Abstract
Nonsyndromic defects in the urinary tract are the most common cause of end-stage renal failure in children and account for a significant proportion of adult nephropathy. The genetic basis of these disorders is not fully understood. We studied seven multiplex kindreds ascertained via an index case with a nonsyndromic solitary kidney or renal hypodysplasia. Systematic ultrasonographic screening revealed that many family members harbor malformations, such as solitary kidneys, hypodysplasia, or ureteric abnormalities (in a total of 29 affected individuals). A genomewide scan identified significant linkage to a 6.9-Mb segment on chromosome 1p32-33 under an autosomal dominant model with reduced penetrance (peak LOD score 3.5 at D1S2652 in the largest kindred). Altogether, three of the seven families showed positive LOD scores at this interval, demonstrating heterogeneity of the trait (peak HLOD 3.9, with 45% of families linked). The chromosome 1p32-33 interval contains 52 transcription units, and at least 23 of these are expressed at stage E12.5 in the murine ureteric bud and/or metanephric mesenchyme. These data show that autosomal dominant nonsyndromic renal hypodysplasia and associated urinary tract malformations are genetically heterogeneous and identify a locus for this common cause of human kidney failure.
The human kidney is composed of 500,000 to 1.8 million independent functional units, called “nephrons.”1,2 Kidney failure occurs when acquired or hereditary disorders cause progressive nephron loss and reduce their numbers to below the threshold required for maintenance of fluid-electrolyte homeostasis and elimination of nitrogenous wastes. Congenital abnormalities that impair nephron development comprise 30%–50% of congenital anomalies detected on prenatal screening.3,4 There is wide interindividual variability in the anatomy and clinical course of these malformations, but, altogether, they account for up to 50% of pediatric and 10% of adult end-stage renal disease worldwide.5,6 Renal agenesis/adysplasia (MIM %191830) is one of the most severe forms of malformations; bilateral renal agenesis occurs in ∼1 in 2,000 births and is nearly always fatal because of oligohydramnios and consequent pulmonary hypoplasia.3,4,7,8 Other related phenotypic variants, such as unilateral agenesis and unilateral or bilateral renal hypodysplasia, occur more frequently (1 in 1,000 births).3,4 Affected kidneys frequently exhibit dysplastic parenchyma, reduced nephron mass, and impaired function, predisposing the individual to renal failure.8 Kidney malformations are also commonly accompanied by anatomic abnormalities in the lower urinary tract, such as ureteropelvic junction obstruction (UPJO) or vesicoureteral reflux (VUR).8–12 Impairment of urinary drainage due to these lower-tract defects further contributes to nephron injury and augments the risk of chronic renal failure.
The etiology of the majority of nonsyndromic forms of urinary tract malformations remains unknown. The overlap in phenotypic expression, together with the limitations of morphological classification, has complicated efforts to understand the primary pathogenesis of these disorders and to formulate clear diagnostic categories. For example, it is not known whether the observation of a congenital solitary kidney in an adult represents a primary failure of kidney development (agenesis) or the end-result of a dysplastic kidney that underwent involution.3,8 Several risk factors for human urinary tract malformations are recognized, including maternal diabetes13 or intrauterine exposure to drugs such as ACE inhibitors.14 In addition, the genetic bases for several rare, syndromic disorders featuring renal developmental defects have been elucidated (e.g., renal-coloboma syndrome [MIM #120330], Fraser syndrome [MIM #219000], and branchiootorenal syndrome [MIM #113650]). Gene-targeting studies have shown that alteration in gene dosage or disruption in the temporospatial sequence of gene expression in intermediate mesodermal tissues (the metanephric mesenchyme [MM] or the ureteric bud [UB]) can result in developmental defects that affect both the upper and lower urinary system.8,12,15–22 Moreover, because of interdependence of developmental pathways, similar renal and urologic phenotypes arise by inactivation of different genes.8,12,15–22 These findings are consistent with the “bud theory” of Mackie and Stephens, which proposes that abnormalities in the kidney and ureter stem from the same common mechanism—namely, abnormal ureteral budding from the Wolffian duct into the bladder results in malposition of the ureteral orifice.12 These data together with the phenotypic overlap observed in human renal and urologic disorders emphasize the potential for genetic heterogeneity of disease. Candidate-gene screening has confirmed heterogeneity, revealing mutations in the Uroplakin 3A gene in a subset of patients with renal dysplasia.23,24 Other recent surveys of patients with renal hypodysplasia, many of whom had extra-renal defects suggestive of syndromic disease, identified mutations in PAX2, TCF2, EYA1, SIX1, or SALL1 in up to 17%–25% of patients.25,26 Thus, mutations identified to date account for a fraction of nonsyndromic renal developmental defects, leaving the majority of cases unexplained.
Several studies have reported familial aggregation of nonsyndromic renal malformations. For example, ultrasonographic screening of first-degree relatives of 41 index patients with bilateral renal agenesis/dysgenesis identified asymptomatic renal malformations (mostly unilateral renal agenesis) in 9% of relatives.9 Additional malformations were also documented, including UPJO and VUR. This increased recurrence risk among relatives has been confirmed in several other studies and is estimated at 4%–20%.9–11,27 Multigenerational occurrence of disease has suggested multifactorial or dominant inheritance in most kindreds,9–11 but families with probable recessive inheritance have also been reported.27 Altogether, these data strongly argue in favor of genetic causation and suggest that familial aggregation is under-recognized because anatomical defects in many family members are often silent. No linkage studies in such families have been reported to date.
We set out to investigate the genetic basis of nonsyndromic urinary tract malformations. Our previous studies of primary VUR had suggested that gene mapping for such heterogeneous disorders would necessitate large numbers of medium-sized pedigrees or uniquely large kindreds.28 In the situation of uncertainty about clinical classification and the potential for genetic heterogeneity, we strived to assemble a cohort with a strong genetic contribution by concentrating on patients with the most severe clinical phenotype—namely, solitary kidneys and/or renal hypodysplasia. We identified seven Italian pedigrees ascertained through an index patient with solitary kidney/hypodysplasia documented by a renal sonogram and/or isotopic scintigraphy (fig. 1). Solitary kidney was diagnosed when one kidney only was seen in the imaging study. Hypodysplasia was defined as kidney length below the 95% tolerance limit based on height- and weight-adjusted sonographic nomograms.29,30 A pediatric nephrologist and clinical geneticist with expertise in diagnosis of developmental disorders of the urinary tract evaluated all the patients. Histories and physical examinations were conducted to search for evidence of multiorgan malformations, which would suggest syndromic forms of renal hypodysplasia. In particular, we focused on the possibility of disease associated with PAX2 or TCF2 mutations, which may masquerade as nonsyndromic malformations.25,26 We performed ophthalmologic exams to search for retinal colobomas and investigated the presence of deafness or genital malformations, which would be indicative of renal-coloboma syndrome. In addition, we searched for abnormalities suggestive of TCF2 mutations, such as presence of renal cysts, diabetes, and elevated liver enzymes or uric acid levels. The exclusion of PAX2 and TCF2 by clinical criteria was further confirmed by linkage analysis in all pedigrees and by direct sequencing of TCF2 exons in one kindred (K101) that had a minimally positive LOD score at the TCF2 locus. There was also no history of intrauterine exposure to teratogenic agents, such as ACE inhibitors and angiotensin receptor antagonists, and no history of maternal diabetes.
