Abstract
In this article, we describe a novel autosomal recessive ichthyosis with hypotrichosis syndrome, characterized by congenital ichthyosis associated with abnormal hair. Using homozygosity mapping, we mapped the disease locus to 11q24.3-q25. We screened the ST14 gene, which encodes matriptase, since transplantation of skin from matriptase−/−-knockout mice onto adult athymic nude mice has been shown elsewhere to result in an ichthyosislike phenotype associated with almost complete absence of erupted pelage hairs. Mutation analysis revealed a missense mutation, G827R, in the highly conserved peptidase S1–S6 domain. Marked skin hyperkeratosis due to impaired degradation of the stratum corneum corneodesmosomes was observed in the affected individuals, which suggests that matriptase plays a significant role in epidermal desquamation.
Autosomal recessive congenital ichthyoses are clinically and genetically heterogeneous genodermatoses characterized by abnormal epidermal differentiation. They can be classified into two groups: (i) primary ichthyoses limited to the skin and (ii) syndromic ichthyoses, associated with extracutaneous features.
Six genes are known to be associated with nonsyndromic autosomal recessive congenital ichthyoses: TGM1 (MIM *190195),1,2 ALOXE3 (MIM *607206), ALOX12B (MIM *603741),3,4 ABCA12 (MIM *607800),5,6 ICHTHYIN (MIM +609383),7 and FLJ39501.8 In addition, linkage to chromosomal loci on 12p11.2-q139 and 19p13.1-p13.210 has been reported.
Several autosomal recessive ichthyoses with hair abnormalities have been described.11 Netherton syndrome (NTS [MIM #256500]) is characterized by congenital ichthyosiform erythroderma, trichorrhexis invaginata, and atopic manifestations. Mutations in SPINK5, which encodes LEKTI, an inhibitor of serine proteases, were identified as the cause of NTS.12 Congenital ichthyosis, follicular atrophoderma, hypotrichosis, and hypohidrosis syndrome (MIM 602400) is characterized by diffuse congenital ichthyosis, patchy follicular atrophoderma, generalized and diffuse nonscarring hypotrichosis, and marked hypohidrosis.13,14 Trichothiodystrophy with congenital ichthyosis—that is, ichthyosis, brittle hair, intellectual impairment, decreased fertility, and short stature (IBIDS [MIM #601675])—a rare autosomal recessive disorder characterized by sulfur-deficient brittle hair and nails, mental retardation, impaired sexual development, and ichthyosis, is caused by mutations in the ERCC3/XPB15 and ERCC2/XPD16 genes. Ichthyosis, split hairs, and amino aciduria syndrome (MIM 242550) is characterized by lamellar ichthyosis, splitting of the hair shaft, amino aciduria, and mental retardation.17
In this study, we describe a novel autosomal recessive ichthyosis with hypotrichosis syndrome (ARIH) characterized by congenital ichthyosis associated with abnormal hair. We report on the identification of the disease-causing mutation in the ST14 gene that encodes the serine protease matriptase.
Subjects and Methods
Patients
We ascertained a consanguineous Israeli-Arab family with ARIH, including three affected and five unaffected individuals. All the patients were siblings and were the offspring of parents who were first cousins. We obtained informed consent from all family members or their legal guardians, according to a protocol approved and reviewed by the National Committee for Genetic Studies, Israel Ministry of Health, and the Rabin Medical Center.
Homozygosity Mapping and Linkage Analysis
Genomic DNA for genotyping was extracted from leukocytes from peripheral venous blood in EDTA by standard procedures.18 SNP genotyping was performed on Affymetrix Human Mapping 50k Xba 240 arrays. All procedures and methods were performed according to the manufacturer’s instructions. In brief, for each sample, 250 ng of genomic DNA were digested using Xbal. Primer sequences were ligated to the digested fragments, and a single-primer PCR was performed on a PE9700 thermal cycler (Applied Biosystems). The PCR products were purified using Qiagen MinElute 96 UF plates. After purification, 40 μg of each amplified sample was fragmented, end-labeled, and hybridized to a GeneChip array. After washing and staining, the arrays were scanned using the GeneChip scanner 3000 with GeneChip Operating Software (GTYPE) version 1.4. Array CEL files were processed with GeneChip DNA Analysis Software version 4.0. For the examined samples, an average call rate of >95% was obtained.
Microsatellite markers were amplified by multiplex PCR, with use of standard protocols. Amplified markers were electrophoresed on an ABI 3700 DNA capillary sequencer and were analyzed with GENESCAN and GENOTYPER software (Applied Biosystems). For the LOD-score calculation, the two-point linkage analysis for markers D11S4091, D11S4150, and D11S4131 on 11q24.3 was performed using the program SUPERLINK.19 We assumed a susceptibility allele with frequency 0.0001 and a recessive mode of inheritance with penetrance 0.99. For LOD-score calculations, the number of alleles was set as the number observed in the pedigree, rather than the number observed elsewhere, to provide a conservative estimate of the LOD score.
Sequencing of the Candidate Genes
Sequencing of the candidate genes was performed with primer sets designed using the Primer3 program. All exons—including exon-intron junctions, 5′ UTRs, and 3′ UTRs—were amplified from genomic DNA with primers designed from the genomic sequences available from the University of California–Santa Cruz (UCSC) Genome Browser. Both strands of the PCR products were sequenced with BigDye Terminators (Applied Biosystems) on an ABI 3100 sequencer. Sequence chromatograms were analyzed using SeqScape software version 1.1 (Applied Biosystems). We initially tested one affected individual and one heterozygous parent.
Sequencing of the ST14 gene was performed using 28 primer pairs (table 1). ST14 mutation screening was performed on genomic DNA by PCR amplification with primer A, 5′-ATGTGCGTGGGCTTCCTC-3′ (forward), and primer B, 5′-AGTCAGCGGTCCAGTCTCC-3′ (reverse), followed by sequencing.
Table 1. .
