Abstract
X-linked dystonia-parkinsonism (XDP) is a movement disorder endemic to the Philippines. The disease locus, DYT3, has been mapped to Xq13.1. In a search for the causative gene, we performed genomic sequencing analysis, followed by expression analysis of XDP brain tissues. We found a disease-specific SVA (short interspersed nuclear element, variable number of tandem repeats, and Alu composite) retrotransposon insertion in an intron of the TATA-binding protein-associated factor 1 gene (TAF1), which encodes the largest component of the TFIID complex, and significantly decreased expression levels of TAF1 and the dopamine receptor D2 gene (DRD2) in the caudate nucleus. We also identified an abnormal pattern of DNA methylation in the retrotransposon in the genome from the patient’s caudate, which could account for decreased expression of TAF1. Our findings suggest that the reduced neuron-specific expression of the TAF1 gene is associated with XDP.
X-linked dystonia-parkinsonism (XDP [MIM314250]) is characterized by severe progressive torsion dystonia followed by parkinsonism.1 Its prevalence is high (5.24 in 100,000) on Panay Island, Philippines.2 Dystonia is a syndrome of sustained muscle contractions causing twisting and repetitive movements or abnormal postures,3 and its pathogenetic basis is still unclear. XDP has a well-defined pathology of extensive neuronal loss and mosaic gliosis in the striatum (caudate nucleus and putamen),4,5 which appears to resemble that in Huntington disease (MIM 143100). Identification of the disease gene of XDP may contribute to the elucidation of the molecular basis underlying not only XDP itself but also other diseases in which basal ganglia show neurodegeneration, such as Huntington disease and Parkinson disease.
A series of linkage analyses has mapped the disease locus, DYT3, to Xq13.1 (fig. 1).6,7 A linkage disequilibrium study narrowed the DYT3 locus to within a 350-kb interval on Xq13.1.8 Subsequently, Nolte et al.9 used PCR-based sequencing and screening analyses to report four SNPs and five disease-specific sequence changes (DSCs) in the “multiple transcript system” (MTS) within 260 kb of the DYT3 region. However, PCR often fails to detect large sequence variants such as transposons. Moreover, the previous study of MTS did not include any brain specimens from patients with XDP. Therefore, the structure and expression of MTS transcripts are still unclear.
Figure 1. .
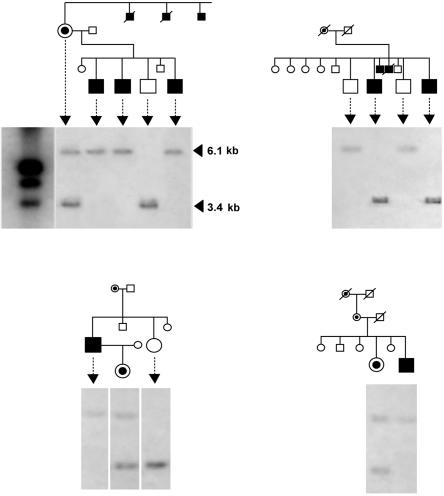
Genomic sequencing analysis of the DYT3 region. a, Physical map of the DYT3 critical region on Xq13.1. Annotated genes in this region that have experimentally verified coding sequences (and their proteins) include NLGN3 (neuroligin 3), GJB1 (gap junction protein, beta-1), ZNF261 (zinc finger protein 261), NONO (non-POU domain–containing octamer-binding protein), ITGB1BP2 (melusin [integrin beta-1 binding protein 2]), TAF1 (TAF-250 [TATA-binding protein-associated factor, 250 kDa]), ING2 (inhibitor of growth 2), OGT (O-linked N-acetylglucosamine transferase), ACRC (acid repeat–containing gene), and CXCR3 (chemokine, CXC motif, receptor 3). The broken arrow indicates MTS. A unique Southern probe was designed for the HindIII-digested fragment containing the SVA retrotransposon insertion. Eight BAC clones comprising a continuous contig map cover 462,651 bp, from 66572462 to 67035112 in NCBI build 30. A total sequence length of 463,567 bp was determined, with 5.7-fold redundancy. Also shown is the distribution of 159 nucleotide variants that were identified by sequencing. None of these variants is located in any exon of these genes, including their alternative splicing forms. A red arrowhead indicates the SVA insertion. b, The SVA insertion in the XDP-related and healthy control populations. Information on the patients with XDP is given in table 1 (patients 1–13). The SVA insertion produces a 6.1-kb fragment, whereas the wild type produces a 3.4-kb fragment. Arrowheads in the gel indicate female individuals. Carriers are defined as mothers or daughters of patients with XDP. Possible carriers are daughters or sons of these carriers. NC = negative control.
We performed the following studies to reveal the disease-causative gene of XDP. To find all disease-specific mutations within the DYT3 region, we first performed genomic sequencing analysis to accurately determine the complete DNA sequence of this region. We also performed detailed expression analysis of the gene in brain specimens obtained from patients with XDP, because the expression of disease genes can be tissue specific. To determine the complete structure of the disease gene, we used a library consisting of “full-length” cDNAs frequently containing their 5′ ends.10
Material and Methods
Subjects and Samples
The study included 67 Filipino individuals (20 affected males) from 16 families residing in Panay. Information on all the patients is listed in table 1. All patients had the disease-specific haplotype between DXS10017 and DXS10018 in the DYT3 region that had been defined elsewhere.8,9 For construction of the BAC contig and portions of the RNA analysis, lymphoblastoid cells were immortalized by infection with the Epstein-Barr virus. This study involved a total of 137 healthy control subjects: 14 unrelated Filipinos, 44 Japanese, 38 African Americans, and 43 European Americans. Materials from the non-Filipino individuals were commercially provided by Coriell Cell Repositories. We used seven postmortem brains from seven male Filipino patients who had XDP. One of the seven brain specimens was frozen, and six were formalin fixed immediately at autopsy. The information on these seven patients with XDP is provided in table 2. We used the frozen brain from a single patient with XDP for long RT-PCR, northern analysis, quantitative RT-PCR, and in situ hybridization. In addition, we used the six formalin-fixed brain specimens for immunohistochemical staining. This study complied with the ethical guidelines of the institutions involved.
Table 1. .
Information on All the Patients with XDP Who Were Studied by Southern Hybridization[Note]
| Signal |
|||||
| Patient Number |
Sample | 6 kb | 3 kb | Age at Onset (years) |
Clinical Description |
| 1 | XD001 | + | − | 31 | Generalized dystonia |
| 2 | XD002 | + | − | 38 | Generalized dystonia |
| 3 | XD011 | + | − | 29 | Muscle atrophy and generalized dystonia |
| 4 | XD013 | + | − | 45 | No atrophy and dystonia |
| 5 | XD015 | + | − | 31 | Limb atrophy and parkinsonism |
| 6 | XD024 | + | − | 41 | Dystonia and parkinsonism |
| 7 | XD027 | + | − | 30 | Dystonia and parkinsonism |
| 8 | XD028 | + | − | 30 | Leg tremor and generalized dystonia |
| 9 | XD033 | + | − | 33 | Parkinsonism (hand tremor) |
| 10 | XD036 | + | − | 50 | Mild dystonia and parkinsonism |
| 11 | XD041 | + | − | 52 | Dystonia and parkinsonism |
| 12 | XD054 | + | − | 30 | Generalized dystonia |
| 13 | XD062 | + | − | 40 | Severe atrophy |
| 14 | XD101 | + | − | 42 | Swelling feet, difficulty walking, and sensory trick |
| 15 | XD103 | + | − | 46 | Dystonia, parkinsonism, difficulty swallowing, and freezing gait |
| 16 | XD111 | + | − | 39 | Dystonia, parkinsonism, and difficulty walking |
| 17 | XD112 | + | − | 33 | Torticollis and parkinsonism |
| 18 | XD115 | + | − | 31 | Jaw-opening dystonia and parkinsonism |
| 19 | XD131 | + | − | 36 | Blephalospasm and writer’s clamp |
| 20 | XD141 | + | − | 40 | Cervical dystonia, axial dystonia, and parkinsonism |
Note.— All patients are male. Plus sign (+) = presence of signal; minus sign (−) = absence of signal.
Table 2. .
Information on All the Patients with XDP Who Were Studied by Expression Analysis[Note]
| Age(years) |
||||||
| Patient Number |
At Onset | At Death | Clinical Description | Cause of Death | Fixation | Examination(s)a |
| 1b | 42 | 52 | Dystonia and parkinsonism | Pneumonia | Frozen; 4% paraformaldehyde | NB, LRT, QRT, ISH |
| 2 | 40 | 54 | Dystonia and parkinsonism | Pneumonia | Formalin | IHC |
| 3 | 23 | 47 | Dystonia and parkinsonism | Pneumonia | Formalin | IHC |
| 4 | 56 | 59 | Dystonia | Suicide | Formalin | IHC |
| 5 | 38 | 52 | Dystonia | Unknown | Formalin | IHC |
| 6 | 35 | 42 | Dystonia | Suicide | Formalin | IHC |
| 7 | 34 | 46 | Dystonia | Pneumonia | Formalin | IHC |
Genome Analysis
We constructed two series of BAC libraries, using genomic DNA from a patient with XDP who was aged 41 years and had generalized torsion dystonia without parkinsonism. Cultured lymphoblastoid cells from a patient with XDP were embedded in agarose plugs, and then high–molecular-weight DNA was partially digested with EcoRI and was size fractionated by pulse-field gel electrophoresis. Size-fractionated DNA was cloned into the CopyControl pCC1BAC vector (Epicentre) and was transformed into DH10B cells by use of an electroporator. The same procedure was repeated with HindIII. We identified BACs covering the DYT3 region by hybridizing 32P-labeled PCR probes to filters generated from these BAC libraries. A shotgun library in pUC118 was constructed from each BAC clone. Cycle sequencing was performed using a BigDye Terminator v3.1 Cycle Sequencing kit (Applied Biosystems) and was analyzed on an ABI 3700 capillary sequencer. The gap clones were sequenced using a GPS-1 Genome Priming System (New England Biolabs). Raw sequence data were analyzed and assembled by ATGC software (Genetyx).