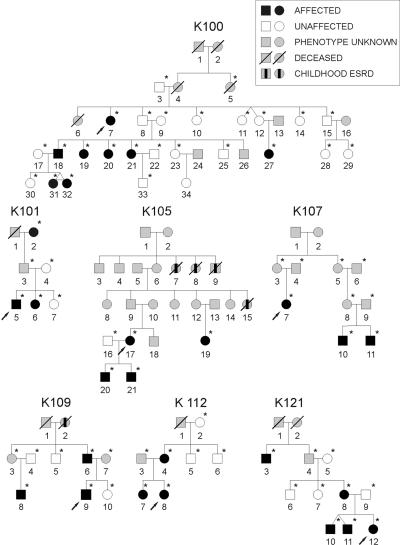
Figure 1. .
Pedigree structure of the seven families studied. Patients with childhood end-stage renal disease (ESRD) but no sonographic data available are indicated by a blackened rectangle inside the symbol. Arrows identify the index cases. Asterisks (*) mark the individuals from whom DNA was available for the study. Individual identification numbers correspond to table 1.
To detect additional affected relatives, we performed ultrasonographic screening and chart review of family members. We identified 22 family members with anatomic abnormalities (table 1). Most family members had congenital solitary kidneys/hypodysplasia, but other abnormalities were also identified, such as UPJO (11 individuals), VUR (6 individuals), infundibulopelvic stenosis (1 individual), and multicystic dysplastic kidney (1 individual). These additional defects mostly presented in association with the hypodysplasia phenotype but also occurred in isolation in a few family members (table 1). One individual (31 in K100, age 10 years) had asymmetric kidney sizes that fell within the normal range, but she was classified as affected because she had an MZ twin with unilateral renal hypodysplasia (this individual and her twin were considered a single affected individual in the genome scan).
Table 1. .
Clinical Data of the Affected Individuals from the Seven Pedigrees[Note]
| Pedigree and Individual |
Sex | Age (years) |
Kidney Phenotype | Kidney Lengtha R/L (cm) |
Associated Defect | Chronic Renal Failure | Dialysis |
| K100: | |||||||
| 7 | F | 55 | Solitary kidney | -/12 | None | No | No |
| 18 | M | 36 | Hypodysplasia (R) | 6/12.3 | None | No | No |
| 19 | F | 38 | Hypodysplasia (R+L) | 7/5.5 | None | Yes | Yes |
| 20 | F | 32 | Hypodysplasia (R+L) | 6/5.8 | UPJO (L) | Yes | Yes |
| 21 | F | 28 | Hypodysplasia (R+L) | 5/5.5 | UPJO (L) | Yes | Yes |
| 27 | F | 12 | Infundibulopelvic stenosis (R) | 10.5/10.8 | Hydro-calix (R) | No | No |
| 31 | F | 10 | Asymmetric sizes | 11/8.5 | None | No | No |
| 32 | F | 10 | Hypodysplasia (R) | 6/10.5 | None | No | No |
| K101: | |||||||
| 2 | F | 82 | Hypodysplasia (R+L) | 5/6 | ND | Yes | Yes |
| 5 | M | 15 | Hypodysplasia (R) | 7/10.9 | UPJO (R) and VUR (R+L) | No | No |
| 6 | F | 23 | Normal kidney size | 11/10.5 | UPJO (L) | No | No |
| K105: | |||||||
| 17 | F | 37 | Solitary kidney | 12/- | None | No | No |
| 19 | F | 35 | Hypodysplasia (R) | 6/11 | None | No | No |
| 20 | M | 16 | Normal kidney size | 10/10.8 | VUR (R+L) | No | No |
| 21 | M | 14 | Normal kidney size | 9.8/10 | MCDK (L) and VUR (L) | No | No |
| K107: | |||||||
| 7 | F | 40 | Hypodysplasia (R) | 6/10.8 | VUR (R) | No | No |
| 10 | M | 10 | Hypodysplasia (R+L) | 7/5.3 | VUR (L) | No | No |
| 11 | M | 13 | Hypodysplasia (R+L) | 8.8/6.4 | VUR (L) | No | No |
| K109: | |||||||
| 6 | M | 48 | Normal kidney size | 12/12.2 | UPJO (L) | No | No |
| 8 | M | 18 | Normal kidney size | 11.5/12 | UPJO (R) | No | No |
| 9 | M | 19 | Hypodysplasia (R) | 6/12 | UPJO (R) | No | No |
| K112: | |||||||
| 4 | F | 60 | Normal kidney size | 12.5/12.3 | UPJO (L) | No | No |
| 7 | F | 28 | Normal kidney size | 11.5/12 | UPJO (R) | No | No |
| 8 | F | 30 | Hypodysplasia (R+L) | 8/7 | UPJO (R) | Yes | No |
| K121: | |||||||
| 3 | M | 80 | Solitary kidney | 12/- | None | No | No |
| 8 | F | 28 | Normal kidney size | 12/12.5 | UPJO (L) | No | No |
| 10 | M | 6 | Hypodysplasia (L) | 10/5 | None | No | No |
| 11 | M | 6 | Solitary kidney | 11/- | None | No | No |
| 12 | F | .3 | Solitary kidney | -/9 | None | No | No |
Note.— R = right; L = left; MCDK = multicystic dysplastic kidney; ND = not determined.
A hyphen (-) indicates that the kidney was not visualized. For reference, the normal range for kidney size in an adult is 10–15 cm; the 95th percentile interval for a 10-year-old is 7–11 cm.
Altogether, 29 individuals were classified as affected on the basis of abnormal urinary anatomy seen on imaging studies (12 males and 17 females, listed in table 1). Of these affected individuals, 10 were asymptomatic and received a diagnosis at the time of screening, and 5 had renal dysfunction (as defined by serum creatinine >1.5 mg/dl) or had developed end-stage renal failure requiring dialysis or transplantation. Twenty-nine family members had normal imaging studies and normal renal function and were classified as unaffected (fig. 1). In addition, chart review revealed that five other family members had died in childhood from end-stage renal failure, but renal imaging studies for these individuals were not available to make a definitive diagnosis (fig. 1). These individuals and all other family members who did not undergo imaging studies were considered to have an unknown phenotype. All phenotypes were assigned prospectively. In these pedigrees, the mode of inheritance was most consistent with multifactorial inheritance or autosomal dominant transmission with incomplete penetrance.
