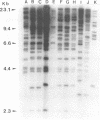
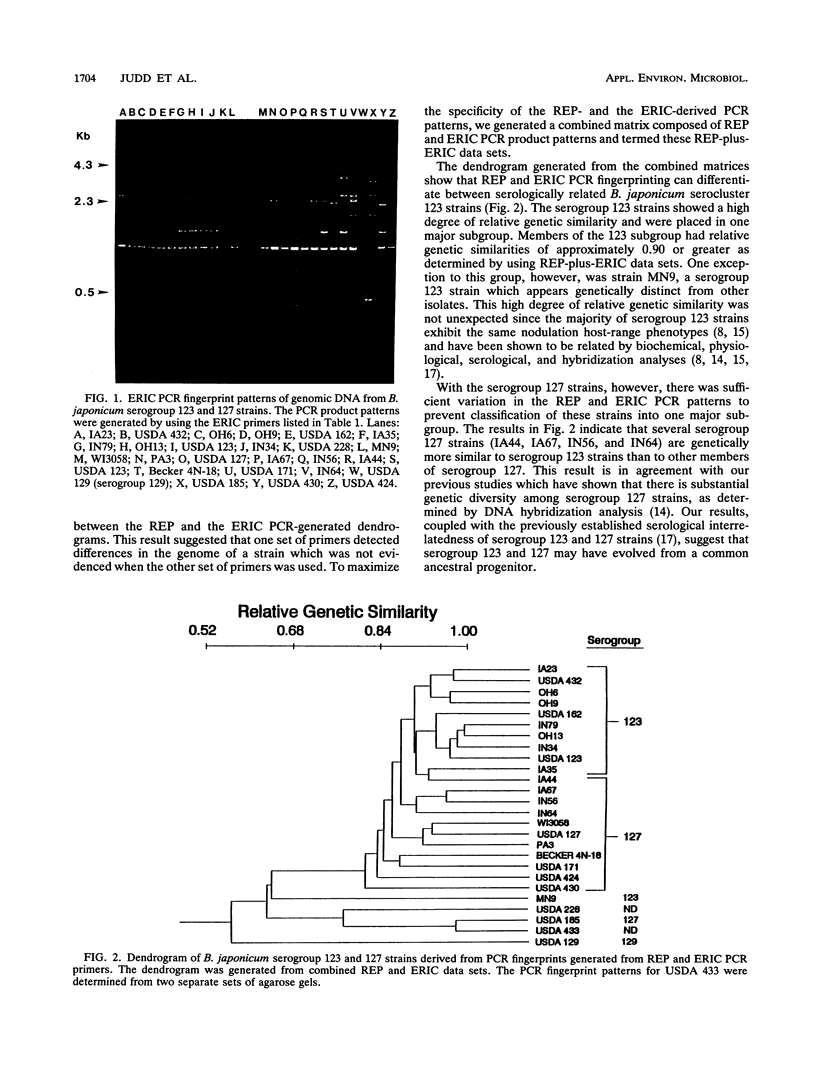
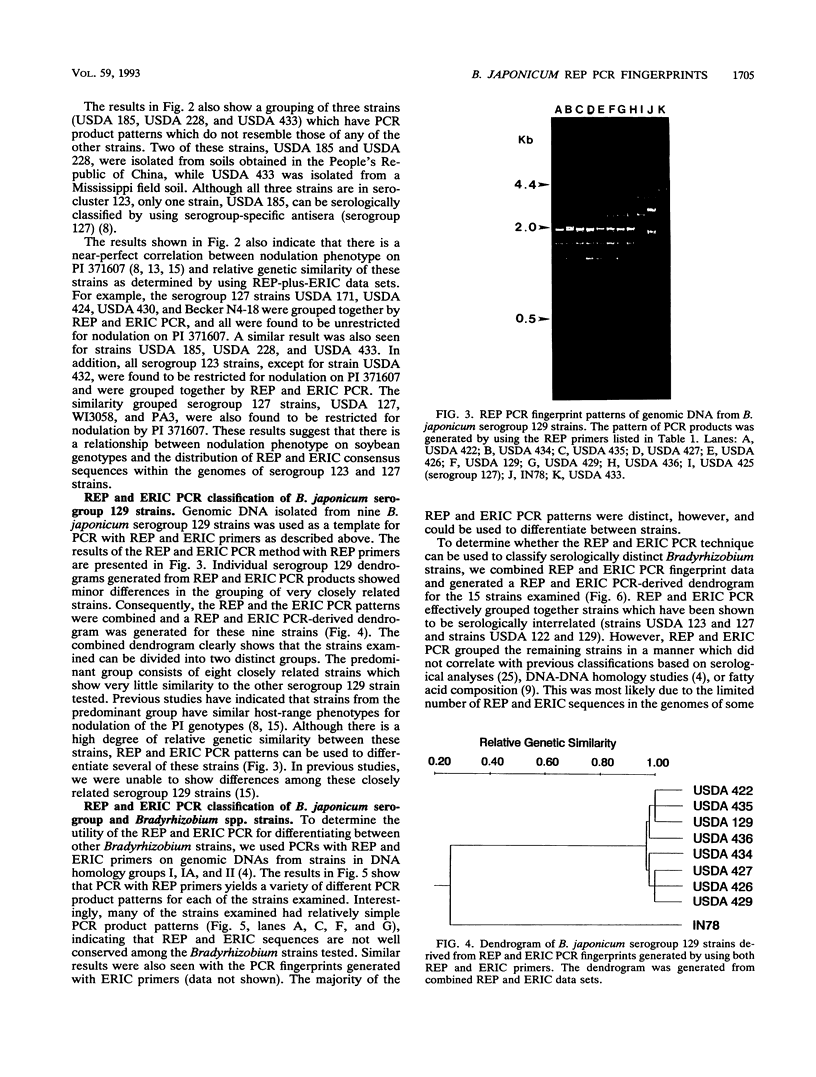
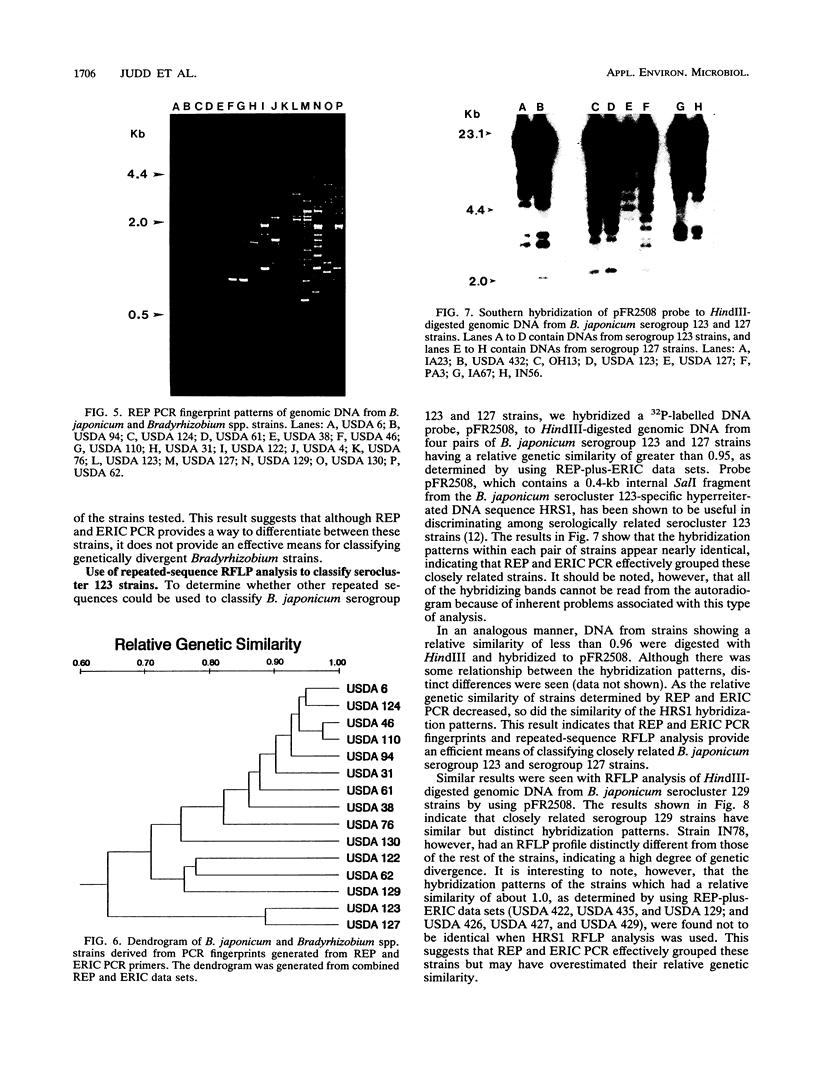
Abstract
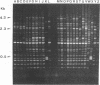
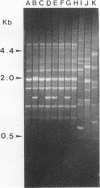
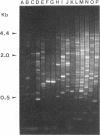
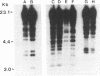
We have determined that repetitive (repetitive extragenic palindromic [REP] and enterobacterial repetitive intergenic consensus [ERIC]) sequences used in conjunction with the polymerase chain reaction technique (REP and ERIC PCR) provide an effective means of differentiating between and classifying genetically related Bradyrhizobium japonicum serocluster 123 strains. Analysis of REP and ERIC PCR-generated dendrograms indicated that this technique can effectively differentiate between closely related strains which were indistinguishable by using other classification methods. To maximize the genomic differences detected by REP and ERIC PCR fingerprint patterns, the REP and the ERIC data sets were combined for statistical analyses. REP-plus-ERIC PCR fingerprints were also found to provide a method to differentiate between highly diverse strains of Bradyrhizobium spp., but they did not provide an effective means for classifying these strains because of the relatively low number of REP and ERIC consensus sequences found in some of the bradyrhizobia. Our results also suggest that there is a relationship between nodulation phenotypes and the distribution of REP and ERIC consensus sequences within the genomes of B. japonicum serogroup 123 and 127 strains. Results obtained by restriction fragment length polymorphism hybridization analyses were correlated with the phylogenetic classification of B. japonicum serocluster 123 strains obtained by using REP and ERIC PCR.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- DATE R. A., DECKER A. M. MINIMAL ANTIGENIC CONSTITUTION OF 28 STRAINS OF RHIZOBIUM JAPONICUM. Can J Microbiol. 1965 Feb;11:1–8. doi: 10.1139/m65-001. [DOI] [PubMed] [Google Scholar]
