Abstract
Craniofacial abnormality is one of the major clinical manifestations of Smith-Magenis syndrome (SMS). Previous analyses in a mixed genetic background of several SMS mouse models—including Df(11)17/+ and Df(11)17-1/+, which have 2-Mb and 590-kb deletions, respectively, and Rai1−/+—revealed that the penetrance of the craniofacial phenotype appears to be influenced by deletion size and genetic background. We generated an additional strain with a 1-Mb deletion intermediate in size between the two described above. Remarkably, the penetrance of its craniofacial anomalies in the mixed background was between those of Df(11)17 and Df(11)17-1. We further analyzed the deletion mutations and the Rai1−/+ allele in a pure C57BL/6 background, to control for nonlinked modifier loci. The penetrance of the craniofacial anomalies was markedly increased for all the strains in comparison with the mixed background. Mice with Df(11)17 and Df(11)17-1 deletions had a similar penetrance, suggesting that penetrance may be less influenced by deletion size, whereas that of Rai1−/+ mice was significantly lower than that of the deletion strains. We hypothesize that potential trans-regulatory sequence(s) or gene(s) that reside within the 590-kb genomic interval surrounding Rai1 are the major modifying genetic element(s) affecting the craniofacial penetrance. Moreover, we confirmed the influence of genetic background and different deletion sizes on the phenotype. The complicated control of the penetrance for one phenotype in SMS mouse models provides tools to elucidate molecular mechanisms for penetrance and clearly shows that a null allele caused by chromosomal deletion can have different phenotypic consequences than one caused by gene inactivation.
A characteristic craniofacial phenotype is presented in many genetic syndromes, which enables an initial diagnosis by experienced clinical geneticists and dysmorphologists.1 The major facial features in Smith-Magenis syndrome (SMS [MIM 182290]) include midface hypoplasia, a broad nasal bridge, prognathia, a down-turned mouth, and bulky philtral pillars.2–4 Both anthropometric measurements and three-dimensional facial morphology analyses quantitatively confirmed these features.2,5 The characteristic facial appearance enables discrimination between patients with SMS and unaffected controls and between SMS and other syndromes, with the ocular and nasal regions revealing the most-significant differences.5 These findings are consistent with the abnormalities observed in the SMS mouse models, indicating that studies in the mouse models may reveal insights into the pathogenesis of the SMS craniofacial phenotype, as has recently been suggested for studies of Williams-Beuren syndrome.6
SMS is a multiple congenital anomaly and mental retardation condition due to a heterozygous 3.7-Mb interstitial deletion on chromosome 17p11.2 in the majority (70%–80%) of patients.7–10 Prominent SMS features include developmental delay, mental retardation, craniofacial and skeletal defects, and neurobehavioral anomalies.3,4 Patients with deletions of unusual size have enabled the refinement of the SMS critical region (SMCR) to an ∼1.1-Mb interval.9,10 The identification of frameshift and nonsense mutations in RAI1 (MIM 607642), a retinoic acid–inducible gene that maps within the SMCR, in several patients with SMS without detectable deletions suggests that it is the major gene responsible for the SMS phenotype, through haploinsufficiency.11–14 The variation of phenotypes, even in patients with the same deletion size,8 and the small number of patients with RAI1 point mutations make it challenging to determine whether RAI1 is the only gene responsible for the phenotype.
To take advantage of the high conservation between the human SMS deletion interval and the mouse syntenic region,9,15 several mouse models have been constructed, including Df(11)17, a strain with an ∼2-Mb chromosome-engineered deletion; Df(11)17-1, with a 590-kb deletion; Df(11)17-2 and -3, both with a 595-kb deletion; and Rai1-targeted disruption.16–18 Craniofacial abnormalities—short, concave, and/or curved snouts and a broader distance between the eyes (hypertelorism)—are manifested in each of these mouse models, but the penetrance is a function of the size of the deletion interval, as well as the genetic background. These results indicate that Rai1 is a major gene responsible for the craniofacial phenotype; however, genetic elements inside and outside the Df(11)17-1 deletion also contribute to the penetrance of the phenotype. The molecular mechanistic basis of penetrance is essentially unknown.
Previous analyses16–18 have been performed in F1 and N2 mixed genetic backgrounds. Because of the variations between the littermates in these backgrounds, we sought to control the confounding variable of background strain by making isogenic strains in a pure C57BL/6 background. This enabled systematic evaluation of the effects of chromosome-engineered deletion size on penetrance of the craniofacial phenotype. In addition, we created Df(11)17-4 mice with an ∼1-Mb chromosome-engineered deletion intermediate in size between the largest 2-Mb deletion (Df(11)17) and the smallest 590-kb deletion (Df(11)17-1), to further investigate the effects of deletion size on the penetrance of the craniofacial phenotype. Our results confirmed the influence of the genetic background on the craniofacial phenotype and revealed that the major modifying genetic element(s) for penetrance in the SMS syntenic region is located within the genomic interval included in the Df(11)17-1 deletion; this region includes Rai1 and surrounding sequences. The multiple contributions to the penetrance of the craniofacial phenotype in the SMS mouse models suggest that subtle changes in Rai1 expression levels and/or developmental timing of expression may underlie penetrance. Such studies will not only enable better understanding of phenotypic variation in patients with SMS but may also elucidate the mechanisms of penetrance.