We digested 4 μg of genomic DNA in 300 μl of a standard reaction mixture containing 125 units of HindIII for 3 h at 37°C. For methylation detection, an aliquot of the HindIII-digested DNA was digested under the same conditions with a CpG methylase–sensitive enzyme, HpaII. As a control reaction, an aliquot was also digested with MspI, which is a CpG methylase–insensitive isoschizomer of HpaII. After ethanol precipitation, the digested DNA was loaded onto a 0.8% agarose gel in 0.1× Tris-borate-EDTA buffer for electrophoresis. After denaturation and the subsequent neutralization steps, the DNA was transferred to a positively charged nylon membrane (Roche Diagnostics). The filter was prehybridized for 1 h in DIG Easy Hyb buffer (Roche Diagnostics) and then was hybridized overnight (16–18 h) at 40°C. The PCR probe was amplified using primers XD_probe-F (5′-AGCTTTGCTGCCATTG-3′) and XD_probe-R (5′-AAGACCCTTATTATTCATGAGTG-3′). After washing the filter for 30 min at 68°C in DIG Wash and Block buffer, drops of 1/100-diluted disodium 3-(4-methoxyspiro {1,2-dioxetane-3,2′-(5′-chloro) tricyclo [3.3.1.13,7]decan}-4-yl) phenyl phosphate (CSPD [Roche Diagnostics]) were added and the filter was exposed to x-ray film for 1–5 h.
RNA Isolation
Total RNA was isolated from caudate, cortex, and accumbens of a frozen brain by use of an RNeasy Lipid Tissue Midi kit (QIAGEN) with a DNaseI treatment step, after homogenization with a Polytron PT1300D (Kinematica). Total RNA from lymphoblastoid cell lines was also isolated by the same procedure. RNA sources from six Japanese brains were used as neurologically healthy controls by the same procedure. Also, two commercial human brain RNA sources were used as controls—cerebral cortex total RNA (catalog number 636561) and caudate nucleus total RNA (63566)—along with human tissue total RNA sources from heart (64100), spleen (64093), lung (64092), liver (64099), and thymus (64107), provided by BD Bioscience Clontech, and stomach (735038), provided by Stratagene. The quality and quantity of total RNAs were assessed using an Agilent 2100 Bioanalyzer (Agilent). The 2:1 ratio of 28S:18S rRNA was employed as a threshold for intact RNA. The quantity was confirmed by the RiboGreen RNA fluorescence assay (Molecular Probes).
Northern Analysis
For synthesis of riboprobes, we performed PCR and cloning into the TA-vector (Promega), using the three primer sets in table 3: TA_f_2333 and TA_r_2690 for probe 1, TA_f_4786 and TA_r_5185 for probe 2, and TA_f_5637 and TA_r_6065 for probe 3. Total RNA samples of 10 μg were loaded into a 1% agarose gel denatured by 2% formaldehyde gel. The gel was run in 1× 3[N-Morpholino]propanesulfonic acid buffer. After electrophoresis, the RNA was transferred to a positively charged nylon membrane. After prehybridization for 1 h in DIG Easy Hyb buffer at 68°C, hybridization was performed overnight (16–18 h) at 68°C. The filters were washed twice for 5 min with 100 ml of 2× saline sodium citrate (SSC) and 0.1% SDS at room temperature and then were washed twice for 15 min with 0.1× SSC and 0.1% SDS at 68°C. We employed the standard conditions and procedure provided by Roche Diagnostics.
Table 3. .
PCR Primer Sets for Fragment Amplification from the TAF1 cDNAs[Note]
| Forward Primer |
Reverse Primer |
||||||
| Fragment Name |
Namea | Sequence (5′→3′) |
Tmb | Namea | Sequence (5′→3′) |
Tmb | Amplicon Size (bp) |
| TA01 | TAF_f_3 | GGGAGCTCAGTAFAGTCACTTCTG | 60.6 | TAF_r_400 | CCACCTCATTGATGTCTGAATAG | 59.6 | 398 |
| TA02 | TAF_f_377 | ACTAFTTCAGACATCAATGAGGTGG | 60.4 | TAF_r_772 | TGGCATCATGCTGCATAATC | 61.8 | 396 |
| TA03 | TAF_f_755 | TTAFTGCAGCATGATGCCAC | 60.4 | TAF_r_1175 | CCCAGCATATCATACCACAGTC | 60.4 | 421 |
| TA04 | TAF_f_1152 | CCGACTGTGGTAFTGATAFTGCTG | 61.5 | TAF_r_1550 | CGTCCATATACCAGATCCTCATTG | 62.7 | 399 |
| TA05 | TAF_f_1528 | AATGAGGATCTGGTAFTAFTGGACG | 60.2 | TAF_r_1910 | CGTAATTCCACAGCAGGAATTG | 62.7 | 383 |
| TA06 | TAF_f_1889 | CAATTCCTGCTGTGGAATTAFCG | 62.7 | TAF_r_2273 | CCATATTTACAATCTGGTGCTCC | 60.7 | 385 |
| TA07 | TAF_f_2251 | GGAGCACCAGATTGTAFAATAFTGG | 60.7 | TAF_r_2598 | TTCCATTCGTATCCTCCGTG | 62 | 348 |
| TA08 | TAF_f_2580 | ACGGAGGATAFCGAATGGAAG | 60.1 | TAF_r_2979 | ATCTGCCACCCCAGTCAC | 60.8 | 400 |
| TA09 | TAF_f_2945 | GCAAGTGTCTGCTAFGAGGTGAC | 60.3 | TAF_r_3331 | GTAGGTCAAAGATGCGCTGAC | 61 | 387 |
| TA10 | TAF_f_3311 | GTCAGCGCATCTTTGACCTAFC | 61 | TAF_r_3710 | GTCCGTATGCGCACATAGG | 61.3 | 400 |
| TA11 | TAF_f_3689 | ATGCCTAFTGTGCGCATAFCG | 61.8 | TAF_r_4089 | CAAGACAATTTTGGTCCCTTC | 59.6 | 401 |
| TA12 | TAF_f_4069 | GAAGGGACCAAAATTGTCTTG | 59.6 | TAF_r_4467 | TAGCTCCAGATGCTCTCTGAAC | 59.9 | 399 |
| TA13 | TAF_f_4413 | CGTGCGTAFAACGCCTCTAFC | 60.6 | TAF_r_4808 | CGACTCTGATACTTGTGCTTGG | 61 | 396 |
| TA14 | TAF_f_4780 | AACATCTCCAAGCACAAGTAFTCAG | 60.7 | TAF_r_5164 | GCTTTTCTGGAGTGGCACTG | 62.1 | 385 |
| TA15 | TAF_f_5130 | TGTCTTGGATAFTTCCCAGTGC | 61.1 | TAF_r_5505 | GTTGCTATCCTCCAAACCATG | 60.5 | 376 |
| TA16 | TAF_f_5482 | TCCCATGGTTTGGAGGATAFG | 60.9 | TAF_r_5862 | AAGGCTAAGGTGTTAGTTAATTTCATG | 60.3 | 381 |
| TA17 | TAF_f_5834 | ATCATGAAATTAFACTAFACACCTTAFGCC | 60.1 | TAF_r_6231 | TTAGTAGAGATGCGGTTTCGC | 60.5 | 398 |
| TA18 | TAF_f_6073 | GTAFACTTTAFTGTCCTCTTGATGTAFTTAFGG | 59.1 | TAF_r_6469 | CAGGAGGGCTCTCATCTGC | 62.3 | 397 |
| TA19 | TAF_f_6451 | GCAGATGAGAGCCCTCCTG | 62.3 | TAF_r_6855 | AAGGATCTGATAGAGTGCTTATCATG | 60.2 | 405 |
| TA20 | TAF_f_6831 | ATGATAFAGCACTCTAFTCAGATCCTTG | 60.2 | TAF_r_7164 | CACACTCTGCCATTTCTAGACTG | 60.1 | 334 |
| TA21 | TAF_f_7140 | CACAGTCTAFGAAATGGCAGAGTG | 60.1 | TAF_r_7553 | GGGTTATCATTGTGAACAGTTAGC | 59.9 | 414 |
| TA22 | TAF_f_7277 | GGCAGGAAGTAFTAFTCATCACAAGC | 62.1 | TAF_r_7606 | CCAGCATACATAACAAACACAGAAG | 60.9 | 330 |
| MT23c | TAF_f_7277 | GGCAGGAAGTAFTAFTCATCACAAGC | 62.1 | pMTS-r | GTGAGAGCTCAAAGACCAATAAG | 58.3 | 779 |
| Probe 1 | TA_f_2333 | TGCAAGCATTTGAGAACAACCT | 62 | TA_r_2690 | GAGTCCATCCCTGTGCGTT | 61.1 | 358 |
| Probe 2 | TA_f_4786 | TCCAAGCACAAGTATCAGAGTCG | 61.7 | TA_r_5185 | CTTCACCTTCCTGTGTTACCTGCT | 63 | 400 |
| Probe 3 | TA_f_5637 | GAGCGTACTAAGCCAGGTCCA | 61.7 | TA_r_6065 | TTCATAATTTCCCCTCCTTCCC | 62.4 | 393 |
Note— The PCR mixture contained 2 μl of the first-strand cDNA, 0.2 mM of each dNTP, 1 μM of each primer, 1× GeneAmp PCR buffer, and 2.5 units of AmpliTaq Gold DNA polymerase, in a 50-μl total volume. The PCR conditions were 9 min at 95°C, followed by 35 cycles at 95°C for 45 s, 60°C for 45 s, and 72°C for 60 s.
