Abstract
The alcohol dehydrogenase (ADH) family of enzymes catalyzes the reversible oxidation of alcohol to acetaldehyde. Seven ADH genes exist in a segment of ∼370 kb on 4q21. Products of the three class I ADH genes that share 95% sequence identity are believed to play the major role in the first step of ethanol metabolism. Because the common belief that selection has operated at the ADH1B*47His allele in East Asian populations lacks direct biological or statistical evidence, we used genomic data to test the hypothesis. Data consisted of 54 single-nucleotide polymorphisms (SNPs) across the ADH clusters in a global sampling of 42 populations. Both the Fst statistic and the long-range haplotype (LRH) test provided positive evidence of selection in several East Asian populations. The ADH1B Arg47His functional polymorphism has the highest Fst of the 54 SNPs in the ADH cluster, and it is significantly above the mean Fst of 382 presumably neutral sites tested on the same 42 population samples. The LRH test that uses cores including that site and extending on both sides also gives significant evidence of positive selection in some East Asian populations for a specific haplotype carrying the ADH1B*47His allele. Interestingly, this haplotype is present at a high frequency in only some East Asian populations, whereas the specific allele also exists in other East Asian populations and in the Near East and Europe but does not show evidence of selection with use of the LRH test. Although the ADH1B*47His allele conveys a well-confirmed protection against alcoholism, that modern phenotypic manifestation does not easily translate into a positive selective force, and the nature of that selective force, in the past and/or currently, remains speculative.
The metabolism of alcohol can significantly influence human drinking behaviors and the development of alcoholism (also called “alcohol dependence” [MIM %103780]), alcohol use disorder, and other alcohol-induced organ damage.1 Most ethanol digestion occurs through a two-step oxidation: alcohol to acetaldehyde and acetaldehyde to acetate. These steps are catalyzed mainly by alcohol dehydrogenase and acetaldehyde dehydrogenase 2, respectively. Various geographic regions have different frequencies for the genetic polymorphisms in the genes (ADH1B [MIM %103720], ADH1C [MIM %103730], and ALDH2 [MIM %100650]) for the primary enzymes.2–6
Alcoholism is a multifactorial disorder. It has been clear for some time that ADH variants common in East Asia affect the risk of developing alcoholism.7–12 Two genomewide linkage studies—Collaborative Studies on Genetics of Alcoholism of populations of European ancestry13,14 and National Institute on Alcohol Abuse and Alcoholism studies of Amerindian populations15—both support the genetic linkage between alcoholism and a region on chromosome 4 encompassing the ADH genes.
Human ADH genes are located in an ∼370-kb cluster on the long arm of chromosome 4. On the basis of gene expression and sequence alignment, the seven distinct genes relate to the five enzyme classes as follows: class I, ADH1A (MIM %103700), ADH1B, and ADH1C; class II, ADH4 (MIM %103740); class III, ADH5 (MIM %103710); class IV, ADH7 (MIM %600086); and class V, ADH6 (MIM %103735). The three class I ADH genes are closely clustered on an ∼77-kb region on chromosome 4 (4q21), flanked upstream (telomeric) ∼60 kb by ADH7 and ∼50 kb downstream (centromeric) by ADH6 (fig. 1).
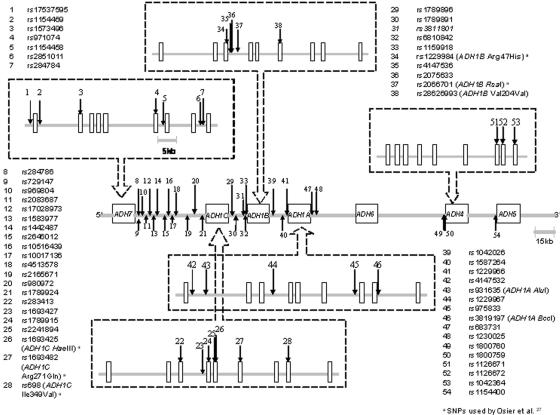
Figure 1. .
Map of the 54 SNPs that cover the ADH7, ADH1C, ADH1B, ADH1A, and ADH4 genes on chromosome 4. SNPs within each locus are shown in an enlarged box with segmented border, whereas SNPs in intergenic regions are listed beside the chromosome segment. The different scales of distance measurement are shown. SNPs are numbered as mentioned in the text.
The protective effect against alcoholism of the ADH1B*47His (previously named “ADH2*2”) allele in East Asian populations is one of the most studied and confirmed associations of a genetic polymorphism and a complex behavior.16 In fact, three functional polymorphisms at class I ADH genes—ADH1B Arg47His and ADH1C Arg271Gln and Ile349Val—are in strong linkage disequilibrium (LD),17 and the variants ADH1B*47His and ADH1C*271Gln&349Val (previously named “ADH3*2”) produce enzymes with higher Vmax enzyme activity for alcohol oxidation. The haplotype with these three variants shows higher frequency in nonalcoholics than in alcoholics in many East Asian populations, including Han Chinese,4,7,17,18 Japanese,19,20 and Koreans,10 making it difficult to attribute the effect to any single site. In addition, the evidence that supports the protective role of ADH1B*47His is not limited to East Asian populations; it has been extended to European,21 Jewish,22 and European Australian23 populations, in which it is much less frequent than in East Asian populations. The observed protective effect of the ADH1C*349Ile allele is attributable to its strong LD with the ADH1B*47His allele in East Asian populations9,17 but appears to have an association with alcoholism in other populations in the absence of the ADH1B*47His allele.24–26
There has been a general belief that selection has operated on these alleles,27–31 because (1) mutations affecting the two sequential steps in ethanol metabolism are both common only in East Asia, and (2) the mutations have a reinforcing effect of increased acetaldehyde that is believed to be responsible for the flushing response to ethanol intake. Although suggestive, this evidence is hardly proof that selection has operated in East Asia.
Osier et al.27 studied nine SNPs in the class I region of ADH. To provide a better understanding of both genetic diversity and the nature of LD in the class I ADH cluster, we examined additional populations for the SNPs they studied (fig. 1) and additional SNPs in all populations. We genotyped individuals from 42 global populations, for a total of 54 SNPs across most of the ADH region.
Positive selection can leave various detectable signatures in the genome. As many have argued,6,32–34 an unusually high Fst can be the signature of selection operating in one region of the world. The LRH test determines whether there has been a rapid rise in haplotype frequency, which can also be a signature of evolutionarily recent positive selection.35 Given the general belief that the Arg47His polymorphism in the ADH1B polypeptide has been the subject of selection in East Asia, we applied both tests to our data, to search for a genomic signature of selection at ADH1B, with a focus on that polymorphism and East Asian populations. The analyses of these data provide strong evidence of selection, but the selective force is still not known.
Material and Methods
Population Samples
We typed 2,250 individuals from a global sample of 42 populations. According to population ancestry and geographic locations, these 42 populations are categorized into nine groups: 9 African, 3 southwest Asian, 9 European, 2 northwestern Asian, 8 East Asian, 2 Pacific Island, 1 eastern Siberian, 4 North American, and 4 South American. The naming convention and geographic categorization of these populations is shown in table 1. Sample descriptions and sample sizes can be found in the Allele Frequency Database (ALFRED) by searching on the population names.
Table 1. .
