Abstract
Although the role of H3K9 methylation in rice (Oryza sativa) is unclear, in Arabidopsis thaliana the loss of histone H3K9 methylation by mutation of Kryptonite [also known as SU(VAR)3-9 homolog] reduces genome-wide DNA methylation and increases the transcription of transposable elements. Here, we report that rice SDG714 (for SET Domain Group Protein714) encodes a histone H3K9-specific methyltransferase. The C terminus of SDG714 confers enzymatic activity and substrate specificity, whereas the N terminus localizes it in the nucleus. Loss-of-function mutants of SDG714 (SDG714IR transformants) generated by RNA interference display a mostly glabrous phenotype as a result of the lack of macro trichomes in glumes, leaves, and culms compared with control plants. These mutants also show decreased levels of CpG and CNG cytosine methylation as well as H3K9 methylation at the Tos17 locus, a copia-like retrotransposon widely used for the generation of rice mutants. Most interestingly, loss of function of SDG714 can enhance transcription and cause the transposition of Tos17. Together, these results suggest that histone H3K9 methylation mediated by SDG714 is involved in DNA methylation, the transposition of transposable elements, and genome stability in rice.
INTRODUCTION
The organization of eukaryotic genomes can be classified into two distinct structural and functional domains, known as euchromatin and heterochromatin, referring, respectively, to the permissive and repressive potential for gene transcription within these regions. This transcriptional regulation is often correlated with distinct posttranslational modification at the N-terminal tails of histones (Strahl and Allis, 2000), including acetylation, methylation, phosphorylation, ubiquitination, ADP-ribosylation, and sumoylation (Jenuwein and Allis, 2001; Fischle et al., 2003). In recent years, significant progress has been achieved in understanding the functional importance of histone methylation in regulating genome organization (Peters et al., 2001; Zhang and Reinberg, 2001). In general, methylation of H3K4, H3K36, and H3K79 primarily correlates with transcriptionally active chromatin, whereas methylation of H3K9 and H3K27, as well as H4K20, is associated with transcriptionally silenced regions (Fischle et al., 2003). In addition to the different Lys residues, each Lys residue can have an added monomethyl, dimethyl, and trimethyl group (Bannister et al., 2001; Czermin et al., 2002; Peters et al., 2003; Tamaru et al., 2003; Xiao et al., 2003). As a result, the net effect of the combination of methylated Lys residues, level of methylation, and potential interaction with other types of modifications creates a complex epigenetic code for transcriptional repression or activation (Fischle et al., 2005).
All identified histone Lys methyltransferases contain a SET domain except for Dot1, which is specific for H3K79 (Fang et al., 2004). The SET domain is composed of ∼130 amino acid residues, named after three Drosophila genes, Su(var), E(z), and TRX (for Trithorax), which are involved in regulating position-effect variegation. SU(VAR)3-9 from Drosophila was the first identified histone methyltransferase specific for K9 of histone H3 (Rea et al., 2000). Thereafter, multiple H3K9 histone methyltransferases, such as ESET, G9a, Suv39 h1, Suv39 h2, and Eu-HMTase, have been identified in mammals and were shown to be of great importance in the control of gene expression and development (Sims et al., 2003; Dodge et al., 2004). For example, suv39 h–defective mice display severely impaired viability and increased risk of tumorigenesis resulting from chromosomal instability (Peters et al., 2001).
In Arabidopsis thaliana, 39 SET domain proteins have been identified (Baumbusch et al., 2001) (http://www.chromdb.org/), and some of them play important roles in developmental processes, such as MEA in seed development (Grossniklaus et al., 1998), ATX1 in flower organ formation (Alvarez-Venegas et al., 2003), and SDG8/EFS in preventing the transition from vegetative to reproductive growth (Kim et al., 2005; Zhao et al., 2005). Overexpression of SUVH2 leads to a dwarf phenotype in Arabidopsis (Naumann et al., 2005). In tobacco (Nicotiana tabacum), overexpression of SET1 delays root and leaf growth (Shen and Meyer, 2004). In rice (Oryza sativa), SET1 represses plant growth, as overexpression of rice SET1 in Arabidopsis leads to retarded plant growth (Liang et al., 2003).
Besides their important roles in the regulation of normal growth and development, SET proteins encoding H3K9 methyltransferases also play pivotal roles in heterochromatin formation and genome integrity. In Neurospora crassa, DIM-5, encoding an H3K9 methyltransferase, is essential for all DNA methylation (Tamaru and Selker, 2001). In Arabidopsis, the H3K9 methyltransferase Kryptonite [also known as SU(VAR)3-9 homolog] (KYP/SUVH4) is required for efficient CNG methylation (Jackson et al., 2002; Malagnac et al., 2002). In addition, kyp/suvh4 mutants also reactivate the transcription of some retrotransposons, such as TSI, At COPIA4, and At SN1 (Jackson et al., 2002; Lippman et al., 2003; Mathieu et al., 2005). Two additional SU(VAR)3-9 homologs, SUVH5 and SUVH6, also encode H3K9 methyltransferases that are required for the maintenance of heterochromatin formation through H3K9 methylation and CMT3-mediated non-CpG DNA methylation in a locus-specific manner (Jackson et al., 2004; Ebbs et al., 2005; Ebbs and Bender, 2006).
Rice, as one of the most important crop species in the world, has become the model monocot species for functional genomic analysis. The rice genome is 430 Mb in size, and >40% of the genome is composed of repetitive sequences or transposable elements (Hirochika et al., 1992; Wang et al., 1999; Goff et al., 2002). Based on the mechanism of transposition, the transposable elements are classified into two groups, DNA transposable elements (class II) and retrotransposons (class I). The movement of a DNA transposon is primarily mediated through a cut-and-paste mechanism, whereas retrotransposons undergo mobilization through an RNA intermediate, resulting in an increase of copy number (Hirochika, 1997; Wang et al., 1999; Jiang et al., 2003). Tos17 is a copia-like retrotransposon, one of only a few retrotransposons that can mobilize under prolonged cell culture conditions (Hirochika et al., 1996). It has been shown that Tos17 has heavy DNA methylation under normal growth conditions, and transposition only occurs during prolonged tissue culture. The activated Tos17 is resilenced during plant regeneration from explants (Hirochika et al., 1996; Hirochika, 1997; Han et al., 2004; Liu et al., 2004; Cheng et al., 2006). A recent study showed that an increase in Tos17 transcripts is accompanied by a loss of Tos17 DNA methylation (Cheng et al., 2006). Although Tos17 mobilization has been widely used as a tool to generate rice mutants for functional genomic analysis, the mechanism of its transposition remains unclear (Hirochika, 1997; Izawa et al., 1997; Kumar and Hirochika, 2001).
Here, we report that SDG714, a histone H3 methyltransferase specific for H3K9, is involved in histone H3K9 methylation, DNA methylation, and transposition of Tos17 in rice.
RESULTS
SDG714 Encodes a Conserved SET Domain Protein in Rice
There are 38 SET domain proteins, SDG701 to SDG741, in the plant chromatin database (http://www.chrombd.org). Phylogenetic analysis suggests that SDG714 is the most similar to KYP/SUVH4, the major Su(var)3-9 class of histone H3K9 methyltransferase in Arabidopsis (Jackson et al., 2002; Malagnac et al., 2002) (see Supplemental Figure 1 online). Full-length cDNA for SDG714 (accession number AK106700) was obtained by RT-PCR.
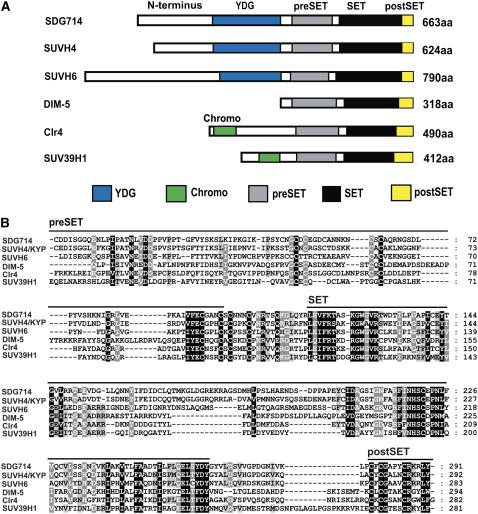
SDG714 encodes a protein of 663 amino acids and contains an YDG domain and a conserved catalytic domain including the SET domain and two Cys-rich motifs, pre-SET and post-SET, critical for histone methyltransferase activity in the C terminus (Rea et al., 2000) (Figure 1A).
Figure 1.
Structures of SDG714 and Its Homologous Proteins.
(A) Diagram of the domain structures of SDG714, KYP/SUVH4, SUVH6, DIM-5, Clr4, and SUV39H1. Proteins all contain pre-SET (gray boxes), SET (black boxes), and post-SET (yellow boxes) domains. The YDG domain (blue boxes) from SDG714, KYP/SUVH4, and SUVH6 and the chromo domain (green boxes) from Clr4 and SUV39H1 are indicated. aa, amino acids.
(B) Alignment of the pre-SET, SET, and post-SET domains of SDG714 with other closely related histone methyltransferases using ClustalW. The amino acid sequence similarities of the catalytic domains from rice SDG714 (372 to 663 amino acids), Arabidopsis thaliana KYP/SUVH4 (332 to 642 amino acids) and SUVH6 (507 to 790 amino acids), Neurospora crassa DIM-5 (64 to 318 amino acids), Schizosaccharomyces pombe Clr4 (208 to 490 amino acids), and Homo sapiens SUV39H1 (131 to 412 amino acids) are shown. The pre-SET, SET, and post-SET domains are indicated by lines.
