Abstract
We used reverse transcription–coupled PCR to produce a high-resolution temporal map of fluctuations in mRNA expression of 112 genes during rat central nervous system development, focusing on the cervical spinal cord. The data provide a temporal gene expression “fingerprint” of spinal cord development based on major families of inter- and intracellular signaling genes. By using distance matrices for the pair-wise comparison of these 112 temporal gene expression patterns as the basis for a cluster analysis, we found five basic “waves” of expression that characterize distinct phases of development. The results suggest functional relationships among the genes fluctuating in parallel. We found that genes belonging to distinct functional classes and gene families clearly map to particular expression profiles. The concepts and data analysis discussed herein may be useful in objectively identifying coherent patterns and sequences of events in the complex genetic signaling network of development. Functional genomics approaches such as this may have applications in the elucidation of complex developmental and degenerative disorders.
The complexity of living organisms poses a challenge to biologists: considering the rapid accumulation of vast amounts of data in the fields of molecular and cell biology, how can we begin to organize these data into a coherent functional whole? To understand the nature of complex biological processes, such as development, we must determine the specific gene expression patterns and biochemical interactions within an organism but, equally important, seek out the organizing principles that allow them to function in a coherent way. Herein, we present a practical experimental-computational strategy that may allow us to advance our understanding of the nature of the complex self-organizing process underlying mammalian central nervous system (CNS) development.
As a first step in this approach, we have addressed the question of whether the temporal expression patterns of large numbers of genes exhibit some degree of order across a tissue, in this case, the developing cervical spinal cord. Further, we are interested in forming hypotheses concerning possible functional relationships between gene families, by examining their patterns of expression over the course of development.
The differentiation and maintenance of a cell phenotype may be viewed as the product of a system of well-coordinated interactions, with some cell types influencing the development of others. Therefore, we have taken a systems approach to CNS development in which the tissue is treated as a whole. In vivo gene expression patterns characteristic of stem cells, pluripotent progenitor cells, and mature neurons and glia should be reflected in the patterns of gene expression obtained at different developmental time points.
Ongoing genome sequencing projects are based on the concept that proteins mediating the functions of organisms are strictly determined by the structure and activity of the genes that encode them. Data from gene-knockout experiments and molecular analysis of individual genes reflect combinatorial regulation, as well as various degrees of redundancy of gene function (1, 2). These characteristics imply a complex network, the underlying principles of which have not yet been explained. We conceptualize this extended genetic network as consisting of two subsystems (3): the proximal genetic network, operating through cis (control regions of DNA) and trans (gene products that regulate cis regions) elements, and the distal genetic network, involving protein–protein and protein–signaling factor interactions governing intra- and extracellular communication (again determined by genes encoding the participating proteins).
Herein, we demonstrate the systematic measurement of multiple gene expression time series, producing a temporal map of developmental gene expression. We have clustered the genes into related expression patterns as a step toward drawing inferences about regulatory origins and interactions among gene families. In regard to large-scale genomics, this study emphasizes temporal patterns (nine time points) and measurement precision [triplicate reverse transcription–coupled PCR (RT-PCR) assays], rather than numbers of genes (112 genes; total 3,024 expression assays), as a strategy for drawing inferences concerning gene interactions and functions. When combined with studies of gene expression in individual cell types, this strategy should be particularly useful in understanding the changes in gene expression underlying the transition of the CNS from a primarily proliferative to a highly differentiated state.