Primers Used to Amplify the ST14 Gene
| Primer Sequence(5′→3′) |
|||
| Primer Pair |
Exon | Forward | Reverse |
| 1 | 1 | GAGGCCACACCCTGAAACTA | GTGCCGGGAGTTGTACTTGA |
| 2 | 1 | GGTGATGGTGAGGGCCTTAG | GGTCTCACAGGCGTCGTC |
| 3 | 1 | AGCCGGAGAAAGAGGAAGAG | ACAGGAAATTCCACCTGTGC |
| 4 | 1 | GACGACGCCTGTGAGACC | ACAGGAAATTCCACCTGTGC |
| 5 | 2 | TTCTCCAAGCTGGACCTCAC | GGCCAAGAGATCCTCGTGTA |
| 6 | 3 | AGTGATGGGAAGCAGTCAGG | GGTCCCCTTTGCAGATCAC |
| 7 | 4 | ATGGGGGTGATCTGCAAAG | AGGACCTCCAGCACCATCT |
| 8 | 5 | GTGTCCTGGAGTCGCTCTG | GTCTTCTCTCTGGGGCACTG |
| 9 | 5 | GCCCATGTGTCCTGGAGT | GGCCAGCACGTAGTCTTCTC |
| 10 | 6 | CAGTGCCCCAGAGAGAAGAC | GGCCTGGAAAGATCGTCATA |
| 11 | 7 | AGGCCTGAGGTCCACACC | TTGTACACCGTCACCAGGTC |
| 12 | 7 | CTTCCCTGACAGCCCCTAC | GCGGCCAAGAGTCATAAACT |
| 13 | 8 | CTGTGGATGGGACCAGAGTT | TCTAGCCTGTGGCCTCCTAA |
| 14 | 9 | ACCTGCTGTGCAGCATCC | CATCTGCACAGGGAGACACA |
| 15 | 10 | AACAAACACGCTGGGAGAAC | CCTCTCTCCGCAGTATCTGG |
| 16 | 11 | GGAGGGGTCCCACCAGAC | CAGCAAAGCACGATGCTAAC |
| 17 | 12 | CTGCTCCTGTGTGTTTGTGG | GGGCCAGAGGTCTCTACACA |
| 18 | 13 | GGTCCCCAGGTAGCTCTGA | CTGGAGAGGCACCAGGACT |
| 19 | 14 | GAGCAGGGGTGCAGTGAGT | GTCCTGCATGTGCGTGTG |
| 20 | 15 | GCCATTGGTGGTTTCTGG | ACCTCCAGGTGCACCCTCT |
| 21 | 16 | GGTCTCCTGGCACACACTG | CCTCGGTGCCAACCTATC |
| 22 | 17 | ATCGTCTTCTCGTAGCAGCA | GTACTCTGCCGGTTTCTCCA |
| 23 | 17 | CCACCCCTTCTTCAATGACT | GACACGCAGAGGAAACACG |
| 24 | 18 | TCTGGGCTGTTCCAGAGTTT | CACACCGGCCTGGAAGAT |
| 25 | 19 | ATGTGCGTGGGCTTCCTC | AGTCAGCGGTCCAGTCTCC |
| 26 | 19 | TTCGGGACTGGATCAAAGAG | TTAGAGACAGGGGAGGCAGA |
| 27 | 19 | CAAAGTGGAGCTGGGAGGTA | CTCAGACCCGTCTGTTTTCC |
| 28 | 19 | CCTCCTCAGTGAAGGTGGTG | CTGACTCCAGGGTCTGTGCT |
Mutation Detection by Restriction Analysis
Amplification of a 379-bp fragment from genomic DNA was performed using primers A and B for 37 cycles at an annealing temperature of 55°C. The mutation introduces a BpmI restriction site, which digests the 379-bp fragment into 215-bp and 164-bp fragments. The fragment was digested with 2 U/μg BpmI (New England Biolabs), and the reaction products were separated by electrophoresis on 3% NuSieve/1% agarose gels.
Bioinformatics Methods
The gene and the protein were described using the UCSC Genome Browser (March 2006 assembly), the Ensembl Genome Browser (June 2006 assembly), and bioinformatics servers. To analyze conservation of the variation site, 10 vertebrate (from fish to human) sequence homologues to the human ST14 protein were collected using BLink (“BLAST Link”). Comparative sequence analysis (multiple sequence analysis [MSA]) was performed by ClustalW with a Java viewer (Clustal Alignment), as described by Higgins et al.20 and Clamp et al.21 The ConSeq Server was used to validate sequence conservation and to predict functionality.22 The SIFT server sorts intolerant from tolerant amino acid substitutions and predicts phenotypic variations, as described by Ng and Henikoff.23 Human ST14 protein was used as the input for SIFT analysis.
Immunohistochemistry
Formaldehyde-fixed 5-μm paraffin-embedded sections were treated with 3% H2O2 in methanol for 15 min at room temperature, were warmed in a microwave oven in citrate buffer for 15 min at 90°C, and were stained with antibodies directed against filaggrin (Novocastra), loricrin (Covance Research Products), involucrin (Covance Research Products), and cytokeratin 1 (BioGenex) or against preimmune rabbit antiserum for 1 h at room temperature. After extensive washings in PBS, the antibodies were revealed using the ABC technique (Zymed Laboratories), and the slides were counterstained using hematoxylin.
Electron Microscopy and Immunoelectron Microscopy
Postembedding electron microscopy with Lowicryl HM20 resin was performed as described elsewhere.24 For immunoelectron microscopy, ultrathin sections were incubated with antibodies against LEKTI (Zymed Laboratories) and corneodesmosin (CDSN).25
Results
Clinical and Laboratory Evaluation
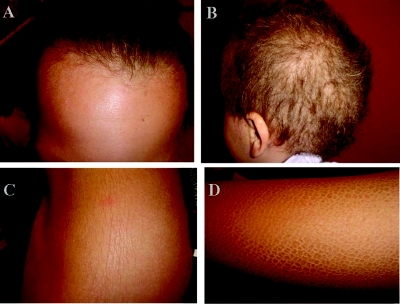
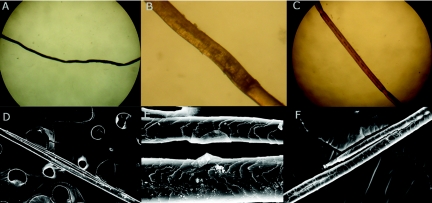
The patients were all born, after full-term, normal pregnancies, with normal birth weights. There was no collodion membrane. Ichthyosis and abnormal hair were present at birth. Patients III-5 and III-8 had a vernixlike layer covering the entire body, which was progressively shed during the first month of life. Hypotrichosis was generalized and diffuse (fig. 1A and 1B). Scaling was diffuse, including on the scalp, but the face was unaffected (fig. 1A, 1C, and 1D). Clinically, the hair of all the patients appeared curly, sparse, fragile, brittle, dry, and lusterless and showed slow growth. Light microscopy and scanning electron microscopy (SEM) examination of hair obtained from affected patients disclosed a number of abnormalities, including dysplastic hair (fig. 2A and 2B), pili torti (fig. 2C), pili bifurcati (fig. 2D and 2E), and central pili mono bifurcati (fig. 2F). Over time, scalp hair growth and appearance improved, and the scalp hair darkened with age. All three affected individuals had photophobia, and one had corneal opacities. The clinical features of the patients are summarized in table 2.