The value in the third part of the primer name represents position in a major form of the TAF1 transcript.
Tm = annealing temperature (°C). The calculation conditions were 1,000 nM of each primer and 50 mM potassium ions.
The primer set was used only for first-strand cDNA from MTS.
Long RT-PCR Analysis
Long RT-PCR analysis was performed in two steps: (1) first-strand synthesis from RNAs of the control and XDP caudates by long reverse transcription (RT) and (2) fragment PCR by use of the long RT products (i.e., cDNA) as a template. Such a long RT-PCR method is known to be effective for defining the extent of a large transcript.11 The two parts revealed not only the extent of the transcripts but also alternative exons included in the transcripts. Long RT was performed using TAF_r_7621 for the TATA-binding protein-associated factor 1 gene (TAF1 [MIM 313650]) and MTS_r for MTS as the long RT primer.
Quantitative RT-PCR
cDNA was synthesized from the total RNA by use of random hexamers with a TaqMan Reverse Transcription Reagents kit (Applied Biosystems). All the primers and probes are listed in table 4. We also employed a control probe for 18S rRNA (4319413E) and the glyceraldehyde-3-phosphate dehydrogenase gene (GAPDH) (4310884E) and examined a probe for the dopamine receptor D2 gene (DRD2) (Hs00156514_m1). We used a final concentration of 250 nM probe, 900 nM primers, and 50 ng cDNA in a 50-μl reaction volume in a 96-well reaction plate on an ABI PRISM 7000 in accordance with the standard procedure. The conditions of real-time PCR consisted of a holding step for 2 min at 50°C and 1 min at 95°C followed by 50 cycles for 15 s at 95°C and 1 min at 60°C. Quantity was calculated every time by use of a standard curve for each well. All quantitative data normalized by adjustment for 18S rRNA were tested by Smirnov’s test with a 5% significance level. We used Student’s t test to test the difference in means of the expression levels between patients with XDP and healthy control caudate nuclei, after checking the acceptance of the quality of variances between the two groups by the F test.
Table 4. .
Primers and Probes for the TaqMan Expression Assay
| Primer Sequence(5′→3′) |
||||
| Assay Name | Forward | Reverse | Reporter 1 Sequencea (5′→3′) |
Amplicon Size (bp) |
| TAF1-5′ | GACTGACGGTGCCTTGGT | GTCTGAATAGTCCACAGCATCTTCT | ACCCACCCTTCATCATTT | 70 |
| TAF1-3′ | ACCTTATTCTGGCCAACAGTGTT | ACAATCTCCTGGGCAGTCTTAGTAT | ACTCTCAGGTCCATTATAC | 73 |
| TA02-334 | TGGGACCAATGAAGAAGGATAAGGA | ATTCTGAGTCAGAGGAACTACTGAAGT | CCACTTTCTCACCAGTAATAG | 81 |
| TA08-269 | CTCTGCGCTGACTTCAAACG | TTCCTCATTTTCTTCTTCTGGAGCAA | CCATAGCCAGCATCCTGT | 79 |
| TA09-391 | GTGGTGAAGGATTCTCCTATGTGAA | ATCTGTTCCTGTCACTGTCTTCTTC | CAGCAGAAGGTAGGATGATAA | 105 |
| TA09-693 | TGCCAAGCAACTTCTACGTAAATTTG | TCCAACTCTATACAAAGCCCCTAGT | CACACTCACCCTCTTCC | 99 |
| TA14-317 | ACCTTATTCTGGCCAACAGTGTT | CCTCCAAAGCTGCTTCTTTAGCA | TTGAGTCAAATGTTCATCATACATT | 97 |
| TA14-389 | GGAGATTGTGAACGTCTGTTACCA | TCCTTCTCAAGTTGAGTCAAATGTTCAT | CATACTACCTCAGTCAATGTC | 73 |
| TA14-391 | CCCCAGGGCCCTACAC | CTGAGGGATGTGTTGGTATCATACAA | CCTCAGGCTAAGCCTC | 64 |
| TA14-407 | GGGAGAGCTTTCTGGATGATGTAAA | ACAATCTCCTGGGCAGTCTTAGTAT | CTGACTCTCAGGTCCTGCACG | 119 |
| TA15-477 | AGTGCCACTCCAGAAAAGCA | CCTCGCCCAGCCTACCT | CCTGGCGCATCTGTGT | 61 |
| TA18-261 | GTGGCTCACACCTGTAATCTCA | CCTCCCGGGTTCAAGTAATTCTC | CAGCCTCCCAGAGTGC | 67 |
| TA14-385Nb | CCCCAGGGCCCTACAC | CTGAGGGATGTGTTGGTATCATACAA | CCTCAGCCTCCTGATT | 58 |
| TAF1-M | GCTAAAGCTCTGCGCTGACT | TTAAGCACCCACCAGTTTGAGT | CATCCCTGTGCGTTTGA | 60 |
| TAF1-3′N | CCCAGTGCCACTCCAGAAAA | CAGTTCCTTCCTCTTCATCTGCAA | ACCTTCCTGTGTTACCTGC | 82 |
| MTS-V4 | TTGCTCTTGGGCTCTGCATT | CTGGCACGGATTTTCACTATCTT | ACTCCTTACAGGTACCAATGA | 251 |
| MTS-37/1 | CCCATGGTTTGGAGGATAGCA | CATCTCTGAATGGCTTGTTCATTG | TCAGGTGATGAACTTCAATA | 166 |
| MTS-37/3 | CCCATGGTTTGGAGGATAGCA | TCGTTGCTCCAGAGATCTTTGTG | ACATCAGGTACCAATGAAC | 83 |
| MTS-2/3 | GGCCTCCGGCATTGCT | TCGTTGCTCCAGAGATCTTTGTG | TCCACTGTACCAATGAA | 112 |
| MTS-32′/34′ | CAACAGTGTTAAGTATAATGGGTACATGTG | CTGAGGGATGTGTTGGTATCATACAA | TCAGGCTAAGCCTCCT | 268 |
| MTS-3/4 | GTGTCCGGAGTTGGTTCCT | TGGCACGGATTTTCACTATCTTCA | CCTTCGCGCTTCAGGC | 105 |
For the reporter 1 sequences, the dye was FAM.
This probe is designed to detect all isoform sequences not containing exon 34′.
In Situ Hybridization
Synthesis of DIG-labeled riboprobes for TAF1 (probe 3 in fig. 2a), as well as for β-actin (ACTB) and glial fibrillary acidic protein (GFAP) as controls, was performed according to the procedure used for northern analysis. The caudate blocks were fixed with 4% paraformaldehyde. After cryoprotection, serial 20-μm sections were cut in a cryostat. The sections were reacted with alkaline phosphatase–labeled anti-DIG antibody (diluted 1:200 [Roche Diagnostics]) in 1% skim milk in sodium Tris (hydroxymethyl)-aminomethane (NT) buffer. After washing with NT buffer, the positive signals were detected by nitroblue tetrazolium chloride (Roche Diagnostics) and 187 μg/ml 5-bromo-4-chloro-3-indolyl-phosphate (Roche Diagnostics).
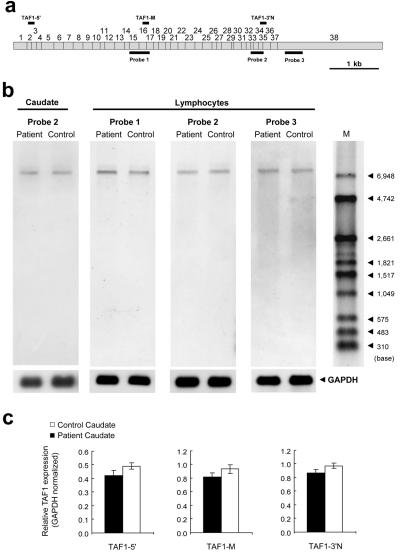
Figure 2. .
Northern analysis. a, Three probes for northern hybridization to TAF1 (detailed information on these probes is given in table 3). b, Total RNA samples from the caudate and lymphoblastoid tissues. The hybridization signal seen at ∼7 kb, which represents TAF1, was observed in every lane, but a signal was never seen at the smaller sizes corresponding to MTS transcripts reported elsewhere. This result suggests that the sequences of TAF1 isoforms have such small differences in size, probably less than a few hundred base pairs, that standard northern analysis cannot discriminate between them in 1% agarose gels denatured by 2% formaldehyde. A probe for GAPDH was also hybridized to each membrane as a loading control. M = DIG-labeled molecular-weight marker. c, Relative TAF1 expression. The hybridization signal seen at ∼7 kb, representing TAF1, had a tendency toward slight reduction in patient caudate, so, to confirm this, TaqMan assays were performed.