MATERIALS AND METHODS
We have used an established RT-PCR protocol (4) to measure the expression of 112 genes in CNS development. Cervical spinal cord tissue was dissected from triplicate animals or litters (Sprague–Dawley albino rats), in accordance with National Institutes of Health guidelines, from embryonic days 11 through 21 (E11–E21; determined by crown–rump length), postnatal days 0–14 (P0–P14), and adult (P90 or adult). Gene-specific primers were designed from GenBank sequences by using the oligo software (National Biosciences, Plymouth, MN). RNA isolated from tissue samples by using RNAstat 60 (Tel-Test, Friendswood, TX) was adjusted to 200 ng/μl according to absorption at 260 nm, before RT-PCR (Perkin–Elmer GeneAmp RNA PCR kit, Applied Biosystems); PCR involved preheating a mixture of Taq antibody (TaqStart, CLONTECH), primers, cDNA, and PCR components to 97°C for 90 sec before amplification. The PCR cycle was 30 sec at 95°C (dissociation), 45 sec at 60°C (annealing), and 60 sec at 72°C (extension). Amplification was within the exponential range (4). PCR product identities were confirmed by restriction enzyme digestion. All RT reactions and PCRs contained control RNA (transcribed from PAW 108 plasmid DNA; Applied Biosystems) to allow ratiometric quantitation and eliminate inefficient reactions from the analysis. Data were obtained by densitometry (nih image) of PCR products resolved with PAGE (ratios of sample bands to corresponding control bands). For every gene, ratiometric data for each group of triplicate animals were averaged for each time point and normalized to maximal expression level among the nine time points (Fig. 1).
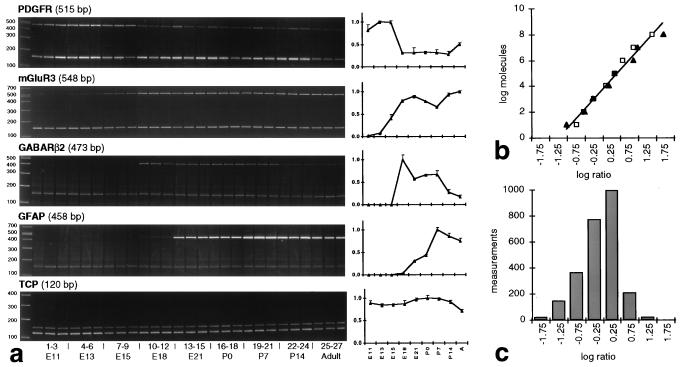
Figure 1.
RT-PCR/PAGE assay. (a) Analysis of representative ethidium bromide-stained polyacrylamide gels. Bands at 150 bp are the PAW 108 internal control PCR product. Every PCR band is from a different animal. Time series of normalized ratiometric densitometry data (averaged ± SEM) are graphed to the right of each gel. (b) Dynamic range of RT-PCR assay. The relationship between the log(starting molecules) and the log(product/control) ratio is linear between 10 and 108 molecules for GAD65 (squares) and GAD67 (triangles; for details, see ref. 4). (c) Range of ratiometric values. The histogram shows the distribution of densitometrically determined product ratios. The measurement values (c) are comfortably within the linear range of the log–log assay (b), far removed from potential saturation.
We clustered the gene expression time series according to the Euclidean distance measure (square root of the sum of the squared differences in each dimension) by using the fitch software (5). We determined the 112 × 112 gene Euclidean distance matrix from the combined 17 dimensional vectors of nine expression values (ranging between 0 and 1) and eight slopes (ranging between −1 and +1; slopes were calculated based on a reduced time interval of 1, not taking into account the variable time intervals). We included slopes to take into account offset but parallel patterns. We used the default parameters for fitch, except that we set the P parameter to zero, to implement the least-squares method appropriate for data with expected linearly proportional error. Cluster boundaries were determined by visual inspection of the Euclidean distance tree. Principal component analysis was performed according to standard routines implemented in the s-plus statistical software package.
RESULTS
Included in the assay were major gene families deemed important for spinal cord development because of their recognized roles in intercellular signaling and a smaller number that are known for major roles in intracellular signaling or transcriptional regulation: neurotransmitter synthesizing and metabolizing enzymes [relating to γ-aminobutyric acid (GABA), acetylcholine, catecholamines, and nitric oxide], ionotropic neurotransmitter receptors [GABAA, N-methyl-d-aspartate (NMDA), nicotinic acetylcholine (nAChR), and serotonin (5HT) receptors], metabotropic neurotransmitter receptors (metabotropic glutamate, muscarinic acetylcholine, and 5HT receptors), neurotrophins and their receptors, heparin-binding growth factors and their receptors, insulin and insulin-like growth factor (IGF) family and their receptors, intracellular inositol 1,4,5-trisphosphate receptors, cell cycle proteins, transcriptional regulatory factors, expressed sequence tags, and other (housekeeping) genes. We included genes for established developmental “marker” proteins as well, to correlate expression time series to indicators of phenotypic differentiation.
