Abstract
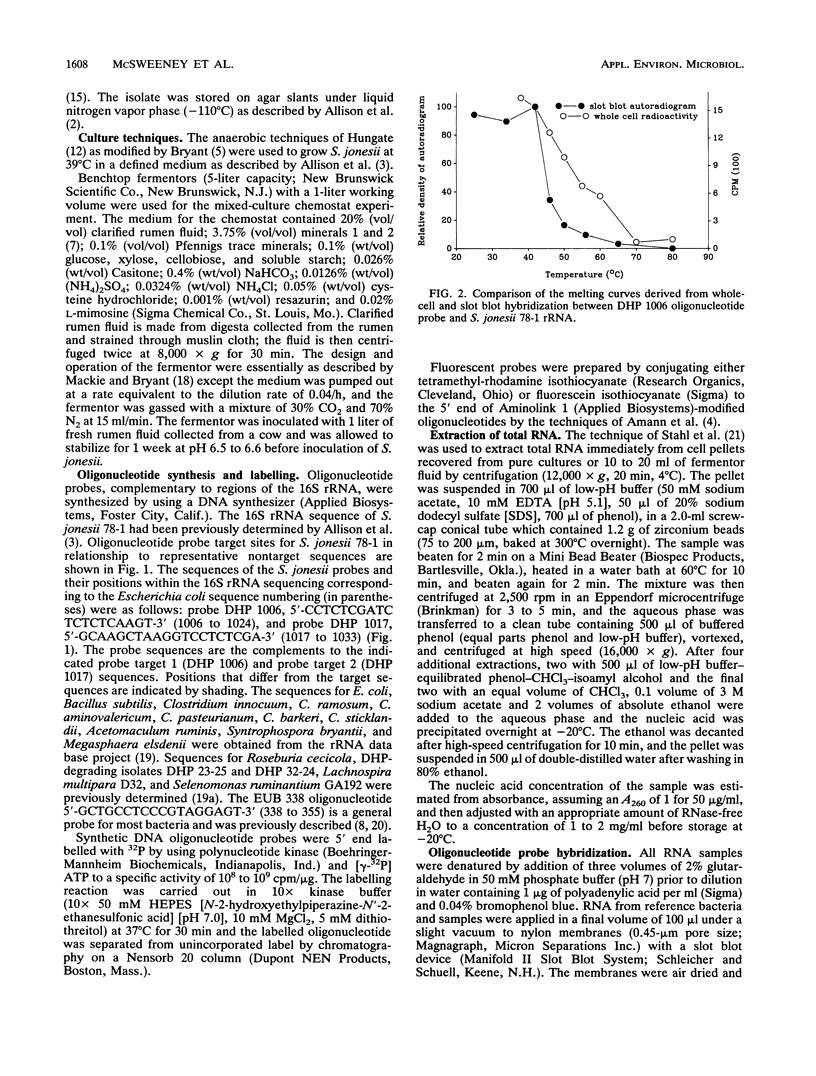
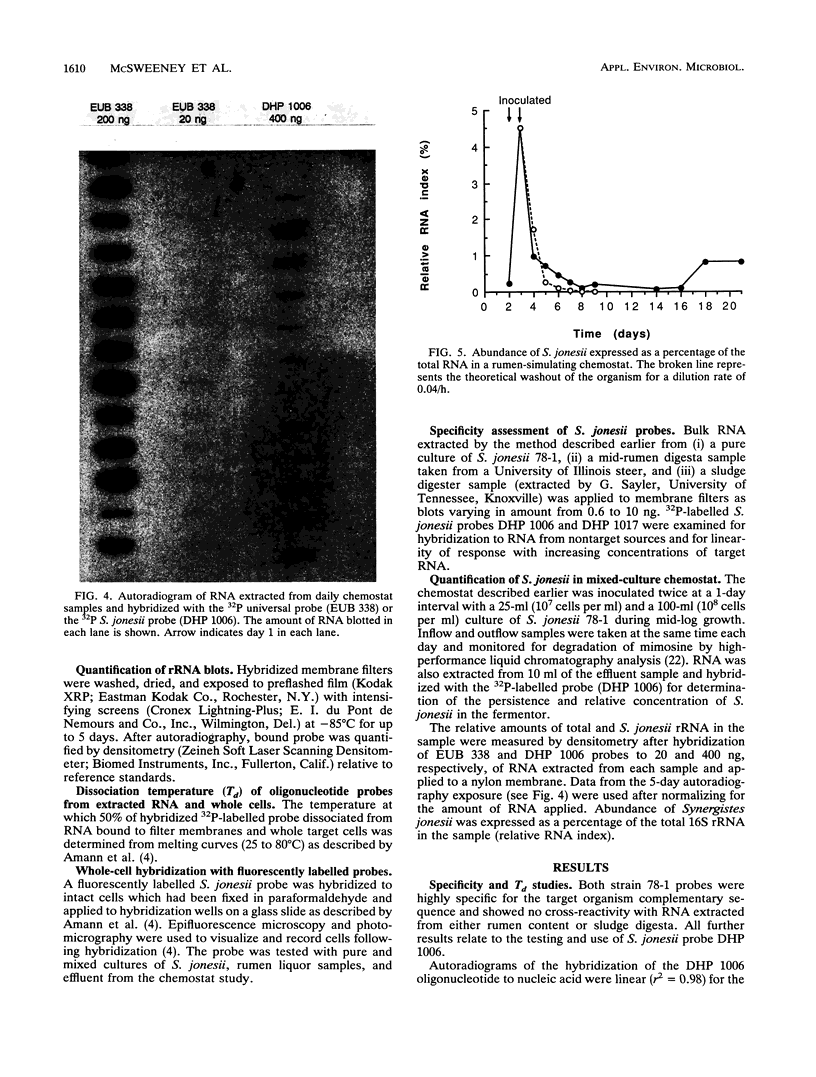
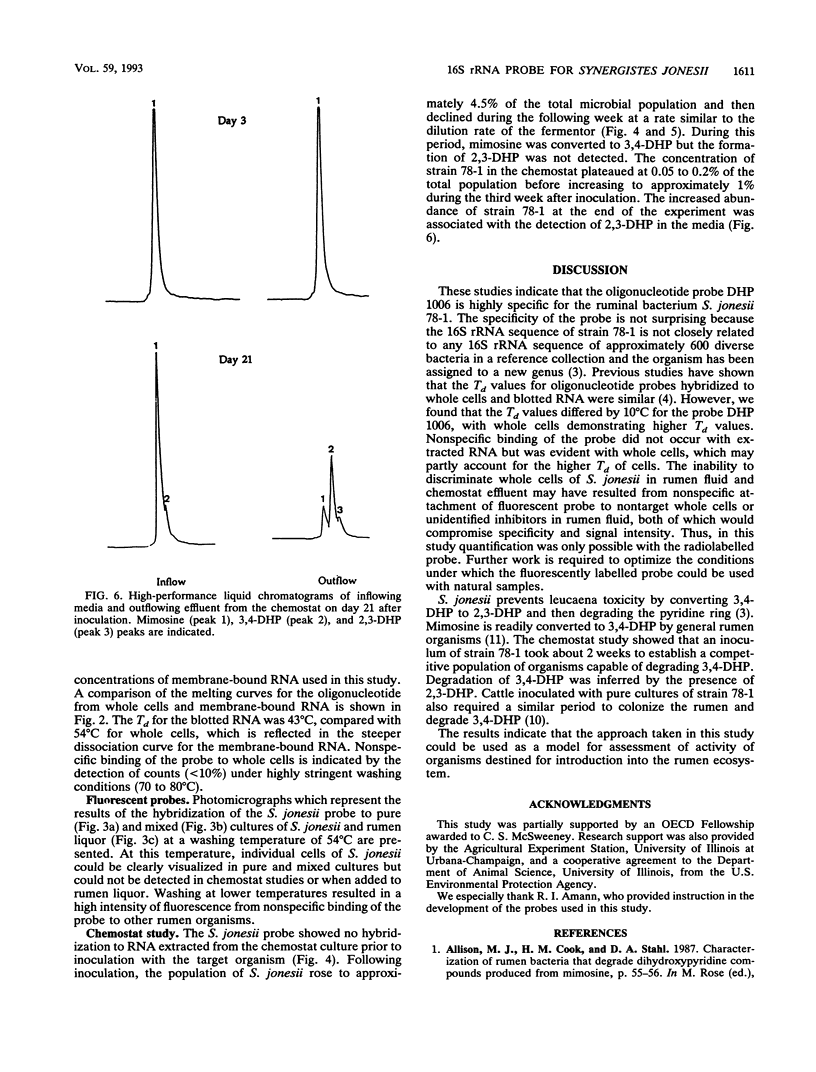
Radiolabelled and fluorescent-dye-conjugated oligonucleotide probes which targeted rRNA sequences were developed for the enumeration of the ruminal bacterium Synergistes jonesii 78-1 in mixed culture. Two probes were tested, and both were highly specific for the respective complementary sequences of the target organism. Individual cells of S. jonesii in pure and mixed cultures were clearly visualized in situ by hybridization with the fluorescent-dye-conjugated probe but could not be detected in natural samples. Therefore the radiolabelled probe was used to monitor the population of S. jonesii introduced into a chemostat which simulated the rumen ecosystem. The S. jonesii probe did not hybridize to RNA extracted from the culture prior to inoculation with the target organism. After inoculation, S. jonesii rRNA represented 4.5% of the total bacterial rRNA and then rapidly declined to < 0.2% before increasing to about 1% of the total bacterial rRNA during the following 3 weeks. This study demonstrates that rRNA-targeted probes could be used for tracking organisms introduced into the rumen ecosystem.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allison M. J., Hammond A. C., Jones R. J. Detection of ruminal bacteria that degrade toxic dihydroxypyridine compounds produced from mimosine. Appl Environ Microbiol. 