Figure 1. .
Clinical features of the patients. A and B, Hair of patients III-1 and III-8, respectively. Sparse and curly hair is observed. C and D, Ichthyotic skin of patient III-1 on the trunk and of patient III-5 on the upper limb, respectively.
Figure 2. .
Light microscopy (A–C) and SEM (D–F) examination of hair obtained from affected patients. A and B, Dysplastic hair. Hair shafts are irregular, flaky, and undulating (A). B, Magnified undulation of panel A. C, Pili torti. Affected shaft is twisted on its own axis. D and E, Pili bifurcati. Each bifurcation produces two separate parallel branches that fuse again to form a single shaft; each branch is covered with its own cuticle. F, Central pili mono bifurcati: bifurcation that does not fuse again to the hair shaft and is located in the middle of the hair shaft. A minimum of 50 hairs from each patient were examined.
Table 2. .
Clinical Characteristics of the Patients[Note]
| Patient |
|||
| Characteristic | III-1 | III-5 | III-8 |
| Age (in years) | 17 | 8 | 2.5 |
| Skin: | |||
| Abnormalities at birth | Scaling | Vernixlike | Vernixlike |
| Color | Gray | Gray and brown | Gray and brown |
| Hair: | |||
| Scalp hair | Dark brown | Uneven length, light brown | Uneven length, light brown |
| Eyebrows | Sparse and curly | Sparse, more lateral parts, light colored | Sparse, more lateral parts, light colored |
| Body hair | Sparse | Sparse | Sparse |
| Eyes: | |||
| Corneal abnormality | No | No | Corneal opacity |
| Photophobia | Yes | Yes | Yes |
| Other | Pingueculum from age 11 years | ||
| Other: | |||
| Teeth | Normal | Notching, pitting | Conical primary teeth |
| Infections | No | Chronic sinusitis | No |
| Itching | Yes | Yes | Yes |
Note.— All patients had skin abnormalities of the scalp, neck, abdomen, back, and limbs (extensor surfaces) and had long, curly, dark upper eyelashes and curly, sparse, fragile, brittle, dry, lusterless scalp hair, with receding frontal hairline.
Follicular atrophoderma, palmoplantar keratoderma, and hyperlinearity were not seen. The nails and mucosa were normal. No erythroderma, cardiac abnormalities, digital contractures, loss of pulp volume, ectropion, or hypoplasia of the nasal/auricular cartilage was observed. Sweating was normal, and there were no episodes of hypernatremic dehydration or hyperthermia. There were no atopic manifestations or photosensitivity. Complete blood count, urea, creatinine, liver enzymes, T-cell count, and immunoglobulins IgG, IgM, IgA, and IgE were all normal.
Patients III-2, III-3, III-6, and III-8 also have Hirschsprung disease, which is unrelated to the ichthyosis. Patient III-2 died at age 2.5 years from gastrointestinal complications of severe Hirschsprung disease (total colonic aganglionosis).
ARIH Mapping on 11q24.3-q25
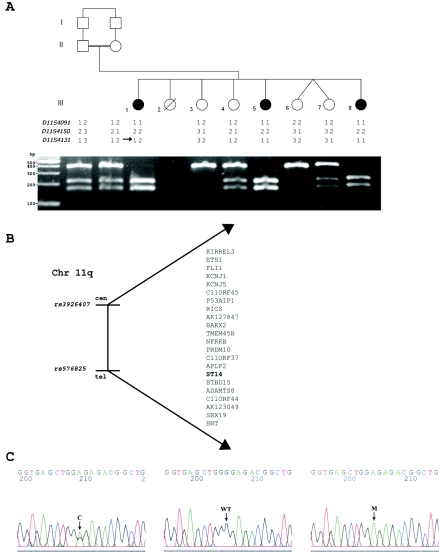
A visual inspection of the SNP data showed that all the patients exhibited a large continuous segment of homozygosity for 53 consecutive SNPs, encompassing 5 Mb between markers rs3926407 and rs576825 on 11q24.3-q25 (table 3 and our tab-delimited txt file). These data were supportive of homozygosity by descent. Other, smaller genomic regions showing homozygosity in consecutive SNPs in all the patients on chromosomes 2, 5, 8, 9, 13, 14, and 17 were excluded by genotyping microsatellite markers D2S2375, D2S2360, D5S1384, D5S1505, D8S549, D8S1731, D9S52, D9S104, D9S43, D9S251, D13S286, D13S1311, D13S173, D13S1265, D13S801, D14S592, D14S1429, D17S947, D17S917, and D17S808 in all nine family members (data not shown). Linkage to the locus on chromosome 11q24.3-q25 was confirmed by genotyping microsatellite markers D11S4091, D11S4150, and D11S4131 in all nine family members (fig. 3A). LOD-score analysis for the marker D11S4150 yielded a Zmax score of 2.21 at recombination fraction (θ) 0.00 (table 4). A higher LOD score could not be obtained because of the small family size. The candidate region contains 22 known or predicted genes (fig. 3B).
Table 3. .