The YDG domain is conserved in the SUVH class of histone methyltransferases in Arabidopsis but is not found in other histone H3K9 methyltransferases, such as DIM-5 from the fungus N. crassa, Clr4 from yeast, and SUV39H1 from human. DIM-5 lacks an N-terminal conserved domain, whereas Clr4 and SUV39H1 contain a chromo domain at their N termini (Rea et al., 2000) (Figure 1A). Alignment of SDG714 with KYP/SUVH4, SUVH6, DIM-5, Clr4, and SUV39H1 shows very high similarity in the pre-SET, SET, and post-SET motifs, indicating that SDG714 could be an active histone methyltransferase (Figure 1B).
SDG714 Specifically Methylates Histone H3K9
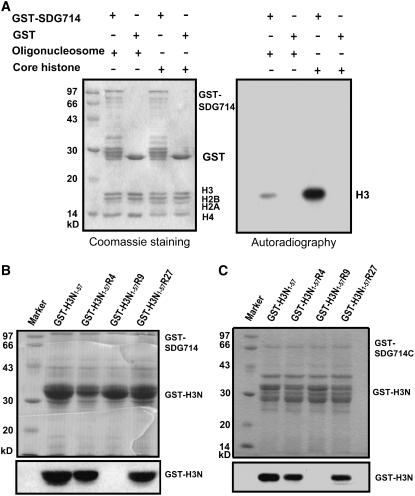
To test whether SDG714 is an active histone methyltransferase, full-length SDG714 fused to glutathione-S-transferase (GST-SDG714) was expressed and purified from Escherichia coli. It has been shown that different histone methyltransferases have different substrate specificities (Nishioka and Reinberg, 2003). For example, SET8 preferentially methylates nucleosomal substrates (oligonucleosomes) at H4K20 (Fang et al., 2002). Therefore, we performed in vitro methylation assays of SDG714 with different substrates, including oligonucleosomes and core histones (Figure 2A). GST-SDG714 strongly methylated histone H3 from core histones but had only weak activity on histone H3 from oligonucleosomes (Figure 2A). As a negative control, GST alone did not methylate any of the substrates. Therefore, we conclude that core histones are the optimal substrate for SDG714 in vitro.
Figure 2.
Methyltransferase Activity and Site Specificity of SDG714 in Vitro.
(A) Methyltransferase activity of SDG714. GST and GST-SDG714 fusion proteins were tested for their ability to methylate oligonucleosomes or core histones. Enzymes and substrates used are indicated at top. The left panel shows the Coomassie blue–stained gel. The positions of GST and GST-SDG714 fusion proteins and different histones are indicated at right. Autoradiography in the right panel indicates methyltransferase activity and specificity.
(B) and (C) Site specificity of SDG714 (B) and the C-terminal part with pre-SET, SET, and post-SET domains of SDG714 (SDG714C; 319 to 663 amino acids) (C). Methyltransferase assays were performed using substrates, as GST-NH3 contains the N-terminal 1 to 57 amino acids of histone H3 fused with GST and point mutations at histone H3K4, H3K9, or H3K27 to Arg (R), named GST-H3N1-57R4, GST-H3N1-57R9, and GST-H3N1-57R27 respectively. Substrates used are indicated at top. The top panels show Coomassie blue–stained gels, and the positions of GST and GST-SDG714C fusion proteins are indicated at right. Autoradiography in the bottom panels indicates methyltransferase activity and specificity.
Because SDG714 is similar to KYP/SUVH4, we wondered whether it also has similar enzymatic specificity. The N terminus of histone H3 was GST-tagged, and GST-H3N1-57 and GST-H3N1-57 with a K-to-R replacement at K4 (GST-H3N1-57R4), K9 (GST-H3N1-57R9), or K27 (GST-H3N1-57R27) were used to test the site specificity of SDG714. As expected, SDG714 methylated GST-H3N1-57, GST-H3N1-57R4, and GST-H3N1-57R27 but not GST-H3N1-57R9, suggesting that SDG714 is a methyltransferase specific for histone H3 at K9 (Figure 2B).
The YDG domain is highly conserved in a group of histone methyltransferases, and we asked whether it is required for enzymatic activity or substrate recognition. The GST fusion protein containing just the pre-SET, SET, and post-SET domains of SDG714 (GST-SDG714C without the N terminus or YDG domain) showed histone methyltransferase activity and specificity similar to full-length GST-SDG714 (Figure 2C). Thus, we conclude that the C-terminal part of SDG714, composed of the pre-SET, SET, and post-SET domains, is sufficient for enzymatic activity and substrate specificity.
SDG714 Localizes to Heterochromatin
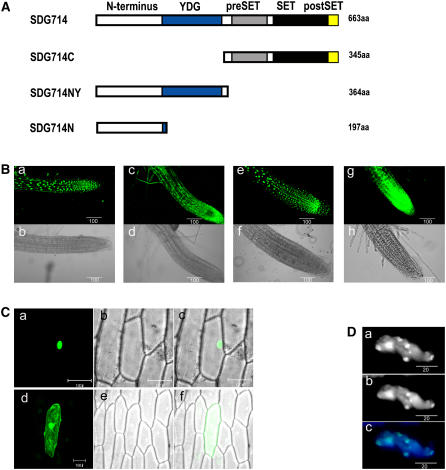
Because SDG714 methylates histones, we would expect it to localize to the nucleus, although no obvious nuclear localization signal was predicted by in silico analysis at http://psort.nibb.ac.jp/. First, we generated stable transgenic Arabidopsis with full-length SDG714, C-terminal SDG714 (SDG714C), or N-terminal SDG714 with or without the YDG domain (SDG714NY or SDG714N) fused to Green Fluorescent Protein (GFP). Schemes of these proteins are shown in Figure 3A. Expression of these fusion proteins in Arabidopsis roots was analyzed by confocal microscopy. GFP fusion proteins of either the full-length SDG714 or the N terminus plus the YDG domain preferentially localized to the nucleus (Figure 3B, panels a, b, e, and f), whereas GFP-SDG714C was observed everywhere (Figure 3B, panels c and d). Thus, SDG714 was localized to the nucleus through its N terminus or YDG domain (Figure 3B, panels g and h). Similar nuclear localization was observed in onion (Allium cepa) epidermal cells, which transiently expressed only the N-terminal SDG714 (GFP-SDG714N) (Figure 3C, panels a to c). Therefore, we conclude that SDG714 nuclear localization is mediated by its N terminus.
Figure 3.
Subcellular Localization of SDG714.
(A) Diagram showing the structures of full-length and truncated SDG714 fused to GFP. Full-length SDG714 (1 to 663 amino acids [aa]) contains the N terminus (white), YDG (blue), and the C-terminal catalytic pre-SET (gray), SET (black), and post-SET (yellow) domains, as indicated at top. SDG714C (319 to 613 amino acids) represents the C-terminal catalytic pre-SET, SET, and post-SET domains; SDG714NY (1 to 364 amino acids) represents the N terminus and YDG domain, and SDG714N (1 to 197 amino acids) represents the N terminus only.
(B) Nuclear localization of SDG714. Subcellular localization of GFP fusion proteins was observed using 7-d-old Arabidopsis seedlings that stably expressed GFP or GFP fusion proteins. Root tip cells overexpressing GFP-SDG714 (panels a and b), GFP-SDG714C (panels c and d), GFP-SDG714NY (panels e and f), and GFP (panels g and h) were observed by confocal laser scanning microscopy (top panels) and bright-field microscopy (bottom panels). Bars = 100 μm.
(C) The N terminus of SDG714 localized in the nucleus. GFP-SDG714N fusion protein (panels a to c) and GFP (panels d to f) were transiently expressed in onion epidermal cells. Green fluorescence was observed by confocal microscopy (panels a and d) and bright-field microscopy (panels b and e). Merged images are shown panels c and f. Bars = 100 μm.
(D) SDG714 was associated with heterochromatin. (a) Seven-day-old Arabidopsis roots containing 35S∷GFP-SDG714 show the distribution of SDG714. (b) The same cell was stained with DAPI, showing heterochromatin of the nucleus. (c) A merged image of panels a and b. Bars = 20 μm.
In Arabidopsis, heterochromatin is located primarily around the centromeres and forms brightly staining chromocenters when stained with 4′,6-diamidino-2-phenylindole (DAPI), whereas heterochromatin in rice is less distinct than in Arabidopsis, being dispersed throughout the genome (Houben et al., 2003). Therefore, we introduced GFP-SDG714 into Arabidopsis to determine whether SDG714 was enriched in heterochromatin. We found a strong correlation between GFP fluorescence and DAPI staining in nuclei, indicating that GFP-SDG714 was highly enriched in chromocenters and hence heterochromatin (Figure 3D, panels a to c).
