Methylation, the attachment of a methyl group in place of a hydrogen atom, is a common modification to numerous biological substrates. Almost all classes of plant metabolite are known to be methylated, including amino acids, alkaloids, phenylpropanoids, sugars, sterols, thiols, flavonoids, and purines (D'Auria et al., 2003). Methylation affects structural characteristics of these metabolites that impact their mobility, activity, and/or interactions with other molecules and therefore is often an important regulatory modification. The most widely used methyl donor in plant systems is S-adenosyl-l-methionine (SAM), and hundreds of SAM-dependent methyltransferases (MTs) have been identified. O-methylation (transfer of the methyl group to an oxygen atom) is the most common reaction that occurs among plant-specialized (i.e., secondary) metabolites, and O-methyltransferases (OMTs) constitute a large superclass of enzyme that contains numerous subgroups or families. Ibrahim et al. (1998) proposed a classification scheme for OMT depending on the substrate (e.g., whether they act on phenylpropanoids, flavonoids, alkaloids, or aliphatic substrates). However, a number of MTs have since been identified that do not fall into any of these classes, and it has been recognized that substrate is not necessarily a reliable predictor of MT provenance or phylogeny (D'Auria et al., 2003).
SABATH METHYLTRANSFERASES ACT ON A NUMBER OF PLANT HORMONES
Among the plant MTs identified recently is a family of enzymes that includes OMTs that methylate an oxygen in the carboxyl group of several plant hormones, including salicylic acid (SA), jasmonic acid (JA), and indole-3-acetic acid (IAA). Some of the first genes to be characterized in this family were those encoding SAM:salicylic acid carboxyl MT (SAMT) from Clarkia brewerii (Ross et al., 1999), SAM:benzoic acid carboxyl MT (BAMT) from snapdragon (Murfitt et al., 2000), and a theobromine synthase (which is an N-methyltransferase) from coffee (Ogawa et al., 2001). This family was therefore given the name SABATH, incorporating letters from SAMT, BAMT, and theobromine synthase. Additional SABATH enzymes have been identified from Arabidopsis, including a SAM:jasomonic acid carboxyl MT (Seo et al., 2001) and a SAM:IAA carboxyl MT (Zubieta et al., 2003; Qin et al., 2005). The SABATH family in the Arabidopsis genome includes 24 genes, and Eran Pichersky and his colleagues have undertaken the task to determine the functions of all 24 genes (Chen et al., 2003).
In this issue of The Plant Cell, Varbanova et al. (pages 32–45) report that two members of the Arabidopsis SABATH methyltransferase gene family, designated GAMT1 and GAMT2, encode SAM-dependent MTs that methylate the 7-carboxyl group of gibberellins (GAs), resulting in the corresponding methyl esters (MeGAs). Both genes were found to be expressed most highly in the siliques during seed development. The authors found that siliques of mutant plants with reduced levels of GAMT1 and GAMT2 showed an increase in concentrations of free GAs, and mature seeds of mutants appeared to have higher concentrations of active GAs, indicating the GAMT activity reduces the level of active GA in these plant tissues. Thus, methylation of GAs may constitute an additional mechanism for modulating cellular GA concentrations in addition to the previously described oxidative pathways.
GAMTs AFFECT GA HOMEOSTASIS IN ARABIDOPSIS
As a first step, the authors expressed full-length cDNAs of the two genes in Escherichia coli, purified the proteins, and tested their activity with a large number of potential substrates using a high-throughput assay. In this test, both enzymes showed MT activity with GA3, the one GA included in the initial high-throughput assay, and no activity was observed with any other substrates tested. At the time of this writing, 136 GAs have been identified in higher plants, fungi, and bacteria (http://www.plant-hormones.info/gibberellins.htm), but only a small number have been found to be biologically active (e.g., GA1, GA3, GA4, and GA7) (Hedden and Phillips, 2000). Varbanova et al. tested numerous additional GAs and found that both enzymes showed activity with all but a few. GAMT1 showed highest activity with GA9 and GA20, followed by GA3 and other GAs, whereas GAMT2 showed highest activity with GA4, followed by GA34, GA9, GA3, and others.
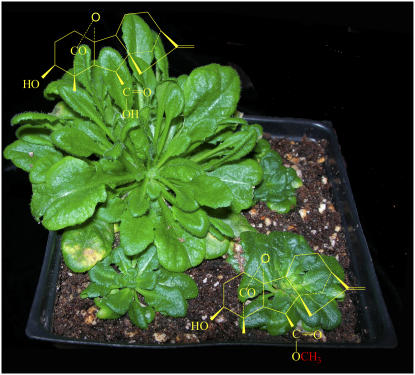
The group then conducted experiments using Arabidopsis, tobacco, and petunia plants overexpressing GAMT1 and GAMT2 and found that overexpression of the genes resulted in dwarf phenotypes similar to those exhibited by GA-deficient mutants (see figure). Measurement of endogenous levels of precursor and bioactive GAs in GAMT-overexpressing Arabidopsis plants just after flowering confirmed that the levels of GA4, the major bioactive form at this stage, as well as a number of precursor GAs, were significantly lower in GAMT-overexpressing plants than in the wild type. They therefore concluded that the dwarfed and semidwarfed phenotypes of plants overexpressing GAMT1 and GAMT2 could be attributed to reduced levels of bioactive GA.