Gene expression levels among independently run triplicate samples were generally uniform (Fig. 1a). Two of the primers sets used herein were tested previously in calibration reactions under similar conditions (4), to demonstrate that the dynamic range of this method covers eight orders of magnitude (Fig. 1b). The distribution of the ratiometric values in the present work (Fig. 1c) suggests that we are well within the linear range of this assay. We determined that it was not practical to perform calibration reactions for every set of PCR primers because of the large numbers of genes assayed and because the absolute quantity of mRNA is not necessarily an indication of its efficacy within the genetic network. Therefore, we have focused on comparing temporal expression patterns. We have diagrammed our measurements as a temporal gene expression map (Fig. 2) and tabulated the raw data (http://rsb.info.nih.gov/mol-physiol/PNAS/GEMtable.html).
Figure 2.
Temporal gene expression map of the developing spinal cord. Expression patterns are labeled according to gene name and general functional class. Densitometry data from ratiometric RT-PCR and PAGE for triplicate animals are shown for each time point. Darkest, maximal expression; white, undetectable expression. MAP2, microtubule-associated protein 2; GAP43, growth-associated protein 43; L1, neural cell adhesion molecule (NCAM); NFL, NFM, NFH, neurofilament light, medium, and heavy; neno, neuron-specific enolase; GFAP, glial fibrillary acidic protein; MOG, myelin-oligodendrocyte glycoprotein; GAD65, glutamate decarboxylase 65; GAD67, G67I80/86, and G67I86, glutamate decarboxylase 67 splice variants; GAT1, GABA transporter 1; ChAT, choline acetyltransferase; AChE, acetylcholinesterase; ODC, ornithine decarboxylase; TH, tyrosine hydroxylase; NOS, nitric oxide synthase; GR, GABA receptor; mGluR, metabotropic glutamate receptor; NMDA, NMDA receptor; nAChR, nicotinic acetylcholine receptor; mAChR, muscarinic acetylcholine receptor; 5HT, serotonin (5-hydroxytryptamine) receptor; NGF, nerve growth factor; NT3, neurotrophin 3; BDNF, brain-derived neurotrophic factor; CNTF, ciliary neurotrophic factor (CNTFR, receptor); trk, NGF receptor; MK2, midkine 2; PTN, pleiotrophin; GDNF, glial-derived neurotrophic factor; EGF and EGFR, epidermal growth factor and its receptor; bFGF, aFGF, basic and acidic fibroblast growth factor; PDGF and PDGFR, platelet-derived growth factor and its receptor; FGFR, fibroblast growth factor receptor; TGFR, transforming growth factor receptor; Ins, insulin; InsR, insulin receptor; IGF, insulin-like growth factor; IGFR, IGF receptor; CRAF, c-raf protooncogene; IP3R, inositol 1,4,5-trisphosphate receptor; H2AZ, H2A.Z histone protein; Brm, brahma; TCP, t-complex protein; SOD, superoxide dismutase; CCO1 and CCO2, cytochrome c oxidase, subunits 1 and 2.
The construction of a temporal gene expression map by using a single RT-PCR protocol allowed us to analyze spinal cord development as a pattern of potentially functionally related genes and gene clusters, all processed in parallel. In this context, we used two objective methods to correlate gene expression time series.