All individuals gave informed consent, and the study protocol was approved by the Western Institutional Review Board for Columbia University and the ethics committee at the Gaslini Institute. Total genomic DNA was isolated from peripheral white blood cells by use of standard procedures. Genomewide screens were performed with two independent methodologies. In the largest kindred (K100), we performed an ∼10-cM microsatellite screen of autosomes (382 markers from Linkage Mapping Set v2.5 MD10 [Applied Biosystems]), using seven affected and seven unaffected individuals. The seven unaffected individuals (8, 10, 11, 12, 14, 15, and 23) were included, to increase inheritance information. During that time, genomewide SNP genotyping technology became readily available. We therefore genotyped all affected individuals from our seven kindreds for 10,204 SNPs, using the GeneChips Mapping 10K 2.0 Arrays (Affymetrix). DNA processing and gene-chip hybridization were performed as suggested by Affymetrix. This provided a uniform data set across all pedigrees.
We performed pairwise analysis of linkage, using FASTLINK 4.1,31 and multipoint analysis, using ALLEGRO 1.2c32 and Simwalk 2.89.33 Parametric analysis was performed under a model of autosomal dominant transmission with reduced penetrance (85%, estimated from the pedigrees), disease-gene frequency of 0.001, and phenocopy rate of 0.001. We computed parametric LOD scores under genetic homogeneity and heterogeneity (calculation of HLODs). Nonparametric statistics were concurrently computed with Allegro (the NPL score and associated exact P value). Allele frequencies were calculated on the basis of the frequencies observed in the data set for the microsatellites; for the SNP data, frequencies were based on public, ethnically matched, frequencies provided by Affymetrix. Published thresholds for suggestive and significant linkage were used.34,35 To detect our study power, 200 simulations under the assumption of linkage with 85% penetrance were performed using the SLINK program.36 The simulated pedigrees were next tested for linkage by use of the autosomal dominant model delineated above. This analysis demonstrated a maximal expected LOD score of 4.1 (average 3.1±1.3) in K100.
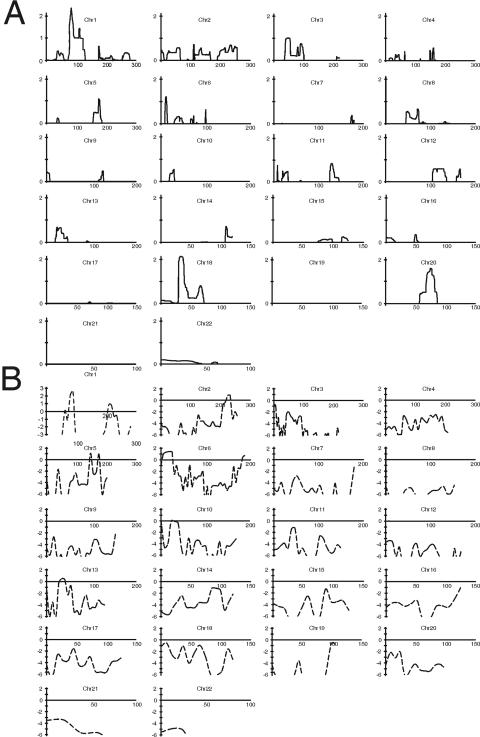
We therefore initially focused on K100, because this pedigree was sufficiently large to localize a trait locus independently. With analysis of affected individuals only (hereafter, “affected-only analysis”), several loci showed LOD scores >1 in K100 for both microsatellite and SNP analyses: 1p32-33 (LOD=1.4), 1q31-33 (LOD=1.5), and 6p24-25 (LOD=1.5) (fig. 2). When the phenotype of unaffected individuals was introduced into the analysis (with the microsatellite scan), the LOD increased to 2.7 at the chromosome 1p32-33 interval (D1S2770) but decreased at the other two intervals. Together, these data suggested that the linkage peak on chromosome 1p32-33 was unlikely to be an artifactual finding produced by linkage disequilibrium between SNP markers. We genotyped 42 additional microsatellite markers across these three promising intervals and genotyped all 16 available family members to extract full inheritance information. After the incorporation of microsatellite markers, we achieved inheritance information >0.9 at these loci. Evidence of linkage to chromosomes 1q31-33 and 6p24-25 became less significant (LOD⩽1.2). However, the LOD score on chromosome 1p32-33 improved significantly, resulting in a peak parametric LOD score of 3.5 between D1S2652 and rs1341347 (fig. 3). The likelihood of linkage to 1p32-33 was 200-fold greater than the next most likely interval at 1q31-33 (LOD=1.2). As shown in table 2, these results were robust to changes in estimates of disease-allele frequencies (0.001–0.01), penetrances (65%–95%), and phenocopy rates (0.001–0.01). This analysis therefore achieved genomewide significance on the basis of published criteria34,35 and established localization of a gene for nonsyndromic renal hypodysplasia on chromosome 1p32-33 (fig. 3).
Figure 2. .
LOD score distribution across the genome. The continuous lines (A) represent the HLOD scores obtained from the SNP scan with affected-only analysis of all seven pedigrees (minimum HLOD=0). The dashed lines (B) represent the parametric LOD scores obtained from microsatellite scan in pedigree K100 alone. Values <−6 are not shown. Chr = chromosome.
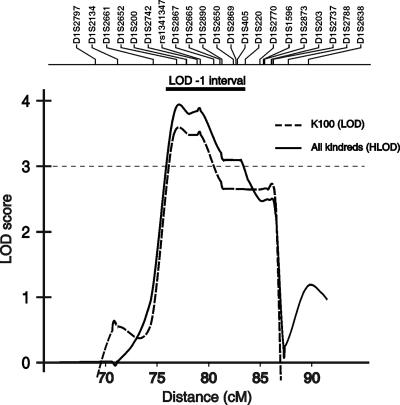
Figure 3. .
LOD plot of chromosome 1p32-33 locus. The multipoint LOD score in K100 and the HLOD in all seven pedigrees combined are shown. The X-axis shows genetic distance based on the deCODE map. The location of microsatellite markers genotyped is shown above the graph. The LOD-1 support interval is indicated by the thick horizontal bar above the LOD curve.
Table 2. .