Naming Conventions and Geographic Categorization of 42 Populations
| Group | Population(s) in Each Group |
| Africa | Biaka (BIA), Mbuti (MBU), Yoruba (YOR), Ibo, Hausa (HAS), Chagga (CGA), Masai (MAS), Ethiopian Jews (ETJ), and African American (AAM) |
| Southwestern Asia | Yemenite Jews (YMJ), Druze (DRU), and Samaritans (SAM) |
| Europe | Adygei (ADY), Chuvash (CHV), Russians from Archangelsk (RUA), Russians from Vologda (RUV), Ashkenazi Jews (ASH), Finns (FIN), Danes (DAN), Irish (IRI), and European Americans (EAM) |
| Northwestern Asia | Komi Zyriane (KMZ), and Khanty (KTY) |
| East Asia | Chinese from San Francisco (CHS), Chinese from Taiwan (CHT), Hakka (HKA), Koreans (KOR), Japanese (JPN), Ami, Atayal (ATL), and Cambodians (CBD) |
| Pacific Islands | Nasioi (NAS) and Micronesians (MIC) |
| Siberia | Yakut (YAK) |
| North America | Cheyenne (CHY), Pima-Arizona (PMA), Pima-Mexico (PMM), and Maya (MAY) |
| South America | Quechua (QUE), Ticuna (TIC), Rondonian Surui (SUR), and Karitiana (KAR) |
DNA samples were extracted from lymphoblastoid cell lines that have been established and/or grown in the Yale University laboratory of J.R.K. and K.K.K. The methods of transformation, cell culture, and DNA purification have been described elsewhere.36,37 For most samples, all volunteers were apparently normal and healthy, with no diagnosis of alcoholism or related disorders. Taiwan samples contained some subjects with an alcoholism diagnosis.17 All samples were collected after receipt of appropriate informed consent and relevant institutional review board approval.
Polymorphic Sites
The 54 SNPs studied extend across ∼350 kb and cover five ADH genes (ADH7, ADH1C, ADH1B, ADH1A, and ADH4) and the intergenic regions. We selected those 54 SNPs on the basis of two major criteria: (1) they have sufficient (⩾0.1) heterozygosities to be used as informative DNA markers for our haplotype analyses, and (2) the SNP density can reach at least 1 SNP per 6 kb, especially in and around the class I ADH cluster. We obtained the information for most of the 54 SNPs from the dbSNP database and the UCSC Genome Browser. Ten SNPs came from the Applied Biosystems (ABI) TaqMan Drug Metabolism Enzyme genotyping assays, and four SNPs in the class I ADH cluster—ADH1B Arg47His (rs1229984), ADH1B RsaI (rs2066701), ADH1C Ile349Val (rs698), and ADH1C HaeIII (rs1693425)—were already included in the earlier studies in our laboratory.27 The dbSNP numbers, the ALFRED numbers, and relative locations of all SNPs are listed in table 2 and figure 1. TaqMan was the main genotyping method, with a small subset of SNPs genotyped with fluorescence polarization (FP) and PCR-based RFLP methods, as noted; some were from the series of Drug Metabolism Assays from ABI. SNP heterozygosity in all populations was checked using HAPLOT.38 See appendix A for details of marker-typing information.
Table 2. .
Spacing, Typing Method, and Reference Numbers for Allele Frequencies for the 54 SNPs in the ADH Region
| SNPa | dbSNP Number | Distance to Next SNP (bp) |
TaqMan Assay Number or Typing Method | ALFRED Number |
| 1 | rs17537595 | 286 | E_ADH7TATA | SI001742O |
| 2 | rs1154469 | 6,510 | C_8934015 | SI000879Y |
| 3 | rs1573496 | 7,808 | E_rs1573496 | S1001660N |
| 4 | rs971074 | 2,265 | C_11942306 | SI000881R |
| 5 | rs1154458 | 3,613 | RFLP | SI000231G |
| 6 | rs2851011 | 109 | C_16129902 | SI001204H |
| 7 | rs284784 | 1,897 | C_1492617 | SI000878X |
| 8 | rs284786 | 710 | C_714911 | SI000877W |
| 9 | rs729147 | 3,574 | FP | SI001207K |
| 10 | rs969804 | 2,356 | C_8933988 | SI001466R |
| 11 | rs2083687 | 4,551 | C_11349421 | S1001661O |
| 12 | rs17028973 | 3,024 | E_rs17028973 | S1001662P |
| 13 | rs1583977 | 5,224 | E_rs1583977 | SI001741N |
| 14 | rs1442487 | 6,750 | E_rs1442487 | S1001663Q |
| 15 | rs2646012 | 4,630 | E_rs2646012 | S1001664R |
| 16 | rs10516439 | 4,477 | C_11349382 | S1001665S |
| 17 | rs10017136 | 3,266 | E_rs10017136 | S1001666T |
| 18 | rs4513578 | 10,267 | E_rs4513578 | S1001667U |
| 19 | rs2165671 | 5,874 | E_rs2165671 | S1001669W |
| 20 | rs980972 | 4,988 | C_2688547 | SI001465Q |
| 21 | rs1789924 | 6,096 | C_2688538 | S1001670O |
| 22 | rs283413 | 1,363 | C_26457440 | SI001429Q |
| 23 | rs1693427 | 456 | C_2688511 | SI001273N |
| 24 | rs1789915 | 238 | C_2688509 | SI001435N |
| 25 | rs2241894 | 21 | C_2688508 | SI001440J |
| 26 | rs1693425 | 2,147 | RFLP | SI000227L |
| 27 | rs1693482 | 3,176 | RFLP | SI000735P |
| 28 | rs698 | 3,805 | RFLP | SI000228M |
| 29 | rs1789896 | 6,565 | C_2688487 | SI001464P |
| 30 | rs1789891 | 6,100 | C_8829540 | S1001671P |
| 31 | rs3811801 | 874 | C_27519856 | S1001672Q |
| 32 | rs6810842 | 436 | E_rs6810842 | S1001673R |
| 33 | rs1159918 | 3,690 | C_2688471 | SI001212G |
| 34 | rs1229984 | 207 | RFLP | SI000229N |
| 35 | rs4147536 | 114 | E_rs4147536 | S1001674S |
| 36 | rs2075633 | 585 | E_rs2075633 | S1001675T |
| 37 | rs2066701 | 3,219 | RFLP | SI000002C |
| 38 | rs2862993 | 6,728 | C_25939834 | SI001451L |
| 39 | rs1042026 | 10,683 | C_2688455 | S1001676U |
| 40 | rs1587264 | 4,350 | E_rs1587264 | S1001677V |
| 41 | rs1229966 | 1,647 | C_8829451 | SI001272M |
| 42 | rs4147532 | 939 | E_rs4147532 | S1001678W |
| 43 | rs931635 | 3,269 | RFLP | SI000737R |
| 44 | rs1229967 | 5,839 | E_rs1229967 | S1001679X |
| 45 | rs975833 | 1,230 | C_2688428 | SI001271L |
| 46 | rs3819197 | 5,236 | FP | SI000738S |
| 47 | rs683731 | 8,897 | C_2688425 | S1001680P |
| 48 | rs1230025 | 120,750 | C_8829387 | S1001681Q |
| 49 | rs1800760 | 117 | C_276457248 | SI001443M |
| 50 | rs1800759 | 17,095 | C_8829281 | SI001444N |
| 51 | rs1126671 | 602 | C_11941799 | SI001447Q |
| 52 | rs1126672 | 2,238 | C_11941798 | SI001448R |
| 53 | rs1042364 | 35,564 | C_9523707 | SI001450K |
| 54 | rs1154400 | … | C_11349123 | SI001449S |
As numbered in figure 1.
Ancestral Allele Inference
The ancestral states of several sites were described elsewhere.27,41 We determined the ancestral states of the remaining SNPs by using the same methods, same primers, or TaqMan probes to genotype genomic DNA for nonhuman primates: three chimpanzees (Pan troglodytes), three gibbons (Hylobates), three gorillas (Gorilla gorilla), three orangutans (Pongo pygmaeus), and three bonobos (Pan paniscus).