Homozygosity for SNP Markers on 11q24.2-q25[Note]
| Genotype for Patient |
||||
| Chromosome and Location |
SNP | III-1 | III-5 | III-8 |
| 11q24.2: | ||||
| 126135641 | rs2226935 | BB | BB | BB |
| 126153350 | rs1946052 | AA | AA | AA |
| 126259661 | rs1940007 | BB | BB | BB |
| 126259725 | rs1940006 | AA | AA | AA |
| 126314224 | rs1939990 | AA | AA | AA |
| 126314287 | rs1939991 | BB | BB | BB |
| 126314915 | rs1939992 | AA | AA | AA |
| 126357648 | rs631562 | AA | AA | AA |
| 126369604 | rs488054 | BB | BB | BB |
| 126369798 | rs541783 | BB | BB | BB |
| 126397423 | rs1944627 | AA | AA | AA |
| 126401733 | rs642387 | AA | AA | AA |
| 126402187 | rs503397 | BB | BB | BB |
| 126553569 | rs1944828 | BB | BB | BB |
| 126553743 | rs1944827 | BB | BB | BB |
| 126861051 | rs749514 | AA | AA | AA |
| 126861482 | rs1729076 | AA | AA | AA |
| 126936044 | rs356248 | BB | BB | BB |
| 126944448 | rs356262 | BB | BB | BB |
| 126971973 | rs763300 | BB | BB | BB |
| 127004821 | rs10490847 | BB | BB | BB |
| 127004961 | rs7928097 | AA | AA | AA |
| 11q24.3: | ||||
| 127344152 | rs1364780 | AA | AA | AA |
| 127525578 | rs1477577 | AA | AA | AA |
| 127562693 | rs476994 | AA | AA | AA |
| 127584911 | rs1940356 | AA | AA | AA |
| 127585108 | rs1940357 | BB | BB | BB |
| 127653338 | rs510628 | AA | AA | AA |
| 127654275 | rs722345 | BB | BB | BB |
| 127774235 | rs2323123 | BB | BB | BB |
| 127920339 | rs4128561 | AA | AA | AA |
| 127921152 | rs4129229 | BB | BB | BB |
| 128134145 | rs631647 | AA | AA | AA |
| 128134871 | rs670809 | BB | BB | BB |
| 128416643 | rs2155177 | BB | BB | BB |
| 128466905 | rs657820 | BB | BB | BB |
| 128624506 | rs4129583 | BB | BB | BB |
| 128904021 | rs2008849 | AA | AA | AA |
| 128983138 | rs2172487 | BB | BB | BB |
| 128984385 | rs1873873 | BB | BB | BB |
| 129012244 | rs952131 | BB | BB | BB |
| 129513314 | rs879780 | BB | BB | BB |
| 129860441 | rs3898490 | BB | BB | BB |
| 11q25: | ||||
| 129879261 | rs1272496 | BB | BB | BB |
| 130306606 | rs1893225 | AA | AA | AA |
| 130679085 | rs1793584 | BB | BB | BB |
| 130680535 | rs1114296 | BB | BB | BB |
| 130697593 | rs1793783 | BB | BB | BB |
| 130729658 | rs4128590 | BB | BB | BB |
| 130729798 | rs4128594 | AA | AA | AA |
| 130853281 | rs1355982 | BB | BB | BB |
| 130897898 | rs411280 | AA | AA | AA |
| 131091392 | rs796873 | AA | AA | AA |
Note.— SNPs are shown on the dbSNP Web site.
Figure 3. .
Family tree, candidate region, and mutation analysis. A, Pedigree of the family and segregation analysis of the mutation. Segregation was assayed by PCR amplification of the genomic DNA and restriction by BpmI. The wild-type allele is uncut. Blackened symbols represent affected individuals. An arrow shows informative recombination. B, Candidate region. C, Mutation analysis of ST14. Sequence chromatogram from wild-type (WT), carrier (C), and affected (M) genotypes on genomic DNA.
Table 4. .
Two-Point LOD Scores for Chromosome 11q24 Markers
| LOD at θ = |
|||||||
| Locus | .00 | .01 | .05 | .10 | .20 | .30 | .40 |
| D11S4091 | 2.0047 | 1.9569 | 1.7651 | 1.5245 | 1.0425 | .5725 | .1736 |
| D11S4150 | 2.2122 | 2.1657 | 1.9776 | 1.7382 | 1.2481 | .7525 | .2912 |
| D11S4131 | −∞ | .1701 | .6990 | .7853 | .6568 | .4177 | .1692 |
Mutation Analysis
All exons—including promoter region, exon-intron junctions, and 5′ and 3′ UTRs—of the BARX2 gene were sequenced, and no pathogenic sequence changes were found. BARX2-knockout mice display short hair.26
Sequencing of the ST14 gene revealed missense mutation c.2672G→A in exon 19 (according to “Recommendations for the Description of DNA Sequence Variants” [Ensembl Genome Browser]) (fig. 3C). The mutation causes a glycine→arginine change at residue 827 (G827R) of the protein, in the peptidase S1–S6 domain.
The ST14 mutation segregated with the disease; affected patients displayed homozygous mutations, whereas parents displayed heterozygosity for a normal and a disease allele, consistent with autosomal recessive inheritance (fig. 3A). The mutation was not observed in 434 chromosomes from unrelated control individuals of Arab origin.
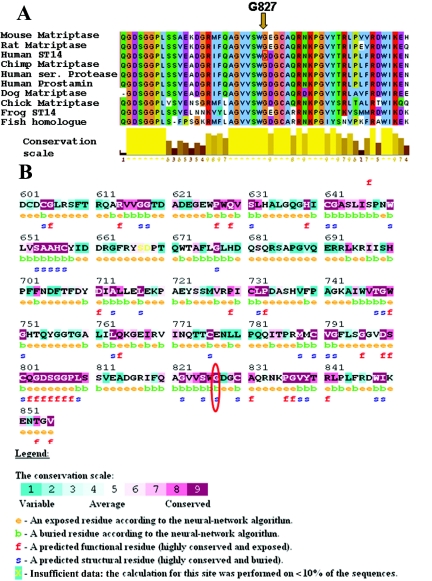
MSA was performed using 10 protein-sequence homologues for human ST14, including vertebrates from fish to human. A highly conserved region is present at the C-terminal region of the protein, harboring the Tryp_SPc (trypsinlike serine protease) domain, which includes the G827 residue of the human protein (fig. 4A). The ConSeq Server was used to further support these data. The peptidase C-terminal domain is shown (fig. 4B). Predicted results for residue G827 in the human protein showed that no change in this amino acid is tolerated (results not shown). Structural analysis of matriptase (1EAW in the Protein Data Bank) showed that the G827 variation is only two residues away from an important catalytic residue (serine 825).
Figure 4. .
Multiple alignment and conservation analysis of human ST14 with its orthologues. A, Multiple sequence alignment, performed by ClustalW (Clustal Alignment), with use of 10 protein sequences of matriptase, including vertebrates from fish to human: Homo sapiens (human) ST14, (GenBank accession number NP_068813), Homo sapiens (human) prostamin (GenBank accession number BAB20376), Homo sapiens (human) serine protease (GenBank accession number AAG15395), Pan troglodytes (chimpanzee) homologue (GenBank accession number XP_508863), Mus musculus (mouse) matriptase (GenBank accession number BAE38987), Rattus norvegicus (rat) matriptase (GenBank accession number AAH97271), Gallus gallus (chicken) matriptase (GenBank accession number XP_417872), Canis familiaris(dog) matriptase (GenBank accession number XP_546396), Xenopus laevis (frog) matriptase (GenBank accession number AAH71077), and Danio rerio (zebrafish) hypothetical protein (GenBank accession number NP_001035441). G827 is marked with an arrow. Residue colors represent amino acid biochemical properties according to Zappo colors. Conservation scale is shown below the alignment (yellow bars). B, Sequence conservation analysis, performed using the ConSeq Server, with internal MSA alignment serving as input. Residue G827 is circled in red. Conserved regions are marked with dark magenta, and variable regions are marked with green. Exposed (e) and functional (f) residues can be seen.
