Abstract
To investigate host and viral mechanisms determining hepadnaviral persistence and hepatocarcinogenesis, we developed a mouse model by transplanting woodchuck hepatocytes into the liver of mice that contain the urokinase-type plasminogen activator transgene (uPA) and lack mature B and T lymphocytes due to a recombination activation gene 2 (RAG-2) gene knockout. The woodchuck hepatocytes were transplanted via intrasplenic injection and were found to integrate into the recipient mouse liver cord structure. Normal adult woodchuck hepatocytes proliferated and reconstituted up to 90% of the uPA/RAG-2 mouse liver. uPA/RAG-2 mice containing woodchuck hepatocytes were infectable with woodchuck hepatitis virus (WHV) and showed WHV replication for at least 10 months with titers up to 1 × 1011 virions per ml in the peripheral blood. WHV-infected hepatocytes from chronic carrier woodchucks also established a persistent infection in uPA/RAG-2 mice after an 8- to 12-week lag period of viremia. Although WHV envelope, core, and X proteins were produced in the uPA/RAG-2 mice, no inflammatory host immune response was observed in the liver of WHV-replicating mice. A first antiviral test demonstrated a greater than four orders of magnitude drop in WHV titer in response to interferon α treatment. WHV replication was up-regulated by dexamethasone treatment. Comparison of precancerous lesions in donor woodchucks versus recipient uPA/RAG-2 mice revealed an enrichment of dysplastic precancerous hepatocytes in transplanted mice. Clonal amplification of hepatocytes from a woodchuck with hepatocellular carcinomas was demonstrated by the detection of unique WHV DNA integration patterns in hepatocellular carcinomas that arose in uPA/RAG-2 mice. In the absence of B or T cell-mediated immune responses, WHV establishes a persistent noncytotoxic infection of woodchuck hepatocytes in uPA/RAG-2 chimeric mouse livers. Further studies of the kinetics of hepadnavirus infection and replication in quiescent and proliferating hepatocytes should increase our understanding of hepadnavirus spread and aid in the design of therapies to block or cure persistent infection.
Hepatitis B virus (HBV) infection remains a major health problem with more than 350 million chronic HBV carriers worldwide, who are at risk for developing liver cirrhosis and hepatocellular carcinoma (HCC) (1–4). The development of effective therapies for eradicating HBV in chronic carriers has been limited by our incomplete understanding of mechanisms of viral persistence (5).
Both humoral and cellular elements of the host immune response are important for HBV clearance. The humoral response to HBV antigens (i.e., antibodies to hepatitis B surface antigen) helps clear circulating virions and confers protection against reinfection, whereas T cell-mediated responses eliminate infected cells (6, 7). Recent work using HBV transgenic mice has shown that this process can be mediated by cytokines, such as tumor necrosis factor α and interferon γ. These cytokines have the capacity to down-regulate HBV replication in a noncytopathic manner (8, 9).
Our aims were to determine whether hepadnavirus replication will become persistent in the absence of B and T cells in the host and whether acute infection of hepatocytes in such an environment would effect the course of hepadnaviral persistence. To analyze this, we took advantage of recent advances in liver repopulation with transplanted hepatocytes (10). The discovery of a hepatocyte-lethal phenotype in urokinase-type plasminogen activator (uPA) transgenic mice and the replacement of livers from those mice with xenografted hepatocytes (11–13) suggested to us that if the mouse liver could be replaced by hepatocytes amenable to hepadnaviral infection, a variety of studies would be possible in mice containing hepatocytes from a single donor. Furthermore, the potential proliferation of hepadnavirus-infected transplanted xenogenic hepatocytes would allow us to study hepadnaviral replication and persistence in proliferating hepatocytes during liver regeneration and in quiescent hepatocytes after completion of liver regeneration. uPA-transgene-expressing hepatocytes are at a growth disadvantage compared with nontransgenic hepatocytes, because transgene expression compromises their function (11), and thus transplanted hepatocytes are selectively amplified in a mixed polyclonal pattern.
To generate the uPA/recombination activation gene 2 (RAG-2) mice, we crossed uPA transgenic mice with RAG-2 knockout mice, which lack mature B and T lymphocytes (14). Woodchuck hepatocytes, transplanted via splenic injection (10) into the liver of uPA/RAG-2 mice, became integrated in the uPA/RAG-2 liver parenchyma, replacing up to 90% of the uPA/RAG-2 mouse hepatocytes, and supported woodchuck hepatitis virus (WHV) replication indefinitely. WHV replication in uPA/RAG-2 mice could be manipulated by up-regulatory (dexamethasone) or down-regulatory (interferon α) pharmacological interventions. In addition, clonal potential of liver tumor cells derived from chronic WHV carrier woodchucks could be tested in the uPA/RAG-2 mice.
METHODS
Animals.