Statistical Analyses
Genotypes and allele frequencies for each individual site were calculated by direct gene counting, under the assumption of codominant inheritance. Fst values, as
 |
were calculated with the program DISTANCE. Maximum-likelihood estimates of haplotype frequencies were calculated from the individual multisite typing results of individuals in each population, with the program HAPLO.42
We examined the extended haplotype homozygosity (EHH)35 and relative EHH (REHH)35 for all 54 SNPs, using two core regions defined by the ADH1B*47His allele and incorporating SNPs flanking both sides, on the basis of the Fst results. We initially examined the EHH and REHH for all 54 SNPs, using SNPs within the ADH1B gene (SNPs 34–38 in fig. 1) as the core region. On the basis of those results and the Fst results, we then examined the upstream region as the core (SNPs 31–34). EHH is defined as the probability that two randomly chosen chromosomes carrying a tested core haplotype are homozygous at all SNPs for the entire interval from the core region to the distance x. REHH is defined as the ratio of the EHH of the tested core haplotype to the EHH of the grouped set of core haplotypes at the region not including the tested core haplotype.35
To determine whether the high REHH observed in East Asian populations is significant, simulations with different parameter inputs were performed to provide reference REHH values. The three models for simulations were a population that experienced a bottleneck and a sudden expansion, a population that experienced a bottleneck and an exponential growth, and a population with a constant size. The schemas for the simulations are illustrated in figure 2. After simulated REHH data points had been obtained, they were categorized into 20 bins on the basis of the haplotype frequency, and then 50th, 75th, and 95th percentile lines were drawn. Observed REHH values were thus compared with percentile lines for evidence of selection. P values were obtained by first binning the simulated data into 20 bins on the basis of the core-haplotype frequency, then by log-transforming the REHH values in each bin to approach normality, and then by calculating the mean and SD. Observed values <.05 were considered significant.
Figure 2. .
Flow charts illustrating the demographic model used for the simulations. Top, Population constant at size 10,000 until it experienced a brief bottleneck 3,000 generations ago, which dropped the population size to 2,000.29 Then the population was constant at size 2,000 until 500 generations ago (on the basis of the rough estimates that the Neolithic period started 9,000–10,000 years ago in East Asia and that the generation length is 20 years, the upper bound of 500 generations was used in this simulation), when it expanded suddenly by a factor of 50. The Ne (effective population size) for the entire period (3,000 generations) for this model is ∼2,400. Middle, Population constant at size 10,000 until it experienced a brief bottleneck 3,000 generations ago, which dropped the population size to 2,000.29 Then the population was constant at size 2,000 until 500 generations ago (the same estimation as for the first model), when it expanded exponentially to the current size of 100,000. The Ne for the entire period (3,000 generations) for this model is ∼2,300. Bottom, Model of the unlikely demographic of a population with a constant size of 10,000.
Results
Allele Frequency
The allele frequencies for all polymorphisms for all 42 populations are available on the ALFRED Web site and are retrievable with use of the numbers listed in table 2. Of the 42×54=2,268 allele frequencies evaluated, 10.6% were fixed and 5.5% had heterozygosity <0.05. Of the Hardy-Weinberg equilibrium (HWE) tests, 1.7% resulted in a P value of .01–.05, and 0.4% fell below the .01 significance level. These percentages are below the expectation of chance deviation from HWE. To provide an overview of allele-frequency similarity among and within each geographic region, pairwise correlations between different populations were examined. Allele-frequency similarity, as pairwise r2 between populations, was examined for four subsections of the ADH region. Only for the intergenic region upstream of the class I cluster and especially for the class I cluster (at r2>0.90) does one see great similarity among the East Asian populations and distinct differences from populations in all other parts of the world (fig. 3). This supports our focus on the ADH1B locus and the ADH1B*47His allele in East Asian populations.
Figure 3. .
The pairwise comparison of allele frequencies in four ADH subregions among all 42 populations. The color scheme is based on the correlation of allele frequencies between each pair of populations, with bright red representing complete correlation (r2=1) and dark blue representing no correlation (r2=0). Both horizontal and vertical axes represent the same 42 populations in the same order as in figures 5 and 10. Generally speaking, the correlation level among populations within the same geographic location tends to be strong. Occasionally, the strong correlation can extend across geographic regions, such as in the intergenic region ADH7–class I ADH (strong correlation extends through Africa, southwestern Asia, and Europe) and downstream of class I ADH (strong correlation extends through southwestern Asia, Europe, and East Asia). Class I ADH, which is of particular interest to our positive-selection study, shows an allele-frequency correlation pattern that makes East Asian populations distinct from those of the rest of the world. Populations are ordered from Africa (1–9), southwestern Asia (10–12), Europe (13–21), northwestern Asia (22–23), East Asia (24–31), Pacific Islands (32–33), northeastern Siberia (34), North America (35–38), and South America (39–42).
Fst Value
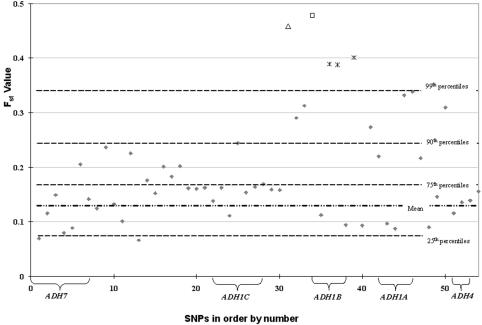
Figure 4 plots the Fst values of the 54 SNPs calculated for the 42 populations. To provide a better context for the different Fst values, we calculated Fst values on the same 42 population samples for 382 presumably neutral sites at other loci not linked to the ADH cluster. This set of 382 sites in the same 42 populations has a mean (SD) Fst value of 0.143 (0.074). The highest Fst value among the 54 ADH region sites was for ADH1B Arg47His (square in fig. 2), which is 4.53 SDs above the mean. The second highest Fst value was observed for the SNP rs3811801 (triangle in fig. 4), which is 4.26 SDs above the mean. Three other highly significant values were observed for three other nearby SNPs—rs2075633, rs2066701, and rs1042026 (asterisk in fig. 4)—on the other side of the Arg47His site.
Figure 4. .
Average Fst values of 42 populations for 54 SNPs, ordered as in table 1 (not to scale). The Mean Fst value of 382 reference sites in 42 populations is represented with a discontinuous dotted line. The 25th, 75th, 90th, and 99th percentiles based on those data are represented with dotted lines. The bracket for each ADH gene includes all SNPs within each gene. SNP 34, ADH1B Arg47His, has the highest Fst value (unblackened square); SNP 31, rs3811801, has the second highest Fst value (unblackened triangle); SNPs 36, 37, and 39, which also have an Fst value >99th percentile, are represented by an asterisk (*).
An East Asian–Prominent Haplotype at ADH1B
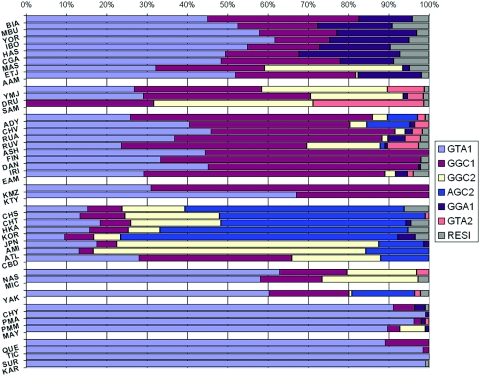
Among the 54 SNPs studied, there are 5 SNPs within the extent of ADH1B: ADH1B Arg47His, rs4147536, rs2075633, rs2066701 (RsaI), and Val204Val (SNPs 34–38 in fig. 1). We analyzed the 5-SNP haplotype-distribution pattern in 42 populations. Of 32 (25) possible haplotypes, 18 were estimated to have nonzero values, and 10 of those 18 haplotypes were observed at a frequency of at least 5% in at least one population in our samples. Those 10 haplotypes account for >96% of all chromosomes in all 42 studied populations. Frequencies for each population are given on the ALFRED Web site and are graphed in figure 5. The ancestral haplotype, based on typing the primates, is 1CA1G (data not shown) and has a frequency of at least 27% in all non–East Asian populations except Samaritans (SAM) (8%) and Micronesians (MIC) (12%). There are two other haplotypes that are frequent in most populations: 1CG2G and 1AA1G. Haplotype 1CG2G is rare in Native American and African populations, but it occurs at ∼20% in Europeans and varies from 3% to 24% in East Asians; haplotype 1AA1G is nearly globally frequent, except in some East Asian populations. Haplotype 2CG2G is the dominant haplotype in East Asia, whereas it is very rarely observed in the rest of the world. Among all eight East Asian populations, haplotype 2CG2G has a minimum frequency of 62%, except in Cambodians (CBD) (34%). Therefore, we consider 2CG2G to be an East Asian–prominent haplotype and our initial focus in LRH analyses.