Figure 1.
Arabidopsis Overexpressing GAMT1 (Foreground and Rear Right) Show Dwarf Phenotypes Similar to GA-Deficient Mutants.
A wild-type plant is shown in the rear left-hand corner. The structures of the free acid of GA4 and of MeGA4 are shown at the top left and bottom right, respectively.
The authors obtained T-DNA insertional mutants of GAMT1 and GAMT2, which were verified as null mutants, and crossed the single mutant lines to obtain the homozygous double mutant. The null mutants all grew normally and had no obvious visible morphological differences compared with wild-type plants. The levels of bioactive GA4 and GA1 were measured and found to be significantly higher in siliques of the gamt1 gamt2 double mutant than in wild-type plants. Analysis of gene expression of GAMT1 and GAMT2 in wild-type plants showed that both genes were highly expressed in siliques and not in any other tissue examined (e.g., leaves, stems, flowers, and roots), except for weak but detectable levels in germinating seeds. These results supported the conclusion that GAs are endogenous substrates for GAMT1 and GAMT2 in wild-type siliques and suggest that methylation of GA plays a role in or has a function during seed development.
THE ROLE OF GA AND MeGA IN SEED DEVELOPMENT
GA is known to function in promoting seed germination, but its role in early seed development is unclear. Many plants accumulate high levels of GA during seed development, and in many cases, the GAs produced are inactive forms that exhibit a range of unusual modifications (a large number of the 136 known GAs are found in seeds). It remains unclear what role GAs play during seed development per se, although they could function in promoting growth and development of the early embryo (Finkelstein, 2004). However, it appears essential that bioactive forms of GA are removed or inactivated during seed maturation; otherwise, they could affect seed dormancy, as well as germination and early seedling growth. For example, the phenotype of the slender mutant of pea is due to loss of GA2 oxidase activity, resulting in the accumulation of GA20 in the seed, which upon germination is converted to GA1 and results in an abnormally elongated seedling (Lester et al., 1999; Martin et al., 1999).
To examine the effect of GA methylation on Arabidopsis seed germination, Varbanova et al. placed homozygous single and double gamt1 gamt2 mutant seeds on medium supplemented with ancymidol, an inhibitor of GA biosynthesis. Germination was inhibited by ancymidol to a lesser extent in the mutant seed compared with wild-type seed and to a greater extent in seed of plants overexpressing GAMT1 and GAMT2 compared with the wild type. This suggests that an important function of GAMTs in seed is in the deactivation or removal of active GA during seed maturation.
WHAT IS THE FATE OF MeGA?
Methyl esters are more hydrophobic than the free acid form of a compound and as a result can more easily pass through membranes and are more highly mobile in planta. For example, MeIAA and MeJA are often used experimentally because, due to their higher rate of mobility, a given concentration applied exogenously is more potent than the same concentration of the corresponding free acid. It has been widely assumed that after the methylated hormone enters a cell, it is hydrolyzed and the free acid is the active form. MeSA and MeJA have been found in many plants and are thought to function as volatile signals that function in plant defense (e.g., Shulaev et al., 1997; Baldwin et al., 2006). However, it is believed that the free acids of SA and JA are released inside the cell and function as the active compounds in eliciting defense response pathways. Until recently, however, there has been very little study of the hydrolysis of these methylated hormones.
Forouhar et al. (2005) recently showed that salicylic acid binding protein 2 (SABP2) from tobacco is a MeSA esterase, and Kumar et al. (2006) have shown that this protein is required for systemic acquired resistance. This work supports the conclusion that MeSA is not the active compound but may be involved in in planta transport of SA, and hydrolysis of MeSA is required to release the active compound. An MeJA esterase has been identified in tomato and implicated in jasmonate signaling (Stuhlfelder et al., 2004), a further indication that plants have specific enzymes to hydrolyze methylated hormones.