UPA mice were purchased from The Jackson Laboratories, RAG-2 knockout mice were from Taconic Farms, and adult woodchucks were from North Eastern Wildlife (South Plymouth, NY) or Cornell University (Ithaca, NY). Animals were housed and maintained under specific pathogen-free conditions in accordance with institutional guidelines under approved protocols. We used one uninfected woodchuck, which was negative for all WHV markers, and three woodchucks (nos. 2765, 3543, and 4940) with persistent WHV infections that were woodchuck hepatitis virus surface antigen (WHsAg)-positive and anti-woodchuck hepatitis virus core antibody (anti-WHc)-positive. The results from nine randomly chosen uPA/RAG-2 mice containing transplanted cells are shown in detail. Partial hepatectomies in mice were performed under tribromoethanol anesthesia (Aldrich) with approved protocols (15).
Generation of Tolerant uPA/RAG-2 Mice.
uPA transgenic mice were crossed with RAG-2 knockout mice. The uPA transgene was identified by PCR of mouse-tail DNA with the following nucleotide sequences: primer 1, 5′-CATCCCTGTGACCCCTCC-3′, and primer 2, 5′-CTCCAAACCACCCCCCTC-3′. Homozygous uPA transgenic mice were distinguished from hemizygous mice by PCR as reported (13). For the experiments described, only hemizygous uPA mice were used. The RAG-2 knockout mutant gene was identified by PCR of tail DNA as described (16).
Isolation and Transplantation of Woodchuck Hepatocytes.
Woodchuck hepatocytes were isolated by the two-step in situ collagenase perfusion method followed by differential centrifugation (17). Hepatocyte viability was >95%, as measured by trypan blue dye exclusion. From 5 × 105 to 1 × 106 hepatocytes were transplanted into a number of 10- to 18-day-old uPA/RAG-2 mice by intrasplenic injection (17).
Histological Studies.
Serial cryostat sections of woodchuck and uPA/RAG-2 mouse livers were examined by hematoxylin/eosin staining, dipeptidyl peptidase IV (DPPIV) enzyme activity (18) and immunohistochemistry with a rabbit antiserum (WHc antiserum) against woodchuck hepatitis core antigen (WHcAg). For immunolabeling, sections were fixed in 4% paraformaldehyde, incubated with WHc antiserum (diluted 1:250 in PBS containing 5% sheep serum) at room temperature, and subsequently incubated with the Cy3-conjugated sheep anti-rabbit IgG antibody (Sigma) diluted 1:200 in PBS with 5% sheep serum at room temperature. Finally, slides were mounted in 40 mg of n-propyl gallate per ml in 90% glycerol/10% 0.5 M sodium carbonate, pH 8, for viewing under a fluorescence microscope.
Immunoprecipitation and Immunoblot Analyses.
The woodchuck hepatitis virus X (WHx) protein was immunoprecipitated from cell extracts with a rabbit WHx antiserum and subjected to SDS/PAGE as described (19). For the detection of WHV core protein, 20 μg of total cell extracts was solubilized (20), boiled, and separated by SDS/PAGE. The transblots were probed with either WHx antiserum (1:1,000 dilution) or WHc antiserum (1:5,000 dilution), and binding was detected by the enhanced chemiluminescence (ECL) system (19) (Amersham).
DNA Analysis.
Genomic DNAs were extracted from frozen liver and used for Southern blot analysis as described (21, 22). For detecting woodchuck genomic DNA in transplanted uPA/RAG-2 mouse livers, 150 ng of PvuII-digested woodchuck DNA was used for a 32P-labeled random genomic probe. Blots were hybridized under high-stringency conditions (23) at 45°C for 2 h. WHV DNA in woodchuck and uPA/RAG-2 mouse liver tissues was detected with a genome-length 3.3-kb WHV DNA 32P-labeled probe (23).
Detection of WHV DNA, Woodchuck Albumin, and WHsAg in Woodchuck and uPA/RAG-2 Mouse Sera.
Woodchucks were anesthetized with ketamine (Fort Dodge Laboratories, Fort Dodge, IA) and xylazine (Bayer, Shawnee Mission, KS) and blood was collected from the femoral vein. Blood was drawn from the tail vein in mice, both woodchuck and mouse sera were dot-blotted onto a nylon membrane (24) and hybridized with a 32P-labeled WHV DNA probe (23), and the number of WHV DNA molecules was quantitated by scanning densitometry. Serial dilutions of known amounts of a plasmid containing one copy of WHV DNA served as a standard. For woodchuck serum albumin, 5 μg of total serum proteins was solubilized (20), boiled, and subjected to SDS/PAGE. Proteins resolved in 7.5% gels were fixed and stained with Coomassie blue. For immunoblotting of WHsAg, proteins were resolved in SDS/PAGE, electrotransferred, probed with a rabbit antiserum against WHsAg (WHs antiserum; 1:1,000 dilution), and visualized by ECL.
Blood Chemistry in uPA/RAG-2 Mice.
Sera were analyzed for total protein, albumin, bilirubin, alanine aminotransferase activity, and aspartate aminotransferase activity in a standard automated clinical analyzer (Technicon).
Drugs.
Interferon α-2b (Schering) at 135 international units/g (body weight) and dexamethasone (Fujisawa, Deerfield, IL) at 27 ng/g (body weight) were given to mice intramuscularly daily for 15 consecutive days.
RESULTS
Repopulation of uPA/RAG-2 Mouse Liver with Xenogenic Woodchuck Hepatocytes.