Consequences of G827R in ST14
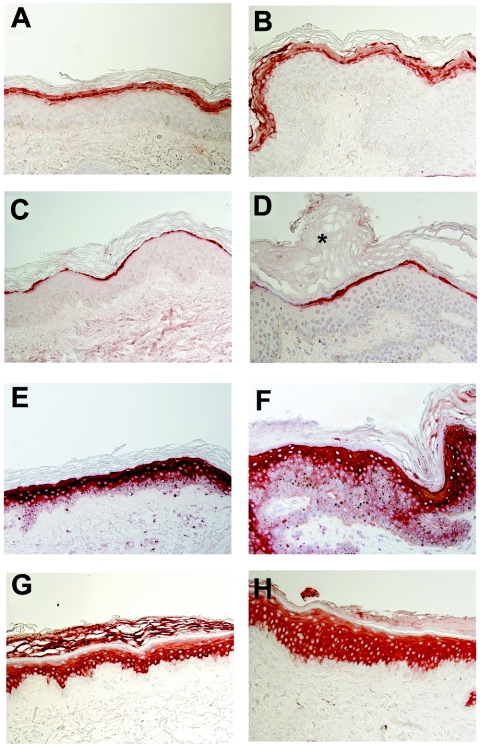
Marked acanthosis and a much thickened stratum corneum were demonstrated in affected skin by light microscopy (fig. 5). Since it has been suggested that proteases in general and matriptase in particular play an important role in the formation of the epidermal barrier,27 we compared the expression of a number of elements of the cornified cell envelope in skin-biopsy samples obtained from patients and healthy control individuals. No significant differences were noticed in the intensity or pattern of expression of filaggrin, loricrin, involucrin, and keratin 1 between unaffected and affected skin (fig. 5).
Figure 5. .
Cornified cell envelope–protein expression (original magnification ×400). Skin-biopsy sections obtained from a control individual (A, C, E, and G) and a patient (B, D, F, and H) were stained with antibodies directed against filaggrin (A and B), loricrin (C and D), involucrin (E and F), and keratin 1 (G and H) and were examined by light microscopy. Note the marked acanthosis (H) and thickened stratum corneum (indicated by an asterisk [*] in panel D) in affected skin.
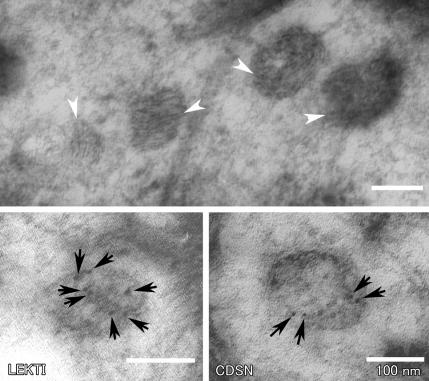
We then examined the ultrastructural consequences of the G827R mutation in the ST14 gene. We first examined the granular layers of the epidermis from a patient. Lamellar granules and keratohyaline granules displayed normal features. Immunoelectron microscopic examination of the skin-biopsy samples of a patient revealed normal distribution of LEKTI and CDSN (fig. 6).
Figure 6. .
Normal-looking lamellar granules with immunoreactivity to LEKTI and CDSN in the skin of a patient. Skin-biopsy samples from a patient were examined by electron microscopy. The top panel is an unlabeled Lowicryl HM20-embedded section stained with uranyl acetate and lead citrate, showing laminated internal structures within lamellar granules (arrowheads). The lower panels are 5-nm immunogold-labeled (arrows) sections with antibodies against LEKTI and CDSN.
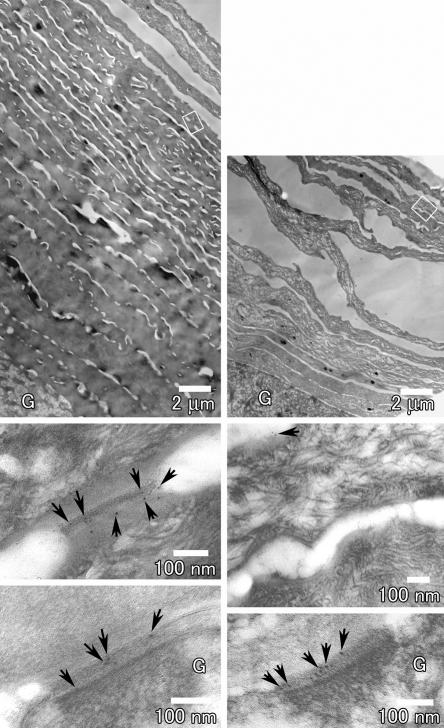
The most remarkable and consistent abnormal finding was the conspicuous presence of intact corneodesmosomes in the upper cornified layers of affected epidermis, in contrast with the normal degradation of these structures in control skin (fig. 7). This finding suggests a major physiological role for matriptase during epidermal desquamation.
Figure 7. .
Impaired degradation of corneodesmosomes in affected skin. Skin-biopsy samples from a patient (left panels) and a healthy individual (right panels) were examined by electron microscopy. Stratum corneum is greatly thickened in the patient’s skin compared with that of the control individual (top panels, with the same magnification). The rectangular areas marked in the top panels are shown at higher magnifications in the middle panels. Corneodesmosomes are intact in the upper layer, with many CDSN labels (arrows) in the patient (left, middle), whereas they are degraded with only a few CDSN labels remaining in the upper stratum corneum in normal skin (right, middle). The lower panels show desmosomes between the uppermost stratum granulosum (G) and the first cornified cells positively labeled with CDSN in both patient and control skin.