Generation of SDG714-Deficient Plants by RNA Interference
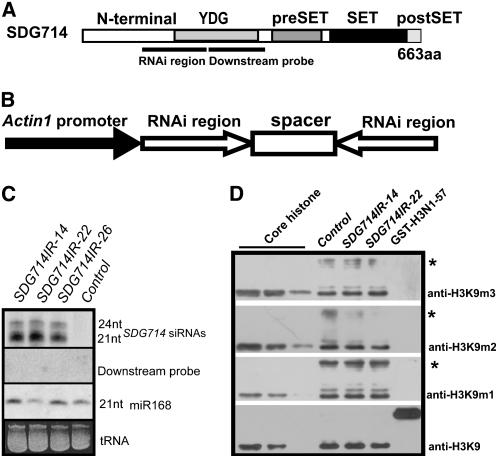
To understand the roles of SDG714 in rice, we used an RNA interference (RNAi) approach to knock down SDG714. Phylogenetic analysis showed that SDG714 was very close to another SET domain–containing protein, SDG727, with 36% identity and 52% similarity. Therefore, amino acids 238 to 362, a less conserved region at the N terminus, and the YDG domain of SDG714 were selected to ensure the specificity of RNAi (Figure 4A). A hairpin construct was driven by the rice Actin1 promoter, and transgenic rice explants were selected and regenerated (Figure 4B). Control transgenic plants in which the vector alone was transformed were also generated. A total of 13 transgenic plants containing SDG714 inverted repeats (SDG714IR) transgenes were obtained, as verified by PCR amplification of the kanamycin resistance gene and detection of small interfering RNA (siRNA) production corresponding to the inverted repeats region of the RNAi construct (Figure 4C; data not shown). By RT-PCR analysis, we observed significant reduction of SDG714 expression in SDG714IR transformants, indicating that we obtained SDG714-deficient plants by RNA interference (data not shown). To rule out the possibility of off-target effects from inverted repeats, the same membrane was blotted using a downstream region as a probe (Figure 4A). We did not detect any siRNAs corresponding to regions downstream of the SDG714IR target site, indicating that off-target effects did not occur in our RNAi progeny (Figure 4C). SDG714 methylates histone H3K9 in vitro; therefore, we first determined whether the global methylation status of histone H3 at K9 was affected by a loss of function of SDG714. Protein immunoblot analysis was performed using antibodies against monomethylated, dimethylated, or trimethylated histone H3K9. However, we detected no significant differences between the control and SDG714IR lines (Figure 4D). There are 38 SET domain–containing proteins in rice, and they could be functionally redundant. Alternatively, SDG714 preferentially acts at specific sequences but not globally in rice.
Figure 4.
SDG714IR Transformants and Protein Gel Blot Analysis.
(A) Domain structure of the SDG714 protein. The relative region used in the RNAi knockdown and downstream probe regions are indicated by lines. aa, amino acids.
(B) Diagram of SDG714 RNAi constructs. Fragments containing the SDG714 fragment in the sense and antisense orientations separated by an unrelated spacer were cloned under the control of the rice Actin1 promoter.
(C) siRNA accumulation in SDG714IR transformants. Results from three independent SDG714IR transformants (SDG714IR-14, SDG714IR-22, and SDG714IR-26) and a control plant transformed with the vector alone (control) are shown. Small RNAs from leaf tissue were probed with the SDG714 inverted repeats region, downstream probe, or oligonucleotides (nt) complementary to microRNA168 (miR168) used as an internal control. tRNA bands visualized by ethidium bromide were used as internal loading controls.
(D) Protein blot analysis of in vivo histone H3K9 methylation status. Histone extracts were isolated from a control plant and SDG714IR-14 and SDG714IR-22 transformants. Core histones (8, 6, and 4 μg) from calf thymus and E. coli–expressed GST-H3N1-57 fusion protein were used as positive and negative controls for histone modifications. Histone extracts were calibrated using H3 modification–insensitive antibody (anti-H3K9). Antibodies against monomethylated (anti-H3K9m1), dimethylated (anti-H3K9m2), and trimethylated (anti-H3K9m3) histone H3K9 are shown at right. Asterisks denote possible nonspecific proteins.
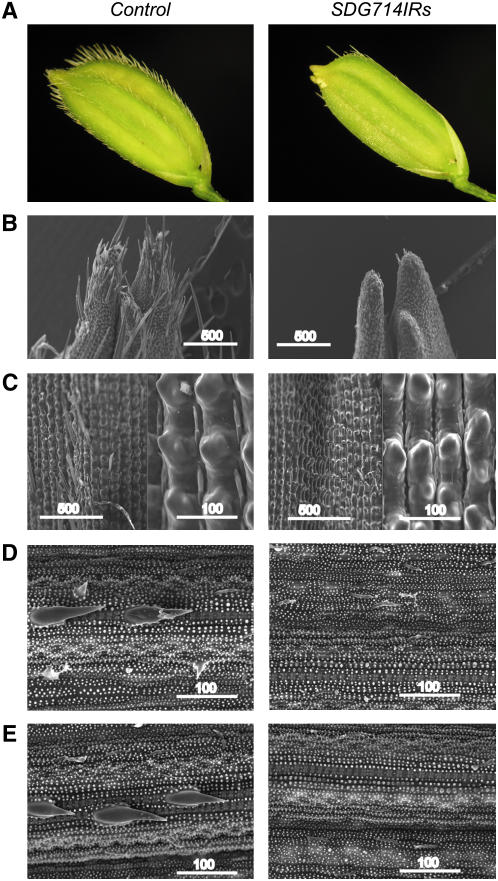
SDG714IR Transformants Are Defective in Macro Trichome Development
After regeneration from explants, 12 of 13 SDG714IR transformants were observed with glabrous glumes. However, 14 control plants that contained the vector alone produced normal trichomes in glumes (Figure 5A). We further examined the glumes with a scanning electron microscope and found that the apical hairs on glumes were lost in SDG714IR transformants (Figure 5B). In rice, there are two types of trichomes, macro and micro hairs. As shown in Figure 5B, only macro hairs disappeared in SDG714IR transformants; the micro hairs were not affected. To confirm this finding, greater magnification was applied to the middle part of the glumes. We found that micro hairs developed normally, whereas no macro hairs were observed in SDG714IR glumes (Figure 5C).
Figure 5.
Macro Trichomes Are Lost in SDG714IR Transformants.
(A) Microscopic analysis of seeds from controls (left) and SDG714IR transformants (right).
(B) to (E) Scanning electron micrographs show loss of macro trichomes on the apical glumes (B), the middle region of glumes with greater magnification (C), the leaves (D), and the culms (E) in controls (left) and SDG714IR transformants (right). Bars = 500 or 100 μm, as indicated.
In wild-type rice, trichomes are present not only on glumes but also on leaves and culms, and we tested whether trichome development on leaves and culms was also affected. Micro hairs were located along the stomata cells on the adaxial side of leaves of both SDG714IR and control plants (Figure 5D). The macro hairs were completely lost on SDG714IR leaves, whereas they were located on silica cells over a thin vascular bundle on control mature leaves. Similarly, macro but not micro hairs were lost on SDG714IR culms (Figure 5E). These observations indicated that development of macro hairs in rice at glumes, leaves, and culms was affected when SDG714 expression was knocked down.
Histone Methylation and DNA Methylation at Tos17 Loci Are Impaired in SDG714IR Transformants
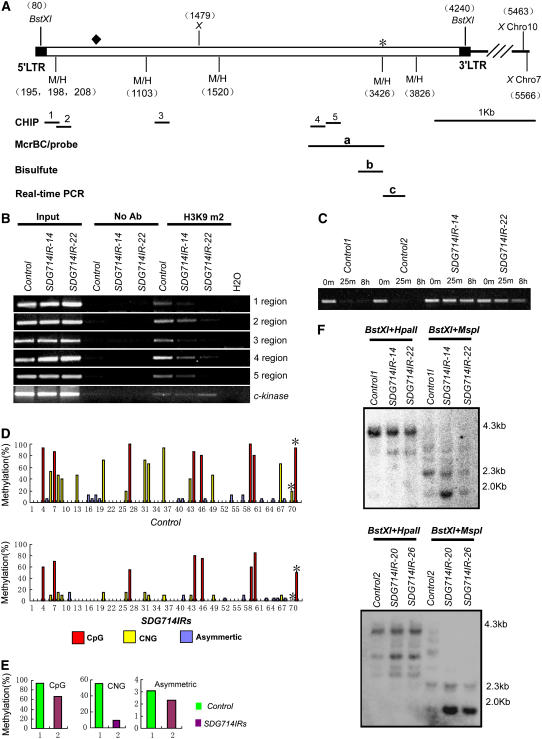
Tos17 is a copia-like retrotransposon belonging to class I transposable elements in wild-type cv Nipponbare rice (Hirochika et al., 1996). Under normal growth conditions, it is associated with large amounts of DNA methylation (Liu et al., 2004; Cheng et al., 2006). Because of its heterochromatic nature, Tos17 was chosen to study the role of SDG714 in histone methylation and DNA methylation. Using the Tos17 sequence as the query (Hirochika et al., 1996), we searched the newly annotated rice genome and identified two copies of Tos17 located at 80,982 to 85,185 of BAC AC087545 on chromosome 10 (renamed Tos17A) and 26,641,215 to 26,645,328 of BAC AP008213 on chromosome 7 (Tos17B). These two copies have identical sequences except for an additional 90-bp insertion in Tos17A (from 569 to 659 bp). A diagram of the Tos17A structure is shown in Figure 6A.
Figure 6.
Decreased Histone H3K9 Methylation and DNA Methylation at Tos17 Loci in SDG714IR Transformants.
(A) Structure of Tos17 on chromosome 10 (Chro10) and chromosome 7 (Chro7). 5′ and 3′ long terminal repeat (LTR) regions are indicated with black boxes. Numbers at top and bottom represent relative positions of the indicated restriction sites to the beginning of the 5′ long terminal repeat on chromosome 10 and chromosome 7, respectively. The black diamond indicates a 90-bp deletion on chromosome 7. Restriction maps of Tos17 were labeled. M/H indicates MspI and HpaII restriction enzymes, and X indicates the XbaI restriction enzyme. Different regions for the chromatin immunoprecipitation (CHIP) assay (regions 1 to 5), McrBC-PCR analysis (a), bisulfite sequencing (b), DNA methylation–sensitive DNA gel hybridization probe (a), and real-time PCR analysis (c) are shown. The asterisk represents the HpaII/MspI site, which was also determined by bisulfite sequencing analysis in (D).