To screen for survival and growth of transplanted woodchuck hepatocytes in uPA/RAG-2 mice, we analyzed serum albumin profiles from all recipients. SDS/PAGE showed that mouse and woodchuck serum albumin migrated differently (Fig. 1). In uPA/RAG-2 mice, 3 months after woodchuck hepatocyte transplantation, this assay demonstrated the presence of both woodchuck and mouse serum albumin (Fig. 1). uPA/RAG-2 mice containing woodchuck hepatocytes were clinically healthy and the livers appeared normal in respect to color, size, and liver weight to body weight ratios at sacrifice. Serum concentrations of total protein, albumin, bilirubin, and transaminases were similar in uPA/RAG-2 mice containing woodchuck hepatocytes compared with control uPA/RAG-2 mice (Table 1).
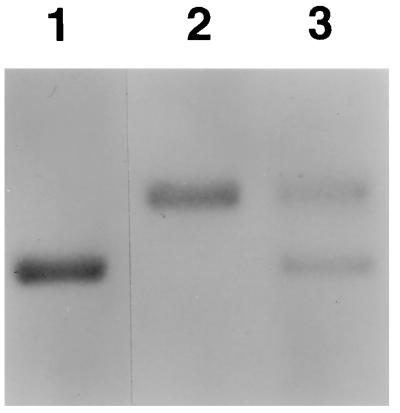
Figure 1.
Migration pattern of mouse (lane 1) and woodchuck (lane 2) serum albumin in a Coomassie blue-stained gel. The migration pattern of serum albumin from a representative uPA/RAG-2 mouse containing woodchuck hepatocytes shows both mouse and woodchuck serum albumin (lane 3).
Table 1.
Serum parameters in representative uPA/RAG-2 mice
| Parameter | Animal
|
|||
|---|---|---|---|---|
| 351 | 457 | 496 | 969 | |
| Woodchuck hepatocytes | − | − | + | + |
| Total protein, g/dl | 4.4 | 4.0 | 4.6 | 4.2 |
| Albumin, g/dl | 2.6 | 2.4 | 2.8 | 2.6 |
| ALT, units/liter | 118 | 104 | 112 | 124 |
| AST, units/liter | 140 | 174 | 139 | 154 |
| Total bilirubin, mg/dl | 0.2 | 0.2 | 0.3 | 0.2 |
uPA/RAG-2 mice containing woodchuck hepatocytes (no. 496 and 969) were compared with untransplanted uPA/RAG-2 mice (no. 351 and 457). ALT, alanine aminotransferase; AST, aspartate aminotransferase.
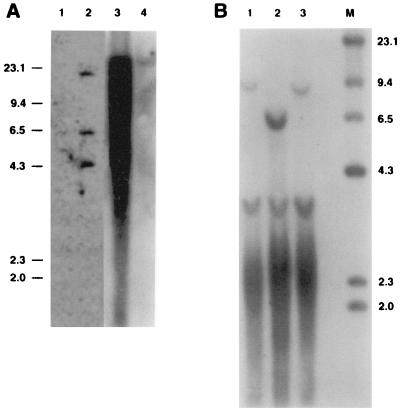
To directly demonstrate the presence of woodchuck hepatocytes in transplanted uPA/RAG-2 mouse livers, we hybridized DNAs extracted from recipients with a total woodchuck genome probe, which detects highly repeated sequences in the woodchuck genome (Fig. 2A). To estimate uPA/RAG-2 mouse liver repopulation with woodchuck hepatocytes, we hybridized test mixtures of woodchuck and mouse genomic DNAs in various proportions (Fig. 2A, lanes 1–5) and DNAs from five liver lobes of a uPA/RAG-2 mouse transplanted with WHV-positive woodchuck hepatocytes (no. 496; Fig. 2A, lanes 7–11). Woodchuck hepatocytes were present in abundance in all liver lobes from approximately 30% to >95% (Fig. 2A). Because intrasplenic injection results in uniform cell distribution to the liver in normal rats (25), our finding could have indicated differences in the proliferative ratios and development of clonally derived islands of woodchuck hepatocytes in uPA/RAG-2 mouse liver. Alternatively, in the disrupted microenvironment of the diseased uPA liver, cell distributions may not have been uniform.
Figure 2.