Discussion
In this study, we describe a novel autosomal recessive ichthyosis syndrome featuring both ichthyosis and abnormal hair distribution and structure, which we have named “autosomal recessive ichthyosis with hypotrichosis syndrome.” This disorder is clinically distinct from other genodermatoses involving skin and hair abnormalities. Compared with subjects affected with NTS, our patients have a milder disease, no erythrodermatous skin changes, no atopic manifestations, and no increased IgE. Congenital ichthyosis, follicular atrophoderma, hypotrichosis, and hypohidrosis syndrome is distinguished by the presence of hypohydrosis and follicular atrophoderma, which were not observed in our family. The ichthyosis, split hairs, and amino aciduria syndrome is characterized by mental retardation, whereas our patients have intact cognition. Mental retardation, brittle nails, and impaired sexual development are present in trichothiodystrophy with congenital ichthyosis syndrome (Tay or IBIDS syndrome); these were absent in our patients. The syndrome of ichthyosis follicularis with atrichia and photophobia (IFAP syndrome [MIM %308205]) is characterized by severe follicular hyperkeratosis, complete baldness, and photophobia; however, our patients have no atrichia or ichthyosis follicularis. Most importantly, IFAP syndrome is considered an X-linked condition.28
We have demonstrated that this new syndrome (i.e., ARIH) is caused by a mutation in ST14 that encodes matriptase, a type II transmembrane serine protease of the S1 trypsinlike family.29 Taking into account strict protease domain conservation and residue-change properties, in addition to its critical location near an important catalytic residue (serine 825), we predict that the amino acid change G827R leads to a severe change in protein function. This protein contains an N-terminal transmembrane domain, two tandem repeats of a CUB (C1r/s, Uegf and bone morphogenetic protein-1) domain, four tandem repeats of a low-density lipoprotein receptor domain, and a conserved extracellular serine protease catalytic domain.30 Matriptase is expressed predominately in the epithelial cells of the surface-lining epithelium.31 In the interfollicular epidermis, matriptase expression is restricted to the postmitotic transitional layer of keratinocytes undergoing terminal differentiation.32 Similar to some other members of the type II transmembrane serine-protease group, matriptase exists in a soluble and membrane-bound form.33,34 The activation of matriptase requires proteolytic cleavage that converts the enzyme from a one-chain zymogen to an active two-chain protease.35
Several proteases and their inhibitors—such as cathepsin L, cathepsin D, cathepsin C, cystatin M/E, the stratum corneum chymotryptic enzyme (KLK7), the stratum corneum tryptic enzyme (KLK5), and LEKTI—are important in epidermal homeostasis and/or hair follicle formation.36,37 Two serine proteases of the kallikrein family—KLK7 and KLK5—have been implicated in the degradation of corneodesmosomal proteins in the epidermis.37
The present study suggests that matriptase may also be involved in the desquamation process, since degradation of corneodesmosomes in ARIH is impaired. The lack of inhibition of KLK7 and KLK5 by the serine-protease inhibitor LEKTI may be related to the phenotypic manifestations of NTS.38 In NTS, corneodesmosome degradation is accelerated,25 suggesting that matriptase may be an additional substrate for LEKTI. Interestingly, similar to ARIH, the persistence of corneodesmosomes in the stratum corneum was observed in autosomal dominant ichthyosis vulgaris and X-linked ichthyosis.39
No human diseases associated with mutations in the ST14 gene have been described elsewhere, although matriptase overexpression has been implicated in cancer metastasis.40 Matriptase−/−-knockout mice have been generated27,41; these mice die shortly after birth as a result of deficient epidermal-barrier function in the skin in newborns. They also demonstrate abnormal hair-follicle development and disturbed thymic homeostasis that results in increased lymphocyte apoptosis in the thymus of the newborn mice.41 Our patients did not display any signs suggestive of a T cell–related immunological defect. Interestingly, 3 wk after transplantation of whole skin from matriptase−/− mice to adult athymic nude mice, the skin from the knockout mice showed severe epidermal thickening, the formation of ichthyosislike epidermal scales, and almost complete absence of erupted pelage hairs.42 Therefore, systemic expression of matriptase does not correct the epidermal defects in matriptase-deficient skin.
The relatively mild manifestations of ARIH syndrome contrast with the severe phenotype of knockout mice. This may be because the identified missense mutation has a milder effect as compared with total protein deficiency in the knockout mice.
It has been shown that, in mice, matriptase deficiency correlates with abnormal proteolytic processing of the epidermal protein profilaggrin into filaggrin monomer units and the NH2-terminal filaggrin S-100 regulatory protein, resulting in accumulation of profilaggrin and aberrant profilaggrin-processing products in the stratum corneum.42 Although we were unable to directly assess filaggrin processing, the distribution of this protein in the epidermis was normal. Ultrastructural analysis of the granular layer of neonatal matriptase−/− epidermis showed sparse and disorganized lamellar bodies. In our patients, lamellar granules were found to be morphologically normal.
All the patients showed various hair abnormalities, including dysplastic hair, pili torti, pili bifurcati, and central pili mono bifurcati. Interestingly, multiple hair abnormalities—including trichorrhexis invaginata, pili torti, trichorrhexis nodosa, and helical hairs—have been reported in NTS.43,44
In conclusion, we have shown that a mutation in ST14 encoding the type II transmembrane serine protease matriptase causes a novel autosomal recessive genodermatosis characterized by ichthyosis and abnormal hair. The fact that the patients show abnormal degradation of corneodesmosmes in the upper layers of the stratum corneum suggests a role of this protease in epidermal desquamation, possibly via impaired proteolytic degradation of corneodesmosomal proteins.
Supplementary Material
Acknowledgments
We are grateful to the families who participated in this study. We also thank Dr. G. Halpern for her help with editing the manuscript and Rafi Rainshtein for his help with preparing the figures. We are grateful to Dr. Tamy Shohat for her help with the LOD-score calculation. This study was supported by the Yeshaya Horovitz foundation and in part by a grant provided by the Deutsche Forschungsgemeinschaft (to E.S.).