Immunohistochemical Staining
The six tissues from six different patients with XDP were fixed in 10% neutral formalin, were sliced, and were embedded in paraffin, and 3-μm sections were cut on a microtome and were mounted on Matsunami adhesive silane–coated glass slides. After routine deparaffinization, rehydration, and blocking of endogenous peroxidase activity, all sections were processed for microwave-enhanced antigen retrieval. The sections were blocked with 3% BSA in PBS (pH 7.2) for 1 h and then were incubated overnight at room temperature in 3% BSA-PBS containing goat polyclonal antibody against TAF1 (diluted 1:5,000 [Santa Cruz]). Rabbit polyclonal antibody against GFAP (Dako) was also used as a control. For the visualization of bound antibodies, we used Histofine Simplestain Max-PO (G) (Nichirei) and the liquid diaminobenzidine (DAB) substrate chromogen system (Dako Cytomation). The sections were further processed for enhancement of the DAB reaction products by use of a Dako Envision Kit (Dako Cytomation).
Full-Length Cloning and Direct Sequencing of the TA14_391 Isoform
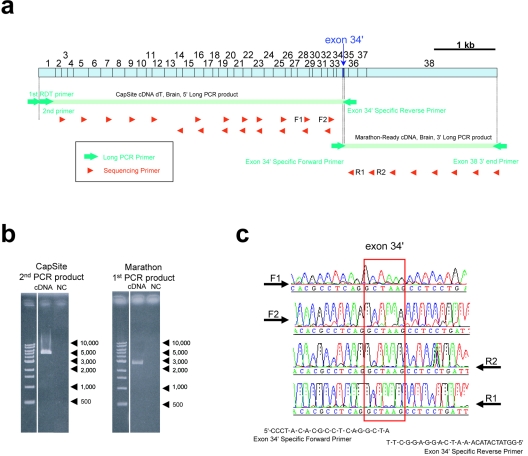
To determine the full-length TAF1 isoform, including the 5′ end of the transcript, a CapSite cDNA library derived from human brain (317-04041 [Nippon Gene]) was used. The CapSite cDNA libraries consist of cDNAs in which the 5′ cap structure (m7Gppp) of eukaryotic mRNA is replaced with a synthetic oligoribonucleotide to label the 5′ end of the cDNA, enabling identification of the 5′ end sequence by PCR.10 For amplification of the 3′ end, a whole Marathon-Ready cDNA library derived from human brain (BD Biosciences Clontech) was used. The reaction mixture contained 1 μl of the library, 200 mM of each primer, 0.16 mM of each deoxyribonucleotide diphosphate (dNTP), 1× BD Advantages2 PCR buffer, and 1× BD Advantages2 Polymerase Mix (639300 [BD Biosciences Clontech]) in a total volume of 50 μl. The PCR consisted of denaturation for 30 s at 94°C, followed by 5 cycles for 5 s at 94°C and 10 min at 70°C and then 20 cycles for 5 s at 94°C and 10 min at 78°C. An aliquot of the first PCR product was used for the second PCR reaction under the same conditions as the first-round PCR but with different primers. We used a primer set for the first PCR—first RDT primer (Nippon Gene) and TA3_r_5070 (5′-GGTATCATACAAATCAGGAGGCTT-3′)—and then used a set for the second heminested PCR—second primer and TA3_r_5070. These primers were purified by PAGE extraction. Alternative exon 34′–specific primers were designed to prevent erroneous amplification due to PCR slippage. For amplification of the 3′ end, a whole Marathon-Ready cDNA library derived from human brain (BD Biosciences Clontech) was used along with the primer set TA6_f_5032 (5′-CCCTACACGCCTCAGGCTA-3′) and TA2_r_7606 (5′-CCAGCATACATAACAAACACAGAAG-3′) under the same conditions as those used for the CapSite cDNA but as a single PCR reaction. Direct sequencing was done on these PCR products. After purification by a PCR Product Pre-Sequencing kit, cycle sequencing was performed using a BigDye Terminator v3.1 Cycle Sequencing kit, with 20 internal primers for a second PCR product from CapSite cDNA and 8 internal primers for the PCR product from Marathon-Ready cDNA. Direct sequencing was done on two long PCR products by use of the following primers: TAF_f_377, TAF_f_755, TAF_f_1152, TAF_f_1528, TAF_f_1889, TAF_f_2580, TAF_f_2945, TAF_f_3311, TAF_f_3689, TAF_f_4069, TAF_f_4413, TAF_f_4780, TAF_r_2273, TAF_r_2598, TAF_r_2979, TAF_r_3331, TAF_r_5164, TAF_r_5505, TAF_r_5862, TAF_r_6231, TAF_r_6469, TAF_r_6855, TAF_r_7164, TAF_r_7553, and TAF_r_7606 (table 3).
Results
Entire Genomic Sequence of the DYT3 Region
Two series of BAC libraries were constructed using DNA from a patient with XDP who had a disease-specific haplotype8,9 in the DYT3 region. From these libraries, a continuous BAC contig consisting of eight BAC clones was then generated to cover the DYT3 region between the GJB1 and CXCR3 genes (fig. 1a). By applying a shotgun sequencing strategy, we accurately determined the complete DNA sequence of the BAC contig, with 5.7-fold redundancy. The total sequence length of the BAC contig was 463,567 bp. A comparison between our sequence from the patient with XDP and a reference sequence from National Center for Biotechnology Information (NCBI) build 30 showed a total of 159 sequence variants: 89 single-nucleotide substitutions, 68 small insertions/deletions (indels), 1 retrotransposal insertion, and 1 large (1,666-bp) deletion (all variants are listed in table 5). Of these variants, 53 were known SNPs, comprising 50 substitutions and 3 indels. Of the 68 indels, 62 were repetitive units of STRs, and the other 6 indels were also located in certain types of STRs. The large deletion was a direct repeat sequence spanning 1,666 bp. The retrotransposal insertion in intron 32 of the TAF1 gene (fig. 1a) was 2,627 bp in length, which is categorized as an SVA (short interspersed nuclear element, VNTR, and Alu composite) retrotransposon.12,13 The SVA retrotransposon insertion in the DYT3 region had been never reported. However, none of these variants was located in any exon or promoter of the annotated genes that have experimentally verified coding sequences, including their alternative splicing exons. In the region between DXS10017 and DXS10018, there were 25 variants, which included the SVA insertion and the 1,666-bp deletion. We confirmed these variants in an ethnic panel (see the “Material and Methods” section) that included patients with XDP who have the disease-specific haplotype. DSCs 12, 10, 1, 3, and 2 were disease-specific among patients with XDP tested here as well as in a previous report.9 The 1,666-bp deletion was detected by PCR among all affected and unaffected Filipinos and in the ethnic panels (data not shown) and was considered to be nonspecific. By contrast, the SVA retrotransposon was clearly disease specific in our data set (figs. 1b and 3). These disease-specific variants, the DSCs and the SVA retrotransposon, were located around the TAF1 and MTS genes.
Table 5. .
