Abstract
We demonstrate that satellite III (SatIII) DNA subfamilies cloned from human acrocentric chromosomes arose in the Hominoidea superfamily. Two groups, distinguished by sequence composition, evolved nonconcurrently, with group 2 evolving 16–23 million years ago (MYA) and the more recent group 1 sequences emerging ∼4.5 MYA. We also show the relative order of emergence of each group 2 subfamily in the various primate species. Our results show that each SatIII subfamily is an independent evolutionary unit, that the rate of evolution is not uniform between species, and that the evolution within a species is not uniform between chromosomes.
Repetitive sequences form a substantial part of the genomes of many eukaryotes.1 Several classes of satellite DNA have been identified, including alpha, beta, and satellites I–IV.2 Satellite III (SatIII) DNA is a family of repetitive sequences that have been localized in humans to the short arms of the acrocentric chromosomes 13–15, 21, and 223–6 and to the heterochromatin of the long arms of chromosomes 1, 9, and Y.7–12 Subfamilies of SatIII DNA have been isolated and divided into two subgroups on the basis of their sequence composition.6,13,14 These two groups show differences in the percentage of two characteristic sequence motifs, GGAAT and GGAGT. The GGAAT motif is predominant in the first group, whereas there is roughly an equal representation of the GGAAT and GGAGT sequence motifs in the second group.14
These subfamilies reside primarily on the short arms of the human acrocentric chromosomes between the centromeric and rDNA sequences. Previous studies of humans have shown that subfamilies pR-1, pR-2, and pR-4 are present on all acrocentric chromosomes; pK-1 and pTRS-47 are present on chromosomes 14 and 22; pE-2 is present on chromosomes 13, 14, and 21; pW-1 is present on chromosomes 13 and 21; and pTRS-63 is present on chromosome 14.14 Although SatIII DNA has been identified by Southern analysis in orangutan, gorilla, and chimpanzee,15,16 the chromosomal locations and identification of particular subfamilies of SatIII in nonhuman primates have not been determined.
We studied 25 nonhuman primate cell lines, four human reference cell lines, and a panel of monochromosomal hybrids of the acrocentric chromosomes and the Y chromosome (table 1), to determine the presence and evolution of SatIII subfamilies in primates. More than one individual was used from each species, with the exception of the pygmy chimpanzee and the white-handed gibbon. DNA extraction was performed on all cell lines and on mouse and hamster controls by standard procedures. Metaphase cells were obtained by standard cytogenetic procedures. We performed PCR with monochromosomal hybrids containing human acrocentric chromosomes, to confirm the specificity of primers for each subfamily. PCR was performed with primers specific to eight subfamilies—three from group 1 (pTRS-63, pW-1, and pK-1) and five from group 2 (pTRS-47, pE-2, pR-1, pR-2, and pR-4)—with use of DNA prepared from the primate panel cell lines, mouse and hamster as controls for the monochromosomal hybrids, monochromosomal hybrids, and a plasmid for each respective subfamily. Primers were designed from the known sequences of each subfamily for pW-1, pR-2, pK-1, pE-2, and pR-4.14 Primers for pR-1, pTRS-63, and pTRS-47 are shown in table 2. For detection of the pTRS-47 subfamily, two sets of primers—pTRS-47[1] and pTRS-47[2]—were designed for the beginning and end of the sequence, respectively.
Table 1. .
Primate Cell Lines and Monochromosomal Cell Lines Used in This Study
| Cell Line | No. of Study Individuals |
| Primates: | |
| Human (Homo sapiens [HSA]) | 4 |
| Pygmy chimpanzee (Pan paniscus [PPA]) | 1 |
| Chimpanzee (Pan troglodytes [PTR]) | 2 |
| Gorilla (Gorilla gorilla [GGO]) | 3 |
| Orangutan (Pongo pygmaeus [PPY]) | 3 |
| Crested gibbon (Nomascus concolor [NCO]) | 2 |
| White-handed gibbon (Hylobates lar [HLA]) | 1 |
| Baboon (Papio hamadryas [PHA]) | 3 |
| Rhesus monkey (Macaca mulatta [MMU]) | 3 |
| African green monkey (Cercopithecus aethiops [CAE]) | 4 |
| Squirrel monkey (Saimiri sciureus [SSC]) | 3 |
| Human monochromosomal hybrids: | |
| Chromosome 13 (GM10898D) | 1 |
| Chromosome 14 (GM10479B, GM11535A, and CP43) | 3 |
| Chromosome 15 (GM11715A) | 1 |
| Chromosome 21 (GM10323A and GM08854A) | 2 |
| Chromosome 22 (GM10888A) | 1 |
| Chromosome Y (GM10898D) | 1 |
Table 2. .