Detailed methods for surface three-dimensional craniofacial scanning have been described elsewhere.17 Mice were anesthetized using Avertin (Sigma), their fur was painted with a mixture of cornstarch and water to create a white, reflective surface for scanning, and they were scanned on a Cyberware Desktop 3D Scanner. Once scanned, the files were edited and converted to .3ds extensions with the use of the Cyberware Mtool software. The .3ds files were opened on free VIScam Solid Viewer software (Marcam Engineering), facial anatomical landmark points were identified and marked, and their three-dimensional coordinates were recorded.
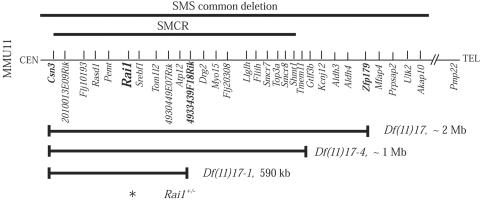
The Df(11)17-4 deletion was created by a retrovirus-mediated chromosome-engineering method used similarly to construct Df(11)17-1.17,19 After characterization with FISH and Southern analysis for detection of the proviral-host junction fragment (data not shown), virus-insertion-site amplification PCR20 was performed to obtain the precise genomic coordinates for the virus insertion site. The sequence was subjected to BLAST analysis against the mouse genomic sequence (National Center for Biotechnology Information [NCBI] mouse genome resources), which located the virus insertion site between the Gtlf3b and Tmem11 genes (fig. 1). The insertion site was ∼1 Mb telomeric to Csn3, which was the starting point for all our deletions.
Figure 1. .
Mouse region on chromosome 11 (MMU11) syntenic to the human SMS deletion interval. The corresponding human deletion regions are shown at the top (bold lines). Shown below are the regions deleted in Df(11)17, Df(11)17-1, and Df(11)17-4 (bold horizontal lines with vertical lines at ends).
The craniofacial phenotype observed in Df(11)17-4/+ mice at the N2 generation was similar to that observed in Df(11)17/+ and Df(11)17-1/+ animals,16,17 as well as to that seen in animals with targeted disruption of one copy of Rai1 (i.e., Rai1−/+).18 The visual observation was confirmed by quantitative analyses with three-dimensional craniofacial scans and skeleton measurements (data not shown). The penetrance was 59%, which, remarkably, was between that of Df(11)17/+ (70%–80%)17 and Df(11)17-1/+ (48%)17 mice (table 1). Because N2 is a mixed background (25% 129/SvEv and 75% C57BL/6), which conveys variation of the craniofacial phenotype between the littermates, we increased the number of Df(11)17-1/+ mice analyzed. The penetrance increased from the 37% reported elsewhere17 to 48%. The penetrance rate difference between Df(11)17/+ and Df(11)17-1/+ mice was statistically significant, whereas the differences between Df(11)17/+ and Df(11)17-4/+ mice and between Df(11)17-4/+ and Df(11)17-1/+ mice were not. The rate differences between each of the three deletion mice and the Rai1−/+ mice were statistically significant (table 1).
Table 1. .
Penetrance of the Craniofacial Phenotype in Different SMS Mouse Models at the N2 Generation (25% 129SvEvBrd and 75% C57BL/6)
| No. of Mice |
||||
| Straina | Deletion Size |
Examined | With Phenotype |
Penetrance (%) |
| Df(11)17/+b | ∼2 Mb | >50 | … | 70–80 |
| Df(11)17-4/+c | ∼1 Mb | 49 | 29 | 59 |
| Df(11)17-1/+ | ∼590 kb | 54 | 26 | 48 |
| Rai1−/+d | NAe | 56 | 10 | 18 |
All strains were maintained by backcrossing to C57BL/6 Tyrc-Brd (B6) wild-type mice. Animals were treated in compliance with relevant animal welfare policies under a protocol approved by the Baylor Institutional Animal Care and Use Committee. For the penetrance comparison, Fisher's exact test was performed. P values <.05 were considered statistically significant.
Data from Yan et al.17
Genotyping was performed with two pairs of primers: 5′ HPRT 2Ty (tyrosinase end) forward 5′-CTGGGAGAAAACATATTTTGAGAGA-3′ and reverse 5′-TTCCTGTTTGGGGTAGAATGTACT-3′ and 3033-2 5′-TCCCGATCAAGGTCAGGAACA-3′ and B1IN-R 5′-AAAAAGAAGTCAGGGGTTGG-3′.