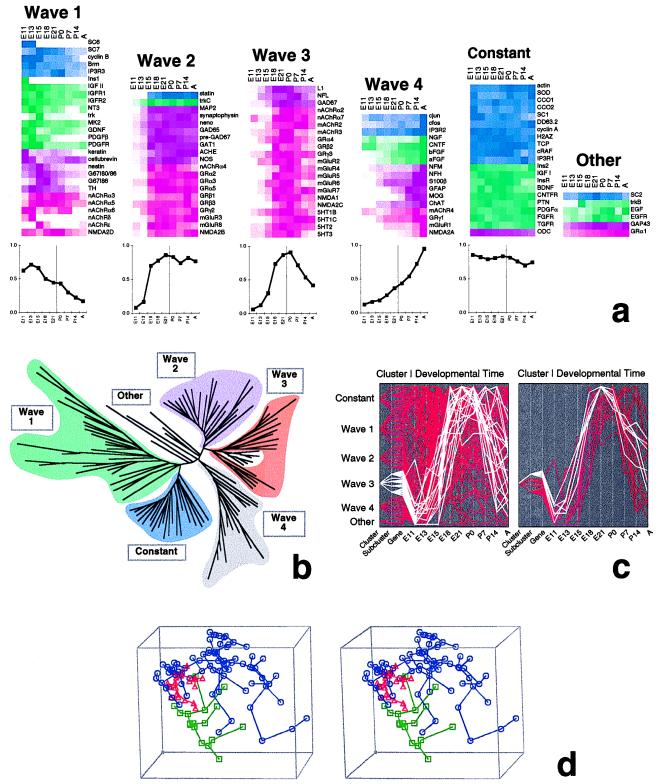
The Euclidean distance matrix and fitch program clustered the 112 genes according to similarities in their temporal expression patterns. This method is similar to grouping according to positive correlations between expression patterns. The genes cluster into four major waves of expression and a largely invariant group (Fig. 3). Wave 1, characterized by high levels of expression occurring only during early developmental stages in 27 genes (E11–E15), includes members of diverse gene families. Two genes, SC6 and SC7, closely cluster to individual members of wave 1, suggesting a functional kinship to known genes. Genes in waves 2 (20 genes) and 3 (21 genes) steeply increase from E13 to E15 and from E15 to E18, respectively. The members of wave 2 remain at their plateau expression level at the completion of development, whereas the members of wave 3 generally have a pattern of transient high expression. Both waves 2 and 3 are notably confined to neurotransmitter signaling. Finally, wave 4 (17 genes) clusters genes that primarily increase during postnatal development, belonging to several functional families. Twenty-one genes showing largely constant expression (constant group) originate from diverse families but strictly exclude neurotransmitter signaling genes and neuroglial markers. Relative contributions of gene sequence families and functional families to the different waves are shown in Table 1. Temporal patterns typical for each Euclidean cluster are shown in Fig. 1; platelet-derived growth factor receptor, metabotropic glutamate receptor mGluR3, GABARβ3, glial fibrillary acidic protein, and t-complex protein correspond to waves 1-4 and constant, respectively.
Figure 3.
Gene expression waves. (a) Normalized gene expression trajectories from Fig. 2 are shown grouped by “waves” determined by Euclidean distance clustering. The graphs below show the average normalized expression pattern or “wave” over the nine time points for all the genes in each cluster (the time of birth is marked by a vertical line). Within each wave, genes are grouped according to gene families, not according to proximity as determined by Euclidean distance. (b) Euclidean distance tree of all gene expression patterns (for annotated tree, see http://rsb.info.nih.gov/mol-physiol/PNAS/tree.html). Major branches correspond to waves in a. (c) Plots of all normalized time series, highlighting wave 3 (Left, white lines) and a subcluster of wave 3 (Right, white lines plotted on top of remaining genes of wave 3 in red). Subclusters (secondary branching) were selected by visual inspection from tree in b; e.g., the plotted time series of the wave 3 subcluster correspond to the branchlet highlighted in white within wave 3 in b. (d) Principal component analysis. Principal components projection viewed as a three-dimensional stereo plot. Each point mapped in three-dimensional space represents an expression time series corresponding to a gene in Fig. 2. Highlighted points correspond to Euclidean distance wave 3 (red triangles), wave 4 (green squares), and the remaining genes (blue octagons). Abbreviations are as in Fig. 2.
Table 1.