Figure 5. .
The haplotype pattern of SNPs 34–38 (ADH1B Arg47His, rs4147536, rs2075633, RsaI, and Val204Val) within the ADH1B gene for 42 populations. Populations are grouped by geographic region, with regions roughly in order of distance from Africa: Africa (including AAM), southwestern Asia, Europe, northwestern Asia, East Asia, Pacific, eastern Siberia, North America, and South America. Haplotype 2CG2G is prominent in East Asian populations (except CBD) but is barely seen in the rest of the world (with a few exceptions, such as NAS, MIC, etc.).
However, on the basis of the initial results and the high Fst values extending upstream of ADH1B, we also examined the core haplotype defined by the ADH1B*47His allele and the three SNPs extending upstream to and including rs3811801. The global pattern of this haplotype also shows an East Asian–prominent haplotype (see the “Discussion” section).
EHH and REHH
Initially, we applied the LRH method to study positive selection on the East Asian–prominent haplotype 2CG2G, which includes the functional variant ADH1B*47His (allele 2 at the first site). The primary rationale of the LRH test is that, under the assumption of neutral evolution, common alleles need an extended period to reach high frequencies in the population; as a function of time, the LD surrounding those alleles will decay because of recombination and mutations. But, under positive selection, we can observe a geographic region–specific high-frequency haplotype that has become common over a short period of time, such that recombination has not had sufficient time to break down the selected haplotype. In this study, according to both the correlation results stated above and the criteria proposed by Yu et al.,43 we defined the core region of 4.1 kb at ADH1B on the basis of the five SNPs (SNPs 34–38 in fig. 1) within the ADH1B gene that defined the East Asian–prominent haplotype (see above). Then, we added increasingly distant SNPs, extending 33 SNPs (∼117 kb) upstream to ADH7 and 15 SNPs (∼225 kb) downstream to ADH5, to study the decay of LD from each core haplotype. We plotted the haplotype-bifurcation diagrams35 for eight East Asian populations (fig. 6). In each haplotype-bifurcation diagram, the root stands for a core haplotype. In general, a diagram with thinner and a greater number of branches from the root visualizes the decay of LD on the core haplotype, and a core haplotype under positive selection has long-range LD and high frequency in some populations. So a core under positive selection will be visualized in the diagram with a large root and a predominant thick line that extends a long distance. At a minimum threshold of 7%, the core region of 54 SNPs defined two haplotypes in Atayal (ATL); three haplotypes in Japanese (JPN), Chinese from Taiwan (CHT), Ami, and Hakka (HKA); and four haplotypes in Koreans (KOR), Chinese from San Francisco (CHS), and CBD. Except CBD, in the seven other East Asian populations, haplotype 2CG2G, which includes the proved protective variant ADH1B*47His at the first SNP of this core region, is visualized with an extended predominance of one thick branch in the haplotype-bifurcation diagram, which clearly suggests long-range LD.
Figure 6. .
Haplotype-bifurcation diagrams for each core haplotype with at least 7% frequency at the ADH1B gene region for eight East Asian populations. The core haplotype 2CG2G shows unusual long-range homozygosity in all East Asia populations except CBD.
The EHH and REHH of major core haplotypes (⩾9%) were plotted against the distance away from the core for all the eight populations (fig. 7A). The EHH of the 2CG2G core haplotype (which has the highest frequency in those populations) decays more slowly than does that of other core haplotypes in HKA, JPN, KOR, CHS, and CHT but not in our Ami, ATL, and CBD populations. We also found that CBD, Ami, and ATL differed from the other East Asian populations in allele frequency and haplotype frequency in ALDH2 studies6 and studies of several other loci44 for the same population samples. The EHH (2CG2G as the core haplotype) upstream of the core extends ∼100 kb at a minimal level of 0.6 in JPN and KOR. In CHS and CHT, the EHH upstream of the core maintains a minimal level of only 0.45, for a distance of 80 kb. The EHH of HKA is between 0.45 and 0.6. However, Ami, ATL, and CBD have EHH that barely stays above 0.4 for an extension of 40 kb. Although some core haplotypes other than 2CG2G do show an even higher level of EHH (upstream of the core), the low core-haplotype frequencies put the results in question. The EHH results downstream of the core are less informative, because those rarer core haplotypes have values either higher than or indistinguishable from the those of haplotype 2CG2G, despite the fact that the EHH of the downstream region seems to decrease at a much slower rate and to extend farther than upstream of the core, as seen in figure 5. Obviously, the REHH values of 2CG2G downstream of the core stay around 1 and are not distinguishable from the results of other core haplotypes. The REHH values upstream of the core suggest that the strongest evidence of selection occurs in KOR, because the REHH continues to increase, and it reaches 2.0 at ∼36 kb, reaches 4.0 at ∼80 kb, and hits 8.0 by 100 kb. JPN also show a signature of selection, since the REHH increases to 2.0 quickly and slowly goes up to 4.0 after 80 kb. Although HKA, CHS, and CHT do not show evidence of selection as strong as JPN and KOR, because their REHH values stay at no more than 1.8 for 80 kb, they do show a slow continuous increase of REHH over distance. Compared with the REHH of other core haplotypes, the REHH of 2CG2G is significantly higher in these three populations. Thus, selection could be considered to have operated in these populations. Consistent with the EHH observations, Ami, ATL, and CBD show no signs of selection, since the REHH values of 2CG2G in these three populations barely exceed 1. In addition, several core haplotypes other than 2CG2G show high REHH levels in these three populations. Thus, in Ami, ATL, and CBD, there is no clear evidence that selection operates uniquely on 2CG2G.
Figure 7. .
A, EHH and REHH plots of core haplotypes covering SNPs 34–38 in all eight East Asian populations. The EHH and REHH values are plotted against the physical distance extending both upstream and downstream of the selected core region. Only core haplotypes with frequency >9% are shown. The EHH and REHH curves based on the core haplotype of interest, 2CG2G, are colored and symbolized in different populations, whereas curves of other core haplotypes are presented in gray. JPN and KOR have the highest EHH and REHH values and the longest extension of high levels upstream of the core, whereas CBD has the lowest values and the shortest extension from the core. The low REHH values of the downstream region seem to negate the possibility of selection operating on variation in that direction, despite the corresponding high EHH levels. B, EHH and REHH plots of core haplotypes covering SNPs 34–38 in the pooled five East Asian populations (JPN, KOR, CHS, CHT, and HKA). The region upstream of the core haplotype 2CG2G shows higher EHH levels over distance (compared with the other core haplotypes) and even significantly higher REHH levels.
From previous studies,29,44,45 we know that JPN, KOR, CHS, CHT, and HKA are very similar genetically. Therefore, it might be possible to pool these five populations for analyses.35 The increased sample size of pooled populations would lead to a more robust statistical inference. Thus, we applied the Fisher’s exact test46 to test the similarity of these five populations; they did not differ significantly with respect to core-haplotype frequencies, in agreement with the very small genetic distances among these samples determined on the basis of large numbers of loci.29,44,45 However, neither Ami, ATL, nor CBD shows similarity in core-haplotype components with the above five populations, also in agreement with large genetic distances between these and the other five.29,44,45 Therefore, we could pool only JPN, KOR, CHS, CHT, and HKA for further analysis. Figure 7B shows the EHH and REHH plots of core haplotypes (minimum threshold of 0.09) for the pooled populations. The results definitely show the footprint of positive selection for 2CG2G.