1990 Mar;56(3):590–594. doi: 10.1128/aem.56.3.590-594.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amann R. I., Krumholz L., Stahl D. A. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J Bacteriol. 1990 Feb;172(2):762–770. doi: 10.1128/jb.172.2.762-770.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant M. P. Commentary on the Hungate technique for culture of anaerobic bacteria. Am J Clin Nutr. 1972 Dec;25(12):1324–1328. doi: 10.1093/ajcn/25.12.1324. [DOI] [PubMed] [Google Scholar]
- Caldwell D. R., Bryant M. P. Medium without rumen fluid for nonselective enumeration and isolation of rumen bacteria. Appl Microbiol. 1966 Sep;14(5):794–801. doi: 10.1128/am.14.5.794-801.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond A. C., Allison M. J., Williams M. J., Prine G. M., Bates D. B. Prevention of leucaena toxicosis of cattle in Florida by ruminal inoculation with 3-hydroxy-4-(1H)-pyridone-degrading bacteria. Am J Vet Res. 1989 Dec;50(12):2176–2180. [PubMed] [Google Scholar]
- Jones R. J. Does ruminal metabolism of mimosine explain the absence of Leucaena toxicity in Hawaii? Aust Vet J. 1981 Jan;57(1):55–56. doi: 10.1111/j.1751-0813.1981.tb07097.x. [DOI] [PubMed] [Google Scholar]