Data from Bi et al.18
NA = not applicable.
Because of the variability of expression of the craniofacial phenotype between littermates in a mixed background, and to remove the confounding effects of strain background from systematic evaluations of engineered deletion mutations and the Rai1 null allele, we analyzed the SMS mouse models in a relatively pure C57BL/6 background (N6 or N8 generations with >98% B6). The penetrance increased for all pure strains in comparison with the mixed N2 background (table 2). Df(11)17/+ and Df(11)17-1/+ mice had a similar penetrance rate (96% and 100%, respectively), whereas that for the Rai1−/+ mice was significantly lower (64%). The P values for comparison of Rai1−/+ with Df(11)17/+ and Rai1−/+ with Df(11)17-1/+ in a pure C57BL/6 background were both .001.
Table 2. .
Penetrance of the Craniofacial Phenotype in Different SMS Mouse Models in a Pure Isogenic C57BL/6 Background
| No. of Mice |
||||
| Strain Genotype |
Generation | Examined | With Phenotype |
Penetrance (%) |
| Df(11)17/+ | N8 | 49 | 47 | 96 |
| Df(11)17-1/+ | N6 | 30 | 30 | 100 |
| Rai1−/+ | N6 | 28 | 18 | 64 |
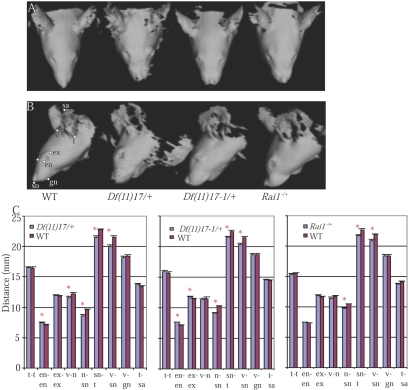
Soft-tissue three-dimensional surface scan analysis provides a rapid, robust, and objective means to assess the craniofacial phenotype without sacrificing the mice.18 The obtained images confirmed our visual observations (fig. 2A and 2B). Several landmarks were selected to obtain quantitative measurements, including the distances between the eyes and the lengths of the snouts (fig. 2B and 2C). Both Df(11)17/+ and Df(11)17-1/+ mice had significantly larger distances between the eyes (i.e., hypertelorism) and shorter snouts (i.e., midface hypoplasia), whereas Rai1−/+ mice had only shorter snouts. This finding persisted even after the exclusion of the Rai1−/+ mice without observable craniofacial phenotype.
Figure 2. .
Three-dimensional craniofacial scan analyses. A and B, Top and side views of the three-dimensional scans of Df(11)17/+, Df(11)17-1/+, and Rai1−/+ mice. C, Statistical analyses of the distances between landmarks. Student’s t test function in Microsoft Excel (Microsoft Office 2001) was used. P values <.05 were considered statistically significant. Mean ± SEM values are presented. Landmarks are as follows: tragion (t), endocanthion (en), exocanthion (ex), vertex (v), subnasale (sn), nasion (n), gnathion (gn), and tip of the ear (sa). WT = wild type; asterisks indicate P<.05. Df(11)17/+ deletion (DEL) n=17, WT n=22; Df(11)17-1/+ DEL n=23, WT n=19; Rai1 −/+ DEL n=28, WT n=17.
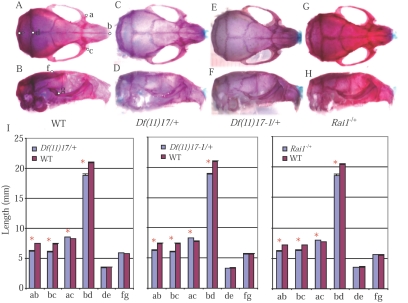
Skeletal analyses during necropsy revealed that the most striking change in the mutants was the deformed nasal bone (fig. 3A–3H). Several distances were measured, including the distance between the anterior notches on the frontal processes and the distance between the nasale and anterior notches on the frontal processes, corresponding to the distance between the eyes and the length of the nasal bone, respectively (fig. 3A and 3B). The measurements of each of these three mutant strains confirmed significant hypertelorism and shorter nasal bones (fig. 3I). Interestingly, by this pathological analysis, the same four measurements differed in a statistically significant way for each of the three isogenic SMS mouse models (fig. 3I). Thus, there was no observable variability of expression among the three different Rai1 haploinsufficiency models (Df(11)17/+, Df(11)17-1/+, and Rai1−/+; the largest and smallest deletion mutations and the knockout allele, respectively), but there was absolutely clear and significantly reduced penetrance in the Rai1−/+ animals.
Figure 3. .