To test further for positive selection within the ADH1B region, for all core haplotypes, we plotted the REHH against their allele frequencies, using the method proposed by Sabeti et al.35 First, we plotted REHH values of each possible core haplotype at the ∼117-kb proximal distance against its allele frequencies for 42 populations and plotted values for the five East Asian populations pooled together (HKA, JPN, KOR, CHS, and CHT) in figure 8A. The REHH of the core haplotype 2CG2G for the pooled five East Asian populations is 7.498, which seems to be an outlier from the distribution of all available data points. To formally test whether our observation is a deviation from evolutionary neutrality, we simulated 1,000 populations under each of three variable neutral assumptions (fig. 2), and we compared the REHH of core haplotype 2CG2G of the pooled five East Asian populations with those simulations in figure 8B. The deviation from the simulation results is highly significant (P values at the 117-kb proximal marker are as follows: for constant-sized population, P=7×10-7; for bottleneck and sudden expansion, P=2.73×10-5; for bottleneck and exponential growth, P=7.64×10-5). The REHH of the core haplotype 2CG2G in those pooled populations is significantly higher than the simulated results at its corresponding haplotype frequency.
Figure 8. .
A, REHH values at the most distant marker, ∼117 kb proximal, plotted against the core haplotype frequencies for 37 populations and the pooled five East Asian populations (HKA, JPN, KOR, CHS, and CHT). The blackened diamond represents the REHH value of core haplotype 2CG2G for the pooled five East Asian populations. B, At the most distant marker, ∼117 kb proximal, REHH values of the pooled five populations and of the simulated data, plotted against the core haplotype frequency. The blackened diamond represents the REHH value of core haplotype 2CG2G for the pooled five East Asian populations, whereas the gray dots are simulated data. The 50th (squares), 75th (*), and 95th (triangles) percentile curves are drawn for visual comparison.
Discussion
Haplotype-Evolution Tree
We observed one East Asian–prominent haplotype for the five SNPs within ADH1B (fig. 5). We are interested, not only in the factors responsible for generating the East Asian–prominent haplotype and whether selection occurred in the ADH1B region, but also in haplotype evolution. We added two additional SNPs (rs6810842 and rs1159918) upstream of ADH1B to the five SNPs within ADH1B, for a total of seven SNPs in a haplotype analysis. Of 128 possible haplotypes, 32 haplotypes were estimated to have nonzero values. Of those 32 haplotypes, 14 were definitely present in at least one individual in our samples (at least one homozygote or one individual heterozygous at only one site), whereas there was strong inferential evidence of the existence of one. We found one East Asian–prominent haplotype, GC2CG2G, in the 42 worldwide human populations. Figure 9 shows a phylogenetic network for eight major haplotypes, with the relative frequencies among the geographic regions for each haplotype. With one exception, all the haplotypes shown (fig. 9) can be explained by sequential accumulation of mutations. That exception, the essentially East Asian–specific haplotype GC2CG2G, requires a crossover of two other haplotypes. One of the nine haplotypes, GC1CG1G, is not common anywhere but is definitely present in some African, European, southwestern Asian, and East Asian samples. Alternatively, the mutations that occurred in the transition from GC1CA1G to GC1CG2G could also be in the other order, first S6 (1→2) then S5 (A→5), but the intermediate haplotype by this order would be GC1CA2G, which was very rare, observed at a frequency of only 1.5%, 0.6%, and 0.9% in Biaka (BIA), African American (AAM), and Pima-Arizona (PMA) populations, respectively. Therefore, this transition was most likely as illustrated in figure 9.
Figure 9. .
Phylogenetic network of eight major haplotypes of seven SNPs for ADH1B. The seven SNPs are rs6810842 (S1), rs1159918 (S2), ADH1B Arg47His (S3), rs4147536 (S4), rs2075633 (S5), RsaI (S6), and Val204Val (S7). All haplotypes in this figure are observed with frequency >5% and are definitely present in at least one individual in our samples. The pie charts represent the haplotypes, and the segments of the pie charts show the proportions of the haplotypes that occurred in each geographic region. This network is started from the ancestral haplotype GA1CA1G. Each arrow represents a single base mutation for the site indicated beside the arrow. The East Asian–specific haplotype GC2GC2G is included in the network; this haplotype was likely generated by recombination between haplotype GC2CA1G, occurring predominantly in southwestern Asia, and haplotype GC1CG2G, occurring much more broadly.
Evidence of Selection at the ADH1B Locus
We initially focused on SNPs within the consensus transcript of the ADH1B gene. As illustrated in the ADH1B 5-SNP haplotype-distribution pattern (fig. 3), we found an East Asian–prominent haplotype, 2CG2G, that includes the functional variant ADH1B*47His, believed to have an effect protective against alcoholism. We conclude that selection is responsible for this haplotype reaching high frequency in East Asian populations, on the basis of two different genomic methods to study selection within the ADH1B region: the Fst statistic and the LRH method.
Among the 54 SNPs, Fst values for only five SNPs are >3 SD above the mean of Fst values of the same 42 populations for 382 presumably neutral sites at other loci: ADH1C-1B intergenic region rs3811801 (0.458), ADH1B Arg47His (0.478), ADH1B rs2075633 (0.389), ADH1B RsaI (0.388), and ADH1B-1A intergenic region rs1042026 (0.401) (fig. 4). The functional variant ADH1B Arg47His has the highest Fst value. Oota et al.6 suggested that selection has operated on the ALDH2 locus and gave evidence of high Fst values (0.30, 0.37, and 0.26) observed for some SNPs around ALDH2. Sakai et al.47 reported a higher Fst value (0.55) in an α-thalassemia polymorphism from Nepal samples and suggested that selection is likely to play a role in allele frequencies at α-thalassemia. Here, we conclude that selection is more likely than genetic drift to be the cause of the high Fst value of ADH1B Arg47His and the high frequency (>60%) of ADH1B*47His in East Asian populations.6,32,34
We also applied the LRH test on the ADH1B region. Three types of results are reported: haplotype-bifurcation diagrams, EHH, and REHH. First, in the ADH1B 5-SNP haplotype-bifurcation diagrams (fig. 6), we observed a large root and a predominant thick line that extends a long distance in East Asians except CBD but not in other geographic groups, which indicates long-range LD uniquely in East Asian populations. Second, in the EHH calculation, the EHH of the East Asian–prominent haplotype 2CG2G tends to decay more slowly than does that of any other identified core haplotypes in five of our eight East Asian samples (HKA, JPN, KOR, CHS, and CHT). This result is consistent with the REHH findings. In the REHH plot, the REHH values of this core haplotype (2CG2G) for the pooled five East Asian populations is statistically distinct from other populations and from our simulation data under neutral assumptions (fig. 8). The REHH value of core haplotype 2CG2G for the pooled five East Asian populations is 7.498 at a frequency of 0.715; the P values of the deviation of this REHH from the simulated data are much more significant than that for the SCA2 haplotype (most common in Utah residents with European ancestry, with ∼39% frequency; REHH ∼13)48 and that for the G6PD haplotype (most common in Africa, with ∼18% frequency; REHH ∼7) (for constant-sized population, P<.0008; for expansion, P<.0006; for bottleneck, P<.0008).35 The latter two loci have been considered to show strong signals of positive selection. We have also done the calculations for the five populations individually and have REHH values ranging from 2.562 to 12.585, with P values all significant, at <.05 (data not shown).
Our observations from the Fst statistic and the LRH test lead to very interesting findings. The SNP rs3811801, which is 5 kb upstream of the functional variant and outside the ADH1B locus, has the second highest Fst value (fig. 4). In addition, the REHH shows a strong increase over distance only upstream of the core we defined in figure 7. Therefore, selection might operate on the upstream part of the gene instead of directly on the core we have selected. Thus, we defined a new core region from rs3811801 to ADH1B*47His. The global haplotype pattern (fig. 10) shows an East Asian–prominent haplotype, AGC2. Within East Asia, five populations (KOR, JPN, CHS, CHT, and HKA) have high frequencies (⩾46%) of this haplotype, whereas Ami, ATL, and CBD have relatively lower frequencies (⩽16%). Outside East Asia, this haplotype occurs only at low frequency in a few populations (Adygei [ADY], Chuvash [CHV], and Ashkenazi Jews [ASH]) or at moderate frequency in one population (Yakut [YAK]). Compared with the haplotype 2CG2G presented in figure 5, this plot illustrates more clearly the difference between the five populations (KOR, JPN, CHS, CHT, and HKA) and the other three (Ami, ATL, and CBD) within East Asia.