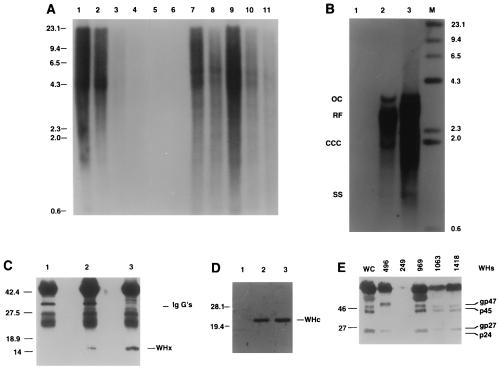
Woodchuck DNA and WHx, WHc, and WHs proteins in uPA/RAG-2 mice transplanted with WHV-positive woodchuck hepatocytes. (A) A Southern blot of genomic DNAs, hybridized with a woodchuck genome DNA probe. Lanes 1–4 present mixtures of genomic woodchuck liver DNA and untransplanted uPA/RAG-2 mouse genomic liver DNA with signals reflecting 100%, 50%, 20%, and 1%, woodchuck hepatocyte DNA, respectively. Lane 5 presents 100% untransplanted uPA/RAG-2 mouse DNA. Woodchuck DNA is undetectable in the spleen of transplanted uPA/RAG-2 mice (lane 6) but present in various amounts in all lobes of the liver of the uPA/RAG-2 mouse transplanted with WHV-positive woodchuck hepatocytes (lanes 7–11). (B) WHV DNA forms, detectable in uPA/RAG-2 mouse genomic liver DNA, hybridized with a WHV DNA probe. Lanes: 1, uPA/RAG-2 mouse liver; 2, uPA/RAG-2 mouse liver transplanted with WHV-positive woodchuck hepatocytes; 3, donor woodchuck genomic liver DNA. OC, open circular DNA; RF, replicative WHV DNA forms; CCC, covalently closed circular DNA; SS, single-stranded DNA. Lane M contains molecular size markers. (C) Immunoprecipitates with WHx antiserum from uPA/RAG-2 mouse liver (lane 1), from uPA/RAG-2 mouse transplanted with WHV-positive woodchuck hepatocytes (lane 2), and from donor woodchuck liver (lane 3). (D) Immunoblotting with WHc antiserum of hepatocyte extracts from uPA/RAG-2 mouse (lane 1), from uPA/RAG-2 mouse transplanted with WHV-positive woodchuck hepatocytes (lane 2), and from donor woodchuck (lane 3). (E) WHs proteins in uPA/RAG-2 mice serum. Immunoblotting with WHs antiserum of uPA/RAG-2 mouse 249 and uPA/RAG-2 mouse sera transplanted with WHV-positive woodchuck hepatocytes (uPA/RAG-2 mice 496, 969, 1063, and 1418). Lane WC was probed with WHV-positive woodchuck serum.
Demonstration of WHV Replication and Persistence in uPA/RAG-2 Mice.
We tested whether transplanted WHV-positive woodchuck hepatocytes supported WHV replication. Genomic liver DNA of a representative woodchuck hepatocyte recipient (no. 496) showed open circular WHV DNA, replicative DNA forms, and covalently closed circular (CCC) WHV DNA that matched the profile of WHV DNA from the donor woodchuck (no. 2765; Fig. 2B, lanes 2 and 3). Also, WHx and WHV core proteins were present in this liver (Fig. 2 C and D, respectively) and the serum contained WHsAg (Fig. 2E). WHV DNA in the serum of uPA/RAG-2 mice became detectable only after completion of liver regeneration with a lag period of viremia of 8 -12 weeks after transplantation. WHV DNA titers stabilized at a level of approximately 5 × 108 viral genomes per ml in transplanted mouse 496 as compared with 1 × 109 WHV genomes per ml in the donor woodchuck (data not shown). In other transplanted uPA/RAG-2 mice, WHV titers of up to 1 × 1011 virions per ml of mouse serum were detected (see Fig. 4). In Dane particles isolated from the serum of transplanted uPA/RAG-2 mice, viral DNA migrated on Southern blots similar to the WHV DNA from the donor woodchuck. Analysis of WHV RNA in mouse 496 liver revealed the expected major mRNA species corresponding to the pregenome mRNA and the major envelope protein mRNAs (data not shown).
Figure 4.
Effect of interferon α and dexamethasone upon WHV secretion in uPA/RAG-2 mice. Each trace shows data from individual uPA/RAG-2 mice containing WHV-secreting woodchuck hepatocytes. Arrows mark starting point and withdrawal of agents. Time points mark the collection of serum samples. The dashed line represents the threshold of sensitivity for the dot blot analysis.
To investigate the growth pattern of woodchuck hepatocytes in uPA/RAG-2 mouse liver, we performed immunohistochemistry with a WHc antiserum. WHV-positive woodchuck hepatocytes had seeded the liver and grown in a nodular pattern within the framework of the preexisting liver with maintenance of the liver cord structure (Fig. 3A). Untransplanted mice did not show any specific staining. Integration of the woodchuck hepatocytes into the liver architecture was evaluated by histochemistry for the enzyme DPPIV, which is localized in bile canaliculi of hepatocytes (18). Mouse hepatocytes appeared smaller and DPPIV-positive areas were, therefore, more compact. In contrast, areas containing woodchuck cells showed greater spacing between DPPIV-positive domains due to larger cell sizes. In chimeric uPA/RAG-2 mouse livers containing woodchuck and mouse hepatocytes, we observed networks of DPPIV-positive bile canaliculi between adjacent mouse and woodchuck hepatocytes (Fig. 3B). The presence of woodchuck hepatocytes in those sections was confirmed by performing immunohistochemistry using a WHc antiserum in serial sections from uPA/RAG-2 mouse liver tissues. Interestingly, despite expression of WHV proteins in transplanted woodchuck hepatocytes, we did not observe any hepatocellular infiltration with inflammatory cells. The uPA/RAG-2 mice are of course deficient in T and B lymphocytes; however, no evidence was found of infiltration with granulocytes or macrophages.