Web Resources
Accession numbers and URLs for data presented herein are as follows:
- BLink, http://www.ncbi.nlm.nih.gov/sutils/blink.cgi?pid=13124575
- Clustal Alignment, http://www.hongyu.org/software/clustal.html (for ClustalW)
- ConSeq Server, http://conseq.bioinfo.tau.ac.il/
- dbSNP, http://www.ncbi.nlm.nih.gov/SNP/
- Ensembl Genome Browser, http://www.ensembl.org/
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for Homo sapiens ST14, (accession number NP_068813), prostamin (accession number BAB20376), and serine protease (accession number AAG15395); P. troglodytes homologue (accession number XP_508863); M. musculus matriptase (accession number BAE38987); R. norvegicus matriptase (accession number AAH97271); G. gallus matriptase (accession number XP_417872); C. familiaris matriptase (accession number XP_546396); X. laevis matriptase (accession number AAH71077); and D. rerio hypothetical protein (accession number NP_001035441)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for TGM1; ALOXE3; ALOX12B; ABCA12; ICHTHYIN; FLJ39501; NTS; congenital ichthyosis, follicular atrophoderma, hypotrichosis, and hypohidrosis syndrome; trichothiodystrophy with congenital ichthyosis; ichthyosis, split hairs, and amino aciduria syndrome; and IFAP syndrome)
- Primer3, http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi
- SIFT, http://blocks.fhcrc.org/sift/SIFT_seq_submit2.html
- UCSC Genome Browser, http://genome.ucsc.edu/
References
- 1.Huber M, Rettler I, Bernasconi K, Frenk E, Lavrijsen SP, Ponec M, Bon A, Lautenschlager S, Schorderet DF, Hohl D (1995) Mutations of keratinocyte transglutaminase in lamellar ichthyosis. Science 267:525–528 10.1126/science.7824952 [DOI] [PubMed] [Google Scholar]
- 2.Russell LJ, DiGiovanna JJ, Rogers GR, Steinert PM, Hashem N, Compton JG, Bale SJ (1995) Mutations in the gene for transglutaminase 1 in autosomal recessive lamellar ichthyosis. Nat Genet 9:279–283 10.1038/ng0395-279 [DOI] [PubMed] [Google Scholar]
- 3.Krebsová A, Küster W, Lestringant GG, Schulze B, Hinz B, Frossard PM, Reis A, Hennies HC (2001) Identification, by homozygosity mapping, of a novel locus for autosomal recessive congenital ichthyosis on chromosome 17p, and evidence for further genetic heterogeneity. Am J Hum Genet 69:216–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jobard F, Lefèvre C, Karaduman A, Blanchet-Bardon C, Emre S, Weissenbach J, Özgüc M, Lathrop M, Prud’homme J-F, Fischer J (2002) Lipoxygenase-3 (ALOXE3) and 12 (R)-lipoxygenase (ALOX12B) are mutated in non-bullous congenital ichthyosiform erythroderma (NCIE) linked to chromosome 17p13.1. Hum Mol Genet 11:107–113 10.1093/hmg/11.1.107 [DOI] [PubMed] [Google Scholar]
- 5.Lefèvre C, Audebert S, Jobard F, Bouadjar B, Lakhdar H, Boughdene-Stambouli O, Blanchet-Bardon C, Heilig R, Foglio M, Weissenbach J, et al (2003) Mutations in the transporter ABCA12 are associated with lamellar ichthyosis type 2. Hum Mol Genet 12:2369–2378 10.1093/hmg/ddg235 [DOI] [PubMed] [Google Scholar]
- 6.Kelsell DP, Norgett EE, Unsworth H, Teh M-T, Cullup T, Mein CA, Dopping-Hepenstal PJ, Dale BA, Tadini G, Fleckman P, et al (2005) Mutations in ABCA12 underlie the severe congenital skin disease Harlequin ichthyosis. Am J Hum Genet 76:794–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lefèvre C, Bouadjar B, Karaduman A, Lefevre C, Bouadjar B, Karaduman A, Jobard F, Saker S, Özgüc M, Lathrop M, et al (2004) Mutations in ichthyin a new gene on chromosome 5q33 in a new form of autosomal recessive congenital ichthyosis. Hum Mol Genet 13:2473–2482 10.1093/hmg/ddh263 [DOI] [PubMed] [Google Scholar]
- 8.Lefèvre C, Bouadjar B, Ferrand V, Tadini G, Megarbane A, Lathrop M, Prud’homme JF, Fischer J (2006) Mutations in a new cytochrome P450 gene in lamellar ichthyosis type 3. Hum Mol Genet 15:767–776 10.1093/hmg/ddi491 [DOI] [PubMed] [Google Scholar]
- 9.Mizrachi-Koren M, Geiger D, Indelman M, Bitterman-Deutsch O, Bergman R, Sprecher E (2005) Identification of a novel locus associated with congenital recessive ichthyosis on 12p11.2-q13. J Invest Dermatol 125:456–462 10.1111/j.0022-202X.2005.23777.x [DOI] [PubMed] [Google Scholar]
- 10.Virolainen E, Wessman M, Hovatta I, Niemi K-M, Ignatius J, Kere J, Peltonen L, Palotie A (2000) Assignment of a novel locus for autosomal recessive congenital ichthyosis to chromosome 19p13.1-p13.2. Am J Hum Genet 66:1132–1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sprecher E (2005) Genetic hair and nail disorders. Clin Dermatol 23:47–55 10.1016/j.clindermatol.2004.09.009 [DOI] [PubMed] [Google Scholar]
- 12.Chavanas S, Bodemer C, Rochat A, Hamel-Teillac D, Ali M, Irvine AD, Bonafe JL, Wilkinson J, Taieb A, Barrandon Y, et al (2000) Mutations in SPINK5, encoding a serine protease inhibitor, cause Netherton syndrome. Nat Genet 25:141–142 10.1038/75977 [DOI] [PubMed] [Google Scholar]
- 13.Lestringant GG, Kuster W, Frossard PM, Happle R (1998) Congenital ichthyosis, follicular atrophoderma, hypotrichosis, and hypohidrosis: a new genodermatosis? Am J Med Genet 75:186–189 [DOI] [PubMed] [Google Scholar]
- 14.Tursen U, Kaya TI, Ikizoglu G, Aktekin M, Aras N (2002) Genetic syndrome with ichthyosis: congenital ichthyosis, follicular atrophoderma, hypotrichosis, and woolly hair: second report. Brit J Derm 147:604–631 10.1046/j.1365-2133.2002.48461.x [DOI] [PubMed] [Google Scholar]
- 15.Weeda G, Eveno E, Donker I, Vermeulen W, Chevallier-Lagente O, Taïeb A, Stary A, Hoeijmakers JHJ, Mezzina M, Sarasin A (1997) A mutation in the XPB/ERCC3 DNA repair transcription gene, associated with trichothiodystrophy. Am J Hum Genet 60:320–329 [PMC free article] [PubMed] [Google Scholar]
- 16.Broughton BC, Berneburg M, Fawcett H, Taylor EM, Arlett CF, Nardo T, Stefanini M, Menefee E, Price VH, Queille S, et al (2001) Two individuals with features of both xeroderma pigmentosum and trichothiodystrophy highlight the complexity of the clinical outcomes of mutations in the XPD gene. Hum Molec Genet 10:2539–2547 10.1093/hmg/10.22.2539 [DOI] [PubMed] [Google Scholar]
- 17.Yesudian P, Srinivas K (1977) Ichthyosis with unusual hair shaft abnormalities in siblings. Brit J Derm 96:199–203 10.1111/j.1365-2133.1977.tb12544.x [DOI] [PubMed] [Google Scholar]