Mapping of expression clusters to functional gene classes
| Cluster | Neurotransmitter receptors
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| General gene class
|
Ligand class | Sequence class
|
||||||||
| % peptide signaling | % neurotr. receptors | % neuroglial markers | % diverse | % ACh | % GABA | % Glu | % 5HT | % ion channel | % G protein coupled | |
| Wave 1 | 37 | 15 | 24 | 24 | 86 | 0 | 14 | 0 | 100 | 0 |
| Wave 2 | 6 | 39 | 48 | 7 | 10 | 65 | 25 | 0 | 69 | 31 |
| Wave 3 | 0 | 79 | 21 | 0 | 17 | 14 | 24 | 45 | 31 | 69 |
| Wave 4 | 23 | 16 | 38 | 23 | 26 | 29 | 45 | 0 | 36 | 64 |
| Constant | 37 | 0 | 4 | 58 | ||||||
| Other | 50 | 11 | 18 | 21 | ||||||
To eliminate the bias caused by gene selection, the percentages reflect the contribution of a gene class to an expression cluster relative to the overall representation of each gene class in the assay. For general gene clusters, expression clusters are enriched for major gene classes (defined in Fig. 2). For neurotransmitter receptors, distinct categories of neurotransmitter (neurotr.) receptors map to selected expression waves. Receptors are classified according to ligand class or functional sequence class. Constant and other clusters are not listed because neurotransmitter receptors are essentially absent from these groups. Boldface type represents dominant class in cluster. ACh, acetylcholine.
Of interest, tyrosine hydroxylase, insulin 1, and IGF-II, whose genes are located on the same human cytogenetic band (11p15.5) (6), are closely clustered in Euclidean distance wave 1. Further, tyrosine hydroxylase and IGF-II are adjacent to each other in the Euclidean distance tree (see http://rsb.info.nih.gov/mol-physiol/PNAS/tree.html). Tyrosine hydroxylase, insulin 2, and IGF-II are also in close proximity to one another on mouse chromosome 7 (Mouse Genome Database, Mouse Genome Informatics, The Jackson Laboratory, Bar Harbor, MA. WWW site: http://www.informatics.jax.org/). However, generally we were not able to correlate patterns of gene regulation with chromosomal location, suggesting that chromosomal proximity is not a major candidate regulatory parameter.
Peptide signaling genes map mainly to Euclidean distance clusters constant and wave 1. Similarly, the neurotransmitter receptor genes map primarily to Euclidean distance waves 2 and 3. Interestingly, all neurotransmitter receptors in wave 1 and most of those in wave 2 (mainly GABAA receptors) are ionotropic, whereas most neurotransmitter receptors in waves 3 and 4 are metabotropic (Fig. 3 and Table 1). The metabotropic receptors are unrelated to ionotropic receptors in terms of sequence kinship; therefore, it is not surprising that these two receptor classes have highly divergent temporal expression patterns. However, comparison of the developmental expression time series for the GABA, nicotinic and muscarinic acetylcholine, and glutamate receptor genes with the phylogenetic trees for these gene families reveals a poor correlation between phylogeny and ontogenetic expression patterns (the same also applies to the data on peptide signaling gene families studied herein). This suggests that sequence homologies within gene families are not tightly coupled to the timing of gene regulation over the course of development.
Further inspection of the data points to deeper relationships between signaling gene families. Note that most of the nAChR genes are found in wave 1 (Fig. 3 and Table 1). In addition to the established developmentally restricted nAChR δ and ɛ subunits, the expression profiles of the α3, α5, and α6 subunits suggest that cholinergic transmission forms the leading edge in neurotransmitter signaling in the cervical spinal cord. The slightly delayed expression of acetylcholinesterase (wave 2) and late expression of choline acetyltransferase (wave 4) suggest that early sources of acetylcholine, if they exist, must originate outside the cervical spinal cord. For the GABAergic signaling system, the relationship between synthesizing enzymes and receptors is more coherent. Glutamate decarboxylase 65 (GAD65) and pre-GAD67 (sum of splicing variants of GAD67) appear with a GABA transporter gene (GAT1) and six of the GABAA receptor subunit genes in wave 2. The remaining four GABAA receptor subunits are found in waves 3 and 4. Wave 3 also shows a preponderance of the metabotropic glutamate receptors (five of eight) and all the analyzed 5HT receptors (Fig. 3 and Table 1).
For peptide growth factor genes, a close correspondence exists between the expression of signal and receptor for insulin 1, IGF-II, IGF receptors 1 and 2, and platelet-derived growth factor β and the platelet-derived growth factor receptor in wave 1. The related neurotrophin pair, nerve growth factor and ciliary neurotrophic factor, and fibroblast growth factor pair (FGF), acidic FGF and basic FGF, are closely clustered in wave 4 (also forming an exception to the general growth factor mapping into wave 1 and constant). However, regulation of receptors for ciliary neurotrophic factor (ciliary neurotrophic factor receptor; constant), nerve growth factor (trk; wave 1), and the FGFs (constant) does not exactly coincide with that of the corresponding ligand genes. Interestingly, epidermal growth factor and its receptor form a unique pair in a separate sixth group (other).
