atlas of Morphology and Development. (Berlin: Springer-Verlag).
- Chen, F., D'Auria, J.C., Tholl, D., Ross, J.R., Gershenzon, J., Noel, J.P., and Pichersky, E. (2003). An Arabidopsis thaliana gene for methylsalicylate biosynthesis, identified by a biochemical genomics approach, has a role in defense. Plant J. 36 577–588. [DOI] [PubMed] [Google Scholar]
- Christmann, A., and Doumas, P. (1998). Detection and identification of gibberellins in needles of silver fir (Abies alba Mill.) by combined gas chromatography-mass spectrometry. Plant Growth Regul. 24 91–99. [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Cowling, R., Kamiya, Y., Seto, H., and Harberd, N.P. (1998). Gibberellin dose–response regulation of GA4 gene transcript levels in Arabidopsis. Plant Physiol. 117 1195–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curaba, J., Moritz, T., Blervaque, R., Parcy, F., Raz, V., Herzog, M., and Vachon, G. (2004). AtGA3ox2, a key gene responsible for bioactive gibberellin biosynthesis, is regulated during embryogenesis by LEAFY COTYLEDON2 and FUSCA3 in Arabidopsis. Plant Physiol. 136 3660–3669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Auria, J.C., Chen, F., and Pichersky, E. (2003). The SABATH family of methyltransferases in Arabidopsis thaliana and other plant species. In Recent Advances in Phytochemistry, J. Romeo, ed (Oxford: Elsevier Science), 253–283.
- Davies, P.G. (1995). Plant Hormones: Physiology, Biochemistry, and Molecular Biology. (Dordrecht, The Netherlands: Kluwer Academic Publishers).
- Eeuwens, C.J., and Schwabe, W.W. (1975). Seed and pod wall development in Pisum sativum L. in relation to extracted and applied hormones. J. Exp. Bot. 26 1–14. [Google Scholar]
- Eriksson, S., Bohlenius, H., Moritz, T., and Nilsson, O. (2006). GA4 is the active gibberellin in the regulation of LEAFY transcription and Arabidopsis floral initiation. Plant Cell 18 2172–2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei, H., Zhang, R., Pharis, R.P., and Sawhney, V.K. (2004). Pleiotropic effects of the male sterile33 (ms33) mutation in Arabidopsis are associated with modifications in endogenous gibberellins, indole-3-acetic acid and abscisic acid. Planta 219 649–660. [DOI] [PubMed] [Google Scholar]
- Forouhar, F., Yang, Y., Kumar, D., Chen, Y., Fridman, E., Park, S.W., Chiang, Y., Acton, T.B., Montelione, G.T., Pichersky, E., Klessig, D.F., and Tong, L. (2005). Structural and biochemical studies identify tobacco SABP2 as a methyl salicylate esterase and implicate it in plant innate immunity. Proc. Natl. Acad. Sci. USA 102 1773–1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gawronska, H., Yang, Y.Y., Furukawa, K., Kendrick, R.E., Takahashi, N., and Kamiya, Y. (1995). Effects of low irradiance stress on gibberellin levels in pea seedlings. Plant Cell Physiol. 36 1361–1367. [Google Scholar]
- Hedden, P., and Kamiya, Y. (1997). Gibberellin biosynthesis: Enzymes, genes and their regulation. Annu. Rev. Plant Phyiol. Plant Mol. Biol. 48 431–460. [DOI] [PubMed] [Google Scholar]
- Hedden, P., and Phillips, A.L. (2000). Gibberellin metabolism: New insights revealed by the genes. Trends Plant Sci. 5 523–530. [DOI] [PubMed] [Google Scholar]
- Huang, S., Raman, A.S., Ream, J.E., Fujiwara, H., Cerny, R.E., and Brown, S.M. (1998). Overexpression of 20-oxidase confers a gibberellin-overproduction phenotype in Arabidopsis. Plant Physiol. 118 773–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen, S.E., and Olszewski, N.E. (1993). Mutations at the SPINDLY locus of Arabidopsis alter gibberellin signal transduction. Plant Cell 5 887–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson, R. (1987). Assaying chimeric genes in plants: The GUS gene fusion system. Plant Mol. Biol. Rep. 5 387–405. [Google Scholar]
- Karssen, C.M., Zagórski, S., Kepczynski, J., and Groot, S.P.C. (1989). Key role for endogenous gibberellins in the control of seed germination. Ann. Bot. (Lond.) 63 71–80. [Google Scholar]