Skeleton analyses. Side and top views of skeleton preparations of a wild-type mouse (A and B), a Df(11)17/+ mouse (C and D), a Df(11)17-1/+ mouse (E and F), and a Rai1−/+ mouse (G and H) are presented. I, Measurement results for Df(11)17/+ mice (DEL n=17, WT n=28), Df(11)17-1/+ mice (DEL n=18, WT n=13), and Rai1 −/+ mice (DEL n=19, WT n=12). Mean ± SEM values are presented. Landmarks used for the measurements are labeled (A and B): a and c = anterior notches on frontal processes situated laterally in relation to the infraorbital fissure, b = nasale, d = intersection of parietal and interparietal bones, e = intersection of interparietal and occipital bones at the midline, f = bregma, and g = intersection of zygoma with zygomatic process of temporal superior aspect.43 WT = wild type; asterisk indicates P<.05. Skeleton preparations were performed as follows. Mice were sacrificed, skinned, eviscerated, and dehydrated in 95% ethanol (EtOH). After the staining of cartilage with 0.05% alcian blue 8GX solution, mice were rinsed in 95% EtOH and were cleared in 2% KOH. Alizarin red (0.015% in 1% KOH) was then used to stain the bones, followed by clearing in 1% KOH and 20% glycerol. Images were obtained with Image Pro Plus software (Media Cybernetics) and were analyzed using Adobe Photoshop version 5.5 (Adobe Systems).
Haploinsufficiency of RAI1 has been suggested to be responsible for the majority of the SMS clinical features, including the craniofacial abnormalities.11,12,21 Rai1 was identified as a gene induced by retinoic acid, which is known to be involved in craniofacial development.22,23 During mouse embryo development, Rai1 is expressed in the craniofacial components derived from branchial arches.18 Furthermore, several lines of evidence indicate that Rai1 is a potential transcription factor containing an extended plant homeodomain zinc finger.12,14 All these observations explain the involvement of Rai1 in craniofacial development. Because of the variation of phenotypes in patients with SMS deletions8 and in those with point mutations,11–14 and because of the ∼50 genes (NCBI human genome resources) present in the common 3.7-Mb deletion, it is challenging to determine whether RAI1 is the only gene responsible for the phenotype. Whether one gene, several contiguous genes, or position effects due to rearrangement contribute to the phenotype is a question pertinent to all microdeletion syndromes.24 With the creation of different mouse models, we were able to demonstrate that genomic regions outside Rai1 apparently contribute to the penetrance of the phenotype or at least to craniofacial abnormalities.16–18 By further analyzing these models in a relatively pure genetic background, we provide apodictic evidence that, within the SMS syntenic region, major modifying genetic element(s) for penetrance of the craniofacial features resides in the Df(11)17-1 deletion, which is an ∼590-kb genomic interval surrounding the 94-kb Rai1 gene with 68 kb of 5′ and 107 bp of 3′ flanking sequences until the next genes, Pemt and Srebf1, respectively (fig. 1).
One possible explanation for the difference in penetrance could relate to the gene content of the smallest 590-kb chromosome-engineered deletion. This 590-kb interval is highly syntenic to the corresponding human region.9,15 There are 11 genes mapped to this region so far. In addition to Rai1, targeted disruptions of Csn3, Rasd1, Pemt, and Srebf1 have been reported.25–28 No heterozygotes of these four strains were reported to manifest an observable craniofacial phenotype, although such a subtle clinical finding might be missed without systematic evaluation, because of the low penetrance rate or the less-severe phenotype. The potential role of the remaining six genes in craniofacial development needs to be further investigated. Nt5m is a mitochondrial deoxyribonucleotidase, which may protect mtDNA replication from overproduction of deoxythymidine triphosphate.29 Flj10193 is a subunit of the multiprotein mediator complex, which is a coactivator for activation of RNA polymerase II transcription.30 Tom1l2, the “target of myb 1–like 2” gene, has been suggested to modulate endosomal functions31 and has been shown to negatively regulate the Src mitogenic signaling induced by platelet-derived growth factor.32 Atp12 is required for assembly of F1-ATPase.33 Nonetheless, chromosome-engineered deletions of different sizes (e.g., the 2-Mb Df(11)17 and the 590-kb Df(11)17-1) (table 2) that have different genes deleted have the same penetrance. Furthermore, the difference in penetrance is observed between the deletion strains and the knockout allele without any variability of expression (table 2 and fig. 3). Thus, if it is a gene in this region that is responsible for the difference in penetrance, this gene must affect the craniofacial development in a very similar way as Rai1. Expression analysis and gene targeting in mice will provide more information about their potential contribution to the SMS phenotype or synergistic effects with Rai1.