PCR Primer Sequences for SatIII Subfamilies
| Primer(5′→3′) |
||
| Clone | Forward | Reverse |
| pTRS-63 | ATTGAAACCTGCTCGATTGG | TCGGTTTGATTCCATTCCAT |
| pR-1 | CTGGACTTTGGTGGAAAGGA | ACAATCTCAGCCCACATTCC |
| pTRS47[1] | CGGAGTGATTGGAGTGGAAT | CCAGTCCCTTCCATTCCTTTAT |
| pTRS47[2] | GGATCAGAACGGAACAGAGC | AGTCCACTGCATTCCATTCC |
PCRs were performed using TAKARA (TAKARA BIO) (for pW-1, pR-2, pK-1, pE-2, pR-1, pTRS-63, and pTRS-47) and Taq DNA (Fermentas Life Sciences) (for pR-4) polymerases. The PCR conditions were different for most subfamilies. The range of annealing temperature was 55°C–64°C; the range of annealing time was 45–90 s. For those subfamilies in which amplification in nonhuman primates could not be obtained, PCR was prepared at a lower annealing temperature (∼53°C). If two or more PCR products were detected, each was cut from the gel before individual purification and preparation for sequencing.
The products for the sequencing reaction were purified using Montage PCR Centrifugal Filter Devices (Millipore). The sequencing reactions were performed in both directions with dye terminators (ABI PRISM BigDye Terminator v3.1 Cycle Sequencing Kit) on an Applied Biosystems 3100 Genetics Analyzer. The analyzed data were edited using Sequencher version 4.1.4 (Gene Codes).
The sizes of the PCR products containing our SatIII subfamilies obtained from primates and human monochromosomal hybrids are shown in table 3. We observed polymorphisms for the PCR product size for the pR-4 subfamily in human chromosomes. Two products, one 500 bp and one 680 bp, were found in human genomic DNA and a hybrid containing a single human chromosome 13. Of three hybrids containing human chromosome 14, one contained the 500-bp product, and two contained the 680-bp product. Monochromosomal hybrids carrying human chromosome 21 or 22 contained the 500-bp product, and the hybrid containing human chromosome 15 contained the 680-bp product. Likewise, the subfamily pE-2 showed two products, 156 bp and 700 bp, from human genomic DNA and monochromosomal hybrids for human chromosomes 13, 14, and 21. Human chromosome 22 showed a slightly larger 170-bp product. pE-2 was not amplified from chromosome 15 (table 3).
Table 3. .
Sizes of PCR Products Containing SatIII Subfamilies[Note]
| PCR Product Size(s)(bp) |
|||||||||||||
| Chromosome |
|||||||||||||
| Group and Clone |
HLA (n=1) |
NCO (n=2) |
PPY (n=5) |
GGO (n=3) |
PPA (n=1) |
PTR (n=4) |
HSA (n=4) |
13 | 14 | 15 | 21 | 22 | Y |
| 1: | |||||||||||||
| pTRS-63 | … | … | … | … | … | … | 207 | … | 207 | … | … | … | 207 |
| pW-1 | … | … | … | … | … | … | 883 | 883 | … | … | 883 | … | 883 |
| pK-1 | … | … | … | … | … | … | 421 | … | 421 | … | … | 421 | … |
| 2: | |||||||||||||
| pTRS-47[1] | … | 234 | … | … | NS | 700 | 234 | … | 234 | … | … | 190 | … |
| pTRS-47[2] | 280 | 280 | 400 | NS | … | … | 190 | … | 190 | … | … | 190 | … |
| pE-2 | … | … | … | … | 156 | 156 | 156 and 700 | 156 and 700 | 156 and 700 | … | 156 and 700 | 170 | … |
| pR-1 | … | … | NS | 1,100 | … | NS | 496 | 496 | 496 | 496 | 496 | NS | 496 |
| pR-2 | 400 | 400 | 1,100 | 900 | 1,100 and 640 | 1,700 | 953 | 953 | 953 | 953 | 953 | 953 | … |
| pR-4 | … | 290 | NS | 400 | 550 | 550 | 500 and 680 | 500 and 680 | 500 or 680 | 680 | 500 | 500 | … |
Note.— PCR products were from primates and human monochromosomal hybrids. NS = fragments could not be sequenced.
