Abstract
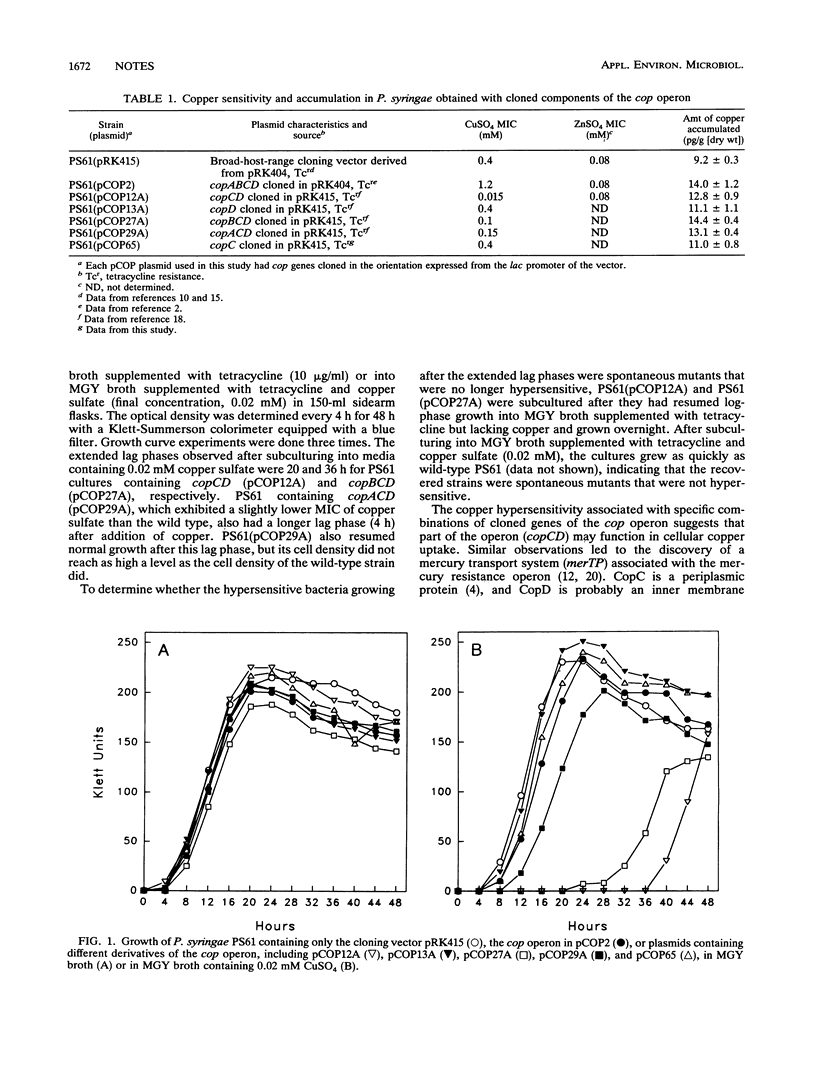
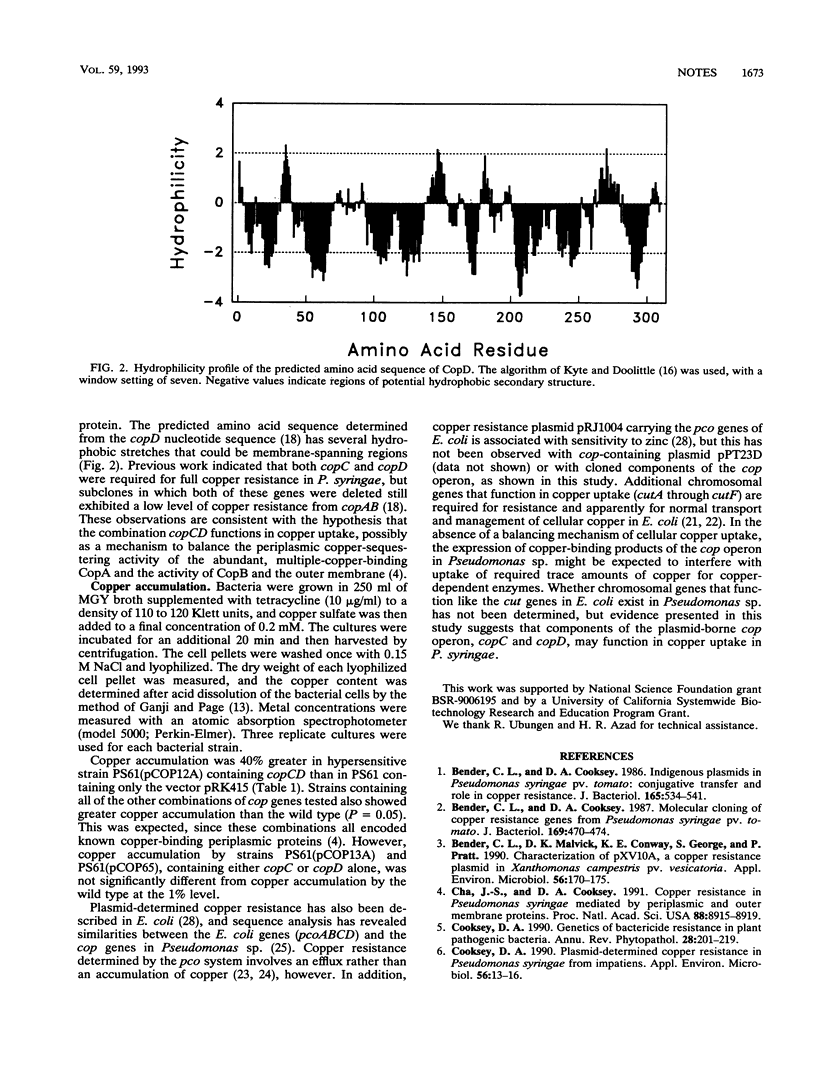
Copper resistance in Pseudomonas syringae carrying the copABCD operon is associated with accumulation of copper in the periplasm and outer membrane, apparently as a function of the copper-binding activities of the copABC gene products. However, no specific function for copD has been determined. In this study, P. syringae cells containing copCD or copBCD cloned behind the lac promoter were hypersensitive to copper. An increased accumulation of copper was measured in cells containing several combinations of cop genes that included copC and copD. Our data suggest that CopC, a periplasmic copper-binding protein, and CopD, a probable inner membrane protein, may function together in copper uptake.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bender C. L., Cooksey D. A. Indigenous plasmids in Pseudomonas syringae pv. tomato: conjugative transfer and role in copper resistance. J Bacteriol. 1986 Feb;165(2):534–541. doi: 10.1128/jb.165.2.534-541.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender C. L., Cooksey D. A. Molecular cloning of copper resistance genes from Pseudomonas syringae pv. tomato. J Bacteriol. 1987 Feb;169(2):470–474. doi: 10.1128/jb.169.2.470-474.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender C. L., Malvick D. K., Conway K. E., George S., Pratt P. Characterization of pXV10A, a Copper Resistance Plasmid in Xanthomonas campestris pv. vesicatoria. Appl Environ Microbiol. 1990 Jan;56(1):170–175. doi: 10.1128/aem.56.1.170-175.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha J. S., Cooksey D. A. Copper resistance in Pseudomonas syringae mediated by periplasmic and outer membrane proteins. Proc Natl Acad Sci U S A. 1991 Oct 15;88(20):8915–8919. doi: 10.1073/pnas.88.20.8915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooksey D. A., Azad H. R. Accumulation of copper and other metals by copper-resistant plant-pathogenic and saprophytic pseudomonads. Appl Environ Microbiol. 1992 Jan;58(1):274–278. doi: 10.1128/aem.58.1.274-278.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooksey D. A., Azad H. R., Cha J. S., Lim C. K. Copper resistance gene homologs in pathogenic and saprophytic bacterial species from tomato. Appl Environ Microbiol. 1990 Feb;56(2):431–435. doi: 10.1128/aem.56.2.431-435.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooksey D. A. Copper uptake and resistance in bacteria. Mol Microbiol. 1993 Jan;7(1):1–5. doi: 10.1111/j.1365-2958.1993.tb01091.x. [DOI] [PubMed] [Google Scholar]