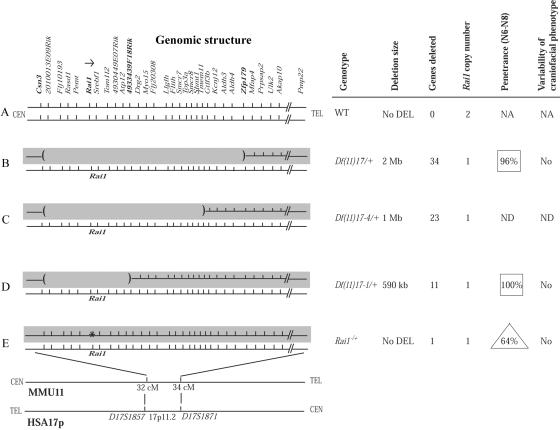
Importantly, all four of our SMS mouse models have one intact wild-type chromosome with one normal Rai1 gene (fig. 4). In isogenic strains, the only genomic differences are either an insertionally inactivated Rai1 gene (fig. 4E) or a complete deletion of one copy of Rai1 and surrounding genomic sequences on the chromosome in trans to the wild-type chromosome (fig. 4B–4D). The craniofacial phenotype results from the remaining wild-type Rai1 gene on the fixed wild-type chromosome, whereas the changes in penetrance most likely relate to the variable changes on the mutated chromosome in trans (fig. 4B–4E). An alternative potential explanation for the reduced penetrance in different SMS models and for the specific observation that Rai1 null alleles caused by deletion can have phenotypic consequences different from a null allele due to gene inactivation is the removal of key control elements for Rai1. Our mouse models have demonstrated that Rai1 is a dosage-sensitive gene,16,18,34 as suggested by human studies involving patients with heterozygous RAI1 point mutations.11,12,14,21 Rai1 was identified as a retinoic acid–induced gene, and a retinoic acid response element was found just upstream of exon 1.22,35 Other than that, the expression control of this gene remains largely unknown.
Figure 4. .
Chromosome-engineered and Rai1-knockout mouse models of SMS. Left, Genomic structure of the critical region on human 17p11.2 and the syntenic interval in mouse. The two chromosome homologues, represented by horizontal lines, are shown for each of the five strains studied, including wild type (WT) (A), three chromosome-engineered deletions with the region deleted demarcated by an absent horizontal line flanked by parenthesis at the break points (B–D), and the Rai1-knockout allele marked by an asterisk (E). Top, Individual genes within the interval examined. The shaded gray background represents the experimentally manipulated chromosome, whereas the fixed or constant chromosome is not shaded. Right, Details of the five strains studied. Note that there is essentially no change in penetrance for the deletion mutants (boxes), whereas there is reduced penetrance (triangle) for the knockout allele. NA = not available. ND = not determined.
Conserved noncoding sequence analysis provides one way to predict potential regulatory modules,36,37 and a regulatory potential (RP) score evaluates the similarity extent to patterns found in alignments of known regulatory elements in comparison to the alignments of neutral DNA.38,39 There is a region ∼8 kb upstream of the mouse Rai1 gene that is highly conserved in human, rat, and chimp and that shows high RP scores (five-way RP: human, chimp, dog, mouse, and rat [UCSC Genome Browser]).
“Position effects” refer to alterations of gene expression due to a change in genomic position and, thus, gain or loss of the regulatory modules acting in cis.40,41 Trans regulation of gene expression is less well studied, particularly in mammalian species. Transvection, the influence of a gene’s expression by the pairing of alleles on homologous chromosomes, is one mechanism for trans regulation.42 As in cis regulation,40,41 the influence of trans regulation is likely to have greater phenotypic consequences and effects in the case of dosage-sensitive genes. The lower penetrance in Rai1−/+ mice could result from the interaction between the remaining control element in the targeted allele and the normal allele, whereas, in deletion mutations, the surrounding sequences are lost. Alternatively, trans-regulatory elements affecting Rai1 expression through other mechanisms could potentially have been removed in different deletions. Different deletion sizes may also influence the phenotype through a position effect by removal or retention of the control element for a phenotype-causing gene located outside the deletions.
The differences in penetrance between two lines with adjacent deletion sizes were small at the N2 generation (Df(11)17/+, 70%−80%; Df(11)17-4/+, 59%; and Df(11)17-1/+, 48%), and there was essentially no difference between Df(11)17/+ (96%) and Df(11)17-1/+ (100%) at the N6 or N8 generations. These latter two deletions differ in the genes deleted—Df(11)17 deletes ∼34 genes, whereas Df(11)17-1 deletes only ∼11—yet they have essentially identical penetrances for the craniofacial phenotype. These findings are most consistent with potential regulatory sequences, rather than the absence of genes included in distinct deletions, being responsible for the penetrance difference. Regardless of genes deleted in the isogenic background, we observed similar penetrance. Reduced penetrance in the isogenic background was observed only for the Rai1 null allele constructed by insertional inactivation and not for deficiency alleles.