(B) Chromatin immunoprecipitation analysis of H3K9 dimethylation in SDG714IR transformants (SDG714IR-14 and SDG714IR-22). Chromatin immunoprecipitation assays were performed with antibodies against dimethyl Lys-9 of histone H3 (H3K9m2). Primers specific for regions 1 to 5 of Tos17 are shown in (A). The euchromatin marker C-Kinase was used as an internal control. No Ab, no antibody.
(C) McrBC-PCR analysis of DNA methylation at Tos17 loci. Equal amounts of genomic DNAs from two controls (control 1 and control 2) and two SDG714IR transformants (SDG714IR-14 and SDG714IR-22) were digested with McrBC for 0 min, 25 min, and 8 h, followed by PCR amplification of region a shown in (A).
(D) Profile of DNA methylation at a 400-bp region (shown in [A]) of Tos17. The numbers on the x axis represent cytosine positions in the analyzed region, and the y axis represents methylation levels in controls (top panel) and SDG714IR transformants (bottom panel). Red, blue, and yellow bars indicate CpG, CNG, and asymmetric methylation, respectively. Asterisks indicate methylation at the CCGG position corresponding to the HpaII/MspI site (3426 bp from the 5′ long terminal repeat on chromosome 10) in control plants and SDG714IR transformants.
(E) Histograms represent the percentage of CpG, CNG, and asymmetric methylation in control plants (green) and SDG714IR transformants (purple).
(F) DNA gel blot analysis of DNA methylation in SDG714IR transformants. Genomic DNAs of two controls or SDG714IR transformants (SDG714IR-14 and SDG714IR-22 in the top panel, SDG714IR-20 and SDG714IR-26 in the bottom panel) were digested with BstXI, followed by methylation-sensitive enzyme HpaII or MspI, and the blot was hybridized with the probes shown in (A). DNA markers are shown at right.
To test whether SDG714 plays a role in maintaining the silenced status of Tos17, chromatin immunoprecipitation analysis was performed. C-Kinase was used as a euchromatin marker in rice, as described previously (Nagaki et al., 2004). We chose five regions of Tos17, two 5′ long terminal repeats and three coding regions of Tos17, as indicated in Figure 6A. We found that H3K9 dimethylation was eliminated or decreased in SDG714IR transformants in all five regions tested compared with those of control plants (Figure 6B). Because histone H3K9 dimethylation is a hallmark for heterochromatin (Lippman et al., 2004), this result indicates that SDG714 is involved in maintaining the heterochromatin status of Tos17.
To examine whether DNA methylation at Tos17 is also dependent on SDG714, DNA methylation status was analyzed with McrBC-PCR. McrBC is an endonuclease that cuts DNA between methylated cytosine residues but not unmethylated DNA. After McrBC treatment, a methylated DNA will be digested and therefore will not be amplified by PCR. The region labeled a in Figure 6A, which was shown previously to be methylated at Tos17, was analyzed (Liu et al., 2004). In control plants with transformed vector alone, this region could not be amplified after only 25 min of McrBC treatment, consistent with a previous report that Tos17 is heavily methylated in wild-type plants (Liu et al., 2004; Cheng et al., 2006). By contrast, the same region could be easily amplified from SDG714IR transformants even after 8 h of incubation with McrBC (Figure 6C). Therefore, Tos17 loci are hypermethylated in two control plants and hypomethylated in SDG714IR transformants. This is consistent with a previous finding that histone methylation controls DNA methylation in Arabidopsis (Jackson et al., 2002), indicating that this mechanism is conserved between monocots and dicots.
In higher plants, DNA methylation is found in all sequence contexts (CpG, CNG, and asymmetric), and kyp/suvh4 shows genome-wide loss of CNG methylation in Arabidopsis (Lindroth et al., 2001; Cao and Jacobsen, 2002; Cao et al., 2003; Tran et al., 2005). To determine which types of DNA methylation are affected in SDG714IR transformants, genomic bisulfite sequencing was used to detect the DNA methylation patterning at the b region, which is part of the a region of Tos17 (Figure 6A). We sequenced 15 individual clones from control plants and 20 from SDG714IR transformants. Compared with control plants, SDG714IR transformants showed reductions of methylation at both CpG (from 94.8 to 66.7%) and at CNG (from 54.9 to 9.1%) sites (Figures 6D and 6E). Very low (<4%) asymmetric methylation was observed in control and SDG714IR transformants (Figures 6D and 6E). Furthermore, DNA methylation–sensitive DNA gel blot analysis was used to verify this result. Genomic DNA was digested with BstXI to give rise to 4.1-kb fragments, followed by digestion with the methylation-sensitive enzyme, either HpaII or MspI, both of which recognize the sequence CCGG. HpaII is sensitive to methylation of either an outer or an inner cytosine, whereas MspI is sensitive to only the outer cytosine (Jackson et al., 2002). Using probe a, corresponding to the middle region of Tos17 (Figure 6A), SDG714IR samples showed a reproducible increased digestion with both HpaII and MspI compared with control plants, indicating that both outer and inner cytosine methylation were affected in SDG714IR plants (Figures 6E and 6F). The DNA gel blot result (Figure 6F, top panel) is also consistent with the bisulfite sequence data analyzed at the HpaII/MspI site, as indicated by the asterisk (Figures 6A and 6D). Collectively, these data suggest that in addition to its conserved role in the control of CNG methylation by KYP/SUVH4 in Arabidopsis, SDG714 also plays a role in maintaining CpG methylation, which may have additional functions in rice.
Active Transcription and Transposition of Tos17 in SDG714 Transformants
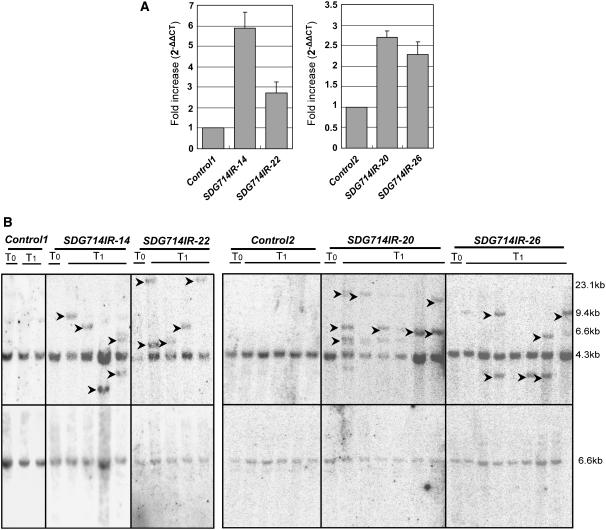
As both histone H3K9 dimethylation and DNA methylation of Tos17 were reduced in SDG714IR transformants, real-time PCR was performed to determine whether transcription of Tos17 was affected. The amplified region is shown in Figure 6A. Ubiquitin was used as an internal control. We found that Tos17 expression levels were increased by twofold to sixfold in SDG714IR plants compared with the control plants (Figure 7A). This result is consistent with the previous finding that loss of DNA methylation and H3K9 methylation mediated by CMT3 and KYP/SUVH4 led to increased expression of TSI (Jackson et al., 2002).
Figure 7.
Transcription and Transposition of Tos17 in SDG714IR Transformants.
(A) Upregulated transcription of Tos17 in SDG714IR transformants. Tos17 transcription was assessed by real-time PCR (top panel) in two control plants (control 1 and control 2) and four independent SDG714IR transformants (SDG714IR-14, SDG714IR-20, SDG714IR-26, and SDG714IR-22) as indicated. The amplified PCR region of Tos17 corresponds to the c region in Figure 6A. The mean ΔΔCt was converted to a relative value by the equation 2−ΔΔCt. Data are presented as 2−ΔΔCt. The value of 2−ΔΔCt indicates the fold change in gene expression relative to the control. Data represent four independent experiments, and error bars represent sd.
(B) Transposition of Tos17 in SDG714IR transformants. Genomic DNA isolated from control plants transformed with the vector alone (control 1 and control 2) or SDG714IR transformants (SDG714IR-14, SDG714IR-22, SDG714IR-20, and SDG714IR-26) from the T0 or T1 generation as indicated at top was digested with the methylation-insensitive enzyme XbaI and sequentially probed with Tos17 (top panel) and Tubulin (bottom panel). Arrowheads indicate the positions of newly transposed Tos17. DNA size markers are shown at right.
It has been shown that loss of both CpG and CNG methylation leads to the transposition of the CACTA transposon in Arabidopsis (Miura et al., 2001; Kato et al., 2003). Here, we tested the hypothesis that a defect in a histone H3K9 methyltransferase will cause the transposition of transposable elements, specifically, whether there will be Tos17 transposition in SDG714IR plants, in which histone H3K9 methylation and DNA methylation at Tos17 are lost. We analyzed the copy number of Tos17 in the T0 and T1 generations of SDG714IR transformants. Transgenic plants containing the vector alone were used as controls to avoid the effect of Tos17 transposition caused by the tissue culture process. There were two copies of Tos17 in both T0 and T1 control plants (Figure 7B). In the SDG714IR transformants, two copies of Tos17 were observed in the T0 generation. However, three to four copies appeared in most of the T1 generation of SDG714IR transgenic progeny (Figure 7B, top panel). The increased copy number was not attributable to incomplete digestion, because only one band was observed when the blot was reprobed with Tubulin (Figure 7B, bottom panel). This result shows a direct link between histone methylation, DNA methylation, transcription, and transposition of a retrotransposon.