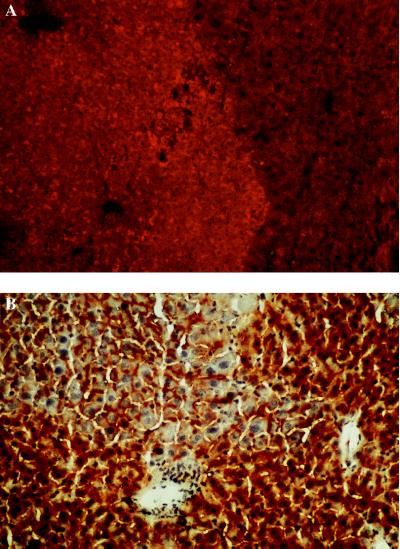
Figure 3.
(A) Detection of WHcAg in a uPA/RAG-2 mouse liver containing WHV-positive woodchuck hepatocytes by immunostaining with a WHc antiserum. A nodule containing transplanted WHV-positive woodchuck hepatocytes (lighter area, rhodamine light) and host mouse hepatocytes that presumably deleted the uPA transgene (darker stained area). (×200.) (B) DPPIV staining of bile canaliculi in uPA/RAG-2 mouse liver containing woodchuck hepatocytes. (×200.) Bile canaliculi are visible between mouse hepatocytes (darker staining) and transplanted woodchuck hepatocytes (lighter staining). Nuclei are counterstained with hematoxylin.
Infection of Woodchuck Hepatocytes in uPA/RAG-2 Mice.
To investigate whether naive woodchuck hepatocytes could be infected with WHV in uPA/RAG-2 mice, hepatocytes from an adult uninfected woodchuck were transplanted into the liver of uPA/RAG-2 mice as described above. After completion of liver regeneration 3 months after transplantation, four uPA/RAG-2 mice were subjected to a liver biopsy and the presence of woodchuck hepatocytes was confirmed by Southern blot analysis as described above. Subsequently, these uPA/RAG-2 mice were infected intramuscularly with either 10 μl of a WHV-positive woodchuck serum, containing approximately 1 × 109 virions per ml or with 10 μl of WHV-containing serum from uPA/RAG-2 mouse 496 (5 × 108 virions per ml). The establishment of productive infection was monitored by serum dot blot analysis for WHV DNA. In quiescent hepatocytes after completion of liver regeneration 3 months after transplantation, WHV DNA became detectable at 4 weeks after infection. Southern blot analysis of uPA/RAG-2 mouse liver DNAs hybridized with WHV DNA demonstrated the presence of open circular and replicative WHV DNA forms. The serum WHV virion levels have remained stable for an additional 10 months in the infected animals (analysis currently ongoing), confirming the persistence of WHV infection in uPA/RAG-2 mice containing woodchuck hepatocytes.
Antiviral Studies in WHV Replicating uPA/RAG-2 Mice.
To confirm the usefulness of the chimeric uPA/RAG-2 mouse model for studying hepadnaviral replication, we investigated modulation of WHV replication with either interferon α or dexamethasone treatment. Three uPA/RAG-2 mice were chosen for this experiment, mouse 1418 containing hepatocytes from a chronic WHV carrier and mice 1063 and 1098 that were transplanted with naive woodchuck hepatocytes and infected with WHV-containing serum as described above. All mice showed a constant level of viral replication before drug administration (Fig. 4). Dexamethasone significantly up-regulated viral replication. After withdrawal of dexamethasone, the level of WHV replication remained at higher levels compared with pretreatment levels. In contrast, treatment with interferon α down-regulated WHV replication in mouse 1063 by greater than four orders of magnitude to nondetectable levels in serum dot blots after 15 days. However, upon withdrawal of interferon α, WHV replication rebounded to levels higher than pretreatment WHV levels. Mouse 1418, with higher WHV pretreatment levels than mouse 1063, showed only a limited response to interferon α treatment but had a rebound effect upon viral replication similar to mouse 1063 after drug withdrawal.
Heterogeneous Precancerous and Malignant Phenotypes of Transplanted Woodchuck Hepatocytes in uPA/RAG-2 Mice.
According to multistep models of carcinogenesis, cells undergo successive cycles of mutation and selection on their path toward malignancy (4). These models predict that a WHV carrier liver will contain hepatocytes that have experienced initiating mutations that predispose them to oncogenic transformation. We reasoned that the regeneration of WHV-positive hepatocytes after transplantation might cause (i) selection of precancerous hepatocytes, (ii) enhanced oncogenic progression of precancerous hepatocytes, or (iii) both.
Therefore, we compared the phenotypes of transplanted woodchuck hepatocytes in relation to hepatocytes present in the donor woodchuck. In chronic carrier woodchuck 2765, precancerous altered hepatic foci (AHFs) were very rare according to hematoxylin/eosin staining of liver sections (Fig. 5A). In contrast, nearly all of the transplanted woodchuck hepatocyte nodules in the corresponding uPA/RAG-2 mouse livers had a distinct AHF phenotype (Fig. 5B, cells to the right of the arrows) that was absent in control nontransplanted uPA/RAG-2 mouse livers. A close up of the border between mouse hepatocytes and woodchuck hepatocytes in the AHFs from Fig. 5B is shown in Fig. 5D. The woodchuck hepatocytes to the right of the arrows in Fig. 5D exhibited an altered phenotype including enlarged nuclei with prominent nucleoli. This phenotype was common to woodchuck hepatocytes in the AHFs from the donor woodchuck (Fig. 5C, to the right of the arrows).