All Nucleotide Variants in the DYT3 Critical Region on Xq13.1
| Variant Number |
Varianta | Positionb | Locationc | Reference Numberd |
| 1 | (C)5→(C)6 | 66573096 | NLGN3 exon 7 + 8,480 bp | rs11399763 |
| 2 | A→T | 66573470 | rs7066438 | |
| 3 | (CAA)6→(CAA)7 | 66573631 | Not verified | |
| 4 | C→A | 66574044 | rs7878993 | |
| 5 | (AAAT)11→(AAAT)8 | 66574555 | Not verified | |
| 6 | (A)17→(A)16 | 66574903 | Not verified | |
| 7 | A→T | 66575368 | rs5981084 | |
| 8 | T→C | 66576795 | rs17174152 | |
| 9 | C→T | 66577311 | Not verified | |
| 10 | T→C | 66579548 | rs4844288 | |
| 11 | T→C | 66579570 | rs6624538 | |
| 12 | (A)14→(A)13 | 66580276 | Not verified | |
| 13 | C→T | 66580741 | rs6624539 | |
| 14 | A→G | 66580815 | rs6625760 | |
| 15 | A→G | 66581634 | rs4844289 | |
| 16 | G→A | 66582609 | rs7891662 | |
| 17 | G→A | 66582761 | rs5981086 | |
| 18 | (TG)8→(TG)9 | 66583578 | Not verified | |
| 19 | G→A | 66583994 | rs7057179 | |
| 20 | C→T | 66584151 | rs4844146 | |
| 21 | C→G | 66584916 | rs6525476 | |
| 22 | (TTGT)6→(TTGT)5 | 66585160 | Not verified | |
| 23 | G→A | 66585224 | rs6625761 | |
| 24 | (ATTTT)8→(A/GTTTT)11 | 66585667 | Not verified | |
| 25 | G→A | 66586595 | rs6624541 | |
| 26 | T→C | 66586686 | rs6624542 | |
| 27 | G→A | 66587216 | Not verified | |
| 28 | (A)11→(A)12 | 66587487 | Not verified | |
| 29 | C→G | 66588293 | rs6624543 | |
| 30 | G→T | 66588505 | Not verified | |
| 31 | (T)23→(T)26 | 66588516 | Not verified | |
| 32 | (T)9→(T)10 | 66588861 | Not verified | |
| 33 | A→G | 66588959 | rs6525478 | |
| 34 | G→C | 66591853 | rs7892772 | |
| 35 | G→C | 66592199 | Not verified | |
| 36 | T→C | 66592559 | rs12396931 | |
| 37 | G→T | 66592688 | Not verified | |
| 38 | (A)16→(A)15 | 66592824 | Not verified | |
| 39 | A→G | 66593416 | rs5981088 | |
| 40 | T→C | 66594092 | rs5980744 | |
| 41 | A→G | 66594674 | rs7891451 | |
| 42 | C→T | 66595654 | rs6525479 | |
| 43 | T→C | 66596926 | rs4844292 | |
| 44 | (TTTC)6→(TTTC)7 | 66597719 | Not verified | |
| 45 | (T)11→(T)12 | 66597742 | Not verified | |
| 46 | (GT)27→(GT)20 | 66597906 | Not verified | |
| 47 | G→A | 66598062 | rs6525480 | |
| 48 | C→A | 66598314 | rs7884258 | |
| 49 | G→A | 66598347 | Not verified | |
| 50 | G→A | 66598626 | rs9698457 | |
| 51 | del(G) | 66598699 | Not verified | |
| 52 | C→T | 66599576 | rs35328964 | |
| 53 | ins(T)→ | 66599603 | Not verified | |
| 54 | T→A | 66599609 | Not verified | |
| 55 | ins(T)→ | 66599615 | Not verified | |
| 56 | C→T | 66599680 | rs34640965 | |
| 57 | (A)15→(A)14 | 66599758 | Not verified | |
| 58 | A→T | 66599766 | rs9698122 | |
| 59 | T→C | 66599820 | rs34030408 | |
| 60 | A→T | 66600629 | rs7051285 | |
| 61 | A→G | 66600840 | rs7878264 | |
| 62 | (CA)21→(CA)17 | 66601155 | Not verified | |
| 63 | A→G | 66601194 | Not verified | |
| 64 | (T)18→(T)21 | 66601314 | Not verified | |
| 65 | T→A | 66601345 | Not verified | |
| 66 | (A)21→(A)20 | 66602510 | Not verified | |
| 67 | (GAAA)4→(GAAA)3 | 66602519 | Not verified | |
| 68 | del(AG) | 66602552 | Not verified | |
| 69 | (GAAA)3→(GAAA)0 | 66602554 | Not verified | |
| 70 | C→G | 66602875 | rs6525481 | |
| 71 | C→A | 66603050 | Not verified | |
| 72 | C→T | 66603721 | Not verified | |
| 73 | A→C | 66605614 | rs7058793 | |
| 74 | (T)20→(T)18 | 66605638 | Not verified | |
| 75 | A→G | 66606071 | GJB1 exon 1 − 2,676 bp | Not verified |
| 76 | (G)4→(G)5 | 66610129 | GJB1 exon 1 + 1,339 bp | Not verified |
| 77 | C→T | 66610170 | rs11094200 | |
| 78 | (A)2→(A)4 | 66611346 | Not verified | |
| 79 | C→T | 66612259 | Not verified | |
| 80 | A→G | 66612280 | Not verified | |
| 81 | A→G | 66612281 | Not verified | |
| 82 | T→G | 66612285 | Not verified | |
| 83 | A→C | 66612532 | Not verified | |
| 84 | A→G | 66612546 | Not verified | |
| 85 | (A)22→(A)24 | 66613275 | Not verified | |
| 86 | C→T | 66613467 | Not verified | |
| 87 | (T)22→(T)18 | 66613644 | Not verified | |
| 88 | del(CT) | 66614391 | Not verified | |
| 89 | G→C | 66615041 | rs5980747 | |
| 90 | A→T | 66615382 | rs11094201 | |
| 91 | (T)19→(T)20 | 66616041 | rs11417492 | |
| 92 | G→A | 66616496 | GJB1 exon 2 − 697 bp | Not verified |
| 93 | T→C | 66619432 | GJB1 exon 2 + 737 bp | rs1997625 |
| 94 | (A)12→(A)13 | 66619843 | Not verified | |
| 95 | (T)14→(T)15 | 66620314 | Not verified | |
| 96 | A→C | 66622409 | Not verified | |
| 97 | (A)26→(A)27 | 66622587 | Not verified | |
| 98 | A→G | 66623453 | rs752081 | |
| 99 | (AGGG)5→(AGGG)4 | 66623969 | Not verified | |
| 100 | A→T | 66624355 | Not verified | |
| 101 | G→A | 66624540 | rs2341629 | |
| 102 | C→A | 66626295 | rs17311899 | |
| 103 | (TTTC)15→(TTTC)12 | 66627870 | DXS10015 | |
| 104 | (TTCC)10→(TTCC)9 | 66629254 | DXS7119 | |
| 105 | (T)15→(T)16 | 66629594 | rs17311899 | |
| 106 | T→A | 66630734 | rs4240822 | |
| 107 | C→A | 66631264 | ZNF261 exon 25 − 1,861 bp | Not verified |
| 108 | ins(AGA) | 66635550 | ZNF261 exon 23 − 121 bp | Not verified |
| 109 | A→G | 66640470 | ZNF261 exon 14 + 278 bp | rs5937076 |
| 110 | G→A | 66642833 | ZNF261 exon 7 − 129 bp | rs2341539 |
| 111 | (TC)32→(T)26 | 66648586 | ZNF261 exon 1 + 513 bp | DXS10016 |
| 112 | G→A | 66656109 | Not verified | |
| 113 | A→T | 66662285 | Not verified | |
| 114 | (A)21→(A)33 | 66666570 | Not verified | |
| 115 | (T)18→(T)19 | 66672301 | NONO exon 1 + 4,852 bp | Not verified |
| 116 | (T)21→(T)22 | 66681621 | NONO exon 3 − 2,509 bp | Not verified |
| 117 | (TATC)10→(TATC)8 | 66703094 | ITGB1 exon 11 + 4,240 bp | DXS0572i |
| 118 | T→A | 66710966 | Not verified | |
| 119 | C→T | 66713140 | rs12845815 | |
| 120 | (T)20→(T)21 | 66714867 | Not verified | |
| 121 | C→T | 66715822 | rs4269695 | |
| 122 | (T)20→(T)21 | 66718066 | rs34793613 | |
| 123 | (ACAA)2→(ACAA)1 | 66721012 | Not verified | |
| 124 | (T)17→(T)18 | 66722840 | Not verified | |
| 125 | G→A | 66733338 | Not verified | |
| 126 | C→T | 66735475 | Not verified | |
| 127 | (GAGGGG)6→(GAGGGG)5 | 66736067 | Not verified | |
| 128 | C→T | 66738056 | Not verified | |
| 129 | (TG)29→(TG)27 | 66746683 | TAF1 exon 1 − 13,082 bp | DXS10017 |
| 130 | (A)26→(A)25 | 66765934 | TAF1 exon 4 − 2,734 bp | ss66974122 |
| 131 | (T)20→(T)19 | 66773098 | TAF1 exon 8 + 567 bp | ss66974125 |
| 132 | T→G | 66784696 | TAF1 exon 19 − 1,374 bp | DSC12 |
| 133 | (TG)21→(TG)22 | 66786726 | TAF1 exon 21 − 76 bp | DXS8030 |
| 134 | A→G | 66791094 | TAF1 exon 23 + 128 bp | rs5980760 |
| 135 | (T)21→(T)22 | 66793069 | TAF1 exon 24 + 832 bp | ss66974128 |
| 136 | (T)26→(T)24 | 66817916 | TAF1 exon 32 + 178 bp | ss66974131 |
| 137 | (T)29→(T)30 | 66824505 | ss66974134 | |
| 138 | ins(SVA retroposon) | 66834003 | Disease specific | |
| 139 | C→T | 66834990 | DSC10 | |
| 140 | (A)21→(A)20 | 66838800 | ss66974137 | |
| 141 | (C)12→(C)11 | 66845611 | TAF1 exon 33 − 2,060 bp | ss66974140 |
| 142 | (T)43→(T)41 | 66850192 | TAF1 exon 35 − 1,550 bp | ss66974143 |
| 143 | C→A | 66877281 | ING2 exon 1 − 7,899 bp | dbSTS143400 |
| 144 | (TA34/TG8)→(TA16/TG2) | 66896816 | −48-bp deletion | |
| 145 | (A)30→(A)33 | 66898243 | ss66974146 | |
| 146 | A→C | 66901589 | ss66974149 | |
| 147 | T→A | 66907161 | DSC1 | |
| 148 | (T)16→(T)17 | 66909738 | ss66974152 | |
| 149 | C→T | 66910841 | SNP5 | |
| 150 | (T)21→(T)22 | 66912194 | ss66974155 | |
| 151 | del(1666bp)e | 66913932 | Polymorphism | |
| 152 | C→T | 66923286 | DSC3 | |
| 153 | G→A | 66924934 | DSC2 | |
| 154 | (TG)19→(TG)16 | 66937236 | DXS8101 | |
| 155 | (TG)23→(TG)24 | 66948636 | OGT exon 3 − 55 bp | DXS10018 |
| 156 | C→T | 66954497 | OGT exon 11 − 779 bp | SNP4 |
| 157 | (T)26→(T)23 | 66963535 | OGT exon 17 + 1,919 bp | Not verified |
| 158 | C→T | 67027072 | Not verified | |
| 159 | (TA)24→(TA)33 | 67034222 | Not verified |
Insertion and deletions are indicated by “ins” and “del,” respectively, against the reference allele.