- Huber T. A., Agarwal A. K., Keister D. L. Extracellular polysaccharide composition, ex planta nitrogenase activity, and DNA homology in Rhizobium japonicum. J Bacteriol. 1984 Jun;158(3):1168–1171. doi: 10.1128/jb.158.3.1168-1171.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulton C. S., Higgins C. F., Sharp P. M. ERIC sequences: a novel family of repetitive elements in the genomes of Escherichia coli, Salmonella typhimurium and other enterobacteria. Mol Microbiol. 1991 Apr;5(4):825–834. doi: 10.1111/j.1365-2958.1991.tb00755.x. [DOI] [PubMed] [Google Scholar]
- Kaluza K., Hahn M., Hennecke H. Repeated sequences similar to insertion elements clustered around the nif region of the Rhizobium japonicum genome. J Bacteriol. 1985 May;162(2):535–542. doi: 10.1128/jb.162.2.535-542.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keyser H. H., Cregan P. B. Nodulation and Competition for Nodulation of Selected Soybean Genotypes among Bradyrhizobium japonicum Serogroup 123 Isolates. Appl Environ Microbiol. 1987 Nov;53(11):2631–2635. doi: 10.1128/aem.53.11.2631-2635.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupski J. R., Weinstock G. M. Short, interspersed repetitive DNA sequences in prokaryotic genomes. J Bacteriol. 1992 Jul;174(14):4525–4529. doi: 10.1128/jb.174.14.4525-4529.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minamisawa K., Seki T., Onodera S., Kubota M., Asami T. Genetic relatedness of Bradyrhizobium japonicum field isolates as revealed by repeated sequences and various other characteristics. Appl Environ Microbiol. 1992 Sep;58(9):2832–2839. doi: 10.1128/aem.58.9.2832-2839.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Quiñones F., Judd A. K., Sadowsky M. J., Liu R. L., Cregan P. B. Hyperreiterated DNA regions are conserved among Bradyrhizobium japonicum serocluster 123 strains. Appl Environ Microbiol. 1992 Jun;58(6):1878–1885. doi: 10.1128/aem.58.6.1878-1885.