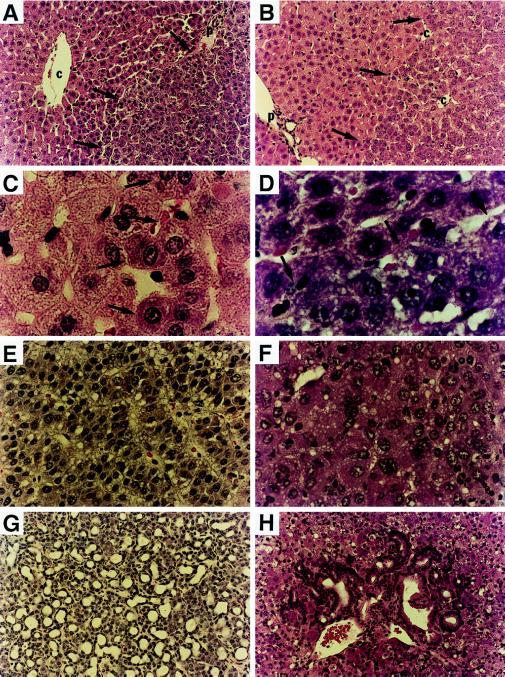
Figure 5.
Hematoxylin/eosin staining of frozen liver sections from donor woodchucks (A, C, E, and G) and uPA/RAG-2 mice recipients (B, D, F, and H) after transplantation of WHV-positive woodchuck hepatocytes. A woodchuck liver (no. 2765) contained altered hepatic foci (A, ×200; C, ×1,000; right side of arrows marks border), with large nuclei and prominent nucleoli that appear similar to transplanted woodchuck hepatocytes in uPA/RAG-2 mouse liver (B, ×200; D, ×1,000; right side of arrows marks border). (E and G) Examples of an HCC and cholangiocarcinoma, respectively, from a woodchuck (no. 4940) chronically infected with WHV. (×200.) (F and H) Presence of an HCC (F) and a cholangiocarcinoma (H) in a uPA/RAG-2 mouse after transplantation of hepatocytes from the donor woodchuck shown in E and G. (×200.)
In another example, we detected both a primary HCC (Fig. 5F) and a cholangiocarcinoma (Fig. 5H) after transplantation of woodchuck cells from woodchuck 4940. Woodchuck 4940 was a chronic WHV carrier that had developed three HCCs and a cholangiocarcinoma (Fig. 5 E and G) in a lobe of it’s liver that was removed before perfusion. The WHV DNA integration pattern in the HCC from transplanted woodchuck cells was different from the integration patterns of the three donor woodchuck HCCs (Fig. 6). Therefore, either the donor liver contained a small tumor that was not detected and was selectively amplified or an infected hepatocyte may have obtained a tumorigenic mutation during growth after transplantation leading to its malignant transformation.
Figure 6.
(A) Southern blot analysis of uPA/RAG-2 mouse liver genomic DNA with a woodchuck genomic DNA probe (lanes 3 and 4). Woodchuck DNA is detectable in tumors arising in uPA/RAG-2 mice (lane 3). Lane 4 is a negative control. Unique WHV DNA integrations in DNA from the tumor of lane 3 are detected with a WHV DNA probe (lane 2, with lane 1 as negative control) that are different from WHV DNA integrations in donor woodchuck tumor DNA samples (B, lanes 1–3).
DISCUSSION
We have developed a hepatitis B mouse model by transplanting xenogenic woodchuck hepatocytes into tolerant uPA/RAG-2 mice. Transplanted woodchuck hepatocytes responded to the mouse liver environment with significant proliferation and replaced more than 90% of the diseased transgenic mouse liver. Transplanted cells grew as microclones in several recipients from common donor sources of cells and their growth pattern generally restored a normal cord structure of the liver.
The experimental system reported herein combined several desirable characteristics of previous animal models for studying hepadnavirus infection and pathogenesis. The woodchuck animal model allows the study of WHV infection in a natural host setting in which complete viral clearance occurs in relatively inaccessible genetically heterogeneous animals (26–28). Although transgenic HBV mice have provided important new information regarding viral pathobiology (6–9), they are tolerant to HBV-encoded antigens and do not entirely reproduce the viral life cycle because CCC DNA cannot be detected in the nuclei of their hepatocytes. The chimeric uPA/RAG-2 mouse model offers a unique chance to study mechanisms of viral persistence in natural host hepatocytes in the absence of B and T cell-mediated immune responses. The absence of B and T cells provide potential opportunities for their replacement with either woodchuck or mouse immune system cells selected for specific B or T cell functions. Furthermore, this model provides the chance to study viral replication in rapidly proliferating hepatocytes during liver repopulation and in quiescent hepatocytes after completion of liver regeneration.