Start position of nucleotide variant in NCBI build 30.
None of the variants listed here are located in any exon of experimentally verified coding sequence, so location indicates a distance (in bp) from an exon to the closest variant.
Reference number in public resources: dbSNP, dbSTS, GenBank, and/or Medline. The variants “Not verified” were not confirmed using our ethnic panel.
The 1,666-bp deletion was observed by PCR in all samples tested here, including all affected and unaffected samples from the Philippines.
Figure 3. .
The SVA insertion in an additional seven patients with XDP and their relatives, from four families. Information on these patients is given in table 1 (patients 14–20).
Previously Proposed MTS Transcripts
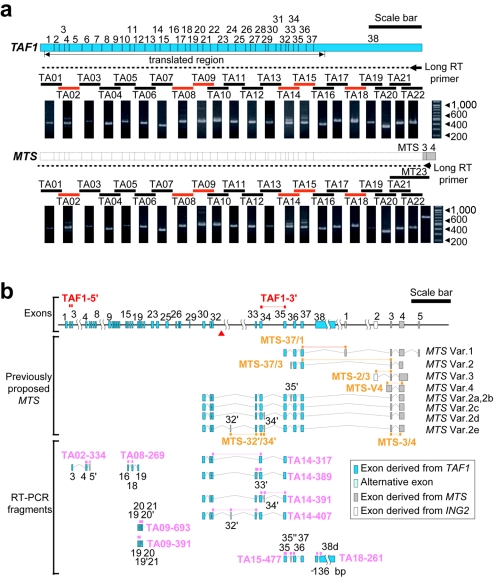
Next, northern hybridization and long RT-PCR analysis were undertaken to confirm the structures and expression of the TAF1 and MTS genes. Northern analysis showed that the hybridization signal seen at ∼7 kb of TAF1 had a tendency toward reduction in patient caudate and that the MTS transcript lengths reported elsewhere9 were not detectable in RNAs from either patient or control tissues (fig. 2b). Long RT-PCR analysis showed that the PCR fragment pattern from MTS was identical to that from TAF1 (fig. 4a). If the MTS transcripts had lacked upstream exons 1–29 and exon 38, spanning >2 kb, as shown in the previous report,9 the northern analysis would have detected the corresponding signals at ∼2–5 kb. Moreover, our long RT-PCR analysis showed that exons 3 and 4 from MTS (gray exons in fig. 4b) are attached at the 3′ end of exon 38 of TAF1. Quantitative RT-PCR analysis by the TaqMan assay with five probes designed to detect the previously proposed MTS transcripts MTS-V4—which was regarded as a candidate transcript for DYT3 because of a deduced amino-acid change9—MTS-37/1, MTS-37/3, MTS-2/3, and MTS-32′/34′ (fig. 4b) yielded very weak and irregular amplification signals that did not allow quantification of expression levels in the caudate nucleus (table 6), whereas the probe MTS-3/4 for exons 3 and 4 attached to TAF1 yielded a relatively weak but regular signal (table 6). These results consistently suggested that the previously proposed MTS transcripts may be extremely rare or unexpressed and that at least exons 3 and 4 of MTS may be just an additional part of the 3′ UTR of TAF1, rather than part of a new distinct short gene.
Figure 4. .
Long RT-PCR and the alternative exons of TAF1. a, Long RT-PCR analysis. The broken line indicates an expected cDNA fragment of TAF1 with the long RT primer on the end of exon 38 (short arrow). By subsequent PCR with the use of the long cDNA, six lanes—TA02, TA08, TA09, TA14, TA15, and TA18 (red bars)—showed multiple bands. Other lanes (black bars) contained single bands. Also shown is the result obtained using the MTS-specific long RT primer on its 3′ end. There was no difference between the TAF1 and MTS primers or between the patient and the control results (patient data not shown). Scale bar=1 kb. b, Ten alternative exons, including two exon skippings and one deletion, were identified by RT-PCR. The detailed sequence information for these exons is annotated in our AB191243 deposition in DNA Databank of Japan (DDBJ). Of 10 alternative exons, 3 were reported elsewhere as exons of MTS, but a form including both exon 32′ and exon 34′ was not detected. For quantitative RT-PCR, 17 TaqMan probes were designed. Two TAF-series probes (red) and 10 TA-series probes (pink) were designed to detect mainly the TAF1 common forms and alternative splicing isoforms, respectively. The other five MTS-series probes (orange) were designed to detect the MTS transcripts reported elsewhere. Var. = variant. Scale bar=5 kb.
Table 6. .
Linearity of Amplification Curve from Threshold Cycle (Ct) = 25 to Ct = 35[Note]
| Assay | Slope (SD) | r (SD) | r2 (SD) |
| MTS-37/1 | .0020 (.0032) | .31 (.641) | .48 (.226) |
| MTS-37/3 | .035 (.0123) | .82 (.036) | .68 (.061) |
| MTS-V4 | .011 (.0098) | .51 (.422) | .43 (.285) |
| MTS-2/3 | .00016 (.0014) | −.10 (.580) | .32 (.213) |
| MTS-32′/34′ | .00073 (.0005) | .77 (.258) | .65 (.292) |
| MTS-3/4 | .054 (.0351) | .81 (.017) | .66 (.032) |
| TA14-385Na | 1.37 (.0424) | .99 (.002) | .97 (.004) |
Note.— Slopes of the resulting lines, correlation coefficient (r) values, and coefficient of determination (r2) values were calculated by curve-fitting with linear function against normalized reporter signal (ΔRn) values from Ct = 25 to Ct = 35 in each amplification curve of the real-time PCRs. We employed the threshold with slope >0.5 and r2>0.6.
The TaqMan probe was employed as a positive control to show standard amplification and linearity from Ct = 25 to Ct = 35.
Decreased Expression of TAF1 in the XDP Brain
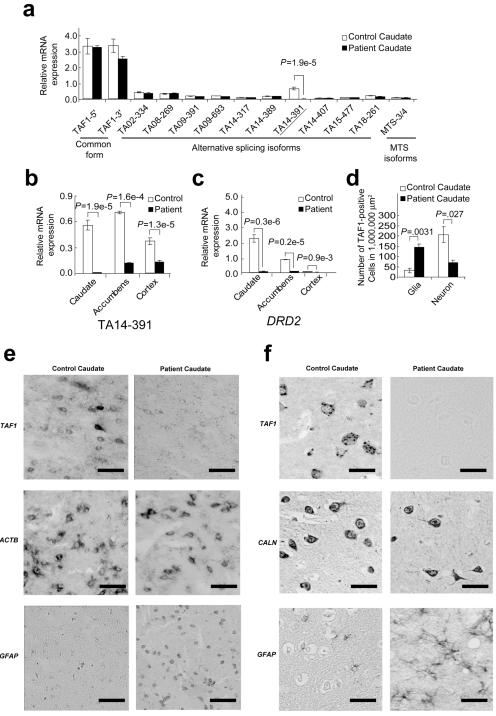
At least 10 new alternative splicing exons around TAF1 were found by long RT-PCR analysis (fig. 4b), but neither the DSCs nor the SVA insertion were located in any known and predictably translated regions of the TAF1 exons. We then examined the expression levels of various forms of TAF1 in the XDP caudate nucleus, using quantitative RT-PCR by designing specific probes for the TaqMan assay (fig. 5a). One of these probes, TA14-391—with an alternative exon of 6 additional bp, named “exon 34′”—showed a highly significant decrease in its expression level in the caudate nucleus of the patient with XDP (fig. 5a), which was less than ∼1/40 of that in the normal control, and its expression was virtually limited to brain and neurons (table 7). TA14-391 was the second-most abundant among all TAF1 species (fig. 5a), and its expression level in the control caudate nucleus was 1/4–1/5 of that of the major form of TAF1 (table 7). TA14-391 also showed a significantly decreased level of expression in the cortex and the nucleus accumbens of the patient with XDP (fig. 5b), although these regions had no neuronal loss. These findings suggest that the decreased level of expression was the cause rather than the result of neuronal loss in the caudate nucleus of the patient with XDP. In addition to the TaqMan assay, in situ hybridization was performed in the caudate by use of a riboprobe common to TAF1 isoforms that are located in exon 38 (probe 3 in fig. 2a). The riboprobe showed decreased expression in the caudate neurons of the patient, although the weak expression was still present in glial cells (fig. 5e). Immunohistochemical examination by use of a polyclonal antibody against the common epitopes of TAF1 also showed decreased immunoreactivity in the XDP neurons in the caudate nucleus from other patients with XDP (fig. 5f). These findings suggest that the deficiency of the neuron-specific isoform of TAF1, TA14-391, reflects these histologically verified neuron-specific decreases of TAF1 expression and that TA14-391 is one of the neuron-specific isoforms, whereas apparently similar levels of mRNA expression for TAF1 or its isoforms as a whole (figs. 2b, 2c, and 5a) can be accounted for by increased glial expression of TAF1 due to intensive astrogliosis4,5 (fig. 5d–5f), obscuring the decreased expression in neurons. The TA14-391 isoform may represent the decreased expression of many TAF1 isoforms in the XDP neurons, because of its original neuron specificity of expression. Finally, to determine the complete structure of the isoform containing the 6 bp of exon 34′, full-length cloning from libraries consisting of cDNAs enriched in their 5′ ends10 was performed. A single cDNA containing exon 34′ was successfully cloned (fig. 6), and this had the complete translation frame of the major form of TAF1 with an insertion of two amino acid residues, alanine and lysine, in the carboxyl terminal kinase domain.