- Kim, Y.C., Nakajima, M., Nakayama, A., and Yamaguchi, I. (2005). Contribution of gibberellins to the formation of Arabidopsis seed coat through starch degradation. Plant Cell Physiol. 8 1317–1325. [DOI] [PubMed] [Google Scholar]
- Lee, M.M., and Schiefelbein, J. (2002). Cell pattern in the Arabidopsis root epidermis determined by lateral inhibition with feedback. Plant Cell 14 611–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lester, D.R., Ross, J.J., Smith, J.J., Elliott, R.C., and Reid, J.B. (1999). Gibberellin 2-oxidation and the SLN gene of Pisum sativum. Plant J. 19 65–73. [DOI] [PubMed] [Google Scholar]
- MacKenzie-Hose, A., Ross, J., Davies, N., and Swain, S. (1998). Expression of gibberellin mutations in fruits of Pisum sativum L. Planta 204 397–403. [Google Scholar]
- Maes, M., and Messens, E. (1992). Phenol as grinding material in RNA preparations. Nucleic Acids Res. 20 4374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall, S.D., Putterill, J.J., Plummer, K.M., and Newcomb, R.D. (2003). The carboxylesterase gene family from Arabidopsis thaliana. J. Mol. Evol. 57 487–500. [DOI] [PubMed] [Google Scholar]
- Martin, D.N., Proebsting, W.M., and Hedden, P. (1999). The SLENDER gene of pea encodes a gibberellin 2-oxidase. Plant Physiol. 121 775–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima, M., et al. (2006). Identification and characterization of Arabidopsis gibberellin receptors. Plant J. 46 880–889. [DOI] [PubMed] [Google Scholar]
- Ogawa, M., Hanada, A., Yamauchi, Y., Kuwahara, A., Kamiya, Y., and Yamaguchi, S. (2003). Gibberellin biosynthesis and response during Arabidopsis seed germination. Plant Cell 15 1591–1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olszewski, N., Sun, T.P., and Gubler, F. (2002). Gibberellin signaling: Biosynthesis, catabolism, and response pathways. Plant Cell 14(Suppl) S61–S80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin, G., Gu, H., Zhao, Y., Ma, Z., Shi, G., Yang, Y., Pichersky, E., Chen, H., Lui, M., Chen, Z., and Qu, L.-J. (2005). Regulation of auxin homeostasis and plant development by an indole-3-acetic acid carboxyl methyltransferase in Arabidopsis. Plant Cell 17 2693–2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rademacher, W. (2000). Growth retardants: Effects on gibberellin biosynthesis and other metabolic pathways. Annu. Rev. Plant Physiol. Plant Mol. Biol. 51 501–531. [DOI] [PubMed] [Google Scholar]
- Roberts, A.V., Blake, P.S., Lewis, R., Taylor, J.M., and Dunstan, D.I. (1999). The effect of gibberellins on flowering in roses. J. Plant Growth Regul. 18 113–119. [DOI] [PubMed] [Google Scholar]
- Ross, J.R., Nam, K.H., D'Auria, J.C., and Pichersky, E. (1999). S-adenosyl-L-methionine:salicylic acid carboxyl methyltransferase, an enzyme involved in floral scent production and plant defense, represents a new class of plant methyltransferases. Arch. Biochem. Biophys. 367 9–16. [DOI] [PubMed] [Google Scholar]
- Schmid, M., Davidson, T.S., Henz, S.R., Pape, U.J., Demar, M., Vingron, M., Scholkopf, B., Weigel, D., and Lohmann, J.U. (2005). A gene expression map of Arabidopsis thaliana development. Nat. Genet. 37 501–506. [DOI] [PubMed] [Google Scholar]
- Schneider, G., Jensen, E., Spray, C.R., and Phinney, B.O. (1992). Hydrolysis and reconjugation of gibberellin A20 glucosyl ester by seedlings of Zea mays L. Proc. Natl. Acad. Sci. USA 89 8045–8048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schomburg, F.M., Bizzell, C.M., Lee, D.L., Zeevaart, J.A.D., and Amasino, R.M. (2003). Overexpression of a novel class of gibberellin 2-oxidases decreases gibberellin levels and creates dwarf plants. Plant Cell 15 151–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo, H.S., Song, J.T., Cheong, J.J., Lee, Y.H., Lee, Y.W., Hwang, I., Lee, J.S., and Choi, Y.D. (2001). Jasmonic acid carboxyl methyltransferase: A key enzyme for jasmonate-regulated plant responses. Proc. Natl. Acad. Sci. USA 98 4788–4793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebert, P.D., Chenchik, A., Kellogg, D.E., Kukyanova, K.A., and Lukyano, S.A. (1995). An improved PCR method for walking in uncloned genomic DNA. Nucleic Acids Res. 23 1087–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuhlfelder, C., Mueller, M.J., and Warzecha, H. (2004). Cloning and expression of a tomato cDNA encoding a methyl jasmonate cleaving esterase. Eur. J. Biochem. 271 2976–2983. [DOI] [PubMed] [Google Scholar]