DISCUSSION
Differences between SDG714 and KYP/SUVH4
It has been shown that tagged KYP/SUVH4, SUVH1, and SUVH2 in Arabidopsis and tobacco SET1 are localized to heterochromatin (Yu et al., 2004; Naumann et al., 2005). Here, we demonstrate that SDG714 is also a nuclear protein that associates with heterochromatic chromocenters in Arabidopsis. Still, the exact mechanism of how SDG714 is targeted to heterochromatin remains to be addressed. The similar enzymatic properties and cellular localization indicate that SDG714 and KYP/SUVH4 gene diversification occurred before the divergence of monocots and dicots. After the monocot and dicot split, rice and Arabidopsis evolved many species-specific sequences in each genome. These unique sequences may require different cofactors along with either SDG714 or KYP/SUVH4 to function. The differences between SDG714 and KYP/SUVH4 were also supported by the observation that DNA methylation patterns maintained by SDG714 and KYP/SUVH4 may not be identical, as SDG714 is required for both CpG and CNG methylation at Tos17 and KYP/SUVH4 acts mainly on CNG methylation. As a result, SDG714 and KYP/SUVH4 may not be orthologous in terms of developmental regulation, because SDG714 could not complement Arabidopsis KYP/SUVH4 in kyp clk-st double mutants (for details, see Supplemental Results online). In addition, we observed that reduction of SDG714 mRNA leads to defects in macro trichome development, whereas there is no obvious developmental defect in plants without KYP, its closest homolog in Arabidopsis. It would be interesting to identify genes causing altered macrohair defects in the loss of function of SDG714 in rice.
Significance of SDG714 in Controlling Tos17 Silencing
In eukaryotes, the presence of 5′ methylcytosine is an important component of transcriptionally silent chromatin and is widely considered to be a mechanism that the genome uses to defend against transposable elements and retroviruses (Constancia et al., 1998; Covey and Al-Kaff, 2000; Lorincz et al., 2001). For example, loss of function of CMT3, a plant-specific DNA methyltransferase that mainly controls non-CpG methylation, causes transcriptional activation of retroelements at Ta3 and TSI in Arabidopsis (Lindroth et al., 2001). Loss of CpG and CNG methylation leads to mobilization of the CACTA transposon in cmt3 and met1 double mutants (Kato et al., 2003). Besides DNA methylation, transcriptionally silenced retroelements are also maintained through histone methylation and siRNA-mediated gene silencing within a gene regulation network (Tran et al., 2005). In the kyp/suvh4 mutant, the retrotransposons TSI, At COPIA4, and At SN1 were actively transcribed, indicating a role for histone and DNA methylation in transcriptional silencing of retrotransposons (Lindroth et al., 2001; Jackson et al., 2002; Lippman et al., 2003; Mathieu et al., 2005). In addition, large-scale DNA methylation analysis also showed that KYP/SUVH4 preferentially regulates transposable elements (Tran et al., 2005). And SUVH5 and SUVH6 were identified to be partially redundant with KYP/SUVH6 in controlling CMT3-mediated non-CpG methylation and repressing the transcription of transposable elements in heterochromatin (Jackson et al., 2004; Ebbs et al., 2005; Ebbs and Bender, 2006).
In rice, Tos17 is a copia-like retrotransposon that is activated and transposed during prolonged cell culture and subsequently remethylated and silenced in regenerated plants (Hirochika et al., 1996; Cheng et al., 2006). Because of its ability to transpose, Tos17 has been widely used as a tool for generating mutants in rice (Hirochika, 1997; Kumar and Hirochika, 2001). However, the exact mechanism of how this transposition is controlled is unknown. Here, we show that decreased expression of SDG714 results in a decrease in histone methylation and DNA methylation and an increase in Tos17 expression. Our work highlights the important connection between histone methylation, DNA methylation, transcription, and the transposition of a retrotransposon.
Surprisingly, our real-time PCR results showed that Tos17 transcript was enhanced by only twofold to sixfold between RNAi and control plants but led to significant differences, between no transposition in the controls and active transposition in the RNAi lines (Figures 7A and 7B). One possibility is that Tos17 transposition requires a certain transcript threshold and that loss of function of SDG714 increases the Tos17 transcript over the threshold, resulting in a high frequency of transposition in SDG714IR plants. A similar phenomenon was observed in CACATA transposition in the met1 and cmt3 backgrounds (Kato et al., 2003). Alternatively, transcript accumulation is not the major barrier to the transposition of retrotransposons. Instead, active transposition might be affected mainly through the epigenetic regulation of chromatin, such as DNA methylation, histone covalent modifications, or ATP-dependent chromatin remodeling. Here, we show that SDG714 targeted to heterochromatin and loss of function of SDG714 decreased histone methylation and DNA methylation at Tos17 loci, which might in turn modulate heterochromatin, resulting in higher levels of transposition. It is reported that Tos17 can be activated and transposed during prolonged cell culture. Although the tissue culture process is required for the generation of SDG714IR plants, no transposition was observed in the control plants transformed with vector only (Figure 7B). Therefore, we proposed that during the transformation of rice, condensed chromatin might be loosened, decreasing the threshold for the activation of Tos17 expression and transposition. However, transformation alone would not reach the threshold for transposition of Tos17, because no transposition of Tos17 was observed in control plants in which the identical transformation process was applied. By contrast, when SDG714 was knocked down, in addition to partially released silencing by transformation, Tos17 was further modulated as a result of the loss of histone H3K9 and DNA methylation, resulting in the upregulation of gene expression and activated transposition. Therefore, a synergic effect between the loss of function of SDG714 and transformation processing led to stronger activation of Tos17 expression and activated transposition.
It has been shown that Tos17 preferentially jumps into euchromatic regions, which would be harmful to the rice genome if it were not properly silenced (Miyao et al., 2003). Here, we find a significant role for SDG714 in rice genome stability, similar to what has been observed in mice. suv39 h–defective mice display severely impaired viability and chromosomal instability, with an increased risk of tumorigenesis (Peters et al., 2001). Practically, loss of function of SDG714 can potentially be used as a tool for the generation of mutants in rice. The advantage of using the SDG714IR system to generate mutants is that prolonged tissue culture is no longer needed, as several extra copies of Tos17 can be observed in the T1 generation. Once the SDG714IR transgene is outcrossed in the T2 generation, Tos17 should be resilenced, and the homozygous progeny of Tos17 insertions will give rise to insertional mutations.
METHODS
Plant Materials and Growth Conditions
Rice plants used in this study were Oryza sativa spp japonica cv Nipponbare. The plants were all regenerated from the callus. T1 generation plants were generated from self-pollinated T0 generation seeds. Plant materials were harvested from leaf tissue grown in the field.
Molecular Cloning of SDG714
cDNAs encoding amino acids 1 to 663 of SDG714 were PCR-amplified from reverse-transcribed rice cDNA by forward primer CX187 (5′-TAGagatctATGGAGGTGATGGATTCGGTGGCCGT-3′; lowercase and underlined letters represent the restriction sites added for subsequent cloning) and reverse primer CX188 (5′-GTCtctagaCTAATAAAGTCGCTTCCGGCAGTAC-3′), and PCR products were cloned into Topo-PCR4.0 (XF00204). SDG714 was then digested with BglII and EcoRI and subcloned into pGEX-4T-1 via BamHI and EcoRI sites, giving GST-SDG714 (XF00206). To generate the C-terminal fragment of SDG714, the cDNA fragments encoding amino acids 319 to 663 of SDG714 were amplified by forward primer CX48 (5′-GAGgaattcTACAAGGTTGTAGATGACTGGGTGCAG-3′) and reverse primer CX47 (5′-GTCgtcgacATCTAATAAAGTCGCTTCCGGCAGTAC-3′). PCR products digested with EcoRI and SalI were further cloned into pGEX-4T-1 at EcoRI and SalI sites, giving GST-SDG714C (XF00163).
Protein Expression and Purification
The plasmids GST-SDG714 and GST-SDG714C, containing either the full length or the C-terminal end of SDG714, respectively, were transformed into BL21(RIL), and single clones were cultured in 2× YT medium plus 100 μg/mL ampicillin at 37°C to OD 0.6, then induced by 0.1 mM isopropylthio-β-galactoside at 18°C for 12 h. GST fusion proteins were purified as reported previously (Jackson et al., 2002).
In Vitro Histone Methyltransferase Assay
The detailed histone methyltransferase assay was performed according to the methods described by Rea et al. (2000) with slight modification. Briefly, 10 μg of oligonucleosomes (containing 2.5 μg of H2A, H2B, H3, and H4 each, wrapped with DNA; kindly provided by Y. Zhang) and core histones (containing 2.5 μg of H2A, H2B, H3, and H4 each) as substrates, 1 μCi of [3H]S-adenosyl-l-[methyl-3H]l-Met (79 Ci/mmol; TRK865; Amersham Biosciences) as methyl donor, and 2 to 5 μg of recombinant protein were used in the histone methyltransferase assay. Histone methyltransferase reactions on GST-NH3 fusion proteins (gifts of Y. Shinkai) were performed using similar reaction conditions. After incubation at 30°C for 1 h, reactions were stopped with SDS loading buffer, and protein was separated by 15 or 12% SDS-PAGE, stained with Coomassie Brilliant Blue R 250, treated with Amplifier (Amersham Biosciences), dried, and fluorographed on x-ray film.