This study further narrowed the region for the major modifying genetic element(s) in the penetrance of the craniofacial phenotype in mouse models of SMS. The construction of isogenic strains for the different Rai1 haploinsufficiency mutation models of SMS apparently uncouples variability of expression from penetrance. Our finding of the lack of variability of phenotypic expression between the Rai1−/+ and deletion models in the context of reduced penetrance suggests that the modifying element(s) either functions in a very similar way as Rai1 in craniofacial development or results in subtle changes in dosage or expression of Rai1, perhaps during specific time intervals of development. In each of the four different mutation models studied, the craniofacial phenotype results from the wild-type Rai1 gene on the fixed or constant wild-type chromosome, whereas the penetrance difference is associated with changes—a knockout allele (fig. 4E) versus chromosome-engineered deletions (fig. 4B and 4D)—on the trans-manipulated chromosome. Future studies to elucidate such potential modifying element(s) will enrich our knowledge of not only the molecular mechanism for SMS but also craniofacial development in general and, perhaps, will reveal insights into the molecular basis of penetrance.
Acknowledgments
We thank Dr. E. O’Brian Smith for assistance with the statistical analyses. This work was supported in part by National Institute for Dental and Craniofacial Research grant RO1 DEO15210 (to W.B. and J.R.L.).
Web Resources
The URLs for data presented herein are as follows:
- NCBI, http://www.ncbi.nlm.nih.gov/ (for the mouse genomic sequence and the 3.7-Mb deletion region)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for SMS and RAI1)
- UCSC Genome Browser, http://genome.ucsc.edu/
References
- 1.Jones KL (ed) (2006) Smith’s recognizable patterns of human malformation, 6th ed. W.B. Saunders, Philadelphia [Google Scholar]
- 2.Allanson JE, Greenberg F, Smith AC (1999) The face of Smith-Magenis syndrome: a subjective and objective study. J Med Genet 36:394–397 [PMC free article] [PubMed] [Google Scholar]
- 3.Bi W, Lupski JR. RAI1, the Smith-Magenis and dup(17)(p11.2p11.2) syndromes. In: Epstein CJ, Erickson RP, Wynshaw-Boris A (eds) Inborn errors of development, 2nd ed. Oxford University Press, New York (in press) [Google Scholar]
- 4.Chen K-S, Potocki L, Lupski JR (1996) The Smith-Magenis syndrome [del(17)p11.2]: clinical review and molecular advances. Ment Retard Dev Disabil Res Rev 2:122–129 [DOI] [Google Scholar]
- 5.Hammond P, Hutton TJ, Allanson JE, Buxton B, Campbell LE, Clayton-Smith J, Donnai D, Karmiloff-Smith A, Metcalfe K, Murphy KC, et al (2005) Discriminating power of localized three-dimensional facial morphology. Am J Hum Genet 77:999–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tassabehji M, Hammond P, Karmiloff-Smith A, Thompson P, Thorgeirsson SS, Durkin ME, Popescu NC, Hutton T, Metcalfe K, Rucka A, et al (2005) GTF2IRD1 in craniofacial development of humans and mice. Science 310:1184–1187 10.1126/science.1116142 [DOI] [PubMed] [Google Scholar]
- 7.Chen K-S, Manian P, Koeuth T, Potocki L, Zhao Q, Chinault AC, Lee CC, Lupski JR (1997) Homologous recombination of a flanking repeat gene cluster is a mechanism for a common contiguous gene deletion syndrome. Nat Genet 17:154–163 10.1038/ng1097-154 [DOI] [PubMed] [Google Scholar]
- 8.Potocki L, Shaw CJ, Stankiewicz P, Lupski JR (2003) Variability in clinical phenotype despite common chromosomal deletion in Smith-Magenis syndrome [del(17)(p11.2p11.2)]. Genet Med 5:430–434 [DOI] [PubMed] [Google Scholar]
- 9.Bi W, Yan J, Stankiewicz P, Park S–S, Walz K, Boerkoel CF, Potocki L, Shaffer LG, Devriendt K, Nowaczyk MJM, et al (2002) Genes in a refined Smith-Magenis syndrome critical deletion interval on chromosome 17p11.2 and the syntenic region of the mouse. Genome Res 12:713–728 10.1101/gr.73702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vlangos CN, Yim DK, Elsea SH (2003) Refinement of the Smith-Magenis syndrome critical region to ∼950kb and assessment of 17p11.2 deletions: are all deletions created equally? Mol Genet Metab 79:134–141 10.1016/S1096-7192(03)00048-9 [DOI] [PubMed] [Google Scholar]