Unlike MeJA and MeIAA, MeGAs are not highly volatile and they do not appear to have activity when applied exogenously. The work of Varbanova et al. suggests that methylation of GA is an irreversible process in planta. The authors attempted to measure endogenous MeGAs in siliques of wild-type plants and found them to be undetectable, although a range of internal standards added to extracts could be detected. These results might indicate that MeGAs are rapidly degraded or converted to other compounds. However, this question needs to be examined more thoroughly before a definitive conclusion can be reached. Interestingly, Varbanova et al. note that the recently identified GID1 family of GA receptors (Ueguchi-Tanaka et al., 2005; Griffiths et al., 2006) are structurally related to MeSA and MeJA hydrolases, although they appear to be inactive as MeGA esterases.
The major route known for GA catabolism is 2β-hydroxylation, which is catalyzed by GA 2-oxidases and results in biologically inactive products that cannot be converted back to active forms (Thomas et al., 1999). GA 2-oxidases are encoded by a small gene family in Arabidopsis and a number of other species (Elliott et al., 2001). The genes are expressed in a range of tissues and are strongly implicated in playing a major role in regulating levels of bioactive GAs (Thomas et al., 1999; Elliott et al., 2001; Schomburg et al., 2003). Deactivation of GAs has also been found to occur via epoxidation by a cytochrome P450 monooxygenase (Zhu et al., 2006) and via conjugation through the formation of glucosyl esters and glucosides (Schneider et al., 1992). Varbanova et al. have shown that methylation of GAs constitutes another route of GA deactivation that may function in regulating levels of bioactive GAs in developing seed.
In addition, the work of Varbanova et al. provides an excellent example of the utility of functional genomics and the importance of the Arabidopsis model system in plant biology. Starting from genome sequence, the authors used high-throughput methods in conjunction with all of the available Arabidopsis tools and resources to identify the function of genes, the activity of gene products, and the possible biological roles of the GA methyl transferases, which had remained elusive to conventional physiological, biochemical, or genetic approaches.
Varbanova et al. have shown that GAMT1 and GAMT2 function in GA homeostasis in developing siliques of Arabidopsis. The authors suggest that methylation of GAs by GAMT1 and GAMT2 serves to deactivate GAs and initiate their degradation in developing seed. This work represents a significant step forward in arriving at a complete understanding of GA metabolism and GA-related processes in plant development.
References
- Baldwin, I.T., Halitschke, R., Paschold, A., von Dahl, C.C., and Preston, C.A. (2006). Volatile signaling in plant-plant interactions “Talking Trees” in the genomics era. Science 311 812–815. [DOI] [PubMed] [Google Scholar]
- Chen, F., D'Auria, J.C., Tholl, D., Ross, J.R., Gershenzon, J., Noel, J.P., and Pichersky, E. (2003). An Arabidopsis thaliana gene for methylsalicylate biosynthesis, identified by a biochemical genomics approach, has a role in defense. Plant J. 36 577–588. [DOI] [PubMed] [Google Scholar]
- D'Auria, J.C., Chen, F., and Pichersky, E. (2003). The SABATH family of methyltransferases in Arabidopsis thaliana and other plant species. In Recent Advances in Phytochemistry, J. Romeo, ed (Oxford, UK: Elsevier Science), pp. 253–283.
- Elliott, R.C., Smith, J.L., Lester, D.R., and Reid, J.B. (2001). Feedforward regulation of gibberellin deactivation in pea. J. Plant Growth Regul. 20 87–94. [Google Scholar]
- Finkelstein, R.R. (2004). The role of hormones during seed development and germination. In Plant Hormones: Biosynthesis, Signal Transduction, Action! P.J. Davies, ed (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 513–537.