Our first antiviral study used interferon α, because this is the only currently approved treatment for persistent HBV infection (5, 29, 30). Besides exhibiting various immunomodulatory effects (31), interferon α induces the release of intracellular enzymes such as 2′,5′-oligoadenylate synthetase and double-stranded RNA-dependent protein kinase, which degrade viral messenger RNAs and inhibit viral protein synthesis (31) in vitro (32) and in vivo (29, 33).
Patients who respond to interferon α therapy show a decrease in circulating HBV DNA levels within the first week (31). In uPA/RAG-2 mice, we observed a transient reduction in WHV DNA in serum after 15 days of interferon α treatment and enhanced WHV replication by stimulation of the glucocorticoid responsive element with dexamethasone. The immediate rebound of viral replication after withdrawal of interferon α strongly suggests that WHV DNA was not cleared from woodchuck hepatocytes and that woodchuck hepatocytes were not eliminated as a result of interferon α treatment. Further studies should allow us to determine whether the sole remaining WHV DNA during interferon α treatment is nuclear CCC DNA. The effectiveness of human interferon α against WHV suggests that other human and murine reagents may cross react with their woodchuck homologues and will allow us to test a wide variety of antiviral agents, including cytokines, for their effectiveness in clearing all molecular species of WHV DNA.
A major objective for hepatocarcinogenesis studies is to define the cellular and molecular phenotype of precancerous hepatocytes to design early diagnostic and intervention protocols. Phenotypic comparison of nodular woodchuck hepatocytes in uPA/RAG-2 mice with hepatocytes present in precancerous lesions in the donor woodchuck revealed an enrichment of AHF phenotypes in transplanted uPA/RAG-2 mice. These data suggest that transplantation led either to a selection for altered hepatocytes that preexisted in the woodchuck liver or progression of initiated hepatocytes from the original woodchuck liver. In either case, the easy identification of amplified precancerous lesions provides a tool for studying genetic changes in those cells.
The presence of unique WHV DNA integrations in an HCC from a uPA/RAG-2 mouse liver demonstrated in vivo selection and clonal expansion of woodchuck hepatocytes, suggesting that tumor progression may have occurred after cell transplantation. Such clonal expansion of hepatocytes should provide sufficient quantities of transformed cells to investigate and better understand the roles of viral genes and WHV DNA integrations in hepadnavirus-associated hepatocarcinogenesis.
Acknowledgments
We thank Leslie E. Rogler for helpful advice during this project and L. Johnson and B. Tennant for providing woodchucks from the experimental woodchuck colony at Cornell University, Ithaca, NY. We also thank J. Gerin, B. Korba, and P. Cote for providing WHs antiserum; W. Mason for providing WHx and WHc antisera; S. Gagandeep and R. Sokhi for technical assistance; and T. Harris for help with computer work. This work was supported by U.S. Public Health Service Grants CA 37232 and DK 46952, Center Grants P30CA13330 and P30DK41296, the Irma T. Hirschl Trust, and the Council for Tob. Res. USA Inc. C.E.R and S.G. are recipients of Irma T. Hirschl-Weiler career scientist awards. J.P. was on leave from the Department of Medicine, University Hospital Eppendorf, University of Hamburg, Germany, and was supported by the Deutsche Forschungsgemeinschaft (Pe/608 1–1), Bonn, Germany.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: WHV, woodchuck hepatitis virus; WHsAg, woodchuck hepatitis virus surface antigen; WHcAg, woodchuck hepatitis virus core antigen; WHx, woodchuck hepatitis virus X protein; uPA, urokinase-type plasminogen activator; RAG-2, recombination activation gene 2; HCC, hepatocellular carcinoma; HBV, hepatitis B virus; DPPIV, dipeptidyl peptidase IV; CCC, covalently closed circular; AHF, altered hepatic focus.
References
- 1.Ganem D, Varmus H E. Annu Rev Biochem. 1987;56:651–693. doi: 10.1146/annurev.bi.56.070187.003251. [DOI] [PubMed] [Google Scholar]
- 2.Buendia M A. Adv Cancer Res. 1992;59:167–226. doi: 10.1016/s0065-230x(08)60306-1. [DOI] [PubMed] [Google Scholar]
- 3.Blumberg B S. Proc Natl Acad Sci USA. 1997;94:7121–7125. doi: 10.1073/pnas.94.14.7121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rogler C E. Curr Top Microbiol Immunol. 1991;168:103–141. doi: 10.1007/978-3-642-76015-0_6. [DOI] [PubMed] [Google Scholar]