With use of the primers derived from the human sequence for the SatIII subfamilies, none of the SatIII subfamilies were detected in squirrel monkey, African green monkey, baboon, or rhesus monkey. To confirm the PCR results, two-color FISH was performed, by standard procedures, on metaphase cells from each of the 25 nonhuman primate cell lines and the four human reference cell lines, with use of unique plasmids containing one SatIII subfamily.14,17,18 Two subfamilies (pTRS-63 and pW-1) from group 1 and four (pTRS-47, pR-4, pR-2, and pR-1) from group 2 were chosen for FISH analysis. DNA from a SatIII subfamily and DNA corresponding to the rDNA genes (pA and pU6.2 simultaneously) were labeled by nick translation with digoxigenin and biotin, respectively.18 At least 10 cells from each cell line were analyzed using microscopic visualization and digital-imaging analysis (Applied Imaging). FISH with plasmids that each correspond to a unique subfamily17 did not hybridize to squirrel monkey, African green monkey, baboon, or rhesus monkey, confirming the PCR results. Thus, these SatIII subfamilies are not present in New World and Old World monkeys and must have evolved later in primate evolution.
Each sequence from our subfamilies was checked for the presence of pentameric GGAGT and/or GGAAT sequence motifs with use of a sequencer program and repeat-masker programs (RepeatMasker at EMBL). Nucleotide alignments were performed using the programs Sequencher version 4.1.4, ClustawW, and Jalview, with standard parameters. To determine and compare the conserved fragments, the Jalview program was used. To observe which nucleotide differences were true differences and which were polymorphisms or transmorphisms, the comparison was performed as follows: first, sequences derived from human cell lines and human monochromosomal hybrids were compared; next, sequences from human and chimpanzee were compared, followed by gorilla, orangutan, and gibbon. We set a threshold of 60% minimum match percentage (MMP) as acceptable for sequence alignment for autosomal SatIII subfamilies and an 80% MMP for the Y chromosome sequences. Pairwise percentage of divergence was calculated as
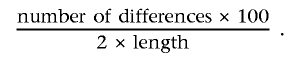 |
Differences between aligned sequences were counted as 1 for each base change and as 1 for each gap, with the length for each comparison adjusted to account for gap lengths.19
PCR analysis of group 1 subfamilies pTRS-63, pK-1, and pW-1 showed amplification in only human cell lines and in human-derived monochromosomal hybrids of the acrocentric chromosomes. At a lower annealing temperature, amplification was achieved with the primer for human pW-1 in gibbon, orangutan, and chimpanzee. However, the sequences obtained at this lower annealing temperature did not contain GGAAT motifs in gibbon, and products obtained from orangutan and chimpanzee contained the repeat motif but did not align at even 60% similarity with human pW-1 sequence and were considered nonspecific amplification. Although FISH analysis showed the presence of hybridization signals from pTRS-63 and pW-1 in gibbon, orangutan, gorilla, and chimpanzee that colocalized with the rDNA (fig. 1 and table 4), on the basis of the sequencing results, this hybridization is considered nonspecific and is likely due to the high homology between group 1 and group 2 subfamilies.14 Thus, group 1 SatIII subfamilies are present only in humans and appeared after the divergence of humans from apes, ∼4.5 million years ago (MYA).
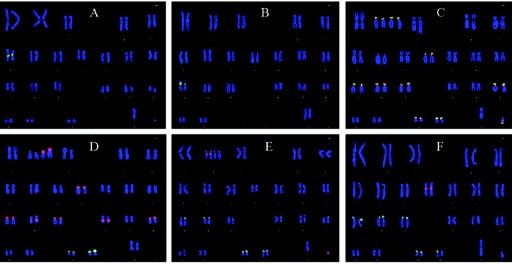
Figure 1. .
Representative FISH with pW-1 SatIII DNA (red) and rDNA (pA and pU6.2) (green) probes for squirrel monkey (A), baboon (B), orangutan (C), gorilla (D), chimpanzee (E), and human (F). Note the lack of hybridization signal in squirrel monkey (A) and baboon (B). Subfamily pW-1 was shown by PCR to be nonspecific in all remaining species studied except humans (see table 4).
Table 4. .