Figure 10. .
Haplotype pattern of SNPs 31–34 (rs3811801, rs6810842, rs1159918, and ADH1B Arg47His) for 42 populations. Abbreviations are shown in table 1.
We therefore applied the LRH test to the new core region (fig. 11). Since Ami, ATL, and CBD have a frequency <16% for haplotype AGC2, the occasional high EHH or REHH values obtained in these populations could be misleading. For example, our sample of CBD is 25 individuals. The 11.1% frequency means that there are 2×25×11.1% (≈6) haplotype sequences containing AGC2. Such a small number brings potentially large sampling errors and thus is not very informative. Therefore, we focus on those five populations (KOR, JPN, CHS, CHT, and HKA) that have a minimum frequency of 0.50 for AGC2. The sample sizes in these populations have a range of 41–60 individuals (or 82–120 chromosomes). Thus, the EHH and REHH calculations in these five populations would be reliable and informative. Compared with haplotype 2CG2G (fig. 5), these five populations show higher consistency in EHH shape (we still focus on the upstream of the core). EHH stays at a higher level and extends farther away from the core (all extend 75 kb above 0.8 and 90 kb above 0.6). The REHH result is consistent with previous findings: KOR shows the strongest footprint of selection, and JPN is similar. HKA, CHS, and CHT show relatively weaker evidence of selection (fig. 11).
Figure 11. .
EHH (left) and REHH (right) plots of core haplotypes covering SNPs 31–34 in all eight East Asian populations. EHH and REHH curves based on the core haplotype of interest, AGC2, are colored and symbolized in different populations, whereas curves of other core haplotypes are presented in gray. Because of the low core-haplotype frequencies, CBD, Ami, and ATL show unexpectedly high EHH (even REHH) levels. The other five populations show similar levels of EHH values (upstream) over distance. KOR and JPN show the highest REHH values (upstream), in agreement with the observations from figure 7A.
Although there seems no doubt that selection does have an effect on the ADH1B region in East Asia, the exact location at which the selective force directly operates is debatable. The SNP rs3811801 is suspected to lie in a regulatory region upstream of ADH1B. If this is true, it could be the primary target of the selective force, and certainly the adjacent functional variant would be affected. Alternatively, the two variants may be operating epistatically. This additional upstream SNP appears to modify the evolutionary scheme in figure 7 by adding an additional G→A mutation deriving from the East Asian–prominent recombinant haplotype at the bottom of the figure.
It seems unlikely that the selection was recent and associated with alcoholism, the modern phenotypic manifestation of the polymorphism. Goldman and Enoch28 suggested that the genetic variations in the ALDH and ADH genes were selectively maintained and suggested two plausible selective forces that could predate the invention of brewing: mycotoxins and infectious disease. Mycotoxins, from toxin-producing fungi found in moldy rice, can be converted by the host ALDH enzymes from protoxin to toxin, and the effects of mycotoxins can be further potentiated by ethanol. If the incidence of hepatic disease found in many East Asians, especially JPN, is related to the consumption of mycotoxins, then individuals carrying the deficient ALDH2*2 variant with lowered alcohol consumption would be selectively favored. The infectious agents—some anaerobes and microaerophiles in several bacterial and protozoan diseases—are susceptible to acetaldehyde levels. Individuals with deficient ALDH2*2 can produce high enough levels of acetaldehyde to inhibit the growth of those anaerobes or another parasite, which thereby confers a selective advantage. Similarly, certain functional polymorphisms in ADH enzymes that cause different efficiency in converting ethanol to acetaldehyde could also be protective, but that has not been sufficiently tested. We have applied various simulation models for the potential magnitude of selection—if the simulation assumes a semidominant effect, the selection coefficient has to be at least 0.03 for the allele frequency of the selected allele (ADH1B*47His) to be promoted from 0.005 initially (approximately assumed) to 0.63 currently (based on our genotyping data) within 500 generations; if the simulation assumes dominant or recessive effect, the selection coefficient would be necessarily weaker or stronger than 0.03. However, there are many uncertainties—for example, we have no information on the initial allele frequency, on how strong the actual selection was or on what kind of selection (dominant, semidominant, or recessive) actually occurred.
In conclusion, these data and analyses provide strong genomic evidence that selection has operated on the ADH1B gene in East Asia populations to increase one haplotype of the gene to high frequency. This provides the first strong evidence supporting the prevalent belief that such selection has operated. However, the nature of the selection force and the time period during which it did operate are both unknown. Biological studies to better understand the broader metabolic consequences of the polymorphisms in regulatory and protein coding sequences of ADH1B are needed to determine the nature of the selection. Once the historical demographies of the relevant populations are better understood, more-sophisticated simulations may better define the magnitude of the timing of the selection. Finally, additional molecular data, including STRPs, about additional populations in East Asia are also needed.
Acknowledgments
This work was funded in part by National Institute of Health grants AA09379 and GM57672 (to K.K.K.) and National Science Foundation grant BCS-9912-28 (to J.R.K.). We thank reviewers for their helpful comments for our revision of the manuscript, and we thank Roy Capper, Andrew R. Dyer, and Valeria Ruggeri for their excellent technical help. We also acknowledge and thank the following people who helped assemble the samples from the diverse populations: F. L. Black, L. L. Cavalli-Sforza, K. Dumars, J. Friedlaender, K. Kendler, W. Knowler, F. Oronsaye, J. Parnas, L. Peltonen, L. O. Schulz, D. Upson, E. L. Grigorenko, S. L. B. Kajuna, N. J. Karoma, J. J. Kim, S. Kungulilo, R. B. Lu, A. Odunsi, F. Okonofua, O. V. Zhukova, and K. Weiss. In addition, some of the cell lines were obtained from the National Laboratory for the Genetics of Israeli Populations at Tel Aviv University, and the AAM samples were obtained from the Coriell Institute for Medical Research. Special thanks are due to the many hundreds of individuals who volunteered to give blood samples for studies such as this. Without such participation of individuals from diverse parts of the world, we would be unable to obtain a true picture of the genetic variation in our species.
Appendix A: Marker Typing and Ascertainment
The discovery and typing method for the ADH1C HaeIII site (SNP 26) was described elsewhere.27 ADH1A AluI (SNP 43) was discovered by comparing the sequences of the contig we assembled with published sequences for ADH1A and then confirming that the observed nucleotide difference is a polymorphism by digestion of PCR products from our standard panel with restriction enzyme AluI. Typing primers were generated using flanking sequences from this contig. Our standard panel consists of 10 individuals: 1 Lisongo, 1 BIA, 1 Yoruba, 2 CHT, 1 Dane, 1 Russian, 1 ADY, 1 Cheyenne, and 1 PMA. The ADH1A BccI site (SNP 46) was discovered by resequencing ADH1A intron 8 with use of two primers (A1IN8UP1 and A1IN8DW2) for the 10 individuals in the standard panel. We designed the PCR primers (A1IN8UP1 and A1BccIDW) appropriate for the FP method.39 The program mfold40 predicted a secondary structure that would likely inhibit the primer-extension reaction. Therefore, we introduced an artificial mismatch in the downstream primer to disrupt the secondary structure. We designed detection primer A1BccITUP for the single-nucleotide base extension giving very tight homo- and heterozygote genotype clusters. Sequences of primers noted above are available from the authors. Most markers were typed by TaqMan with use of standard protocols. The TaqMan assay numbers are listed in table 1. Other markers were selected from dbSNP (the AB catalogue), to provide informative coverage across much of the upstream half of the cluster. These markers have diverse historical discoveries that are largely unknown.