- Swain, S.M., Ross, J.J., Reid, J.B., and Kamiya, Y. (1995). Gibberellins and pea seed development. Planta 195 426–433. [Google Scholar]
- Talon, M., Kornneeff, M., and Zeevaart, J.A.D. (1990). Endogenous gibberellins in Arabidopsis thaliana and possible steps blocked in the biosynthetic pathways of the semidwarf ga4 and ga5 mutants. Proc. Natl. Acad. Sci. USA 87 7983–7987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, S.G., Phillips, A.L., and Hedden, P. (1999). Molecular cloning and functional expression of gibberellin 2-oxidases, multifunctional enzymes involved in gibberellin deactivation. Proc. Natl. Acad. Sci. USA 96 4698–4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueguchi-Tanaka, M., Ashikari, M., Nakajima, M., Itoh, H., Katoh, E., Kobayashi, M., Chow, T.Y., Hsing, Y.I., Kitano, H., Yamaguchi, I., and Matsuoka, M. (2005). GIBBERELLIN INSENSITIVE DWARF1 encodes a soluble receptor for gibberellin. Nature 437 693–698. [DOI] [PubMed] [Google Scholar]
- Wang, H., Caruso, L.V., Downie, A.B., and Perry, S.E. (2004). The embryo MADS domain protein AGAMOUS-Like 15 directly regulates expression of a gene encoding an enzyme involved in gibberellin metabolism. Plant Cell 16 1206–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel, D., Ahn, J.H., Blazquez, M.A., Borevitz, J.O., Christensen, S.K., Fankhauser, C., Ferrandiz, C., Kardailsky, I., Malancharuvil, E.J., and Neff, M.M. (2000). Activation tagging in Arabidopsis. Plant Physiol. 122 1003–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss, D., Vanderluit, A., Knegt, E., Vermeer, E., Mol, J.N.M., and Kooter, J.M. (1995). Identification of endogenous gibberellins in petunia flowers – Induction of anthocyanin biosynthetic gene expression and the antagonistic effect of abscisic acid. Plant Physiol. 107 695–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi, S., and Kamiya, Y. (2000). Gibberellin biosynthesis: Its regulation by endogenous and environmental signals. Plant Cell Physiol. 41 251–257. [DOI] [PubMed] [Google Scholar]
- Yamauchi, T., Oyama, N., Yamane, H., Murofushi, N., Schraudolf, H., Pour, M., Furber, M., and Mander, L.N. (1996). Identification of Antheridiogens in Lygodium circinnatum and Lygodium flexuosum. Plant Physiol. 111 741–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Y., Yuan, J.S., Ross, J., Noel, J.P., Pichersky, E., and Chen, F. (2006). An Arabidopsis thaliana methyltransferase capable of methylating farnesoic acid. Arch. Biochem. Biophys. 448 123–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, F., Park, S., and Rodermel, S.R. (2004). The Arabidopsis FtsH methaloprotease gene family: Interchangeability of subunits in chloroplast oligomeric complexes. Plant J. 37 864–876. [DOI] [PubMed] [Google Scholar]
- Zhu, Y., et al. (2006). ELONGATED UPPERMOST INTERNODE encodes a cytochrome P450 monooxygenase that epoxidizes gibberellins in a novel deactivation reaction in rice. Plant Cell 18 442–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann, P., Hirsch-Hoffmann, M., Hennig, L., and Gruissem, W. (2004). GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 136 2621–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zubieta, C., Koscheski, P., Ross, J.R., Yang, Y., Pichersky, E., and Noel, J.P. (2003). Structural basis for substrate recognition in the salicylic acid carboxyl methyltransferase family. Plant Cell 15 1704–1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.