FISH Results for Four Subfamilies of SatIII DNA and rDNA Probes[Note]
| FISH Results for Species |
|||||||||||
| Group and Clone |
CAE (n=4) |
SSC (n=3) |
PHA (n=3) |
MMU (n=3) |
HLA (n=1) |
NCO (n=2) |
PPY (n=3) |
GGO (n=3) |
PPA (n=1) |
PTR (n=2) |
HSA (n=4) |
| NORa | 1 | 1 | 1 | 1 | 1 | 2 | 8−9+Y | 2 | 5 | 5 | 5 |
| 1: | |||||||||||
| pTRS-63 | 0 | 0 | 0 | 0 | (2/1) | (7+Y/2) | (9+Y/8−9+Y) | (8+Y/1) | (6+Y/2) | (5+Y/2) | 2/1 |
| pW-1 | 0 | 0 | 0 | 0 | (2/1) | (3+Y/2) | (9+Y/8−9+Y) | (8+Y/1) | (6+Y/2) | (7−9+Y/4) | 3/2 |
| 2: | |||||||||||
| pTRS-47 | 0 | 0 | 0 | 0 | 5/1 | 4+Y/2 | 9+Y/8−9+Y | (8+Y/1) | (6+Y/2) | 5+Y/2−3 | 3/2 |
| pR-4 | 0 | 0 | 0 | 0 | (5/1) | 5−7+Y/2 | (9+Y/8−9+Y) | 4,5,7+Y/0−1 | 8+Y/5 | 7+Y/5 | 5+Y/5 |
| pR-2 | 0 | 0 | 0 | 0 | 5/1 | 7−8+Y/2 | 9+Y/8−9+Y | 3−4/0−1 | 8+Y/5 | 7+Y/5 | 5/5 |
| pR-1 | 0 | 0 | 0 | 0 | (5/1) | (7−8+Y/2) | (9+Y/8−9+Y) | 5−7+Y/2 | (8+Y/5) | (7+Y/5) | 5+Y/5 |
Note.— Numbers indicate pairs. Ranges indicate variable signal intensities among different individuals. Numbers after virgules (/) indicate the number of pairs colocalized with the rDNA. Numbers in parentheses indicate cross-hybridization based on sequence analysis. Numbers not enclosed in parentheses and bold indicate signals determined to be true hybridization that correlated with sequencing results.
NOR = nucleolar organizer region.
The evolution of satellite DNA sequences in primates appears to be highly complex; some sequences seem to remain conserved for a long evolutionary time, whereas other sequences show dynamic nucleotide changes even in the same population.20 We made these same observations in our group 2 subfamilies. The pR-1 subfamily was detected in two gorilla individuals, in humans, and in the monochromosomal hybrids for the human acrocentric chromosomes 13–15 and 21. PCR products from orangutans and chimpanzees showed only a smear and could not be sequenced. The gorilla pR-1 PCR product showed ∼4.5% sequence divergence from the human sequence (table 5). Remarkably, among the human chromosomes analyzed, pR-1 showed 4.11% divergence. Thus, this subfamily appears to be relatively dynamic. The absence of the pR-1 subfamily in chimpanzees may be the result of a DNA sequence change in the chimpanzee genome for the regions corresponding to the human primers. Alternatively, these sequences may not be present in chimpanzee, since this sequence may have been lost from the chimpanzee genome after the divergence of gorilla and chimpanzee ancestors or may have evolved independently in human and gorilla.
Table 5. .