The ADH1B Arg47His (SNP 34), ADH1B RsaI (SNP 37), and ADH1C Ile349Val (SNP 28) polymorphisms were typed as described elsewhere.17 For markers not typed by TaqMan, PCR conditions were optimized using gradient PCR in 96-well plates (total volume 25 μl), and amplifications were done in 384-well plates (total volume: 10 μl). The genomic DNA and PCR and restriction enzyme reaction mixtures were dispensed by a TOMTEC Workstation, and the reactions were performed on a PTC-225 Peltier Thermal Cycler (MK Research). The PCR products were digested with appropriate enzymes following the manufacturers’ protocols. The digestion patterns were detected using 2% regular agarose gels. The FP genotyping was read on an LJL BioSystem Analyst. The TaqMan genotyping was read on an ABI PRISM 7900HT Sequence Detection System. We repeated the typing of markers with failed or unclear typings until the proportion of typed individuals was >95% in each population.
Web Resources
The URLs for data presented herein are as follows:
- ALFRED, http://alfred.med.yale.edu/
- dbSNP, http://www.ncbi.nlm.nih.gov/projects/SNP/
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for alcohol dependence, ADH1B, ADH1C, ALDH2, ADH1A, ADH4, ADH5, ADH7, and ADH6)
- UCSC Genome Browser, http://genome.ucsc.edu/cgi-bin/hgGateway
References
- 1.Ramchandani VA, Bosron WF, Li TK (2001) Research advances in ethanol metabolism. Pathol Biol (Paris) 49:676–682 [DOI] [PubMed] [Google Scholar]
- 2.Yoshida A, Hsu LC, Yasunami M (1991) Genetics of human alcohol-metabolizing enzymes. Prog Nucleic Acid Res Mol Biol 40:255–287 [DOI] [PubMed] [Google Scholar]
- 3.Agarwal DP, Goedde HW (1992) Pharmacogenetics of alcohol metabolism and alcoholism. Pharmacogenetics 2:48–62 10.1097/00008571-199204000-00002 [DOI] [PubMed] [Google Scholar]
- 4.Osier MV, Pakstis AJ, Goldman D, Edenberg HJ, Kidd JR, Kidd KK (2002) A proline-threonine substitution in codon 351 of ADH1C is common in Native Americans. Alcohol Clin Exp Res 26:1759–1763 [DOI] [PubMed] [Google Scholar]
- 5.Mulligan CJ, Robin RW, Osier MV, Sambuughin N, Goldfarb LG, Kittles RA, Hesselbrock D, Goldman D, Long JC (2003) Allelic variation at alcohol metabolism genes (ADH1B, ADH1C, ALDH2) and alcohol dependence in an American Indian population. Hum Genet 113:325–336 10.1007/s00439-003-0971-z [DOI] [PubMed] [Google Scholar]
- 6.Oota H, Pakstis AJ, Bonne-Tamir B, Goldman D, Grigorenko E, Kajuna SL, Karoma NJ, Kungulilo S, Lu RB, Odunsi K, et al (2004) The evolution and population genetics of the ALDH2 locus: random genetic drift, selection, and low levels of recombination. Ann Hum Genet 68:93–109 10.1046/j.1529-8817.2003.00060.x [DOI] [PubMed] [Google Scholar]
- 7.Thomasson HR, Edenberg HJ, Crabb DW, Mai XL, Jerome RE, Li TK, Wang SP, Lin YT, Lu RB, Yin SJ (1991) Alcohol and aldehyde dehydrogenase genotypes and alcoholism in Chinese men. Am J Hum Genet 48:677–681 [PMC free article] [PubMed] [Google Scholar]
- 8.Thomasson HR, Crabb DW, Edenberg HJ, Li TK, Hwu HG, Chen CC, Yeh EK, Yin SJ (1994) Low frequency of the ADH2*2 allele among Atayal natives of Taiwan with alcohol use disorders. Alcohol Clin Exp Res 18:640–643 10.1111/j.1530-0277.1994.tb00923.x [DOI] [PubMed] [Google Scholar]
- 9.Chen C-C, Lu R-B, Chen Y-C, Wang M-F, Chang Y-C, Li T-K, Yin S-J (1999) Interaction between the functional polymorphisms of the alcohol-metabolism genes in protection against alcoholism. Am J Hum Genet 65:795–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shen YC, Fan JH, Edenberg HJ, Li TK, Cui YH, Wang YF, Tian CH, Zhou CF, Zhou RL, Wang J, et al (1997) Polymorphism of ADH and ALDH genes among four ethnic groups in China and effects upon the risk for alcoholism. Alcohol Clin Exp Res 21:1272–1277 [PubMed] [Google Scholar]
- 11.Higuchi S, Matsushita S, Murayama M, Takagi S, Hayashida M (1995) Alcohol and aldehyde dehydrogenase polymorphisms and the risk for alcoholism. Am J Psychiatry 152:1219–1221 [DOI] [PubMed] [Google Scholar]
- 12.Muramatsu T, Wang ZC, Fang YR, Hu KB, Yan H, Yamada K, Higuchi S, Harada S, Kono H (1995) Alcohol and aldehyde dehydrogenase genotypes and drinking behavior of Chinese living in Shanghai. Hum Genet 96:151–154 10.1007/BF00207371 [DOI] [PubMed] [Google Scholar]
- 13.Reich T, Edenberg HJ, Goate A, Williams JT, Rice JP, Van Eerdewegh P, Foroud T, Hesselbrock V, Schuckit MA, Bucholz K, et al (1998) Genome-wide search for genes affecting the risk for alcohol dependence. Am J Med Genet 81:207–215 [DOI] [PubMed] [Google Scholar]
- 14.Saccone NL, Kwon JM, Corbett J, Goate A, Rochberg N, Edenberg HJ, Foroud T, Li TK, Begleiter H, Reich T, et al (2000) A genome screen of maximum number of drinks as an alcoholism phenotype. Am J Med Genet 96:632–637 [DOI] [PubMed] [Google Scholar]
- 15.Long JC, Knowler WC, Hanson RL, Robin RW, Urbanek M, Moore E, Bennett PH, Goldman D (1998) Evidence for genetic linkage to alcohol dependence on chromosomes 4 and 11 from an autosome-wide scan in an American Indian population. Am J Med Genet 81:216–221 [DOI] [PubMed] [Google Scholar]
- 16.Uhl GR (2004) Molecular genetic underpinnings of human substance abuse vulnerability: likely contributions to understanding addiction as a mnemonic process. Neuropharmacology Suppl 47 1:140–147 10.1016/j.neuropharm.2004.07.029 [DOI] [PubMed] [Google Scholar]
- 17.Osier M, Pakstis AJ, Kidd JR, Lee J-F, Yin S-J, Ko H-C, Edenberg HJ, Lu R-B, Kidd KK (1999) Linkage disequilibrium at the ADH2 and ADH3 loci and risk of alcoholism. Am J Hum Genet 64:1147–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen WJ, Loh EW, Hsu YP, Chen CC, Yu JM, Cheng AT (1996) Alcohol-metabolising genes and alcoholism among Taiwanese Han men: independent effect of ADH2, ADH3 and ALDH2. Br J Psychiatry 168:762–767 [DOI] [PubMed] [Google Scholar]
- 19.Higuchi S (1994) Polymorphisms of ethanol metabolizing enzyme genes and alcoholism. Alcohol Alcohol Suppl 2:29–34 [PubMed] [Google Scholar]
- 20.Higuchi S, Matsushita S, Imazeki H, Kinoshita T, Takagi S, Kono H (1994) Aldehyde dehydrogenase genotypes in Japanese alcoholics. Lancet 343:741–742 10.1016/S0140-6736(94)91629-2 [DOI] [PubMed] [Google Scholar]
- 21.Borras E, Coutelle C, Rosell A, Fernandez-Muixi F, Broch M, Crosas B, Hjelmqvist L, Lorenzo A, Gutierrez C, Santos M, et al (2000) Genetic polymorphism of alcohol dehydrogenase in Europeans: the ADH2*2 allele decreases the risk for alcoholism and is associated with ADH3*1. Hepatology 31:984–989 10.1053/he.2000.5978 [DOI] [PubMed] [Google Scholar]