Multipoint LOD Scores on Chromosome 1p32-33 with Varying Linkage Parameters[Note]
| LOD for K100 at |
HLOD at |
|||||
| Disease-Allele Frequency .001 |
Disease-Allele Frequency .001 |
|||||
| Penetrances (%) |
Phenocopy Rate .001 | Phenocopy Rate .01 | Disease-Allele Frequency .01 and Phenocopy Rate .001 |
Phenocopy Rate .001 | Phenocopy Rate .01 | Disease-Allele Frequency .01 and Phenocopy Rate .001 |
| 65 | 3.2 | 3.2 | 3.2 | 3.6 | 3.5 | 3.3 |
| 75 | 3.4 | 3.4 | 3.4 | 3.7 | 3.7 | 3.5 |
| 85 | 3.5 | 3.5 | 3.5 | 3.9 | 3.8 | 3.6 |
| 95 | 3.3 | 3.3 | 3.3 | 3.7 | 3.7 | 3.4 |
Note.— Peak parametric HLODs are shown for all seven pedigrees combined (with 45% of families linked).
We next determined whether our six other pedigrees with renal hypodysplasia demonstrate linkage to this interval. After genotyping 24 microsatellites across 1p32-33 in these remaining pedigrees, affected-only analysis provided an HLOD of 1.1 (with 45% of families linked) and an NPL of 2.2 (P=.02) at 1p32-33, demonstrating that some but not all families linked to this interval. Two kindreds (K105 and K109) showed near-maximal expected LOD scores between D1S200 and D1S220, encompassing the same linkage interval as K100. In these two kindreds, the conditional probability of linkage to 1p32-33 was >0.95 (table 3). In K112, a LOD score of 0.3 was achieved by affected-only analysis, but it decreased to −0.6 when the phenotype of unaffected individuals was introduced in the calculations. Results for the other three kindreds (K101, K107, and K121) were negative in the affected-only analysis and after incorporation of unaffected phenotypes. To further verify that the data for the kindreds with positive LOD scores point to the same interval as K100, we combined all seven families and analyzed linkage to 1p32-33 under a model of locus heterogeneity. This analysis resulted in a peak HLOD of 3.9 with 45% of families linked; this HLOD peak coincides perfectly with the LOD curve obtained with K100 alone (fig. 3). As before, varying linkage parameters had little effect on the HLOD results (table 2), exceeding the threshold for genomewide significance in all cases.34,35 Finally, evidence of linkage to 1p32-33 was also supported by nonparametric analysis (NPL=5.3; P=1.5×10-4). Altogether, these data provide genomewide evidence for localization of a gene for renal hypodysplasia on 1p32-33 and confirm that this trait is genetically heterogeneous.
Table 3. .
List of Positional Candidates with Detectable Expression in the Developing Murine Urinary Tract[Note]
| Human Gene Symbol |
Human Gene Description | Mus Gene Symbol | Affymetrix ProbeSet | MMa | UB Tipa | UB Stalka |
| YIPF1 | Yip1 domain family, member 1 | Yipf1 | 1424196_at | 270 | 288 | 284 |
| C1ORF41 | Chromosome 1 ORF 41 | 2900042B11Rik | 1453016_at | 390 | 248 | 398 |
| LRRC42 | Leucine-rich repeat containing 42 | Lrrc42 | 1424049_at | 394 | 442 | 419 |
| TMEM59 | Transmembrane protein 59 | Tmem59 | 1450046_at | 13 | 17 | 13 |
| C1orf83 | Chromosome 1 ORF 83 | 2210012G02Rik | 1418662_at | 141 | 148 | 143 |
| MRPL37 | Mitochondrial ribosomal protein L37 | Mrpl37 | 1423764_s_at | 385 | 494 | 400 |
| SSBP3 | Single-stranded DNA binding protein 3 | Ssbp3 | 1425940_a_at | 217 | 148 | 225 |
| 1427917_s_at | 985 | 805 | 954 | |||
| ACOT11 | Acyl-CoA thioesterase 11 | Acot11 | 1429267_at | 95 | 76 | 104 |
| C1orf179 | Chromosome 1 ORF 179 | BC026682 | 1417443_at | 55 | 52 | 52 |
| TTC4 | Tetratricopeptide repeat domain 4 | Ttc4 | 1451193_x_at | 528 | 523 | 518 |
| 1451192_a_at | 549 | 497 | 539 | |||
| PARS2 | Prolyl-tRNA synthetase (mitochondrial) (putative) | Pars2 | 1427309_at | 203 | 203 | 208 |
| 1418130_at | 132 | 153 | 112 | |||
| PCSK9 | Proprotein convertase subtilisin/kexin type 9 AD hypercholesterolemia | Pcsk9 | 1437453_s_at | 18 | 48 | 27 |
| USP24 | Ubiquitin-specific protease 24 | Usp24 | 1452868_at | 209 | 287 | 263 |
| 1448013_at | 178 | 259 | 244 | |||
| 1448014_s_at | 124 | 171 | 138 | |||
| 1441018_at | 40 | 35 | 35 | |||
| PPAP2B | Phosphatidic acid phosphatase type 2B | Ppap2b | 1429514_at | 673 | 488 | 738 |
| 1448908_at | 545 | 390 | 623 | |||
| PRKAA2 | Protein kinase, AMP-activated, alpha | Prkaa2 | 1429463_at | 28 | 52 | 45 |
| DAB1 | Disabled homolog 1 (Drosophila) | Dab1 | 1427308_at | 31 | 47 | 37 |
| 1427307_a_at | 66 | 90 | 71 | |||
| 1421100_a_at | 11 | 16 | 12 | |||
| 1435578_s_at | 25 | 40 | 38 | |||
| 1435577_at | 15 | 18 | 18 | |||
| OMA1 | OMA1 homolog, zinc metallopeptidase (S. cerevisiae) | Oma1 | 1424299_at | 199 | 246 | 214 |
| TACSTD2 | Tumor-associated calcium signal transducer 2 | Tacstd2 | 1423323_at | 11 | 535 | 523 |
| MYSM1 | Myb-like, SWIRM and MPN domains 1 | Mysm1 | 1437202_at | 118 | 110 | 142 |
| JUN | V-jun sarcoma virus 17 oncogene homolog (avian) | Jun | 1417409_at | 2,347 | 2,774 | 3,758 |
| 1448694_at | 1,137 | 1,119 | 1,866 | |||
| FLJ10986 | Hypothetical protein FLJ10986 | 2310009E04Rik | 1452799_at | 102 | 114 | 108 |
| HOOK1 | Hook homolog 1 (Drosophila) | Hook1 | 1438018_at | 252 | 881 | 446 |
| 1439173_at | 18 | 56 | 27 | |||
| CYP2J2 | Cytochrome P450, family 2, subfamily J, polypeptide 2 | Cyp2j6 | 1417952_at | 51 | 121 | 117 |
Note.— Genes reported to be present in at least one compartment are shown.