Pairwise Percentage of Divergence for Sequences from Nonhuman Primates Compared with Human Consensus Sequence
| Pairwise Percentage of Divergence |
|||||||
| Group 2 Clone |
HLA (n=1) |
NCO (n=2) |
PPY (n=3) |
GGO (n=3) |
PPA (n=1) |
PTR (n=2) |
HSA (n=11) |
| pTRS-47[1] | … | 13.35 | … | … | … | 3.49 | 2.65 |
| pTRS-47[2] | 17.98 | 13.49 | 11.78 | … | … | .52 | |
| pE-2 | … | … | … | … | 4.38 | 1.47 | |
| pR-1 | … | … | … | 4.49 | … | … | 4.11 |
| pR-2 | 14.7 | 13.44 | 12.62 | 10.05 | 10.38 | 9.12 | 1.68–11.56 |
| pR-4 | … | 12.69 | … | 7.81 | 5.61 | 6.81 | .65–2.2 |
The group 2 subfamilies pTRS-47, pR-2, and pR-4 were detected in gibbon, orangutan, gorilla, chimpanzee, and bonobo by both sequencing (table 3) and FISH analysis (table 4). Sequence analysis showed that the pairwise percentage of divergence ranged from ∼3.5% to ∼18% (table 5) for these subfamilies in the various primates compared with humans, and, interestingly, the divergence among human acrocentric chromosomes ranged from ∼0.5% to ∼11.5%. Sequence comparisons among acrocentric chromosomes showed that the pR-2 subfamily has high variation among humans and among chromosomes, as analyzed in the monochromosomal hybrids, as compared with other subfamilies. Although each chromosome has a slightly different sequence for this subfamily, differences were also found between two different hybrids containing human chromosomes 21. Divergence between human pR-2 sequences had a range of 1.68%–11.56%. When we compared human pR-2 sequences with those of primates, we observed similar variation between human and chimpanzee (9.12%) and between human and gorilla (10.05%). Between humans and the gibbon subspecies, we observed 13.44% divergence with white-handed gibbon and 14.7% divergence with crested gibbon. This high level of variation in the pR-2 subfamily cannot be explained by rapid spreading alone, because pR-4, by comparison, is highly stable in sequence composition (fig. 2), and both pR-2 and pR-4 are found in the same chromosomal distributions among species. More interesting, though, is the discrepancy among pR-1, pR-2, and pR-4 in chromosomal distribution and species emergence. All three subfamilies are present on all human acrocentric chromosomes, suggesting that they evolved together and spread with rDNA sequences.14 However, pR-1 did not emerge until the divergence of gorillas, but pR-2 and pR-4 are present in gibbons.
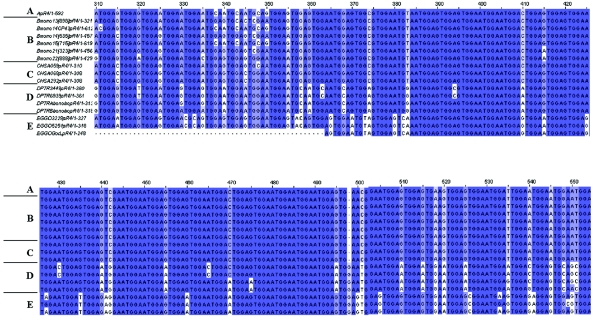
Figure 2. .
Sequence alignment, prepared using Jalview multiple-alignment editor, for the plasmid containing human pR-4 (A), human monochromosomal hybrids (B), human cell lines (C), chimpanzee (D), and gorilla (E). Dark blue indicates perfect sequence alignment across all comparisons. Light blue indicates perfect sequence alignment for some comparisons. White indicates a base change from consensus. Regions with the most-significant homology are shown.
A pE-2 product was detected in gibbon, orangutan, and chimpanzee. However, we found several nonspecific sequences that did not have the characteristic motifs found in human for gibbon and orangutan. Thus, the subfamily pE-2 is present only in chimpanzee and humans. In chimpanzee, we observed only one (156 bp) of the two products (156 bp and ∼700 bp) found in humans.
Thus, our results for group 2 subfamilies show that sequences homologous to pR-2, pR-4, and pTRS-47 are present in gibbon, which suggests that these subfamilies appeared suddenly in the Hominoidea lineage ∼16–23 MYA. In addition, pR-1 did not appear until the divergence of orangutan and gorilla, ∼7–10 MYA. pE-2 is the youngest group 2 subfamily, occurring after the divergence of bonobo and chimpanzee, ∼4.5–5 MYA.
The sequences on the human acrocentric short arms are thought to have evolved through interchromosomal recombination between repeats. Further understanding of the chromosomal evolution in primates may be gained by observing differences in repetitive DNA distribution. The loci to the 18S and 28S ribosomal RNA genes reside on the acrocentric short arms in humans and chimpanzee, with one pair in chimpanzee residing on chromosome 18 instead of chromosome 15.21–23 Orangutan has nine pairs of chromosomes (2p, 2q, 9, 13–15, 18, 21, and 22) with rDNA.23,24 The exception to the characteristic multiple distributions among pongidae is gorilla, which has only two pairs of chromosomes (21 and 22) with rDNA.22,23 The white-handed gibbon, similar to baboon and rhesus monkey, has only one pair of chromosomes (12 and 15 in baboon and rhesus monkey, respectively) with rDNA, and they are metacentric.23,25,26 The crested gibbon has three pairs of acrocentric chromosomes, but only two of them (in addition to the Y chromosome) contain rDNA.27 Two-color FISH analysis with use of the subfamily-specific plasmids17 and 18S/28S rDNA sequences showed the number of chromosomes containing both SatIII sequences and rDNA (table 4). Our SatIII subfamilies were found primarily on the acrocentric chromosomes and the Y chromosome in orangutan, gorilla, and chimpanzee. In white-handed gibbon, our SatIII subfamilies were found in the pericentromeric regions on 2–5 pairs of metacentric chromosomes, whereas, in crested gibbon, our subfamilies were found on two pairs of acrocentric chromosomes, in the pericentromeric regions on 2–5 pairs of nonacrocentric chromosomes, and on the Y chromosome. This colocalization of SatIII and rDNA increased in number from gibbon to orangutan, with a dramatic reduction in the number of rDNA-bearing chromosomes in gorilla and a moderate reduction in more-recent primates in the number of chromosomes that colocalize SatIII DNA with rDNA.