- 5.Fried M W. Med Clin N Am. 1996;80:957–971. doi: 10.1016/s0025-7125(05)70475-2. [DOI] [PubMed] [Google Scholar]
- 6.Chisari F V, Ferrari C. Annu Rev Immunol. 1995;13:29–60. doi: 10.1146/annurev.iy.13.040195.000333. [DOI] [PubMed] [Google Scholar]
- 7.Chisari F V. Curr Top Microbiol Immunol. 1996;206:150–173. doi: 10.1007/978-3-642-85208-4_9. [DOI] [PubMed] [Google Scholar]
- 8.Guidotti L G, Ando K, Hobbs M V, Ishikawa T, Runkel L, Schreiber R D, Chisari F V. Proc Natl Acad Sci USA. 1994;91:3764–3768. doi: 10.1073/pnas.91.9.3764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guidotti L G, Ishikawa T, Hobbs M V, Matzke B, Schreiber R, Chisari F V. Immunity. 1996;4:25–36. doi: 10.1016/s1074-7613(00)80295-2. [DOI] [PubMed] [Google Scholar]
- 10.Gupta S, Rajvanshi P, Bhargava K K, Kerr A. Prog Liver Dis. 1996;14:199–222. [PubMed] [Google Scholar]
- 11.Sandgren E P, Palmiter R D, Heckel J L, Daugherty C C, Brinster R L, Degen J L. Cell. 1991;66:245–256. doi: 10.1016/0092-8674(91)90615-6. [DOI] [PubMed] [Google Scholar]
- 12.Rhim J A, Sandgren E P, Degen J L, Palmiter R D, Brinster R L. Science. 1994;263:1149–1152. doi: 10.1126/science.8108734. [DOI] [PubMed] [Google Scholar]
- 13.Rhim J A, Sandgren E P, Palmiter R D, Brinster R L. Proc Natl Acad Sci USA. 1995;92:4942–4946. doi: 10.1073/pnas.92.11.4942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shinkai Y, Rathbun G, Lam K P, Oltz E M, Stewart V, Mendelsohn M, Charron J, Datta M, Young F, Stall A M, Alt F W. Cell. 1992;68:855–867. doi: 10.1016/0092-8674(92)90029-c. [DOI] [PubMed] [Google Scholar]
- 15.Vemuru R P, Aragona E, Gupta S. Hepatology. 1992;16:968–973. doi: 10.1002/hep.1840160419. [DOI] [PubMed] [Google Scholar]
- 16.Horton R M, Karachunski P I, Conti-Fine B M. BioTechniques. 1995;19:690–691. [PubMed] [Google Scholar]
- 17.Gupta S, Vemuru R P, Lee C D, Yerneni P, Aragona E, Burk R D. Hum Gene Ther. 1994;4:249–257. doi: 10.1089/hum.1994.5.8-959. [DOI] [PubMed] [Google Scholar]
- 18.Rajvanshi P, Kerr A, Bhargava K K, Burk R D, Gupta S. Hepatology. 1996;23:482–496. doi: 10.1002/hep.510230313. [DOI] [PubMed] [Google Scholar]
- 19.Dandri M, Schirmacher P, Rogler C E. J Virol. 1996;70:5246–5254. doi: 10.1128/jvi.70.8.5246-5254.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laemmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 21.Gong S S, Jensen A D, Rogler C E. J Virol. 1996;70:2000–2007. doi: 10.1128/jvi.70.3.2000-2007.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Petersen J, Buerkle A, Zhang L, Rogler C E. J Virol. 1997;71:5455–5463. doi: 10.1128/jvi.71.7.5455-5463.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ogston C W, Jonak G J, Rogler C E, Astrin S M, Summers J. Cell. 1982;29:385–394. doi: 10.1016/0092-8674(82)90155-6. [DOI] [PubMed] [Google Scholar]
- 24.Scotto J, Hadchouel M, Hery C, Yvart Y, Tiollais P, Brechot C. Hepatology. 1983;3:279–284. doi: 10.1002/hep.1840030301. [DOI] [PubMed] [Google Scholar]
- 25.Gupta S, Lee C D, Vemuru R P, Bhargava K K. Hepatology. 1994;19:750–757. doi: 10.1002/hep.1840190330. [DOI] [PubMed] [Google Scholar]
- 26.Roggendorf M, Tolle T K. Intervirology. 1995;38:100–112. doi: 10.1159/000150418. [DOI] [PubMed] [Google Scholar]
- 27.Korba B E, Cote P J, Wells F N, Baldwin B, Popper H, Purcell R H, Tennant B C, Gerin J L. J Virol. 1989;63:1360–1370. doi: 10.1128/jvi.63.3.1360-1370.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tennant B C, Gerin J L. In: The Liver. Biology and Pathobiology. Arias I M, Boyer J L, Fausto N, Jakoby W B, Schachter D, Shafritz D A, editors. New York: Raven; 1994. pp. 1455–1469. [Google Scholar]
- 29.Perillo R P, Mason A L. Gastroenterol Clin N Am. 1994;23:581–601. [PubMed] [Google Scholar]
- 30.Perillo R P, Schiff E R, Davis G L, Bodenheimer H C, Lindsay K, et al. N Engl J Med. 1990;323:295–301. doi: 10.1056/NEJM199008023230503. [DOI] [PubMed] [Google Scholar]
- 31.Haria M, Benfield P. Drugs. 1995;50:873–896. doi: 10.2165/00003495-199550050-00007. [DOI] [PubMed] [Google Scholar]
- 32.Yamashita Y, Koike K, Takashi M, Matsuda S. Microbiol Immunol. 1988;32:1119–1126. doi: 10.1111/j.1348-0421.1988.tb01476.x. [DOI] [PubMed] [Google Scholar]
- 33.Nagahata T, Araki K, Yamamura K I, Matsubara K. Antimicrob Agents Chemother. 1992;36:2042–2045. doi: 10.1128/aac.36.9.2042. [DOI] [PMC free article] [PubMed] [Google Scholar]