The last three columns show raw expression levels across metanephric compartments (from Schmidt-Ott et al.39).
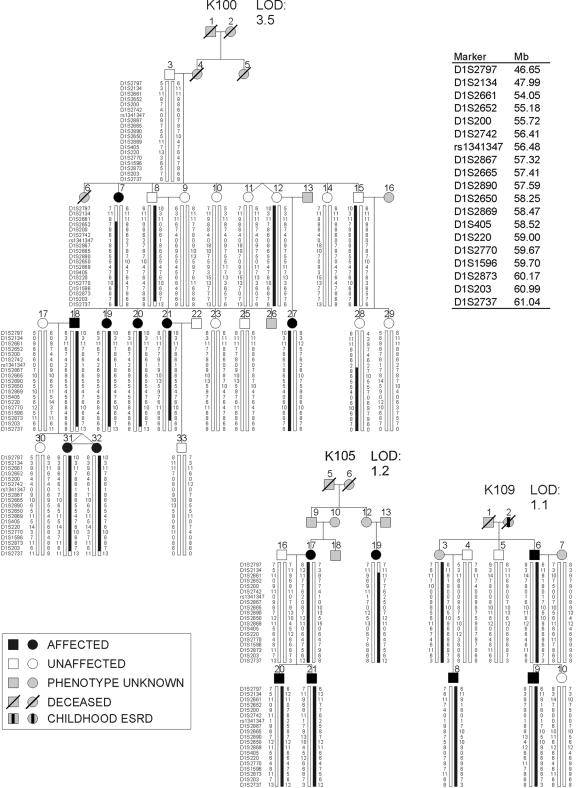
We next analyzed the 1p32-33 haplotypes, which are defined by 51 SNP and microsatellite markers across our linked interval. This demonstrated that all affected members and obligate carriers of K100, K105, and K109 inherited the linked haplotypes at 1p32-33 (fig. 4). Consistent with incomplete penetrance of the trait, one unaffected member in pedigree K100 (individual 15) had inherited the linked haplotype, and his unaffected daughter (individual 28) had inherited the distal portion of the haplotype. The recombination interval, inferred from affected-only analysis, localizes the disease gene within the 6.9-Mb region between D1S2661 and D1S203, corresponding to the region with the maximal LOD scores from SNP and microsatellite analysis. Comparison of haplotypes showed no evidence of shared segments between the three families with positive LOD scores on 1p32-33, suggesting independent mutations. This is not surprising, because K100 originates from Sardinia, whereas the other two kindreds belong to other regions of Italy.
Figure 4. .
Haplotype structure of the three pedigrees that show linkage to the 1p32-33 locus. Genotypes for the most informative markers spanning the linkage interval are shown (19 of 51 SNPs and microsatellite loci genotyped are shown). The vertical bars highlight the linked (black) and the wild-type (white) haplotypes. The physical locations of the markers are indicated in a list at the top right. Patients with childhood end-stage renal disease (ESRD) but no sonographic data available are indicated by a blackened rectangle inside the symbol.
Examination of genes located within the LOD-1 interval on 1p32-33 (by use of the NCBI Map Viewer, build 36.1; Ensembl, v39; and University of California–Santa Cruz [UCSC] Genome Browser, March 2006 assembly) reveals 52 transcription units, of which 23 encode hypothetical or predicted proteins. None of the positional candidates have been previously implicated in structural abnormalities of the kidney or urologic tract. Consultation of Online Mendelian Inheritance of Man (OMIM) revealed that mutations in six positional candidates have been implicated in human traits (PCSK9, BSDN, C8A, C8B, TACSTD2, and DHCR24). Mice carrying null alleles for seven other genes (HOOK1, DAB1, JUN, PRKAA2, PPAP2B, DHCR24, and SSBP3) have also been reported, but none feature urogenital defects. Two other genes on 1p32-33 had been associated with kidney developmental defects (FOXD2 and CPT2) but were located outside our recombinant interval.37,38
A powerful method to annotate positional candidates is to determine their expression pattern within relevant tissues and developmental time points. Nephron development is a recursive process that starts in the 2nd month of gestation, when the UB invades the MM, initiating branching morphogenesis.21,22 Genetic manipulations in mice have shown that inactivation of genes expressed in either compartment can produce the renal hypodysplasia phenotype.12–20 Hence, one can predict that the phenotype observed in our families originates from dysregulation of a gene that is normally expressed early in nephrogenesis but could be localized to the MM, the UB, or both. Accordingly, we performed annotation of positional candidates with the results of a recently published study that characterized gene expression in the murine MM, UB tips, and UB stalk.39 This expression profiling was performed at the E12.5 time point, corresponding to the 2nd and 3rd round of cleavage in branching morphogenesis, among the earliest time points at which the MM can be anatomically differentiated from the UB in the mouse. We particularly focused on genes that show differential expression in UB tips, which coordinate UB branching and secrete factors necessary for nephron induction in the MM. Examination of the data revealed that 30 positional candidates had murine homologs and corresponding probe sets on the arrays; of these, 23 had detectable expression in the developing MM, UB tip, or UB stalk (table 3). Among positional candidates, three genes initially stood out because they had high expression levels across metanephric compartments (JUN and HOOK1) or displayed UB-specific expression in mouse (TACSTD2). JUN, a component of the AP-1 transcription factor complex, is essential for cell proliferation and organogenesis after midgestation.40 It acts downstream of RET signaling and may modify UB branching and nephrogenesis.41 HOOK1 is a microtubule-binding protein involved in endocytosis in Drosophila and is responsible for the abnormal spermatozoon head shape phenotype in mice.42 TACSTD2 encodes a cell-surface receptor recognized as a carcinoma-associated antigen, with mutations associated with gelatinous drop-like corneal dystrophy.43 TACSTD2 and TACSTD1 were identified as UB-specific secreted proteins in multiple studies; alteration in TACSTD2 may therefore modify UB branching and nephrogenesis.39,44 We performed direct sequencing of the coding exons of the JUN, HOOK1, TACSTD2, FOXD2, and CPT2 genes, using the index patients from our linked families, but found no evidence of pathogenic mutations.
We also tested whether the 1p32-33 interval may be relevant to inherited disorders that affect the ureters but not the kidneys. This search was motivated by the possibility that primary ureteric defects can produce chronic renal parenchymal injury and subsequent involution of the kidneys, resulting in the finding of small kidney size on sonogram. We tested linkage to 1p32-33 in 10 families segregating isolated ureteric abnormalities (7 kindreds with primary VUR,28 2 multiplex kindreds with isolated UPJO, and 1 family with duplicated collecting system). All individuals in this second cohort had normal kidney size. Parametric linkage analysis was performed with 20 microsatellite loci spanning to 1p32-33, under the same model used to map the hypodysplasia pedigrees (affected-only analysis). We found no evidence for linkage to 1p32-33 in these kindreds. Moreover, our 1p32-33 interval is > 90 Mb proximal to, and therefore distinct from, the 1p13 locus reported elsewhere for linkage to primary VUR and reflux nephropathy.45 These data suggest the 1p32-33 interval harbors a gene that may be specific to familial disorders that feature the renal hypodysplasia phenotype.