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadowsky M. J., Bohlool B. B., Keyser H. H. Serological Relatedness of Rhizobium fredii to Other Rhizobia and to the Bradyrhizobia. Appl Environ Microbiol. 1987 Aug;53(8):1785–1789. doi: 10.1128/aem.53.8.1785-1789.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadowsky M. J., Tully R. E., Cregan P. B., Keyser H. H. Genetic Diversity in Bradyrhizobium japonicum Serogroup 123 and Its Relation to Genotype-Specific Nodulation of Soybean. Appl Environ Microbiol. 1987 Nov;53(11):2624–2630. doi: 10.1128/aem.53.11.2624-2630.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt E. L., Zidwick M. J., Abebe H. M. Bradyrhizobium japonicum Serocluster 123 and Diversity among Member Isolates. Appl Environ Microbiol. 1986 Jun;51(6):1212–1215. doi: 10.1128/aem.51.6.1212-1215.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobral B. W., Sadowsky M. J., Atherly A. G. Genome analysis of Bradyrhizobium japonicum serocluster 123 field isolates by using field inversion gel electrophoresis. Appl Environ Microbiol. 1990 Jun;56(6):1949–1953. doi: 10.1128/aem.56.6.1949-1953.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern M. J., Ames G. F., Smith N. H., Robinson E. C., Higgins C. F. Repetitive extragenic palindromic sequences: a major component of the bacterial genome. Cell. 1984 Jul;37(3):1015–1026. doi: 10.1016/0092-8674(84)90436-7. [DOI] [PubMed] [Google Scholar]
- Versalovic J., Koeuth T., Lupski J. R. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 1991 Dec 25;19(24):6823–6831. doi: 10.1093/nar/19.24.6823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bruijn F. J. Use of repetitive (repetitive extragenic palindromic and enterobacterial repetitive intergeneric consensus) sequences and the polymerase chain reaction to fingerprint the genomes of Rhizobium meliloti isolates and other soil bacteria. Appl Environ Microbiol. 1992 Jul;58(7):2180–2187. doi: 10.1128/aem.58.7.2180-2187.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]