Construction of Binary Vectors
To generate pCAMBIA1300-35S-GFP, 35S-GFP from pAVA321 vector was cloned into pCAMBIA1300 vector as a GFP control and named pCAMBIA1300-35S-GFP (XF00183). To generate GFP fusion proteins, primers used for the amplification of each fragment were as follow: CX187 and CX188 for full-length SDG714 (SDG714-GFP; 1 to 663 amino acids); CX48 and CX47 for the C terminus of SDG714 (SDG714C-GFP; 319 to 663 amino acids); CX187 and reverse primer CX317 (5′-GACtctagaATCGTGGTGGGAGCTTCAGCACGAG-3′) for the NY terminus of SDG714 (SDG714NY-GFP; 1 to 364 amino acids); and CX187 and reverse primer CX1366 (5′-AGCtctagaTGATCCCCAACATCAACACCAGGTA-3′) for the N terminus of SDG714 (SDG714N-GFP; 1 to 197 amino acids). Each fragment was subcloned into binary vector pCAMBIA1300-35S-GFP (XF00183) with appropriate restriction enzymes. The detailed process will be provided upon request. Binary vectors of pCAMBIA1300-35S-GFP, SDG714-GFP, SDG714NY-GFP, and SDG714C-GFP were transferred into Agrobacterium tumefaciens strain AGL0 and used to transform Arabidopsis thaliana ecotype Columbia plants by vacuum infiltration. T1 seeds were selected on half-strength Murashige and Skoog medium supplemented with hygromycin (25 μg/mL). To generate the SDG714 RNAi vector, the fragment was amplified with primers CX52 (5′-ATTctcgagGCAACTTGTATTGTCATGTCGGG-3′) and CX53 (5′-AGCagatctGGGAGCTTCAGCACGAGTAA-3′), then sequentially cloned into pUCRNAi vector with XhoI/BglII and BamHI/SalI. Finally, the full stem-loop fragment was cloned into pCAMBIA2300-Actin vector (Liu et al., 2005).
Transient Expression of SDG714N-GFP
The SDG714N-GFP construct was coated onto gold particles (0.1 μm) and delivered into onion (Allium cepa) epidermal cells with a Helios Gene Gun System (Bio-Rad Laboratories). The bombardment parameters were as follows: discharge pressure of 150 p.s.i.; and distance to target tissue of 3.5 cm. Onion epidermal cells were placed onto half-strength Murashige and Skoog agar plates before bombardment, incubated at 23°C for 24 h, and analyzed with a Zeiss LSM 510 confocal microscope.
Protein Immunoblot Analysis
Leaves were homogenized in histone extraction buffer (0.25 M sucrose, 60 mM KCl, 15 mM NaCl, 5 mM MgCl2, 1 mM CaCl2, 15 mM PIPES, pH 6.8, 0.8% Triton X-100, 0.1 mM Pefablock, and protease inhibitor cocktail), as modified from Tariq et al. (2003). After centrifugation at 10,000g for 10 min, the pellet was resuspended with 0.4 M H2SO4 and centrifuged again; the soluble proteins were precipitated with 12 volumes of acetone, left at −20°C overnight, and then collected by centrifugation at 8000g for 15 min. The pellet was resuspended with 4 M urea (100 μL), separated on a 15% PAGE gel, detected by monomethylated, dimethylated, or trimethylated specific H3K9 and methylation-insensitive H3 antibodies (catalog Nos. 07-450, 07-441, 07-442, and 06-755; Upstate). Peroxidase-conjugated goat anti-rabbit antibodies (Pierce) were used as secondary antibody, and the SuperSignal Western Pico detection system (Pierce) was used for signal detection. The same blot was stripped before probing the next antibody. Escherichia coli–expressed GST-H3 (1 to 57 amino acids) fusion protein and diluted core histones were used as loading controls.
Microscopy
Seven-day-old seedlings were used for GFP analysis. The GFP fluorescence of protein was visualized by confocal laser scanning microscopy (Olympus). Heterochromatin sublocation was analyzed with Arabidopsis roots. The roots were stained with 2 μg/mL DAPI, and GFP images were captured with the Olympus BX61 fluorescence microscope in conjunction with a micro CCD camera. Gray-scale images were captured for each color channel and then merged using Image-Pro Plus software.
Scanning Electron Microscopy
Scanning electron microscopy was performed as described previously with slight modifications (Li et al., 2003). Rice tissue was placed into 5% formaldehyde dissolved in 1× PBS for 18 h, dried with 30, 50, 70, 90, and 100% ethanol, each for 10 min, and then placed into glutaraldehyde for 12 h. The material was then point-dried with a CO2 critical point dryer (Hitachi HCP-2), spotted with gold powder with ion sputter (Hitachi E-1010), and observed with a scanning electron microscope (Hitachi S-3000N).
Reverse Transcription and Real-Time PCR
Total RNA isolation and RT-PCR were performed as described previously (Liu et al., 2005). Real-time PCR analysis was performed using the Chromo4 real-time PCR instrument (PTC-200; Bio-Rad) and SYBR Green I (Invitrogen S-7567). The relative expressions of specific genes were quantitated using 2−ΔΔCt calculation, where ΔΔCt is the difference in the threshold cycles of the test and housekeeping gene Ubiquitin. ΔCt is the threshold cycle of the target gene subtracted from the threshold cycle of the housekeeping gene. The mean threshold cycle values for the genes of interest were calculated from three experiments. Primers used were as follows: for Ubiquitin, CX2137 (5′-GCCCAAGAAGAAGATCAAGAAC-3′) and CX2138 (5′-AGATAACAACGGAAGCATAAAAGTC-3′); for Tos17, CX2141 (5′-TGTCAGTGCTCCGATTTCAGTTCA-3′) and CX2142 (5′-AAATACAATAGCCAGTGACAGAGCG-3′).
Small RNA Gel Blot Analysis
Small RNA gel blot analysis was performed as described by Liu et al. (2005). Probes corresponding to the inverted repeated and downstream region of SDG714 were PCR-amplified with CX52 and CX53 or with CX2095 (5′-CACGATTTCGGAATTACCTG-3′) and CX2096 (5′-AAGCGATACTGCAGACCTTT-3′), respectively. The PCR products were then cloned into pGEM-T easy vector (Promega), transcribed with T7 polymerase in vitro, and labeled with [α-32P]ATP. Probe corresponding to microRNA168 (CX161, 5′-GTCCCGATCTGCACCAAGCGA-3′) was end-labeled with [γ-32P]ATP.
Chromatin Immunoprecipitation Assay
Chromatin immunoprecipitation was performed as described previously (Johnson et al., 2002). Briefly, 2 g of leaves was fixed with formaldehyde, and the DNA/protein complex was immunoprecipitated with α-dimethyl H3K9. After reverse cross-linking and proteinase K treatment, the immunoprecipitated DNA was extracted with phenol/chloroform. All PCR procedures were performed in 30-μL volumes, started with 2 min at 94°C, and followed with 30 to 40 cycles. The PCR product was electrophoresed on a 2 to 3% agarose gel. The primers were as follows: for region 1, CX1675 (5′-TGTACTGTATAGTTGGCCCATGTCC-3′) and CX1676 (5′-GATGGGGAATTGGCAGCTAG-3′); for region 2, CX1250 (5′-GTTAGGTTGCAAGTTAGTTAAGA-3′) and CX1251 (5′-GTCAACGACAAATCGCGCTG-3′); for region 3, CX1668 (5′-ACTCAGGAGCCTCCTTTCAT-3′) and CX1669 (5′-GTTTTGGAACAAGTGACACA-3′); for region 4, CX430 (5′-GCTACCCGTTCTTGGACTAT-3′) and CX1665 (5′-ATACATGTCCTGGTGGAGCT-3′); for region 5, CX1692 (5′-GATCTTCATGAGGAGGTATA-3′) and CX1693 (5′-TATGAATGAATAGTGCTGGG-3′); and for C-Kinase, CX1262 (5′-CGACTAAACCACTCCAATCATC-3′) and CX1263 (5′-CCAATCAAAACTTCTCCTGTAA-3′).
McrBC-PCR
Genomic DNA was isolated from leaf tissue. For McrBC-PCR analysis of Tos17, 500 ng of genomic DNA was digested with 20 units of McrBC restriction enzyme (New England Biolabs) for 0 min, 25 min, and 8 h. Equal amounts of McrBC-digested DNAs were used for PCR amplification by primers CX430 and CX431 (5′-CTGAAATCGGAGCACTGACA-3′).
Bisulfite Sequencing
For Tos17 bisulfite sequencing, 3 μg of genomic DNA was digested with EcoRI (New England Biolabs). Samples were extracted once with phenol/chloroform and then precipitated with 3 volumes of ethanol and 30 μg of tRNA. After centrifugation, pellets were dissolved in 40 μL of water, heated at 97°C for 5 min, and quenched on ice. Two microliters of NaOH (6.3 M; freshly prepared) was added and incubated at 39°C for 30 min, followed by the addition of 416 μL of bisulfite solution to the denatured DNA. Bisulfite solution was prepared as follows: 40.5 g of sodium bisulfite (Fisher S654-500) was dissolved in 80 mL of water, adjusted to pH 5.1, 3.3 mL of 20 mM hydroquinone was added (Sigma-Aldrich H-9003), and the volume was adjusted to 100 mL with water. Samples were incubated in a PCR machine for five cycles of 55°C for 3 h and 95°C for 5 min. After bisulfite conversion, the Wizard DNA clean-up system was used to remove extra salt (Promega). Then, NaOH was added to a final concentration of 0.3 M and incubated at 37°C for 15 min. The bisulfite-treated DNA was precipitated with 3 volumes of ethanol plus 20 μg of tRNA. DNA was dissolved in 100 μL of TE (10 mM Tris-HCl and 1 mM EDTA, pH 8.0). PCR was performed, and the PCR products were cloned into pGEM-T easy vector (Promega) and sequenced. The primers used for Tos17 were CX1787 (5′-GTTGATTAtAGGGGATGATTTGGAGTAtATTGtTT-3′) and CX1103 (5′-CaTAAaACACaAAaCAAaTaACCATAaTaAACTa-3′), with lowercase letters representing C-to-T or G-to-A substitution in Tos17, respectively.