In addition, we applied a statistical method, principal component analysis, to confirm independently the cluster analysis results. This method is based on the covariance of the gene expression time series. The first three principal components, shown as x, y, and z axes in the stereo plot in Fig. 3d capture 66% of the variability in the 17 coordinates (nine expression values and eight slopes). One may use a stereo viewer to fuse the images of Fig. 3d; the square dot on the frame appears at the back of the fused image. Minimal spanning trees are included to facilitate image fusion and study (7, 8). The paucity of points in the center of the data cloud suggests strong constraints on gene expression (such as the absence of high–low–high patterns). In contrast, random rearrangement of the data within each time series results in accumulation of the data points in the center of the first three principal components view (data not shown), as expected for random data. The genes clustered by Euclidean distance are grouped together within the three-dimensional view. Wave 3 (red) is a tight cluster but some of the other (blue) points are very close. Wave 4 (green) is spread out but well separated from the other clusters. This suggests that many genes in these clusters would continue to be grouped together by various Euclidean distance clustering algorithms (confirmed by comparison of several alternative clustering algorithms; results not shown).
DISCUSSION
The rapidly progressing genome projects are providing a toolbox that enables us to go beyond the study of individual genes in isolation to the characterization of a network of combinatorial gene interactions (3). A fundamental aspect of this “functional genomics” is a straightforward cataloging of gene expression in different species and tissues (9). In addition to assaying large numbers of genes, extensive perturbation studies, and time series of the appropriate temporal resolution will be essential for “reverse engineering” to produce a gene–gene “interaction diagram” (3, 10–12).
Despite our use of whole cervical spinal cord, we found a high degree of order among most of the 112 selected genes: five basic expression patterns or waves. This demonstrates that fundamental patterns of temporal fluctuations in gene expression can be discerned even without dissecting whole tissue into distinct anatomical subregions. Further, this suggests that each gene in waves 1–4 is not expressed in all anatomical regions at a different time point for each region; if this were the case, all the genes we assayed would exhibit relatively constant expression levels over time. The data therefore suggest the existence of strong constraints on gene regulation on the tissue level. Interestingly, we did not observe gene expression patterns characterized by large amplitude oscillations or a U-shaped time course of high early, low intermediate, and high final developmental expression. It remains to be determined whether the absence of such patterns is a characteristic of gene expression in spinal cord or the result of a bias in our selection of genes.
Beyond grouping functional gene families, the Euclidean clusters identify distinct phases of spinal cord development. Specifically, wave 1 is indicative of an immature proliferative stage. The genes of wave 1 represent indicators for dividing neuroglial progenitor cells (e.g., the ectodermal marker keratin, the cell cycle gene cyclin B, and the progenitor cell marker nestin) and the physiological signals ostensibly important to their activity (growth factors and their receptors). These genes represent a group that should be studied in spinal cord disease and injury, during which the organism may make attempts at reactivation of developmental programs (13). Wave 2 is indicative of neurogenesis, as evidenced by the coexpression of neuronal markers such as synaptophysin, neuron-specific enolase, and a vast group of neurotransmitter metabolizing enzymes and receptors, in particular relating to GABAergic signaling (see also ref. 14).
Wave 3, although exclusively covering neurotransmitter signaling genes and neuronal markers, is distinguished from wave 2 by a characteristic low–high–low pattern of developmental gene expression and a slower rise time. A fundamental phenomenon in CNS development is overproduction of cells, many of which are later eliminated during maturation and cementing of synaptic connections. In spinal cord, 80% of all cells disappear between E15 and E18 (15). Many motoneurons are eliminated during the first weeks of postnatal spinal cord development (16). It has been hypothesized that induction of spinal cord cell death may be mediated by the NMDA class of glutamate receptors (14). Could reduction of the NMDA1 and NMDA2C receptors, as well as metabotropic glutamate and other neurotransmitter receptors, particularly in wave 3, be related to postnatal elimination of cells expressing these genes? Alternatively, genes in wave 3 may have a developmentally restricted role and may be down-regulated independent of cell death. Studies on localization of gene expression and coanalysis of cell-death-related genes may help to elucidate these alternative interpretations. In conclusion, the pattern of gene expression in wave 3 emphasizes that neuronal-signaling gene expression is not a gradual linear process in which genes asymptotically approach their mature tissue levels but that there is a transient phase of high expression, analogous to the transient overabundance of cells in neurodevelopment.