The reduction of SatIII DNA localization in humans can be explained by several chromosomal evolutionary events. First, chromosomes 2p and 2q in higher apes underwent pericentromeric inversion, and, after the divergence of apes and humans, these chromosomes underwent fusion. Second, human chromosome 18 underwent pericentromeric inversion,23 losing both rDNA and SatIII DNA. Third, human chromosome 9, the orthologue of which is acrocentric in orangutan and is submetacentric in gorilla and chimpanzee, retained SatIII subfamilies pW-1, pTRS-63, and pTRS-47 but lost the rDNA and the remaining SatIII subfamilies.
Although the Y chromosome is not an rDNA-bearing chromosome in humans, we investigated the SatIII DNA content in the Y chromosome for those species for which we had male individuals (table 4). From a monochromosomal hybrid with a human Y chromosome, we obtained amplification products with the predicted sizes, with primers specific to pW-1 (883 bp), pR-1 (496 bp), and pTRS-63 (∼200 bp), and several bands for primers specific to pR-2, pTRS-47, pR-4, and pE-2. PCR with pK-1–specific primers did not show any product from the Y chromosome. After sequence analysis, the product from pE-2 did not contain any SatIII consensus sequence. For sequences derived from the human Y chromosome, we established a minimum threshold of 80% similarity. Using these criteria, we discarded sequences from pR-4, pR-2, and pTRS-47, which contain characteristic motifs but insufficient similarity to be characterized as proper SatIII subfamilies. pTRS-47 showed three products, one with 93% similarity to the HSDYZ1 sequence (GenBank accession number X06228) from the Y chromosome. However, the two other fragments could not be found using BLAT (Human BLAT Search), likely because of sequence gaps in the pericentromeric regions of the human genome. By FISH analysis, we determined that SatIII DNA is present on the Y chromosome from gibbons to humans. In humans, the signal on the Y chromosome is weak or absent in the pericentromeric region. The human Y chromosome shows very weak signals for pR-4 and pR-1 (data not shown) and no signals for pW-1, pTRS-63, or pTRS-47. Because hybridization signals were not visible by FISH but sequencing of PCR amplification products showed sequence consistent with pW-1, pR-1, and pTRS-63 SatIII DNA, it is likely that fewer copies of these repetitive sequences are present on the human Y chromosome than on the human acrocentric chromosomes or on the nonhuman primate Y chromosomes. By comparison, the various subfamilies (fig. 1) have strong hybridization signals in the Y pericentromeric region in orangutan, gibbon, gorilla, and chimpanzee and on the q12 band in orangutan, gorilla, and chimpanzee. Thus, the results for the human Y chromosome indicate that the reduction of satellite DNA in the less ancient primates is due not only to the reduction of SatIII DNA in chromosomes or chromosome regions but also to the loss of the number of repeats in some subfamilies.
Our results suggest that SatIII DNA arose suddenly in gibbon 16–23 MYA and underwent amplification, peaking in orangutan and gorilla and then decreasing during more-recent speciation events. The group 2 subfamilies are older than the group 1 subfamilies, with the group 1 subfamilies appearing after the divergence of humans from apes, ∼4.5 MYA. Additionally, our data suggest that the SatIII subfamilies have evolved from an ancient sequence similar to pR-2 or pR-4 (both group 2). Our data support the hypothesis that each satellite can be an independent evolutionary unit, concerning not only the copy number but also the sequence.20 In addition, our data indicate that the rate of evolution within and among species is not uniform,28,29 supporting patchy homogenization during evolution, as proposed for the rDNA genes.19
Recently, it has been shown that SatIII DNA from the human 9q heterochromatic region is transcribed after cellular stress, specifically heat-shock treatment, and that these transcripts bind splicing factors and recruit them to nuclear stress granules.30–32 This process seems to regulate splicing under specific physiologic conditions. The significant alignment of the sequences in some subfamilies among various primates may indicate conservation during evolution. This conservation suggests a selective pressure to remain unchanged, which may support the functional significance of SatIII DNA in response to cellular stress.