- 11.Slager RE, Newton TL, Vlangos CN, Finucane B, Elsea SH (2003) Mutations in RAI1 associated with Smith-Magenis syndrome. Nat Genet 33:466–468 10.1038/ng1126 [DOI] [PubMed] [Google Scholar]
- 12.Bi W, Saifi GM, Shaw CJ, Walz K, Fonseca P, Wilson M, Potocki L, Lupski JR (2004) Mutations of RAI1, a PHD-containing protein, in nondeletion patients with Smith-Magenis syndrome. Hum Genet 115:515–524 10.1007/s00439-004-1187-6 [DOI] [PubMed] [Google Scholar]
- 13.Girirajan S, Vlangos CN, Szomju BB, Edelman E, Trevors CD, Dupuis L, Nezarati M, Bunyan DJ, Elsea SH (2006) Genotype-phenotype correlation in Smith-Magenis syndrome: evidence that multiple genes in 17p11.2 contribute to the clinical spectrum. Genet Med 8:417–427 [DOI] [PubMed] [Google Scholar]
- 14.Bi W, Saifi GM, Girirajan S, Xin S, Szomju B, Firth H, Magenis RE, Potocki L, Elsea SH, Lupski JR (2006) RAI1 point mutations, CAG repeat variation, and SNP analysis in nondeletion Smith-Magenis syndrome. Am J Med Genet A 140:2454–2463 [DOI] [PubMed] [Google Scholar]
- 15.Zody MC, Garber M, Adams DJ, Sharpe T, Harrow J, Lupski JR, Nicholson C, Searle SM, Wilming L, Young SK, et al (2006) DNA sequence of human chromosome 17 and analysis of rearrangement in the human lineage. Nature 440:1045–1049 10.1038/nature04689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Walz K, Caratini-Rivera S, Bi W, Fonseca P, Mansouri DL, Lynch J, Vogel H, Noebels JL, Bradley A, Lupski JR (2003) Modeling del(17)(p11.2p11.2) and dup(17)(p11.2p11.2) contiguous gene syndromes by chromosome engineering in mice: phenotypic consequences of gene dosage imbalance. Mol Cell Biol 23:3646–3655 10.1128/MCB.23.10.3646-3655.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yan J, Keener VW, Bi W, Walz K, Bradley A, Justice MJ, Lupski JR (2004) Reduced penetrance of craniofacial anomalies as a function of deletion size and genetic background in a chromosome engineered partial mouse model for Smith-Magenis syndrome. Hum Mol Genet 13:2613–2624 10.1093/hmg/ddh288 [DOI] [PubMed] [Google Scholar]
- 18.Bi W, Ohyama T, Nakamura H, Yan J, Visvanathan J, Justice MJ, Lupski JR (2005) Inactivation of Rai1 in mice recapitulates phenotypes observed in chromosome engineered mouse models for Smith-Magenis syndrome. Hum Mol Genet 14:983–995 10.1093/hmg/ddi085 [DOI] [PubMed] [Google Scholar]
- 19.Su H, Wang X, Bradley A (2000) Nested chromosomal deletions induced with retroviral vectors in mice. Nat Genet 24:92–95 10.1038/71756 [DOI] [PubMed] [Google Scholar]
- 20.Hansen GM, Skapura D, Justice MJ (2000) Genetic profile of insertion mutations in mouse leukemias and lymphomas. Genome Res 10:237–243 10.1101/gr.10.2.237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Girirajan S, Elsas LJ 2nd, Devriendt K, Elsea SH (2005) RAI1 variations in Smith-Magenis syndrome patients without 17p11.2 deletions. J Med Genet 42:820–828 10.1136/jmg.2005.031211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Imai Y, Suzuki Y, Matsui T, Tohyama M, Wanaka A, Takagi T (1995) Cloning of a retinoic acid-induced gene, GT1, in the embryonal carcinoma cell line P19: neuron-specific expression in the mouse brain. Brain Res Mol Brain Res 31:1–9 10.1016/0169-328X(95)00020-S [DOI] [PubMed] [Google Scholar]
- 23.Padmanabhan R, Ahmed I (1997) Retinoic acid-induced asymmetric craniofacial growth and cleft palate in the TO mouse fetus. Reprod Toxicol 11:843–860 10.1016/S0890-6238(97)00068-3 [DOI] [PubMed] [Google Scholar]
- 24.Shaffer LG, Ledbetter DH, Lupski JR (2001) Molecular cytogenetics of contiguous gene syndromes: mechanisms and consequences of gene dosage imbalance. In: Scriver CR, Beaudet AL, Sly WS, Valle D (eds) The metabolic and molecular bases of inherited disease, 8th edition. McGraw-Hill, New York, pp 1291–1324 [Google Scholar]
- 25.Cheng H-YM, Obrietan K, Cain SW, Lee BY, Agostino PV, Joza NA, Harrington ME, Ralph MR, Penninger JM (2004) Dexras1 potentiates photic and suppresses nonphotic responses of the circadian clock. Neuron 43:715–728 10.1016/j.neuron.2004.08.021 [DOI] [PubMed] [Google Scholar]