Finally, wave 4 coincides with gliogenesis and final maturation of the tissue. This is indicated by the expression patterns of glial fibrillary acidic protein (astrocyte marker), myelin-oligodendrocyte glycoprotein (oligodendrocyte marker), and several coincident peptide and neurotransmitter signaling genes. Questions arise as to whether these gene groups are colocalized in glia or whether these signaling genes may be a response of other cells to glial differentiation or may be themselves regulators of gliogenesis.
Overall, there is a clear mapping of functional gene groups to expression profiles. Although we have also seen several examples of coexpression between receptor–ligand gene pairs, exceptions to this rule suggest outside sources of signals and alternative ligands or receptors.
Our analysis suggests testable hypotheses concerning gene regulation. For example, our data suggest that SC6 and nAChRδ may share regulatory inputs, given their tight clustering in the Euclidean distance tree; the same may be said for SC7, nestin, G67I80/86, and G67I80, which are all found closely clustered within wave 1 (see http://rsb.info.nih.gov/mol-physiol/PNAS/tree.html). Inputs to known genes may, therefore, be tested for their ability to regulate the expression of genes such as SC6 and SC7, whose functions have yet to be determined. Generally, similarities in temporal expression patterns may point to the existence of common regulatory structures and pathways.
We need a simple model for conceptualizing how large numbers of genes interact to generate a complex but robust system. Threshold levels of gene expression are a possible mechanism by which the genetic program makes decisions about the timing of development. Boolean network models are based on a binary idealization of thresholding and exhibit dynamic behaviors such as self-organization, cycling, and maintenance of complex stable structures, referred to as attractors (3, 17). Although this model is oversimplified, abstractions such as this may be useful in conceptualizing the nature of genetic information flow (3).
Additional issues must be addressed in understanding the principles that govern the complex behavior of genetic networks. Positional data on gene expression will be required for a greater understanding of how time series relate to anatomical development. In addition, our present strategy does not account for possible differences between mRNA and protein expression; however, it is reasonable to assume that protein and gene expression patterns are generally well-correlated.
Large-scale gene expression assays may be performed by using RT-PCR, serial analysis of gene expression (SAGE, ref. 18), or DNA chip technology (19). We chose to use RT-PCR because of its exceptional sensitivity and dynamic range, reliability, and flexibility. RT-PCR can be scaled up to cover the same number of genes as SAGE and DNA chips by using robotics and capillary electrophoresis arrays for separating PCR products (20, 21).
Finally, analytical computational techniques are needed to interpret data. Reverse engineering (10–12) will require data from experimental perturbations of tissue (e.g., injury or pharmacological perturbation), combined with analysis of spatial localization and cell-type-specific gene expression patterns. Cluster analysis places constraints on the structure of the genetic network, although short of finding the inputs and regulatory rules themselves. Functional gene families may be recast in terms of gene expression clusters. This may be useful in defining roles for the large numbers of newly sequenced genes with unknown function. A genetic networks approach may provide a better understanding of the flow of genetic information during development and could lead to the generation and testing of new hypotheses for the study of developmental disorders or cancer.
Acknowledgments
We thank David Lange of the Research Services Branch, National Institute of Neurological Disorders and Stroke for help in generating the graphics. We are also grateful to George Gabor Miklos (Neurosciences Institute, San Diego), Harold Morowitz (Krasnow Institute, George Mason University), and Jeffrey Scargle (National Aeronautics and Space Administration, Ames) for helpful comments on the manuscript.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: GAD, glutamate decarboxylase; NMDA, N-methyl-d-aspartate; nAChR, nicotinic acetylcholine receptor; 5HT, serotonin (5-hydroxytryptamine); FGF, fibroblast growth factor; IGF, insulin-like growth factor; RT-PCR, reverse transcription–coupled PCR; CNS, central nervous system; E, embryonic day(s); P, postnatal day(s); GABA, γ-aminobutyric acid.