- Forouhar, F., Yang, Y., Kumar, D., Chen, Y., Fridman, E., Park, S.W., Chiang, Y., Action, T.B., Montelione, G.T., Pichersky, E., Klessig, D.F., and Tong, L. (2005). Structural and biochemical studies identify tobacco SABP2 as a methyl salicylate esterase and implicate it in plant immunity. Proc. Natl. Acad. Sci. USA 102 1773–1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths, J., Murase, K., Rieu, I., Zentella, R., Zhang, Z.-L., Powers, S.J., Gong, F., Phillips, A.L., Hedden, P., Sun, T.-p., and Thomas, S.G. (2006). Genetic characterization and functional analysis of the GID1 gibberellin receptors in Arabidopsis. Plant Cell 18 3399–3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedden, P., and Phillips, A.L. (2000). Gibberellin metabolism: New insights revealed by the genes. Trends Plant Sci. 5 523–530. [DOI] [PubMed] [Google Scholar]
- Ibrahim, R.K., Bruneau, A., and Bantignies, B. (1998). Plant O-methyltransferases: Molecular analysis, common signature and classification. Plant Mol. Biol. 36 1–10. [DOI] [PubMed] [Google Scholar]
- Kumar, D., Gustafsson, C., and Klessig, D.F. (2006). Validation of RNAi silencing specificity using synthetic genes: Salicylic acid-binding protein 2 is required for innate immunity in plants. Plant J. 45 863–868. [DOI] [PubMed] [Google Scholar]
- Lester, D.R., Ross, J.J., Smith, J.J., Elliott, R.C., and Reid, J.B. (1999). Gibberellin 2-oxidation and the SLN gene of Pisum sativum. Plant J. 19 65–73. [DOI] [PubMed] [Google Scholar]
- Martin, D.N., Proebsting, W.M., and Hedden, P. (1999). The SLENDER gene of pea encodes a gibberellin 2-oxidase. Plant Physiol. 121 775–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murfitt, L.M., Kolosova, N., Mann, C.J., and Dudareva, N. (2000). Purification and characterization of S-adenosyl-L-methionine:benzoic acid carboxyl methyltransferase, the enzyme responsible for biosynthesis of the volatile ester methyl benzoate in flowers of Antirrhinum majus. Arch. Biochem. Biophys. 382 145–151. [DOI] [PubMed] [Google Scholar]
- Ogawa, M., Herai, Y., Koizumi, N., Kusano, T., and Sano, H. (2001). 7-Methylxanthine methyltransferase of coffee plants. Gene isolation and enzymatic properties. J. Biol. Chem. 276 8213–8218. [DOI] [PubMed] [Google Scholar]
- Qin, G., Gu, H., Zhao, Y., Ma, Z., Shi, G., Yang, Y., Pichersky, E., Chen, H., Lui, M., Chen, Z., and Qu, L.-J. (2005). Regulation of auxin homeostasis and plant development by an indole-3-acetic acid carboxyl methyltransferase in Arabidopsis. Plant Cell 17 2693–2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross, J.R., Nam, K.H., D'Auria, J.C., and Pichersky, E. (1999). S-adenosyl-L-methionine:salicylic acid carboxyl methyltransferase, an enzyme involved in floral scent production and plant defense, represents a new class of plant methyltransferases. Arch. Biochem. Biophys. 367 9–16. [DOI] [PubMed] [Google Scholar]
- Schneider, G., Jensen, E., Spray, C.R., and Phinney, B.O. (1992). Hydrolysis and reconjugation of gibberellin A20 glucosyl ester by seedlings of Zea mays L. Proc. Natl. Acad. Sci. USA 89 8045–8048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schomburg, F.M., Bizzell, C.M., Lee, D.L., Zeevaart, J.A.D., and Amasino, R.M. (2003). Overexpression of a novel class of gibberellin 2-oxidases decreases gibberellin levels and creates dwarf plants. Plant Cell 15 151–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo, H.S., Song, J.T., Cheong, J.J., Lee, Y.H., Lee, Y.W., Hwang, I., Lee, J.S., and Choi, Y.D. (2001). Jasmonic acid carboxyl methyltransferase: A key enzyme for jasmonate-regulated plant responses. Proc. Natl. Acad. Sci. USA 98 4788–4793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulaev, V., Silverman, P., and Raskin, I. (1997). Airborne signalling by methyl salicylate in plant pathogen resistance. Nature 385 718–721. [Google Scholar]
- Stuhlfelder, C., Mueller, M.J., and Warzecha, H. (2004). Cloning and expression of a tomato cDNA encoding a methyl jasmonate cleaving esterase. Eur. J. Biochem. 271 2976–2983. [DOI] [PubMed] [Google Scholar]
- Thomas, S.G., Phillips, A.L., and Hedden, P. (1999). Molecular cloning and functional expression of gibberellin 2-oxidases, multifunctional enzymes involved in gibberellin deactivation. Proc. Natl. Acad. Sci. USA 96 4698–4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueguchi-Tanaka, M., Ashikari, M., Nakajima, M., Itoh, H., Katoh, E., Kobayashi, M., Chow, T.Y., Hsing, Y.I., Kitano, H., Yamaguchi, I., and Matsuoka, M. (2005). GIBBERELLIN INSENSITIVE DWARF1 encodes a soluble receptor for gibberellin. Nature 437 693–698. [DOI] [PubMed] [Google Scholar]
- Varbanova, M., et al. (2007). Methylation of gibberellins by Arabidopsis GAMT1 and GAMT2. Plant Cell 19: 32–45. [DOI] [PMC free article] [PubMed]
- Zhu, Y., et al. (2006). ELONGATED UPPERMOST INTERNODE encodes a cytochrome P450 monooxygenase that epoxidizes gibberellins in a novel deactivation reaction in rice. Plant Cell 18 442–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zubieta, C., Koscheski, P., Ross, J.R., Yang, Y., Pichersky, E., and Noel, J.P. (2003). Structural basis for substrate recognition in the salicylic acid carboxyl methyltransferase family. Plant Cell 15 1704–1716. [DOI] [PMC free article] [PubMed] [Google Scholar]