Because several families displayed negative LOD scores on 1p32-33, we searched for additional loci across the genome that account for disease. As K100 clearly linked to 1p32-33, we performed genomewide analysis of linkage after removing this kindred from our cohort (affected-only analysis with SNPs). There was no evidence of linkage across the genome under a model of genetic homogeneity. Under a model of locus heterogeneity, we identified several linkage peaks with HLOD >1 on chromosomes 18p11-q11, 20q11-13, 1p32-33, and 3p21-22. We next added 40 additional markers at these four loci and typed all available family members, to yield inheritance information >0.9. After this analysis, only the HLOD signal on chromosome 3p21-22 improved (HLOD 1.5; α=0.35; NPL=2.3; P=.02; maximum between D3S1768 and D3S2409). These data indicate that, other than 1p32-33, there are no significant intervals across the genome that account for the trait in remaining families (table 4).
Table 4. .
Multipoint Parametric LOD Scores at the Most Significant Intervals in the Seven Pedigrees
| LOD |
||||
| Kindred | 1p32-33a | 3p21-22b,e | 18p11-q11c,e | 20q11-13d,e |
| 100 | 3.5 | −4 | −2.6 | −5 |
| 101 | −2.5 | −1.3 | −2.8 | .8 |
| 105 | 1.2 | −1.3 | 1 | −2 |
| 107 | −1.4 | 1.2 | 1.2 | 1.2 |
| 109 | 1.1 | −2.4 | −.5 | −1.9 |
| 112 | −.6 | −.4 | .4 | .6 |
| 121 | −2.3 | 1.8 | −.5 | −3 |
HLOD=3.9 (45% of families linked).
HLOD=1.5 (35% of families linked).
HLOD=1.2 (50% of families linked).
HLOD=1 (35% of families linked).
HLOD was calculated without K100 because this kindred shows significant linkage to chromosome 1p32-33.
In this study, we performed detailed phenotypic characterization of families with nonsyndromic renal hypodysplasia, identified a locus for this trait, and completed the initial steps for functional annotation and prioritization of positional candidates. Because of the complex signaling network involved in urogenital development, renal developmental phenotypes are likely to display a high degree of genetic heterogeneity, necessitating large numbers of pedigrees or uniquely large kindreds for gene mapping.28 We therefore performed systematic sonographic screening of families with renal hypodysplasia, to extend kindreds and maximize study power. We confirmed that this phenotype segregates as an autosomal dominant trait with reduced penetrance. Next, genomewide analysis of linkage demonstrated that this trait is genetically heterogeneous, with about half of families showing linkage to a 6.9-Mb interval on 1p32-33. This linkage assignment is firmly established by a large family (K100), which provided evidence for genomewide significance on its own.
Our present findings can be pursued by several approaches. We can perform systematic sequencing of all positional candidates to identify the pathogenic mutations. However, further prioritization of candidates may help accelerate gene identification. This can be achieved by determining renal gene expression at multiple developmental time points and by examining tissue and cell-specific expression through in situ hybridization. In addition, our largest kindred (K100) is of Sardinian origin, providing the opportunity to refine the interval by disequilibrium mapping. Sardinia is composed of subpopulations that descend from limited sets of ancestors and exhibit strong linkage disequilibrium.46 This characteristic has facilitated gene identification for several traits, such as uric acid nephrolithiasis.47 Hence, comparison of the haplotypes between K100 and Sardinian patients from the same locality may identify chromosomal segments inherited by descent and help refine the interval to a limited set of candidates.
Identification of the gene underlying the 1p32-33 linkage will provide insight into the molecular basis of renal developmental disorders and will help improve diagnostic schemes. Low nephron number at birth and consequent low renal reserve have been proposed as a major susceptibility factor for the development of nephropathy or hypertension.48,49 Some genes regulating kidney development may also participate in tissue repair after renal injury.50 This suggests that identification of the 1p32-33 hypodysplasia gene may also inform us about interindividual variability in nephron number and have additional diagnostic or therapeutic implications.
Acknowledgments
We thank the patients and family members for participating in this study. A.G.G. is supported by the Emerald Foundation, the Irving Clinical Scholar Program, and the National Kidney Foundation (NFK) Clinical Scientist Program. P.L.W. is supported by an NKF research fellowship. G.M.G. was supported by Telethon grant E.1122. We thank Richard Lifton, Cathy Mendelsohn, Juan Oliver, and Qais Al-Awqati for their insightful comments.