- Jones R. J., Megarrity R. G. Successful transfer of DHP-degrading bacteria from Hawaiian goats to Australian ruminants to overcome the toxicity of Leucaena. Aust Vet J. 1986 Aug;63(8):259–262. doi: 10.1111/j.1751-0813.1986.tb02990.x. [DOI] [PubMed] [Google Scholar]
- KISTNER A. An improved method for viable counts of bacteria of the ovine rumen which ferment carbohydrates. J Gen Microbiol. 1960 Dec;23:565–576. doi: 10.1099/00221287-23-3-565. [DOI] [PubMed] [Google Scholar]
- Mackie R. I., Bryant M. P. Metabolic Activity of Fatty Acid-Oxidizing Bacteria and the Contribution of Acetate, Propionate, Butyrate, and CO(2) to Methanogenesis in Cattle Waste at 40 and 60 degrees C. Appl Environ Microbiol. 1981 Jun;41(6):1363–1373. doi: 10.1128/aem.41.6.1363-1373.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen G. J., Larsen N., Woese C. R. The ribosomal RNA database project. Nucleic Acids Res. 1991 Apr 25;19 (Suppl):2017–2021. doi: 10.1093/nar/19.suppl.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl D. A., Flesher B., Mansfield H. R., Montgomery L. Use of phylogenetically based hybridization probes for studies of ruminal microbial ecology. Appl Environ Microbiol. 1988 May;54(5):1079–1084. doi: 10.1128/aem.54.5.1079-1084.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tangendjaja B., Wills R. B. Analysis of mimosine and 3-hydroxy-4(1H)-pyridone by high-performance liquid chromatography. J Chromatogr. 1980 Dec 19;202(2):317–318. doi: 10.1016/s0021-9673(00)81746-x. [DOI] [PubMed] [Google Scholar]