Figure 5. .
Expression of the TAF1 isoforms in the caudate. a, Expressions are shown relative to the expression of 18S rRNA (as an internal control). The label “relative mRNA expression” means relative mRNA expression level to 1/20 × 18S rRNA. Values are expressed as means ± SEM (n=3). The TA14-391 probe showed a significant reduction in the patient’s caudate. Two-sided P values are shown. Also shown is the expression level of TA14-391 (b) and DRD2 (c) in three brain regions: caudate, accumbens, and cortex. All regions showed a significant decrease in TA14-391 expression. d, Morphometry analysis of TAF1-positive cells in XDP and control caudate nuclei. The number of each type of cell was counted in 1,000,000-μm2 areas of the caudate nuclei. These were gliosis and neuronal loss in the XDP caudate nucleus. e, In situ hybridization analysis of TAF1. Although many TAF1-positive neurons were observed in both tissues, the expression level was apparently low in the patient’s caudate neurons, even when a common probe for exon 38 was used. By contrast, the expression level of TAF1 in glial cells was weak in both tissues. ACTB =β-actin; GFAP = glial fibrillary acidic protein. Strong ACTB signals were shown in glial and neuronal cells. The GFAP probe stains active glial cells, especially in the XDP caudate nucleus, because of activation by astrogliosis. In contrast, we observed no signal when using sense probes of these genes (data not shown). Scale bar=25 μm. f, TAF1 immunohistochemical staining. Nearby sections were stained with polyclonal antibodies against TAF1, calcineurin (CALN), and GFAP. The immunoreactivity of TAF1 in the XDP caudate neurons was apparently weak. Moreover, the immunoreactivity of TAF1 in glial cells was originally weak in both tissues. Similar immunoreactivity was observed in three other brain tissues from three different patients with XDP. Scale bar=25 μm.
Table 7. .
Abundance of the Probe TA14-391 in Various Tissues and Cell Lines[Note]
| Mean (SE)[10−5 fmol per 100 ng RNA] |
||
| Tissue or Cell Line | TA14-391 | TAF1 Major Form |
| Lymphocytes | 0 | 3.68 (.08897) |
| Heart | 0 | .0873 (.00484) |
| Spleen | 0 | 3.13 (.06749) |
| Lung | 0 | .133 (.00536) |
| Liver | 0 | 4.20 (.21582) |
| Thymus | 0 | 1.44 (.07147) |
| Stomach | 0 | 1.23 (.03865) |
| Caudate | .699 (.00761) | 3.30 (.50351) |
| Cortex | .463 (.00308) | 3.70 (.57268) |
| Neuroblastoma, SH-SY5Y | .213 (.00694) | 4.68 (.55110) |
| Glioblastoma, HTB15 | 0 | 1.67 (.44595) |
Note.— The abundances for TA14-391 and the TAF1 major form (TA14-385N) were determined using quantitative RT-PCR by use of each clone of known concentration in the plasmid as an internal control.
Figure 6. .
a, Full-length cloning of a TAF1 isoform sequence containing alternative exon 34′. The 5′ end was obtained from CapSite cDNA from brain. The 3′ end was obtained from Marathon-Ready cDNA from brain. The complete DNA sequences of these long products were determined by a PCR direct-sequencing method by use of 28 redundant internal sequencing primers (red triangles). b, From the long PCR products, products of sufficient quantity and quality for PCR direct sequencing were amplified. NC = negative control without DNA template. c, Each exon 34′–specific primer was designed to have only 3 nt of exon 34′, to prevent erroneous amplification due to slippage.
Discussion
SVA retrotransposon insertions are thought to be active in the human genome and to alter the expression level of adjacent genes that cause diseases, such as autosomal recessive hypercholesterolemia (ARH [MIM 605747])14 and Fukuyama-type congenital muscular dystrophy (FCMD [MIM 607440]).15 SVA insertion was found in the 3′ UTR region of the FCMD gene and in an intronic region of the ARH gene, which showed association with the reduced expression level of these genes. SVA retrotransposon has a high degree of GC content (∼70%) and a large number of CpG sites (>150) in its nucleotide sequence, so that it is frequently hypermethylated in its insertion site. In fact, the present study demonstrated that the SVA retrotransposon was hypermethylated in genomic DNA from the caudate nucleus of the patient with XDP (fig. 7), which was also used in northern analysis, quantitative RT-PCR, and in situ hybridization. Such hypermethylated status and high GC content are able to affect dynamics of surrounding nucleotide sequence, such as the cis-regulatory element, so SVA insertions may reduce the expression level of adjacent genes. For instance, many large introns of eukaryotes often contain tissue-specific cis-regulatory elements, such as enhancers or silencers.16–22 Intron 32, the largest intron of TAF1 (29,932 bp), possibly contains a neuron-specific cis-regulatory element. Although involvement of other sequence variants in XDP pathogenesis is still possible, it is most likely that the SVA insertion impairs the function of a hypothetical neuron-specific cis-regulatory element, such as an enhancer, through changes in the methylation content and then substantially reduces the expressions of many isoforms of TAF1, including TA14-391, in the neurons. Because of the original neuron specificity of the expression, the TA14-391 isoform might represent the impairment more remarkably than do other isoforms. On the other hand, the remaining expression, as shown for TA14-391, despite being low, may compensate for complete loss of function and may account for the relatively late disease onset (age 39.5±8.44 years) and the recessive mode of inheritance.
Figure 7. .
Detection of methylation around the SVA insertion. a, Restriction sites around the SVA insertion. The HindIII fragment and Southern probe are identical to those in figure 1a. The SVA insertion created 47 new HpaII sites in the HindIII restriction fragment. MspI is a CpG methylase-insensitive isoschizomer of HpaII. b, Southern hybridization of genomic DNA from the patient’s caudate. The 6.1-kb HindIII fragment was shifted, by additional HpaII digestion, to an ∼4.6-kb fragment. By contrast, the HindIII fragment was completely digested, by additional MspI digestion, to a fragment of ∼2.4 kb.
In summary, our results suggest that the SVA retrotransposon insertion into the TAF1 gene may cause XDP by altering the expression of TAF1 isoforms, including TA14-391, possibly through DNA methylation changes. To our knowledge, our report is the first to reveal the entire genomic sequence of the DYT3 region and to demonstrate at least one whole structure of the neuron-specific isoform of TAF1 and the disease-specific mutation, with a possible mechanism. To establish the disease specificity and the involvement mechanism of the SVA insertion in reduced expression of TAF1 in XDP, further studies, such as an extensive population screening and genetic modification in model organisms, will be necessary and warranted. The TAF1 protein is the largest and the essential component of the TFIID complex in the pathway of RNA polymerase II–mediated gene transcription,23,24 and it regulates transcription of a large number of genes related to cell division and proliferation.23–25 How can a ubiquitous gene such as TAF1 cause a disease that affects a selective part of the nervous system? We hypothesize that the neuron-specific isoforms and/or their enhanced expression level of TAF1 (table 7) may play important roles in the nondividing cell. Sharing similar pathological features in the caudate nucleus, XDP and Huntington disease might result from disorders in the same biochemical pathway of RNA polymerase II–mediated gene transcription. In Huntington disease, for example, the abnormal huntingtin protein has been shown to interfere with the interaction between Sp1 and TAFII130, resulting in reduced expression of DRD2 (MIM 126450) in the brain, including the caudate nucleus.26 In XDP, the decreased expression of the TA14-391 isoform, and probably other TAF1 isoforms, may result in transcriptional dysregulation of many neuronal genes, including DRD2. We believe that the present findings in XDP support the concept of “transcription syndromes”27 in TFIID, which include congenital cataracts facial dysmorphism neuropathy (CCFDN [MIM 604168]) syndrome, caused by a partial deficiency of RNA polymerase II28; Huntington disease26; dentato-rubro-pallidoluysian atrophy (DRPLA [MIM 125370]),29 caused by interference in the signals to TFIID; and spinocerebellar ataxia 17 (SCA17 [MIM 607136]),30 caused by an expanded polyglutamine in the TATA-binding protein (TBP [MIM 600075]).
Acknowledgments
We thank S. Fahn (Neurological Institute of New York, Columbia University) and M. Nakagawa (Kyoto Prefectural University of Medicine) for their critical reading of the manuscript. This study was partly supported by a Grant-in-Aid for Scientific Research on Priority Areas (C) “Medical Genome Science”; by Center of Excellence grant 16101J-1 from the Japanese Ministry of Education, Science, Culture, and Sports; and by Grant-in-Aid for Dystonia Research from the National Center of Neurology and Psychiatry, Japan.