References
- 1.Crossin K L. Perspect Dev Neurobiol. 1994;2:21–32. [PubMed] [Google Scholar]
- 2.Shastry B S. Mol Cell Biochem. 1994;136:171–182. doi: 10.1007/BF00926078. [DOI] [PubMed] [Google Scholar]
- 3.Somogyi R, Sniegoski C A. Complexity. 1996;1(6):45–63. [Google Scholar]
- 4.Somogyi R, Wen X, Ma W, Barker J L. J Neurosci. 1995;15:2575–2591. doi: 10.1523/JNEUROSCI.15-04-02575.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Felsenstein J. phylip (Phylogeny Inference Package), version 3.5c. Univ. of Washington, Seattle: Department of Genetics; 1993. [Google Scholar]
- 6.Lucassen A M, Julier C, Beressi J P, Boitard C, Froguel P, Lathrop M, Bell J I. Nat Genet. 1993;4:305–310. doi: 10.1038/ng0793-305. [DOI] [PubMed] [Google Scholar]
- 7.Carr D B, Littlefield R J. Computer Science and Statistics, Proceedings of the 15th Symposium on the Interface. New York: North Holland; 1983. pp. 295–299. [Google Scholar]
- 8.Carr D B. Stat Comp Graph News. 1993;4:2–7. [Google Scholar]
- 9.Lander E S. Science. 1996;274:536–539. doi: 10.1126/science.274.5287.536. [DOI] [PubMed] [Google Scholar]
- 10.Somogyi, R., Fuhrman, S., Askenazi, M. & Wuensche, A. Proceedings of the Second World Congress of Nonlinear Analysts (WCNA96) (Elsevier Science, Amsterdam), in press.
- 11.Liang, S., Fuhrman, S. & Somogyi, R. (1998) Proceedings of the Pacific Symposium on Biocomputing, 1998, in press. [PubMed]
- 12.Arkin A, Peidong S, Ross J. Science. 1997;277:1275–1279. [Google Scholar]
- 13.Ma W, Chang L, Zhang L, Barker J L. Soc Neurosci Abstr. 1996;22:514.4. [Google Scholar]
- 14.Barker, J. L., Behar, T., Li, Y.-X., Liu, Q.-Y., Ma, W., Maric, D., Maric, I., Schaffner, A. E., Serafini, R., Smith, S. V., Somogyi, R., Vautrin, J. Y., Wen, X. & Xian, H. (1997) Perspect. Dev. Neurobiol. 4, in press. [PubMed]
- 15.Maric D, Maric I, Ma W, Lahjouji F, Somogyi R, Wen X, Sieghart W, Fritschy J-M, Barker J L. Eur J Neurosci. 1996;9:507–522. doi: 10.1111/j.1460-9568.1997.tb01628.x. [DOI] [PubMed] [Google Scholar]
- 16.Jacobson M. Developmental Neurobiology. New York: Plenum; 1991. [Google Scholar]
- 17.Kauffman S A. The Origins of Order: Self-Organization and Selection in Evolution. New York: Oxford Univ. Press; 1993. [Google Scholar]
- 18.Velculescu V E, Zhang L, Vogelstein B, Kinzler K W. Science. 1995;270:484–487. doi: 10.1126/science.270.5235.484. [DOI] [PubMed] [Google Scholar]
- 19.Lockhart D J, Dong H, Byrne M C, Follettie M T, Gallo M V, Chee M S, Mittmann M, Wang C, Kobayashi M, Horton H, Brown E L. Nat Biotechnol. 1996;14:1675–1680. doi: 10.1038/nbt1296-1675. [DOI] [PubMed] [Google Scholar]
- 20.Ueno K, Yeung E S. Anal Chem. 1994;66:1424–1431. [Google Scholar]
- 21.Somogyi R. Federal Register. 1995;60:34544–34545. [Google Scholar]