Acknowledgment
This research was supported in part by a grant from the National Institutes of Health (to L.G.S.).
Web Resources
The accession number and URLs for data presented herein are as follows:
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for the HSDYZ1 sequence [accession number X06228])
- Human BLAT Search, http://genome.ucsc.edu/cgi-bin/hgBlat
- Jalview, http://www.jalview.org/
- RepeatMasker at EMBL, http://woody.embl-heidelberg.de/repeatmask/
References
- 1.John B, Miklos GL (1979) Functional aspects of satellite DNA and heterochromatin. Int Rev Cytol 58:1–114 [DOI] [PubMed] [Google Scholar]
- 2.Choo KH, Vissel B, Earle E (1989) Evolution of alpha-satellite DNA on human acrocentric chromosomes. Genomics 5:332–344 10.1016/0888-7543(89)90066-9 [DOI] [PubMed] [Google Scholar]
- 3.Gosden JR, Lawrie SS, Gosden CM (1981) Satellite DNA sequences in the human acrocentric chromosomes: information from translocations and heteromorphisms. Am J Hum Genet 33:243–251 [PMC free article] [PubMed] [Google Scholar]
- 4.Dale S, Earle E, Voullaire L, Rogers J, Choo KH (1989) Centromeric alpha satellite DNA amplification and translocation in an unusually large chromosome 14p+ variant. Hum Genet 82:154–158 10.1007/BF00284049 [DOI] [PubMed] [Google Scholar]
- 5.Earle E, Dale S, Choo KH (1989) Amplification of satellite III DNA in an unusually large chromosome 14p+ variant. Hum Genet 82:187–190 10.1007/BF00284055 [DOI] [PubMed] [Google Scholar]
- 6.Choo KH, Earle E, Vissel B, Kalitsis P (1992) A chromosome 14-specific human satellite III DNA subfamily that shows variable presence on different chromosomes 14. Am J Hum Genet 50:706–716 [PMC free article] [PubMed] [Google Scholar]
- 7.Cooke HJ, Hindley J (1979) Cloning of human satellite III DNA: different components are on different chromosomes. Nucleic Acids Res 6:3177–3197 10.1093/nar/6.10.3177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moyzis RK, Albright KL, Bartholdi MF, Cram LS, Deaven LL, Hildebrand CE, Joste NE, Longmire JL, Meyne J, Schwarzacher-Robinson T (1987) Human chromosome-specific repetitive DNA sequences: novel markers for genetic analysis. Chromosoma 95:375–386 10.1007/BF00333988 [DOI] [PubMed] [Google Scholar]
- 9.Schwarzacher-Robinson T, Cram LS, Meyne J, Moyzis RK (1988) Characterization of human heterochromatin by in situ hybridization with satellite DNA clones. Cytogenet Cell Genet 47:192–196 [DOI] [PubMed] [Google Scholar]
- 10.Gosden JR, Spowart G, Lawrie SS (1981) Satellite DNA and cytological staining patterns in heterochromatic inversions of human chromosome 9. Hum Genet 58:276–278 10.1007/BF00294922 [DOI] [PubMed] [Google Scholar]
- 11.Nakahori Y, Mitani K, Yamada M, Nakagome Y (1986) A human Y-chromosome specific repeated DNA family (DYZ1) consists of a tandem array of pentanucleotides. Nucleic Acids Res 14:7569–7580 10.1093/nar/14.19.7569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cooke HJ, Schmidtke J, Gosden JR (1982) Characterisation of a human Y chromosome repeated sequence and related sequences in higher primates. Chromosoma 87:491–502 10.1007/BF00333470 [DOI] [PubMed] [Google Scholar]
- 13.Choo KH, Earle E, McQuillan C (1990) A homologous subfamily of satellite III DNA on human chromosomes 14 and 22. Nucleic Acids Res 18:5641–5648 10.1093/nar/18.19.5641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bandyopadhyay R, McQuillan C, Page SL, Choo KH, Shaffer LG (2001) Identification and characterization of satellite III subfamilies to the acrocentric chromosomes. Chromosome Res 9:223–233 10.1023/A:1016648404388 [DOI] [PubMed] [Google Scholar]