- 22.Neumark YD, Friedlander Y, Thomasson HR, Li TK (1998) Association of the ADH2*2 allele with reduced ethanol consumption in Jewish men in Israel: a pilot study. J Stud Alcohol 59:133–139 [DOI] [PubMed] [Google Scholar]
- 23.Whitfield JB, Nightingale BN, Bucholz KK, Madden PA, Heath AC, Martin NG (1998) ADH genotypes and alcohol use and dependence in Europeans. Alcohol Clin Exp Res 22:1463–1469 [PubMed] [Google Scholar]
- 24.Konishi T, Calvillo M, Leng AS, Feng J, Lee T, Lee H, Smith JL, Sial SH, Berman N, French S, et al (2003) The ADH3*2 and CYP2E1 c2 alleles increase the risk of alcoholism in Mexican American men. Exp Mol Pathol 74:183–189 10.1016/S0014-4800(03)00006-6 [DOI] [PubMed] [Google Scholar]
- 25.Tiemersma EW, Wark PA, Ocke MC, Bunschoten A, Otten MH, Kok FJ, Kampman E (2003) Alcohol consumption, alcohol dehydrogenase 3 polymorphism, and colorectal adenomas. Cancer Epidemiol Biomarkers Prev 12:419–425 [PubMed] [Google Scholar]
- 26.Cichoz-Lach H, Partycka J, Nesina I, Celinski K, Slomka M, Wojcierowski J (2006) Genetic polymorphism of alcohol dehydrogenase 3 in alcohol liver cirrhosis and in alcohol chronic pancreatitis. Alcohol Alcohol 41:14–17 [DOI] [PubMed] [Google Scholar]
- 27.Osier MV, Pakstis AJ, Soodyall H, Comas D, Goldman D, Odunsi A, Okonofua F, Parnas J, Schulz LO, Bertranpetit J, et al (2002) A global perspective on genetic variation at the ADH genes reveals unusual patterns of linkage disequilibrium and diversity. Am J Hum Genet 71:84–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goldman D, Enoch MA (1990) Genetic epidemiology of ethanol metabolic enzymes: a role for selection. World Rev Nutr Diet 63:143–160 [DOI] [PubMed] [Google Scholar]
- 29.Kidd KK, Pakstis AJ, Speed WC, Kidd JR (2004) Understanding human DNA sequence variation. J Hered 95:406–420 10.1093/jhered/esh060 [DOI] [PubMed] [Google Scholar]
- 30.Voight BF, Kudaravalli S, Wen X, Pritchard JK (2006) A map of recent positive selection in the human genome. PLoS Biol 4:e72 10.1371/journal.pbio.0040072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Endo T, Ikeo K, Gojobori T (1996) Large-scale search for genes on which positive selection may operate. Mol Biol Evol 13:685–690 [DOI] [PubMed] [Google Scholar]
- 32.Norman PJ, Cook MA, Carey BS, Carrington CV, Verity DH, Hameed K, Ramdath DD, Chandanayingyong D, Leppert M, Stephens HA, et al (2004) SNP haplotypes and allele frequencies show evidence for disruptive and balancing selection in the human leukocyte receptor complex. Immunogenetics 56:225–237 10.1007/s00251-004-0674-1 [DOI] [PubMed] [Google Scholar]
- 33.Hu XS, He F (2005) Background selection and population differentiation. J Theor Biol 235:207–219 10.1016/j.jtbi.2005.01.004 [DOI] [PubMed] [Google Scholar]
- 34.Walsh EC, Sabeti P, Hutcheson HB, Fry B, Schaffner SF, de Bakker PI, Varilly P, Palma AA, Roy J, Cooper R, et al (2006) Searching for signals of evolutionary selection in 168 genes related to immune function. Hum Genet 119:92–102 10.1007/s00439-005-0090-0 [DOI] [PubMed] [Google Scholar]
- 35.Sabeti PC, Reich DE, Higgins JM, Levine HZ, Richter DJ, Schaffner SF, Gabriel SB, Platko JV, Patterson NJ, McDonald GJ, et al (2002) Detecting recent positive selection in the human genome from haplotype structure. Nature 419:832–837 10.1038/nature01140 [DOI] [PubMed] [Google Scholar]
- 36.Anderson MA, Gusella JF (1984) Use of cyclosporin A in establishing Epstein-Barr virus-transformed human lymphoblastoid cell lines. In Vitro 20:856–858 [DOI] [PubMed] [Google Scholar]
- 37.Sambrook J, Fritsch EF, Maniatis T (1989) Quantitation of DNA and RNA. In: Ford N, Nolan C, Ferguson M (eds) Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 38.Gu S, Pakstis AJ, Kidd KK (2005) HAPLOT: a graphical comparison of haplotype blocks, tagSNP sets and SNP variation for multiple populations. Bioinformatics 21:3938–3939 10.1093/bioinformatics/bti649 [DOI] [PubMed] [Google Scholar]
- 39.Chen X, Levine L, Kowk PY (1999) Fluorescence polarization in homogeneous nucleic acid analysis. Genome Res 9:492–498 [PMC free article] [PubMed] [Google Scholar]
- 40.SantaLucia J Jr (1998) A unified view of polymer, dumbbell, and oligonucleotide DNA nearest-neighbor thermodynamics. Proc Natl Acad Sci USA 95:1460–1465 10.1073/pnas.95.4.1460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Han Y, Oota H, Osier MV, Pakstis AJ, Speed WC, Odunsi A, Okonofua F, Kajuna SL, Karoma NJ, Kungulilo S, et al (2005) Considerable haplotype diversity within the 23 kb encompassing the ADH7 gene. Alcohol Clin Exp Res 29:2091–2100 [DOI] [PubMed] [Google Scholar]
- 42.Hawley ME, Kidd KK (1995) HAPLO: a program using the EM algorithm to estimate the frequencies of multi-site haplotypes. J Hered 86:409–411 [DOI] [PubMed] [Google Scholar]
- 43.Yu F, Sabeti PC, Hardenbol P, Fu Q, Fry B, Lu X, Ghose S, Vega R, Perez A, Pasternak S, et al (2005) Positive selection of a pre-expansion CAG repeat of the human SCA2 gene. PLoS Genet 1:e41 10.1371/journal.pgen.0010041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim J, Verdu P, Pakstis AJ, Speed WC, Kidd JR, Kidd KK (2005) Use of autosomal loci for clustering individuals and populations of East Asian origin. Hum Genet 117:511–519 10.1007/s00439-005-1334-8 [DOI] [PubMed] [Google Scholar]
- 45.Tishkoff SA, Kidd KK (2004) Implications of biogeography of human populations for “race” and medicine. Nat Genet Suppl 11 36:S21–S27 10.1038/ng1438 [DOI] [PubMed] [Google Scholar]
- 46.Raymond M, Rousset F (1995) An exact test for population differentiation. Evolution 49:1280–1283 10.2307/2410454 [DOI] [PubMed] [Google Scholar]
- 47.Sakai Y, Kobayashi S, Shibata H, Furuumi H, Endo T, Fucharoen S, Hamano S, Acharya GP, Kawasaki T, Fukumaki Y (2000) Molecular analysis of α-thalassemia in Nepal: correlation with malaria endemicity. J Hum Genet 45:127–132 10.1007/s100380050198 [DOI] [PubMed] [Google Scholar]
- 48.Yu F, Sabeti PC, Hardenbol P, Fu Q, Fry B, Lu X, Ghose S, Vega R, Perez A, Pasternak S, et al (2005) Positive selection of a pre-expansion CAG repeat of the human SCA2 gene. PLoS Genet 1:e41 10.1371/journal.pgen.0010041 [DOI] [PMC free article] [PubMed] [Google Scholar]