- 26.Shimano H, Shimomura I, Hammer RE, Herz J, Goldstein JL, Brown MS, Horton JD (1997) Elevated levels of SREBP-2 and cholesterol synthesis in livers of mice homozygous for a targeted disruption of the SREBP-1 gene. J Clin Invest 100:2115–2124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Walkey CJ, Donohue LR, Bronson R, Agellon LB, Vance DE (1997) Disruption of the murine gene encoding phosphatidylethanolamine N-methyltransferase. Proc Natl Acad Sci USA 94:12880–12885 10.1073/pnas.94.24.12880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yan J, Walz K, Nakamura H, Carattini-Rivera S, Zhao Q, Vogel H, Wei N, Justice MJ, Bradley A, Lupski JR (2003) COP9 signalosome subunit 3 is essential for maintenance of cell proliferation in the mouse embryonic epiblast. Mol Cell Biol 23:6798–6808 10.1128/MCB.23.19.6798-6808.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rampazzo C, Gallinaro L, Milanesi E, Frigimelica E, Reichard P, Bianchi V (2000) A deoxyribonucleotidase in mitochondria: involvement in regulation of dNTP pools and possible link to genetic disease. Proc Natl Acad Sci USA 97:8239–8244 10.1073/pnas.97.15.8239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tomomori-Sato C, Sato S, Parmely TJ, Banks CA, Sorokina I, Florens L, Zybailov B, Washburn MP, Brower CS, Conaway RC, et al (2004) A mammalian mediator subunit that shares properties with Saccharomyces cerevisiae mediator subunit Cse2. J Biol Chem 279:5846–5851 10.1074/jbc.M312523200 [DOI] [PubMed] [Google Scholar]
- 31.Katoh Y, Imakagura H, Futatsumori M, Nakayama K (2006) Recruitment of clathrin onto endosomes by the Tom1-Tollip complex. Biochem Biophys Res Commun 341:143–149 10.1016/j.bbrc.2005.12.156 [DOI] [PubMed] [Google Scholar]
- 32.Franco M, Furstoss O, Simon V, Benistant C, Hong WJ, Roche S (2006) The adaptor protein Tom1L1 is a negative regulator of Src mitogenic signaling induced by growth factors. Mol Cell Biol 26:1932–1947 10.1128/MCB.26.5.1932-1947.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Z-G, White PS, Ackerman SH (2001) Atp11p and Atp12p are assembly factors for the F(1)-ATPase in human mitochondria. J Biol Chem 276:30773–30778 10.1074/jbc.M104133200 [DOI] [PubMed] [Google Scholar]
- 34.Walz K, Paylor R, Yan J, Bi W, Lupski JR (2006) Rai1 duplication cause physical and behavioral phenotypes in a mouse model of dup(17)(p11.2p11.2). J Clin Invest 116:3035–3041 10.1172/JCI28953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Toulouse A, Rochefort D, Roussel J, Joober R, Rouleau GA (2003) Molecular cloning and characterization of human RAI1, a gene associated with schizophrenia. Genomics 82:162–171 10.1016/S0888-7543(03)00101-0 [DOI] [PubMed] [Google Scholar]
- 36.Cooper GM, Sidow A (2003) Genomic regulatory regions: insights from comparative sequence analysis. Curr Opin Genet Dev 13:604–610 10.1016/j.gde.2003.10.001 [DOI] [PubMed] [Google Scholar]
- 37.Frazer KA, Elnitski L, Church DM, Dubchak I, Hardison RC (2003) Cross-species sequence comparisons: a review of methods and available resources. Genome Res 13:1–12 10.1101/gr.222003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.King DC, Taylor J, Elnitski L, Chiaromonte F, Miller W, Hardison RC (2005) Evaluation of regulatory potential and conservation scores for detecting cis-regulatory modules in aligned mammalian genome sequences. Genome Res 15:1051–1060 10.1101/gr.3642605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kolbe D, Taylor J, Elnitski L, Eswara P, Li J, Miller W, Hardison R, Chiaromonte F (2004) Regulatory potential scores from genome-wide three-way alignments of human, mouse, and rat. Genome Res 14:700–707 10.1101/gr.1976004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kleinjan D-J, van Heyningen V (1998) Position effect in human genetic disease. Hum Mol Genet 7:1611–1618 10.1093/hmg/7.10.1611 [DOI] [PubMed] [Google Scholar]
- 41.Kleinjan DA, van Heyningen V (2005) Long-range control of gene expression: emerging mechanisms and disruption in disease. Am J Hum Genet 76:8–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Duncan IW (2002) Transvection effects in Drosophila. Annu Rev Genet 36:521–556 10.1146/annurev.genet.36.060402.100441 [DOI] [PubMed] [Google Scholar]
- 43.Richtsmeier JT, Baxter LL, Reeves RH (2000) Parallels of craniofacial maldevelopment in Down syndrome and Ts65Dn mice. Dev Dyn 217:137–145 [DOI] [PubMed] [Google Scholar]