- Cooksey D. A. Plasmid-Determined Copper Resistance in Pseudomonas syringae from Impatiens. Appl Environ Microbiol. 1990 Jan;56(1):13–16. doi: 10.1128/aem.56.1.13-16.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditta G., Schmidhauser T., Yakobson E., Lu P., Liang X. W., Finlay D. R., Guiney D., Helinski D. R. Plasmids related to the broad host range vector, pRK290, useful for gene cloning and for monitoring gene expression. Plasmid. 1985 Mar;13(2):149–153. doi: 10.1016/0147-619x(85)90068-x. [DOI] [PubMed] [Google Scholar]
- Figurski D. H., Helinski D. R. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster T. J., Nakahara H., Weiss A. A., Silver S. Transposon A-generated mutations in the mercuric resistance genes of plasmid R100-1. J Bacteriol. 1979 Oct;140(1):167–181. doi: 10.1128/jb.140.1.167-181.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keen N. T., Tamaki S., Kobayashi D., Trollinger D. Improved broad-host-range plasmids for DNA cloning in gram-negative bacteria. Gene. 1988 Oct 15;70(1):191–197. doi: 10.1016/0378-1119(88)90117-5. [DOI] [PubMed] [Google Scholar]
- Kyte J., Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982 May 5;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Mellano M. A., Cooksey D. A. Induction of the copper resistance operon from Pseudomonas syringae. J Bacteriol. 1988 Sep;170(9):4399–4401. doi: 10.1128/jb.170.9.4399-4401.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellano M. A., Cooksey D. A. Nucleotide sequence and organization of copper resistance genes from Pseudomonas syringae pv. tomato. J Bacteriol. 1988 Jun;170(6):2879–2883. doi: 10.1128/jb.170.6.2879-2883.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahara H., Silver S., Miki T., Rownd R. H. Hypersensitivity to Hg2+ and hyperbinding activity associated with cloned fragments of the mercurial resistance operon of plasmid NR1. J Bacteriol. 1979 Oct;140(1):161–166. doi: 10.1128/jb.140.1.161-166.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers S. D., Bhave M. R., Mercer J. F., Camakaris J., Lee B. T. Cloning and characterization of cutE, a gene involved in copper transport in Escherichia coli. J Bacteriol. 1991 Nov;173(21):6742–6748. doi: 10.1128/jb.173.21.6742-6748.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouch D., Camakaris J., Lee B. T., Luke R. K. Inducible plasmid-mediated copper resistance in Escherichia coli. J Gen Microbiol. 1985 Apr;131(4):939–943. doi: 10.1099/00221287-131-4-939. [DOI] [PubMed] [Google Scholar]
- Tetaz T. J., Luke R. K. Plasmid-controlled resistance to copper in Escherichia coli. J Bacteriol. 1983 Jun;154(3):1263–1268. doi: 10.1128/jb.154.3.1263-1268.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]