DNA Gel Blot Analysis
Genomic DNA (5 to 10 μg) was digested with 30 units of the appropriate restriction enzymes (New England Biolabs) for 3 h. The digested DNAs were separated on 0.8% agarose gels and transferred onto Hybond N+ (Amersham). The preparation of blots, hybridization, and probes were as described previously (Cao and Jacobsen, 2002). The PCR product used for Tos17 probe in DNA methylation and the copy number assay was amplified with CX430 and CX431 from genomic DNA as a template. The probes for tubulin were amplified with primers CX1201 (5′-GAGAGAGATCCTGCACATCCA-3′) and CX1202 (5′-ACTCCTCCCTGATCTTTGATATC-3′).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: Tos17, D85876; SDG714, AK106700; Arabidopsis thaliana KYP/SUVH4, AAK28969; SUVH6, AAK28971; Neurospora crassa DIM-5, Q8X225; Schizosaccharomyces pombe Clr4, CAA07709; Homo sapiens SUV39H1, CAG46546.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Results. SDG714 Fails to Complement KYP/SUVH4 in Arabidopsis.
Supplemental Figure 1. Phylogenetic Analysis of SET Domain Proteins between Arabidopsis and Oryza sativa.
Supplemental Figure 2. Complementary Analysis of kyp-2 clk-st with SDG714.
Supplementary Material
Acknowledgments
We thank Y. Shinkai for the GST-NH3 expression vector, T. Jenuwein for histone H3K9 dimethylated antibody, Y. Zhang for oligonucleosome, Ying Lan for scanning electron microscopy analysis, and YuXiang Wen for GFP analysis. The confocal images from this research were acquired with the Zeiss LSM 510 META confocal microscope at the Institute of Genetics and Developmental Biology, Chinese Academy of Sciences. We acknowledge Dong-Qiao Shi and Wei-Cai Yang for assistance with confocal microscopy. We thank Steve Jacobsen and Lianna Johnson at the University of California Los Angeles and Falong Lu at the Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, for critically reading the manuscript. This research was supported by National Basic Research Program of China Grants 2005CB522400 and 2005CB120806 and National Natural Science Foundation of China Grants 30430410, 30325015, and 30621001 to X.C. and by Chinese Academy of Sciences Grant CXTD-S2005-2.
The authors responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) are: ChengCai Chu (ccchu@genetics.ac.cn) and XiaoFeng Cao (xfcao@genetics.ac.cn).
Online version contains Web-only data.
References
- Alvarez-Venegas, R., Pien, S., Sadder, M., Witmer, X., Grossniklaus, U., and Avramova, Z. (2003). ATX-1, an Arabidopsis homolog of trithorax, activates flower homeotic genes. Curr. Biol. 13 627–637. [DOI] [PubMed] [Google Scholar]
- Bannister, A.J., Zegerman, P., Partridge, J.F., Miska, E.A., Thomas, J.O., Allshire, R.C., and Kouzarides, T. (2001). Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410 120–124. [DOI] [PubMed] [Google Scholar]
- Baumbusch, L.O., Thorstensen, T., Krauss, V., Fischer, A., Naumann, K., Assalkhou, R., Schulz, I., Reuter, G., and Aalen, R.B. (2001). The Arabidopsis thaliana genome contains at least 29 active genes encoding SET domain proteins that can be assigned to four evolutionarily conserved classes. Nucleic Acids Res. 29 4319–4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, X., Aufsatz, W., Zilberman, D., Mette, M.F., Huang, M.S., Matzke, M., and Jacobsen, S.E. (2003). Role of the DRM and CMT3 methyltransferases in RNA-directed DNA methylation. Curr. Biol. 13 2212–2217. [DOI] [PubMed] [Google Scholar]
- Cao, X., and Jacobsen, S.E. (2002). Locus-specific control of asymmetric and CpNpG methylation by the DRM and CMT3 methyltransferase genes. Proc. Natl. Acad. Sci. USA 99 (suppl. 4): 16491–16498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, C., Daigen, M., and Hirochika, H. (2006). Epigenetic regulation of the rice retrotransposon Tos17. Mol. Genet. Genomics 276 378–390. [DOI] [PubMed] [Google Scholar]
- Constancia, M., Pickard, B., Kelsey, G., and Reik, W. (1998). Imprinting mechanisms. Genome Res. 8 881–900. [DOI] [PubMed] [Google Scholar]
- Covey, S.N., and Al-Kaff, N.S. (2000). Plant DNA viruses and gene silencing. Plant Mol. Biol. 43 307–322. [DOI] [PubMed] [Google Scholar]
- Czermin, B., Melfi, R., McCabe, D., Seitz, V., Imhof, A., and Pirrotta, V. (2002). Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell 111 185–196. [DOI] [PubMed] [Google Scholar]
- Dodge, J.E., Kang, Y.K., Beppu, H., Lei, H., and Li, E. (2004). Histone H3-K9 methyltransferase ESET is essential for early development. Mol. Cell. Biol. 24 2478–2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebbs, M.L., Bartee, L., and Bender, J. (2005). H3 lysine 9 methylation is maintained on a transcribed inverted repeat by combined action of SUVH6 and SUVH4 methyltransferases. Mol. Cell. Biol. 25 10507–10515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebbs, M.L., and Bender, J. (2006). Locus-specific control of DNA methylation by the Arabidopsis SUVH5 histone methyltransferase. Plant Cell 18 1166–1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang, J., Feng, Q., Ketel, C.S., Wang, H., Cao, R., Xia, L., Erdjument-Bromage, H., Tempst, P., Simon, J.A., and Zhang, Y. (2002). Purification and functional characterization of SET8, a nucleosomal histone H4-lysine 20-specific methyltransferase. Curr. Biol. 12 1086–1099. [DOI] [PubMed] [Google Scholar]
- Fang, J., Wang, H., and Zhang, Y. (2004). Purification of histone methyltransferases from HeLa cells. Methods Enzymol. 377 213–226. [DOI] [PubMed] [Google Scholar]
- Fischle, W., Tseng, B.S., Dormann, H.L., Ueberheide, B.M., Garcia, B.A., Shabanowitz, J., Hunt, D.F., Funabiki, H., and Allis, C.D. (2005). Regulation of HP1-chromatin binding by histone H3 methylation and phosphorylation. Nature 438 1116–1122. [DOI] [PubMed] [Google Scholar]
- Fischle, W., Wang, Y., and Allis, C.D. (2003). Binary switches and modification cassettes in histone biology and beyond. Nature 425 475–479. [DOI] [PubMed] [Google Scholar]
- Goff, S.A., et al. (2002). A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science 296 92–100. [DOI] [PubMed] [Google Scholar]
- Grossniklaus, U., Vielle-Calzada, J.P., Hoeppner, M.A., and Gagliano, W.B. (1998). Maternal control of embryogenesis by MEDEA, a polycomb group gene in Arabidopsis. Science 280 446–450. [DOI] [PubMed] [Google Scholar]
- Han, F.P., Liu, Z.L., Tan, M., Hao, S., Fedak, G., and Liu, B. (2004). Mobilized retrotransposon Tos17 of rice by alien DNA introgression transposes into genes and causes structural and methylation alterations of a flanking genomic region. Hereditas 141 243–251. [DOI] [PubMed] [Google Scholar]
- Hirochika, H. (1997). Retrotransposons of rice: Their regulation and use for genome analysis. Plant Mol. Biol. 35 231–240. [PubMed] [Google Scholar]
- Hirochika, H., Fukuchi, A., and Kikuchi, F. (1992). Retrotransposon families in rice. Mol. Gen. Genet. 233 209–216. [DOI] [PubMed] [Google Scholar]
- Hirochika, H., Sugimoto, K., Otsuki, Y., Tsugawa, H., and Kanda, M. (1996). Retrotransposons of rice involved in mutations induced by tissue culture. Proc. Natl. Acad. Sci. USA 93 7783–7788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houben, A., Demidov, D., Gernand, D., Meister, A., Leach, C.R., and Schubert, I. (2003). Methylation of histone H3 in euchromatin of plant chromosomes depends on basic nuclear DNA content. Plant J. 33 967–973. [DOI] [PubMed] [Google Scholar]
- Izawa, T., et al. (1997). Transposon tagging in rice. Plant Mol. Biol. 35 219–229. [PubMed] [Google Scholar]
- Jackson, J.P., Johnson, L., Jasencakova, Z., Zhang, X., PerezBurgos, L., Singh, P.B., Cheng, X., Schubert, I., Jenuwein, T., and Jacobsen, S.E. (2004). Dimethylation of histone H3 lysine 9 is a critical mark for DNA methylation and gene silencing in Arabidopsis thaliana. Chromosoma 112 308–315. [DOI] [PubMed] [Google Scholar]
- Jackson, J.P., Lindroth, A.M., Cao, X., and Jacobsen, S.E. (2002). Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase. Nature 416 556–560. [DOI] [PubMed] [Google Scholar]
- Jenuwein, T., and Allis, C.D. (2001). Translating the histone code. Science 293 1074–1080. [DOI] [PubMed] [Google Scholar]
- Jiang, N., Bao, Z., Zhang, X., Hirochika, H., Eddy, S.R., McCouch, S.R., and Wessler, S.R. (2003). An active DNA transposon family in rice. Nature 421 163–167. [DOI] [PubMed] [Google Scholar]