Web Resources
Accession numbers and URLs for data presented herein are as follows:
- dbSNP, http://www.ncbi.nlm.nih.gov/SNP/ (for newly described SNPs ss66974122, ss66974125, ss66974128, ss66974131,ss66974134, ss66974137, ss66974140, ss66974143, ss66974146, ss66974149, ss66974152, and ss66974155)
- DNA Databank of Japan (DDBJ), http://www.ddbj.nig.ac.jp/Welcome-e.html (for the complete genomic sequence of the DYT3 region [accession number AB191243])
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for XDP, Huntington disease, TAF1, ARH, FCMD, DRD2, CCFDN, DRPLA, SCA17, and TBP)
References
- 1.Lee LV, Pascasio FM, Fuentes FD, Viterbo GH (1976) Torsion dystonia in Panay, Philippines. Adv Neurol 14:137–151 [PubMed] [Google Scholar]
- 2.Lee LV, Munoz EL, Tan KT, Reyes MT (2001) Sex linked recessive dystonia parkinsonism of Panay, Philippines (XDP). Mol Pathol 54:362–368 [PMC free article] [PubMed] [Google Scholar]
- 3.Fahn S, Bressman SB, Marsden CD (1998) Classification of dystonia. Adv Neurol 78:1–10 [PubMed] [Google Scholar]
- 4.Goto S, Lee LV, Munoz EL, Tooyama I, Tamiya G, Makino S, Ando S, Dantes MB, Yamada K, Matusmoto S, et al (2005) Functional anatomy of the basal ganglia in X-linked recessive dystonia-parkinsonism. Ann Neurol 58:7–17 10.1002/ana.20513 [DOI] [PubMed] [Google Scholar]
- 5.Waters CH, Faust PL, Powers J, Vinters H, Moskowitz C, Nygaard T, Hunt AL, Fahn S (1993) Neuropathology of lubag (X-linked dystonia parkinsonism). Mov Disord 8:387–390 10.1002/mds.870080328 [DOI] [PubMed] [Google Scholar]
- 6.Wilhelmsen KC, Weeks DE, Nygaard TG, Moskowitz CB, Rosales RL, dela Paz DC, Sobrevega EE, Fahn S, Gilliam TC (1991) Genetic mapping of “Lubag” (X-linked dystonia-parkinsonism) in a Filipino kindred to the pericentromeric region of the X chromosome. Ann Neurol 29:124–131 10.1002/ana.410290203 [DOI] [PubMed] [Google Scholar]
- 7.Haberhausen G, Schmitt I, Kohler A, Peters U, Rider S, Chelly J, Terwilliger JD, Monaco AP, Muller U (1995) Assignment of the dystonia-parkinsonism syndrome locus, DYT3, to a small region within a 1.8-Mb YAC contig of Xq13.1. Am J Hum Genet 57:644–650 [PMC free article] [PubMed] [Google Scholar]
- 8.Nemeth AH, Nolte D, Dunne E, Niemann S, Kostrzewa M, Peters U, Fraser E, Bochukova E, Butler R, Brown J, et al (1999) Refined linkage disequilibrium and physical mapping of the gene locus for X-linked dystonia-parkinsonism (DYT3). Genomics 60:320–329 10.1006/geno.1999.5929 [DOI] [PubMed] [Google Scholar]
- 9.Nolte D, Niemann S, Muller U (2003) Specific sequence changes in multiple transcript system DYT3 are associated with X-linked dystonia parkinsonism. Proc Natl Acad Sci USA 100:10347–10352 10.1073/pnas.1831949100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maruyama K, Sugano S (1994) Oligo-capping: a simple method to replace the cap structure of eukaryotic mRNAs with oligoribonucleotides. Gene 138:171–174 10.1016/0378-1119(94)90802-8 [DOI] [PubMed] [Google Scholar]
- 11.Lee JT, Davidow LS, Warshawsky D (1999) Tsix, a gene antisense to Xist at the X-inactivation centre. Nat Genet 21:400–404 10.1038/7734 [DOI] [PubMed] [Google Scholar]
- 12.Shen L, Wu LC, Sanlioglu S, Chen R, Mendoza AR, Dangel AW, Carroll MC, Zipf WB, Yu CY (1994) Structure and genetics of the partially duplicated gene RP located immediately upstream of the complement C4A and the C4B genes in the HLA class III region: molecular cloning, exon-intron structure, composite retroposon, and breakpoint of gene duplication. J Biol Chem 269:8466–8476 [PubMed] [Google Scholar]
- 13.Ostertag EM, Goodier JL, Zhang Y, Kazazian HH Jr (2003) SVA elements are nonautonomous retrotransposons that cause disease in humans. Am J Hum Genet 73:1444–1451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wilund KR, Yi M, Campagna F, Arca M, Zuliani G, Fellin R, Ho YK, Garcia JV, Hobbs HH, Cohen JC (2002) Molecular mechanisms of autosomal recessive hypercholesterolemia. Hum Mol Genet 11:3019–3030 10.1093/hmg/11.24.3019 [DOI] [PubMed] [Google Scholar]
- 15.Kobayashi K, Nakahori Y, Miyake M, Matsumura K, Kondo-Iida E, Nomura Y, Segawa M, Yoshioka M, Saito K, Osawa M, et al (1998) An ancient retrotransposal insertion causes Fukuyama-type congenital muscular dystrophy. Nature 394:388–392 10.1038/28256 [DOI] [PubMed] [Google Scholar]
- 16.Gillies SD, Morrison SL, Oi VT, Tonegawa S (1983) A tissue-specific transcription enhancer element is located in the major intron of a rearranged immunoglobulin heavy chain gene. Cell 33:717–728 10.1016/0092-8674(83)90014-4 [DOI] [PubMed] [Google Scholar]
- 17.Halder G, Callaerts P, Gehring WJ (1995) Induction of ectopic eyes by targeted expression of the eyeless gene in Drosophila. Science 267:1788–1792 10.1126/science.7892602 [DOI] [PubMed] [Google Scholar]
- 18.Dirksen WP, Mohamed SA, Fisher SA (2003) Splicing of a myosin phosphatase targeting subunit 1 alternative exon is regulated by intronic cis-elements and a novel bipartite exonic enhancer/silencer element. J Biol Chem 278:9722–9732 10.1074/jbc.M207969200 [DOI] [PubMed] [Google Scholar]
- 19.Donoghue M, Ernst H, Wentworth B, Nadal-Ginard B, Rosenthal N (1988) A muscle-specific enhancer is located at the 3′ end of the myosin light-chain 1/3 gene locus. Genes Dev 2:1779–1790 [DOI] [PubMed] [Google Scholar]
- 20.Annweiler A, Muller-Immergluck M, Wirth T (1992) Oct2 transactivation from a remote enhancer position requires a B-cell-restricted activity. Mol Cell Biol 12:3107–3116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Banerji J, Rusconi S, Schaffner W (1981) Expression of a beta-globin gene is enhanced by remote SV40 DNA sequences. Cell 27:299–308 10.1016/0092-8674(81)90413-X [DOI] [PubMed] [Google Scholar]
- 22.Rippe RA, Lorenzen SI, Brenner DA, Breindl M (1989) Regulatory elements in the 5′-flanking region and the first intron contribute to transcriptional control of the mouse alpha 1 type I collagen gene. Mol Cell Biol 9:2224–2227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hisatake K, Hasegawa S, Takada R, Nakatani Y, Horikoshi M, Roeder RG (1993) The p250 subunit of native TATA box-binding factor TFIID is the cell-cycle regulatory protein CCG1. Nature 362:179–181 10.1038/362179a0 [DOI] [PubMed] [Google Scholar]
- 24.Ruppert S, Wang EH, Tjian R (1993) Cloning and expression of human TAFII250: a TBP-associated factor implicated in cell-cycle regulation. Nature 362:175–179 10.1038/362175a0 [DOI] [PubMed] [Google Scholar]
- 25.Wassarman DA, Sauer F (2001) TAF(II)250: a transcription toolbox. J Cell Sci 114:2895–2902 [DOI] [PubMed] [Google Scholar]
- 26.Dunah AW, Jeong H, Griffin A, Kim YM, Standaert DG, Hersch SM, Mouradian MM, Young AB, Tanese N, Krainc D (2002) Sp1 and TAFII130 transcriptional activity disrupted in early Huntington’s disease. Science 296:2238–2243 10.1126/science.1072613 [DOI] [PubMed] [Google Scholar]
- 27.Vermeulen W, van Vuuren AJ, Chipoulet M, Schaeffer L, Appeldoorn E, Weeda G, Jaspers NG, Priestley A, Arlett CF, Lehmann AR, et al (1994) Three unusual repair deficiencies associated with transcription factor BTF2(TFIIH): evidence for the existence of a transcription syndrome. Cold Spring Harb Symp Quant Biol 59:317–329 [DOI] [PubMed] [Google Scholar]
- 28.Varon R, Gooding R, Steglich C, Marns L, Tang H, Angelicheva D, Yong KK, Ambrugger P, Reinhold A, Morar B, et al (2003) Partial deficiency of the C-terminal-domain phosphatase of RNA polymerase II is associated with congenital cataracts facial dysmorphism neuropathy syndrome. Nat Genet 35:185–189 10.1038/ng1243 [DOI] [PubMed] [Google Scholar]
- 29.Igarashi S, Koide R, Shimohata T, Yamada M, Hayashi Y, Takano H, Date H, Oyake M, Sato T, Sato A, et al (1998) Suppression of aggregate formation and apoptosis by transglutaminase inhibitors in cells expressing truncated DRPLA protein with an expanded polyglutamine stretch. Nat Genet 18:111–117 10.1038/ng0298-111 [DOI] [PubMed] [Google Scholar]
- 30.Nakamura K, Jeong SY, Uchihara T, Anno M, Nagashima K, Nagashima T, Ikeda S, Tsuji S, Kanazawa I (2001) SCA17, a novel autosomal dominant cerebellar ataxia caused by an expanded polyglutamine in TATA-binding protein. Hum Mol Genet 10:1441–1448 10.1093/hmg/10.14.1441 [DOI] [PubMed] [Google Scholar]