Web Resources
The URLs for data presented herein are as follows:
- Ensembl, http://www.ensembl.org/Homo_sapiens/index.html (for v39)
- NCBI Map Viewer, http://www.ncbi.nlm.nih.gov/mapview/ (for build 36.1)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for renal agenesis/adysplasia, renal-coloboma syndrome, Fraser syndrome, and branchiootorenal syndrome)
- UCSC Genome Browser, http://genome.ucsc.edu/cgi-bin/hgGateway (for March 2006 assembly)
References
- 1.Hughson M, Farris AB 3rd, Douglas-Denton R, Hoy WE, Bertram JF (2003) Glomerular number and size in autopsy kidneys: the relationship to birth weight. Kidney Int 63:2113–2122 10.1046/j.1523-1755.2003.00018.x [DOI] [PubMed] [Google Scholar]
- 2.Hughson MD, Douglas-Denton R, Bertram JF, Hoy WE (2006) Hypertension, glomerular number, and birth weight in African Americans and white subjects in the southeastern United States. Kidney Int 69:671–678 10.1038/sj.ki.5000041 [DOI] [PubMed] [Google Scholar]
- 3.Hiraoka M, Tsukahara H, Ohshima Y, Kasuga K, Ishihara Y, Mayumi M (2002) Renal aplasia is the predominant cause of congenital solitary kidneys. Kidney Int 61:1840–1844 10.1046/j.1523-1755.2002.00322.x [DOI] [PubMed] [Google Scholar]
- 4.Helin I, Persson PH (1986) Prenatal diagnosis of urinary tract abnormalities by ultrasound. Pediatrics 78:879–883 [PubMed] [Google Scholar]
- 5.US Renal Data System (2005) USRDS 2005 annual data report: atlas of end-stage renal disease in the United States. National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases, Bethesda, MD [Google Scholar]
- 6.Ardissino G, Dacco V, Testa S, Bonaudo R, Claris-Appiani A, Taioli E, Marra G, Edefonti A, Sereni F (2003) Epidemiology of chronic renal failure in children: data from the ItalKid project. Pediatrics 111:e382–e387 10.1542/peds.111.4.e382 [DOI] [PubMed] [Google Scholar]
- 7.Stroup NE, Edmonds L, O‘Brien TR (1990) Renal agenesis and dysgenesis: are they increasing? Teratology 42:383–395 10.1002/tera.1420420407 [DOI] [PubMed] [Google Scholar]
- 8.Woolf AS, Price KL, Scambler PJ, Winyard PJ (2004) Evolving concepts in human renal dysplasia. J Am Soc Nephrol 15:998–1007 10.1097/01.ASN.0000113778.06598.6F [DOI] [PubMed] [Google Scholar]
- 9.Roodhooft AM, Birnholz JC, Holmes LB (1984) Familial nature of congenital absence and severe dysgenesis of both kidneys. N Engl J Med 310:1341–1345 [DOI] [PubMed] [Google Scholar]
- 10.McPherson E, Carey J, Kramer A, Hall JG, Pauli RM, Schimke RN, Tasin MH (1987) Dominantly inherited renal adysplasia. Am J Med Genet 26:863–872 10.1002/ajmg.1320260413 [DOI] [PubMed] [Google Scholar]
- 11.Carter CO, Evans K, Pescia G (1979) A family study of renal agenesis. J Med Genet 16:176–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mackie GG, Stephens FD (1975) Duplex kidneys: a correlation of renal dysplasia with position of the ureteral orifice. J Urol 114:274–280 [DOI] [PubMed] [Google Scholar]
- 13.Parikh CR, McCall D, Engelman C, Schrier RW (2002) Congenital renal agenesis: case-control analysis of birth characteristics. Am J Kidney Dis 39:689–694 [DOI] [PubMed] [Google Scholar]
- 14.Cooper WO, Hernandez-Diaz S, Arbogast PG, Dudley JA, Dyer S, Gideon PS, Hall K, Ray WA (2006) Major congenital malformations after first-trimester exposure to ACE inhibitors. N Engl J Med 354:2443–2451 10.1056/NEJMoa055202 [DOI] [PubMed] [Google Scholar]
- 15.Vize PD, Woolf AS, Bard JBL (2003) The kidney: from normal development to congenital disease. Elsevier Science, San Diego, CA [Google Scholar]
- 16.Grieshammer U, Le M, Plump AS, Wang F, Tessier-Lavigne M, Martin GR (2004) SLIT2-mediated ROBO2 signaling restricts kidney induction to a single site. Dev Cell 6:709–717 10.1016/S1534-5807(04)00108-X [DOI] [PubMed] [Google Scholar]
- 17.Yu J, McMahon AP, Valerius MT (2004) Recent genetic studies of mouse kidney development. Curr Opin Genet Dev 14:550–557 10.1016/j.gde.2004.07.009 [DOI] [PubMed] [Google Scholar]
- 18.Miyazaki Y, Oshima K, Fogo A, Ichikawa I (2003) Evidence that bone morphogenetic protein 4 has multiple biological functions during kidney and urinary tract development. Kidney Int 63:835–844 10.1046/j.1523-1755.2003.00834.x [DOI] [PubMed] [Google Scholar]
- 19.Durbec P, Marcos-Gutierrez CV, Kilkenny C, Grigoriou M, Wartiowaara K, Suvanto P, Smith D, Ponder B, Costantini F, Saarma M, et al (1996) GDNF signalling through the Ret receptor tyrosine kinase. Nature 381:789–793 10.1038/381789a0 [DOI] [PubMed] [Google Scholar]
- 20.Torres M, Gomez-Pardo E, Dressler GR, Gruss P (1995) Pax-2 controls multiple steps of urogenital development. Development 121:4057–4065 [DOI] [PubMed] [Google Scholar]
- 21.Schmidt-Ott KM, Lan D, Hirsh BJ, Barasch J (2006) Dissecting stages of mesenchymal-to-epithelial conversion during kidney development. Nephron Physiol 104:p56–p60 10.1159/000093287 [DOI] [PubMed] [Google Scholar]
- 22.Vainio S, Lin Y (2002) Coordinating early kidney development: lessons from gene targeting. Nat Rev Genet 3:533–543 10.1038/nrg842 [DOI] [PubMed] [Google Scholar]
- 23.Jenkins D, Bitner-Glindzicz M, Malcolm S, Hu CC, Allison J, Winyard PJ, Gullett AM, Thomas DF, Belk RA, Feather SA, et al (2005) De novo Uroplakin IIIa heterozygous mutations cause human renal adysplasia leading to severe kidney failure. J Am Soc Nephrol 16:2141–2149 10.1681/ASN.2004090776 [DOI] [PubMed] [Google Scholar]
- 24.Schonfelder EM, Knuppel T, Tasic V, Miljkovic P, Konrad M, Wuhl E, Antignac C, Bakkaloglu A, Schaefer F, Weber S (2006) Mutations in Uroplakin IIIA are a rare cause of renal hypodysplasia in humans. Am J Kidney Dis 47:1004–1012 10.1053/j.ajkd.2006.02.177 [DOI] [PubMed] [Google Scholar]
- 25.Weber S, Moriniere V, Knuppel T, Charbit M, Dusek J, Ghiggeri GM, Jankauskiene A, Mir S, Montini G, Peco-Antic A, et al (2006) Prevalence of mutations in renal developmental genes in children with renal hypodysplasia: results of the ESCAPE study. J Am Soc Nephrol 17:2864–2870 10.1681/ASN.2006030277 [DOI] [PubMed] [Google Scholar]