- Johnson, L., Cao, X., and Jacobsen, S. (2002). Interplay between two epigenetic marks. DNA methylation and histone H3 lysine 9 methylation. Curr. Biol. 12 1360–1367. [DOI] [PubMed] [Google Scholar]
- Kato, M., Miura, A., Bender, J., Jacobsen, S.E., and Kakutani, T. (2003). Role of CG and non-CG methylation in immobilization of transposons in Arabidopsis. Curr. Biol. 13 421–426. [DOI] [PubMed] [Google Scholar]
- Kim, S.Y., He, Y., Jacob, Y., Noh, Y.S., Michaels, S., and Amasino, R. (2005). Establishment of the vernalization-responsive, winter-annual habit in Arabidopsis requires a putative histone H3 methyl transferase. Plant Cell 17 3301–3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, A., and Hirochika, H. (2001). Applications of retrotransposons as genetic tools in plant biology. Trends Plant Sci. 6 127–134. [DOI] [PubMed] [Google Scholar]
- Li, Y., Qian, Q., Zhou, Y., Yan, M., Sun, L., Zhang, M., Fu, Z., Wang, Y., Han, B., Pang, X., Chen, M., and Li, J. (2003). BRITTLE CULM1, which encodes a COBRA-like protein, affects the mechanical properties of rice plants. Plant Cell 15 2020–2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang, Y.K., Wang, Y., Zhang, Y., Li, S.G., Lu, X.C., Li, H., Zou, C., Xu, Z.H., and Bai, S.N. (2003). OsSET1, a novel SET-domain-containing gene from rice. J. Exp. Bot. 54 1995–1996. [DOI] [PubMed] [Google Scholar]
- Lindroth, A.M., Cao, X., Jackson, J.P., Zilberman, D., McCallum, C.M., Henikoff, S., and Jacobsen, S.E. (2001). Requirement of CHROMOMETHYLASE3 for maintenance of CpXpG methylation. Science 292 2077–2080. [DOI] [PubMed] [Google Scholar]
- Lippman, Z., et al. (2004). Role of transposable elements in heterochromatin and epigenetic control. Nature 430 471–476. [DOI] [PubMed] [Google Scholar]
- Lippman, Z., May, B., Yordan, C., Singer, T., and Martienssen, R. (2003). Distinct mechanisms determine transposon inheritance and methylation via small interfering RNA and histone modification. PLoS Biol. 1 E67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, B., Li, P., Li, X., Liu, C., Cao, S., Chu, C., and Cao, X. (2005). Loss of function of OsDCL1 affects microRNA accumulation and causes developmental defects in rice. Plant Physiol. 139 296–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Z.L., Han, F.P., Tan, M., Shan, X.H., Dong, Y.Z., Wang, X.Z., Fedak, G., Hao, S., and Liu, B. (2004). Activation of a rice endogenous retrotransposon Tos17 in tissue culture is accompanied by cytosine demethylation and causes heritable alteration in methylation pattern of flanking genomic regions. Theor. Appl. Genet. 109 200–209. [DOI] [PubMed] [Google Scholar]
- Lorincz, M.C., Schubeler, D., and Groudine, M. (2001). Methylation-mediated proviral silencing is associated with MeCP2 recruitment and localized histone H3 deacetylation. Mol. Cell. Biol. 21 7913–7922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malagnac, F., Bartee, L., and Bender, J. (2002). An Arabidopsis SET domain protein required for maintenance but not establishment of DNA methylation. EMBO J. 21 6842–6852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu, O., Probst, A.V., and Paszkowski, J. (2005). Distinct regulation of histone H3 methylation at lysines 27 and 9 by CpG methylation in Arabidopsis. EMBO J. 24 2783–2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura, A., Yonebayashi, S., Watanabe, K., Toyama, T., Shimada, H., and Kakutani, T. (2001). Mobilization of transposons by a mutation abolishing full DNA methylation in Arabidopsis. Nature 411 212–214. [DOI] [PubMed] [Google Scholar]
- Miyao, A., Tanaka, K., Murata, K., Sawaki, H., Takeda, S., Abe, K., Shinozuka, Y., Onosato, K., and Hirochika, H. (2003). Target site specificity of the Tos17 retrotransposon shows a preference for insertion within genes and against insertion in retrotransposon-rich regions of the genome. Plant Cell 15 1771–1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaki, K., Cheng, Z., Ouyang, S., Talbert, P.B., Kim, M., Jones, K.M., Henikoff, S., Buell, C.R., and Jiang, J. (2004). Sequencing of a rice centromere uncovers active genes. Nat. Genet. 36 138–145. [DOI] [PubMed] [Google Scholar]
- Naumann, K., Fischer, A., Hofmann, I., Krauss, V., Phalke, S., Irmler, K., Hause, G., Aurich, A.C., Dorn, R., Jenuwein, T., and Reuter, G. (2005). Pivotal role of AtSUVH2 in heterochromatic histone methylation and gene silencing in Arabidopsis. EMBO J. 24 1418–1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishioka, K., and Reinberg, D. (2003). Methods and tips for the purification of human histone methyltransferases. Methods 31 49–58. [DOI] [PubMed] [Google Scholar]
- Peters, A.H., Kubicek, S., Mechtler, K., O'Sullivan, R.J., Derijck, A.A., Perez-Burgos, L., Kohlmaier, A., Opravil, S., Tachibana, M., Shinkai, Y., Martens, J.H., and Jenuwein, T. (2003). Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol. Cell 12 1577–1589. [DOI] [PubMed] [Google Scholar]
- Peters, A.H., et al. (2001). Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell 107 323–337. [DOI] [PubMed] [Google Scholar]
- Rea, S., Eisenhaber, F., O'Carroll, D., Strahl, B.D., Sun, Z.W., Schmid, M., Opravil, S., Mechtler, K., Ponting, C.P., Allis, C.D., and Jenuwein, T. (2000). Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 406 593–599. [DOI] [PubMed] [Google Scholar]
- Shen, W.H., and Meyer, D. (2004). Ectopic expression of the NtSET1 histone methyltransferase inhibits cell expansion, and affects cell division and differentiation in tobacco plants. Plant Cell Physiol. 45 1715–1719. [DOI] [PubMed] [Google Scholar]
- Sims, R.J., III, Nishioka, K., and Reinberg, D. (2003). Histone lysine methylation: A signature for chromatin function. Trends Genet. 19 629–639. [DOI] [PubMed] [Google Scholar]
- Strahl, B.D., and Allis, C.D. (2000). The language of covalent histone modifications. Nature 403 41–45. [DOI] [PubMed] [Google Scholar]
- Tamaru, H., and Selker, E.U. (2001). A histone H3 methyltransferase controls DNA methylation in Neurospora crassa. Nature 414 277–283. [DOI] [PubMed] [Google Scholar]
- Tamaru, H., Zhang, X., McMillen, D., Singh, P.B., Nakayama, J., Grewal, S.I., Allis, C.D., Cheng, X., and Selker, E.U. (2003). Trimethylated lysine 9 of histone H3 is a mark for DNA methylation in Neurospora crassa. Nat. Genet. 34 75–79. [DOI] [PubMed] [Google Scholar]
- Tariq, M., Saze, H., Probst, A.V., Lichota, J., Habu, Y., and Paszkowski, J. (2003). Erasure of CpG methylation in Arabidopsis alters patterns of histone H3 methylation in heterochromatin. Proc. Natl. Acad. Sci. USA 100 8823–8827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran, R.K., Zilberman, D., de Bustos, C., Ditt, R.F., Henikoff, J.G., Lindroth, A.M., Delrow, J., Boyle, T., Kwong, S., Bryson, T.D., Jacobsen, S.E., and Henikoff, S. (2005). Chromatin and siRNA pathways cooperate to maintain DNA methylation of small transposable elements in Arabidopsis. Genome Biol. 6 R90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, S., Liu, N., Peng, K., and Zhang, Q. (1999). The distribution and copy number of copia-like retrotransposons in rice (Oryza sativa L.) and their implications in the organization and evolution of the rice genome. Proc. Natl. Acad. Sci. USA 96 6824–6828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao, B., Jing, C., Wilson, J.R., Walker, P.A., Vasisht, N., Kelly, G., Howell, S., Taylor, I.A., Blackburn, G.M., and Gamblin, S.J. (2003). Structure and catalytic mechanism of the human histone methyltransferase SET7/9. Nature 421 652–656. [DOI] [PubMed] [Google Scholar]
- Yu, Y., Dong, A., and Shen, W.H. (2004). Molecular characterization of the tobacco SET domain protein NtSET1 unravels its role in histone methylation, chromatin binding, and segregation. Plant J. 40 699–711. [DOI] [PubMed] [Google Scholar]
- Zhang, Y., and Reinberg, D. (2001). Transcription regulation by histone methylation: Interplay between different covalent modifications of the core histone tails. Genes Dev. 15 2343–2360. [DOI] [PubMed] [Google Scholar]
- Zhao, Z., Yu, Y., Meyer, D., Wu, C., and Shen, W.H. (2005). Prevention of early flowering by expression of FLOWERING LOCUS C requires methylation of histone H3 K36. Nat. Cell Biol. 7 1156–1160. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.