- 18.Sambrook J, Fritsch EF, Maniatis T (1987) Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 19.Fishelson M, Geiger D (2002) Exact genetic linkage computations for general pedigrees. Bioinformatics Suppl 18:S189–S198 [DOI] [PubMed] [Google Scholar]
- 20.Higgins D, Thompson J, Gibson T (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680 10.1093/nar/22.22.4673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clamp M, Cuff J, Searle SM, Barton GJ (2004) The Jalview Java alignment editor. Bioinformatics 20:426–427 10.1093/bioinformatics/btg430 [DOI] [PubMed] [Google Scholar]
- 22.Berezin C, Glaser F, Rosenberg J, Paz I, Pupko T, Fariselli P, Casadio R, Ben-Tal N (2004) ConSeq: the identification of functionally and structurally important residues in protein sequences. Bioinformatics 20:1322–1324 10.1093/bioinformatics/bth070 [DOI] [PubMed] [Google Scholar]
- 23.Ng PC, Henikoff S (2002) Accounting for human polymorphisms predicted to affect protein function. Genome Res 12:436–446 10.1101/gr.212802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ishida-Yamamoto A, Simon M, Kishibe M, Miyauchi Y, Takahashi H, Yoshida S, O’Brien TJ, Serre G, Iizuka H (2004) Epidermal lamellar granules transport different cargoes as distinct aggregates. J Invest Dermatol 122:1137–1144 10.1111/j.0022-202X.2004.22515.x [DOI] [PubMed] [Google Scholar]
- 25.Descargues P, Deraison C, Bonnart C, Kreft M, Kishibe M, Ishida-Yamamoto A, Elias P, Barrandon Y, Zambruno G, Sonnenberg A, et al (2005) Spink5-deficient mice mimic Netherton syndrome through degradation of desmoglein 1 by epidermal protease hyperactivity. Nat Genet 37:56–65 [DOI] [PubMed] [Google Scholar]
- 26.Olson LE, Zhang J, Taylor H, Rose DW, Rosenfeld MG (2005) Barx2 functions through distinct corepressor classes to regulate hair follicle remodeling. Proc Natl Acad Sci USA 102:3708–3713 10.1073/pnas.0500519102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.List K, Bugge TH, Szabo R (2006) Matriptase: potent proteolysis on the cell surface. Mol Med 12:1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Konig A, Happle R (1999) Linear lesions reflecting lyonization in women heterozygous for IFAP syndrome (ichthyosis follicularis with atrichia and photophobia). Am J Med Genet 85:365–368 [DOI] [PubMed] [Google Scholar]
- 29.Hooper JD, Clements JA, Quigley JP, Antalis TM (2001) Type II transmembrane serine proteases: insights into an emerging class of cell surface proteolytic enzymes. J Biol Chem 276:857–860 10.1074/jbc.R000020200 [DOI] [PubMed] [Google Scholar]
- 30.Lin CY, Anders J, Johnson M, Sang QA, Dickson RB (1999) Molecular cloning of cDNA for matriptase, a matrix-degrading serine protease with trypsin-like activity. J Biol Chem 274:18231–18236 10.1074/jbc.274.26.18231 [DOI] [PubMed] [Google Scholar]
- 31.Oberst MD, Singh B, Ozdemirli M, Dickson RB, Johnson MD, Lin CY (2003) Characterization of matriptase expression in normal human tissues. J Histochem Cytochem 51:1017–1025 [DOI] [PubMed] [Google Scholar]
- 32.List K, Szabo R, Molinolo A, Nielsen BS, Bugge TH (2006) Delineation of matriptase protein expression by enzymatic gene trapping suggests diverging roles in barrier function, hair formation, and squamous cell carcinogenesis. Am J Pathol 168:1513–1512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin CY, Wang JK, Torri J, Dou L, Sang QA, Dickson RB (1997) Characterization of a novel, membrane-bound, 80-kDa matrix-degrading protease from human breast cancer cells: monoclonal antibody production, isolation, and localization. J Biol Chem 272:9147–9152 10.1074/jbc.272.14.9147 [DOI] [PubMed] [Google Scholar]
- 34.Cho EG, Kim MG, Kim C, Kim SR, Seong IS, Chung C, Schwartz RH, Park D (2001) N-terminal processing is essential for release of epithin, a mouse type II membrane serine protease. J Biol Chem 276:44581–44589 10.1074/jbc.M107059200 [DOI] [PubMed] [Google Scholar]
- 35.Oberst MD, Williams CA, Dickson RB, Johnson MD, Lin CY (2003) The activation of matriptase requires its noncatalytic domains, serine protease domain, and its cognate inhibitor. J Biol Chem 278:26773–26779 10.1074/jbc.M304282200 [DOI] [PubMed] [Google Scholar]
- 36.Zeeuwen P (2004) Epidermal differentiation: the role of proteases and their inhibitors. Eur J Cell Biol 83:761–773 10.1078/0171-9335-00388 [DOI] [PubMed] [Google Scholar]
- 37.Caubet C, Jonca N, Brattsand M, Guerrin M, Bernard D, Schmidt R, Egelrud T, Simon M, Serre G (2004) Degradation of corneodesmosome proteins by two serine proteases of the kallikrein family, SCTE/KLK5/hK5 and SCCE/KLK7/hK7. J Invest Dermatol 122:1235–1244 10.1111/j.0022-202X.2004.22512.x [DOI] [PubMed] [Google Scholar]
- 38.Egelrud T, Brattsand M, Kreutzmann P, Walden M, Vitzithum K, Marx UC, Forssmann WG, Magert HJ (2005) hK5 and hK7, two serine proteinases abundant in human skin, are inhibited by LEKTI domain 6. Br J Dermatol 153:1200–1203 10.1111/j.1365-2133.2005.06834.x [DOI] [PubMed] [Google Scholar]
- 39.Elsayed-Ali H, Barton S, Marks R (1992) Stereological studies of desmosomes in ichthyosis vulgaris. Br J Dermatol 126:24–28 10.1111/j.1365-2133.1992.tb08398.x [DOI] [PubMed] [Google Scholar]
- 40.Satomi S, Yamasaki Y, Tsuzuki S, Hitomi Y, Iwanaga T, Fushiki T (2001) A role for membrane-type serine protease (MT-SP1) in intestinal epithelial turnover. Biochem Biophys Res Commun 287:995–1002 10.1006/bbrc.2001.5686 [DOI] [PubMed] [Google Scholar]
- 41.List K, Haudenschild CC, Szabo R, Chen W, Wahl SM, Swaim W, Engelholm LH, Behrendt N, Bugge TH (2002) Matriptase/MT-SP1 is required for postnatal survival, epidermal barrier function, hair follicle development, and thymic homeostasis. Oncogene 21:3765–3779 10.1038/sj.onc.1205502 [DOI] [PubMed] [Google Scholar]
- 42.List K, Szabo R, Wertz PW, Segre J, Haudenschild CC, Kim SY, Bugge TH (2003) Loss of proteolytically processed filaggrin caused by epidermal deletion of matriptase/MT-SP1. J Cell Biol 163:901–910 10.1083/jcb.200304161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ito M, Ito K, Hashimoto K (1984) Pathogenesis in trichorrhexis invaginata (bamboo hair). J Invest Dermatol 83:1–6 10.1111/1523-1747.ep12261618 [DOI] [PubMed] [Google Scholar]
- 44.Lurie R, Garty BZ (1995) Helical hairs: a new hair anomaly in a patient with Netherton’s syndrome. Cutis 55:349–352 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.