- 26.Ulinski T, Lescure S, Beaufils S, Guigonis V, Decramer S, Morin D, Clauin S, Deschenes G, Bouissou F, Bensman A, et al (2006) Renal phenotypes related to hepatocyte nuclear factor-1beta (TCF2) mutations in a pediatric cohort. J Am Soc Nephrol 17:497–503 10.1681/ASN.2005101040 [DOI] [PubMed] [Google Scholar]
- 27.Pasch A, Hoefele J, Grimminger H, Hacker HW, Hildebrandt F (2004) Multiple urinary tract malformations with likely recessive inheritance in a large Somalian kindred. Nephrol Dial Transplant 19:3172–3175 10.1093/ndt/gfh514 [DOI] [PubMed] [Google Scholar]
- 28.Sanna-Cherchi S, Reese A, Hensle T, Caridi G, Izzi C, Kim YY, Murer L, Scolari F, Ravazzolo R, Ghiggeri GM, Gharavi AG (2005) Familial vesicoureteral reflux: testing replication of linkage in seven new multigenerational kindreds. J Am Soc Nephrol 16:1781–1787 10.1681/ASN.2004121034 [DOI] [PubMed] [Google Scholar]
- 29.Dinkel E, Ertel M, Dittrich M, Peters H, Berres M, Schulte-Wissermann H (1985) Kidney size in childhood: sonographical growth charts for kidney length and volume. Pediatr Radiol 15:38–43 10.1007/BF02387851 [DOI] [PubMed] [Google Scholar]
- 30.Han BK, Babcock DS (1985) Sonographic measurements and appearance of normal kidneys in children. AJR Am J Roentgenol 145:611–616 [DOI] [PubMed] [Google Scholar]
- 31.Cottingham RW Jr, Idury RM, Schaffer AA (1993) Faster sequential genetic linkage computations. Am J Hum Genet 53:252–263 [PMC free article] [PubMed] [Google Scholar]
- 32.Gudbjartsson DF, Jonasson K, Frigge ML, Kong A (2000) Allegro, a new computer program for multipoint linkage analysis. Nat Genet 25:12–13 10.1038/75514 [DOI] [PubMed] [Google Scholar]
- 33.Sobel E, Lange K (1996) Descent graphs in pedigree analysis: applications to haplotyping, location scores, and marker-sharing statistics. Am J Hum Genet 58:1323–1337 [PMC free article] [PubMed] [Google Scholar]
- 34.Faraway JJ (1993) Distribution of the admixture test for the detection of linkage under heterogeneity. Genet Epidemiol 10:75–83 10.1002/gepi.1370100108 [DOI] [PubMed] [Google Scholar]
- 35.Lander E, Kruglyak L (1995) Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet 11:241–247 10.1038/ng1195-241 [DOI] [PubMed] [Google Scholar]
- 36.Ott J (1989) Computer-simulation methods in human linkage analysis. Proc Natl Acad Sci USA 86:4175–4178 10.1073/pnas.86.11.4175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bonnefont JP, Djouadi F, Prip-Buus C, Gobin S, Munnich A, Bastin J (2004) Carnitine palmitoyltransferases 1 and 2: biochemical, molecular and medical aspects. Mol Aspects Med 25:495–520 10.1016/j.mam.2004.06.004 [DOI] [PubMed] [Google Scholar]
- 38.Kume T, Deng K, Hogan BL (2000) Minimal phenotype of mice homozygous for a null mutation in the forkhead/winged helix gene, Mf2. Mol Cell Biol 20:1419–1425 10.1128/MCB.20.4.1419-1425.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schmidt-Ott KM, Yang J, Chen X, Wang H, Paragas N, Mori K, Li JY, Lu B, Costantini F, Schiffer M, et al (2005) Novel regulators of kidney development from the tips of the ureteric bud. J Am Soc Nephrol 16:1993–2002 10.1681/ASN.2004121127 [DOI] [PubMed] [Google Scholar]
- 40.Johnson RS, van Lingen B, Papaioannou VE, Spiegelman BM (1993) A null mutation at the c-jun locus causes embryonic lethality and retarded cell growth in culture. Genes Dev 7:1309–1317 [DOI] [PubMed] [Google Scholar]
- 41.Jijiwa M, Fukuda T, Kawai K, Nakamura A, Kurokawa K, Murakumo Y, Ichihara M, Takahashi M (2004) A targeting mutation of tyrosine 1062 in Ret causes a marked decrease of enteric neurons and renal hypoplasia. Mol Cell Biol 24:8026–8036 10.1128/MCB.24.18.8026-8036.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mendoza-Lujambio I, Burfeind P, Dixkens C, Meinhardt A, Hoyer-Fender S, Engel W, Neesen J (2002) The Hook1 gene is non-functional in the abnormal spermatozoon head shape (azh) mutant mouse. Hum Mol Genet 11:1647–1658 10.1093/hmg/11.14.1647 [DOI] [PubMed] [Google Scholar]
- 43.Tsujikawa M, Kurahashi H, Tanaka T, Nishida K, Shimomura Y, Tano Y, Nakamura Y (1999) Identification of the gene responsible for gelatinous drop-like corneal dystrophy. Nat Genet 21:420–423 10.1038/7759 [DOI] [PubMed] [Google Scholar]
- 44.Caruana G, Cullen-McEwen L, Nelson AL, Kostoulias X, Woods K, Gardiner B, Davis MJ, Taylor DF, Teasdale RD, Grimmond SM, et al (2006) Spatial gene expression in the T-stage mouse metanephros. Gene Expr Patterns 6:807–825 10.1016/j.modgep.2006.02.001 [DOI] [PubMed] [Google Scholar]
- 45.Feather SA, Malcolm S, Woolf AS, Wright V, Blaydon D, Reid CJ, Flinter FA, Proesmans W, Devriendt K, Carter J, et al (2000) Primary, nonsyndromic vesicoureteric reflux and its nephropathy is genetically heterogeneous, with a locus on chromosome 1. Am J Hum Genet 66:1420–1425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Angius A, Bebbere D, Petretto E, Falchi M, Forabosco P, Maestrale B, Casu G, Persico I, Melis PM, Pirastu M (2002) Not all isolates are equal: linkage disequilibrium analysis on Xq13.3 reveals different patterns in Sardinian sub-populations. Hum Genet 111:9–15 10.1007/s00439-002-0753-z [DOI] [PubMed] [Google Scholar]
- 47.Gianfrancesco F, Esposito T, Ombra MN, Forabosco P, Maninchedda G, Fattorini M, Casula S, Vaccargiu S, Casu G, Cardia F, et al (2003) Identification of a novel gene and a common variant associated with uric acid nephrolithiasis in a Sardinian genetic isolate. Am J Hum Genet 72:1479–1491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brenner BM, Chertow GM (1994) Congenital oligonephropathy and the etiology of adult hypertension and progressive renal injury. Am J Kidney Dis 23:171–175 [PubMed] [Google Scholar]
- 49.Keller G, Zimmer G, Mall G, Ritz E, Amann K (2003) Nephron number in patients with primary hypertension. N Engl J Med 348:101–108 10.1056/NEJMoa020549 [DOI] [PubMed] [Google Scholar]
- 50.Mori K, Lee HT, Rapoport D, Drexler IR, Foster K, Yang J, Schmidt-Ott KM, Chen X, Li JY, Weiss S, et al (2005) Endocytic delivery of lipocalin-siderophore-iron complex rescues the kidney from ischemia-reperfusion injury. J Clin Invest 115:610–621 10.1172/JCI200523056 [DOI] [PMC free article] [PubMed] [Google Scholar]