- 15.Fowler JC, Skinner JD, Burgoyne LA, Drinkwater RD (1989) Satellite DNA and higher-primate phylogeny. Mol Biol Evol 6:553–557 [DOI] [PubMed] [Google Scholar]
- 16.Mitchell AR, Gosden JR, Ryder OA (1981) Satellite DNA relationships in man and the primates. Nucleic Acids Res 9:3235–3249 10.1093/nar/9.14.3235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Han JY, Choo KH, Shaffer LG (1994) Molecular cytogenetic characterization of 17 rob(13q14q) Robertsonian translocations by FISH, narrowing the region containing the breakpoints. Am J Hum Genet 55:960–967 [PMC free article] [PubMed] [Google Scholar]
- 18.Shaffer LG, McCaskill C, Han JY, Choo KH, Cutillo DM, Donnenfeld AE, Weiss L, Van Dyke DL (1994) Molecular characterization of de novo secondary trisomy 13. Am J Hum Genet 55:968–974 [PMC free article] [PubMed] [Google Scholar]
- 19.Gonzalez IL, Sylvester JE (2001) Human rDNA: evolutionary patterns within the genes and tandem arrays derived from multiple chromosomes. Genomics 73:255–263 10.1006/geno.2001.6540 [DOI] [PubMed] [Google Scholar]
- 20.Ugarkovic D, Plohl M (2002) Variation in satellite DNA profiles—causes and effects. EMBO J 21:5955–5959 10.1093/emboj/cdf612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Henderson AS, Atwood KC, Warburton D (1976) Chromosomal distribution of rDNA in Pan paniscus, Gorilla gorilla beringei, and Symphalangus syndactylus: comparison to related primates. Chromosoma 59:147–155 10.1007/BF00328483 [DOI] [PubMed] [Google Scholar]
- 22.Hirai H, Taguchi T, Godwin AK (1999) Genomic differentiation of 18S ribosomal DNA and beta-satellite DNA in the hominoid and its evolutionary aspects. Chromosome Res 7:531–540 10.1023/A:1009237412155 [DOI] [PubMed] [Google Scholar]
- 23.Yunis JJ, Prakash O (1982) The origin of man: a chromosomal pictorial legacy. Science 215:1525–1530 10.1126/science.7063861 [DOI] [PubMed] [Google Scholar]
- 24.Henderson AS, Warburton D, Megraw-Ripley S, Atwood KC (1979) The chromosomal location of rDNA in the Sumatran orangutan, Pongo pygmaeus albei. Cytogenet Cell Genet 23:213–216 [DOI] [PubMed] [Google Scholar]
- 25.Henderson AS, Warburton D, Megraw-Ripley S, Atwood KC (1977) The chromosomal location of rDNA in selected lower primates. Cytogenet Cell Genet 19:281–302 [DOI] [PubMed] [Google Scholar]
- 26.Quirk S, Henderson AS (1985) Equivalence of nucleolar organizer activity among primate species. Cytogenet Cell Genet 39:134–135 [DOI] [PubMed] [Google Scholar]
- 27.Koehler U, Bigoni F, Wienberg J, Stanyon R (1995) Genomic reorganization in the concolor gibbon (Hylobates concolor) revealed by chromosome painting. Genomics 30:287–292 10.1006/geno.1995.9875 [DOI] [PubMed] [Google Scholar]
- 28.Hall SE, Kettler G, Preuss D (2003) Centromere satellites from Arabidopsis populations: maintenance of conserved and variable domains. Genome Res 13:195–205 10.1101/gr.593403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mravinac B, Plohl M, Ugarkovic D (2004) Conserved patterns in the evolution of Tribolium satellite DNAs. Gene 332:169–177 10.1016/j.gene.2004.02.055 [DOI] [PubMed] [Google Scholar]
- 30.Metz A, Soret J, Vourc’h C, Tazi J, Jolly C (2004) A key role for stress-induced satellite III transcripts in the relocalization of splicing factors into nuclear stress granules. J Cell Sci 117:4551–4558 10.1242/jcs.01329 [DOI] [PubMed] [Google Scholar]
- 31.Ugarkovic D (2005) Functional elements residing within satellite DNAs. EMBO Rep 6:1035–1039 10.1038/sj.embor.7400558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jolly C, Metz A, Govin J, Vigneron M, Turner BM, Khochbin S, Vourc’h C (2004) Stress-induced transcription of satellite III repeats. J Cell Biol 164:25–33 10.1083/jcb.200306104 [DOI] [PMC free article] [PubMed] [Google Scholar]