Abstract
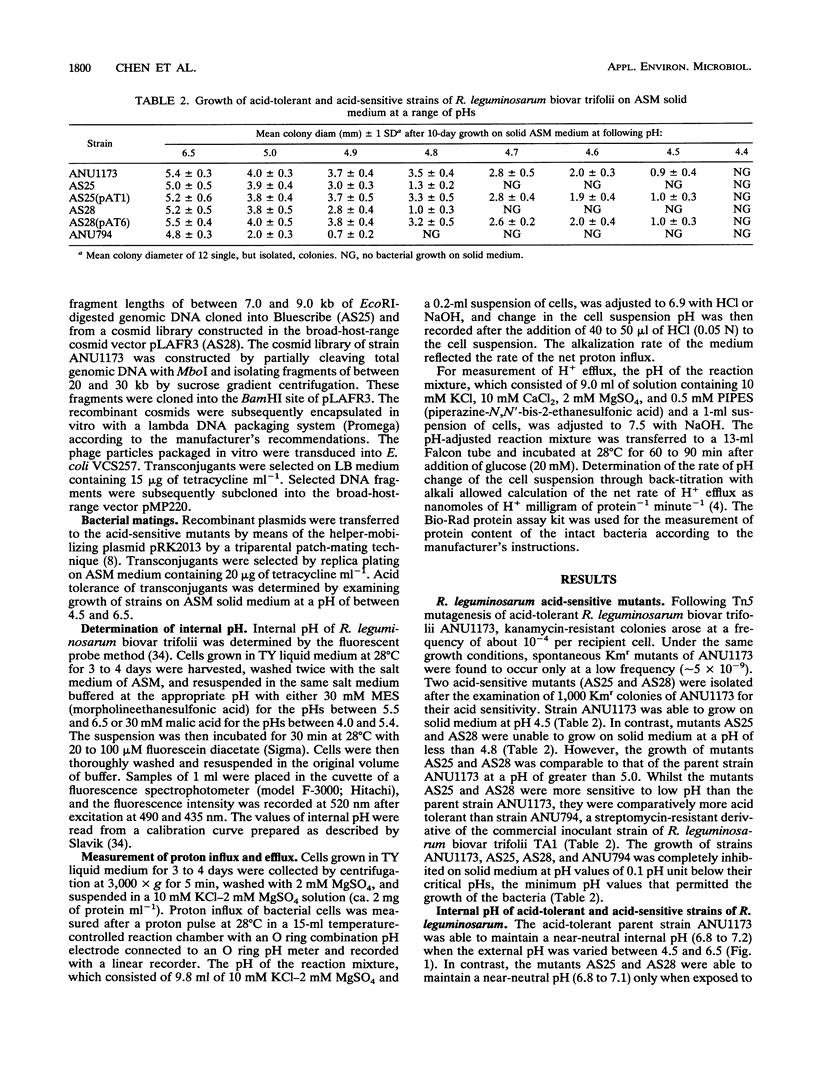
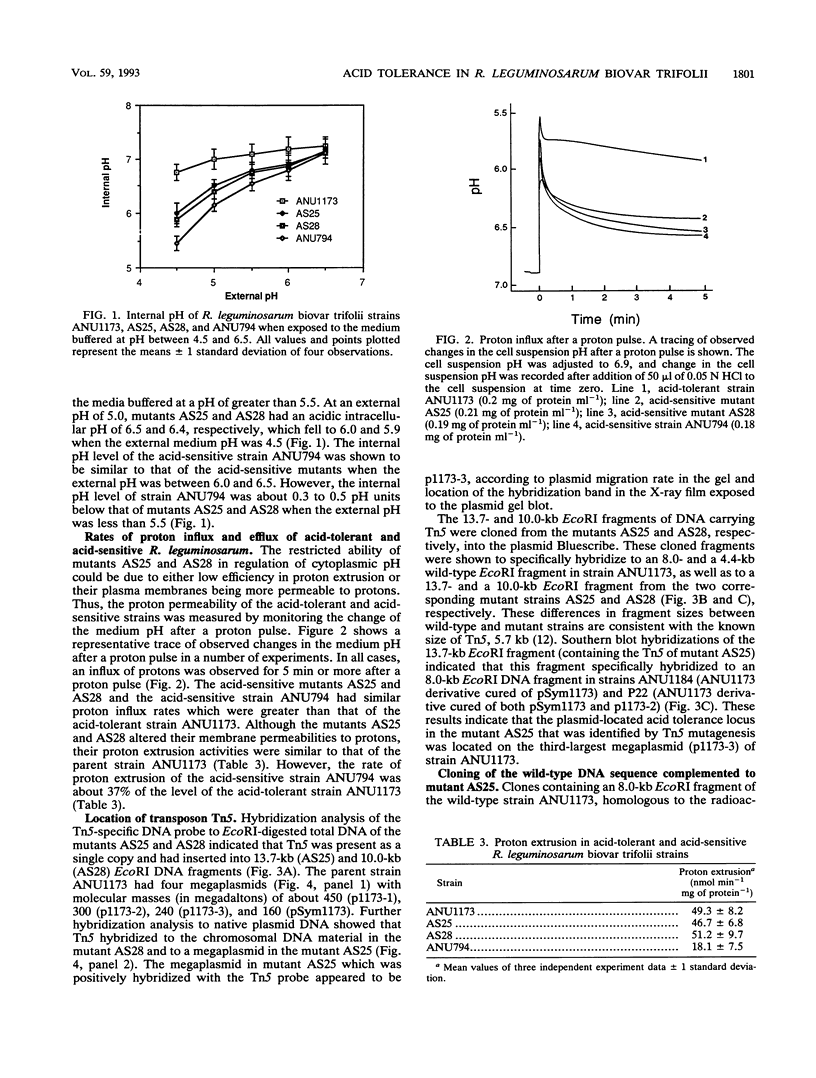
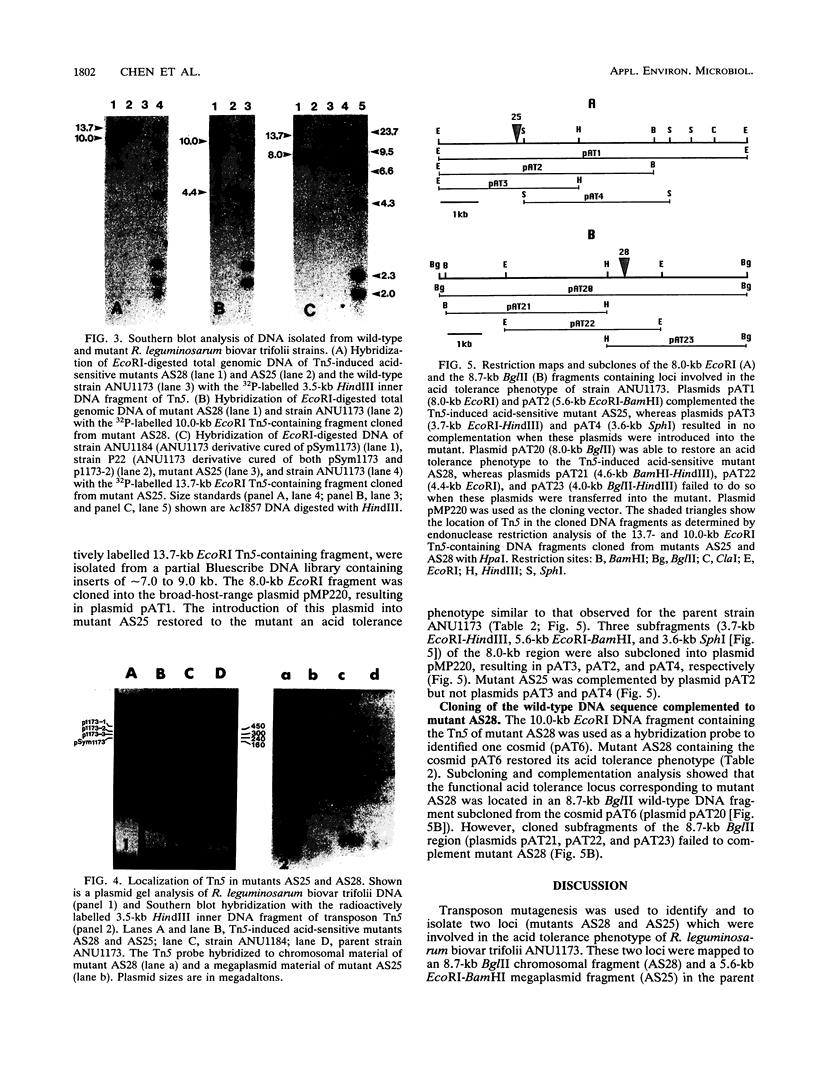
Acid-tolerant Rhizobium leguminosarum biovar trifolii ANU1173 was able to grow on laboratory media at a pH as low as 4.5. Transposon Tn5 mutagenesis was used to isolate mutants of strain ANU1173, which were unable to grow on media at a pH of less than 4.8. The acid-tolerant strain ANU1173 maintained a near-neutral intracellular pH when the external pH was as low as 4.5. In contrast, the acid-sensitive mutants AS25 and AS28 derived from ANU1173 had an acidic intracellular pH when the external pH was less than 5.5. The acid-sensitive R. leguminosarum biovar trifolii ANU794, which was comparatively more sensitive to low pH than mutants AS25 and AS28, showed a more acidic internal pH than the two mutants when the three strains were exposed to medium buffered at a pH of less than 5.5. The two acid-sensitive mutants had an increased membrane permeability to protons but did not change their proton extrusion activities. However, the acid-sensitive strain ANU794 exhibited both a higher membrane permeability to protons and a lower proton extrusion activity compared with the acid-tolerant strain ANU1173. DNA hybridization analysis showed that mutants AS25 and AS28 carried a single copy of Tn5 located in 13.7-kb (AS25) and 10.0-kb (AS28) EcoRI DNA fragments. The wild-type DNA sequences spanning the mutation sites of mutants AS25 and AS28 were cloned from genomic DNA of strain ANU1173. Transfer of these wild-type DNA sequences into corresponding Tn5-induced acid-sensitive mutants, respectively, restored the mutants to their acid tolerance phenotypes. Mapping studies showed that the AS25 locus was mapped to a 5.6-kb EcoRI-BamHI megaplasmid DNA fragment, whilst the AS28 locus was located in an 8.7-kb BglII chromosomal DNA fragment.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Beringer J. E. R factor transfer in Rhizobium leguminosarum. J Gen Microbiol. 1974 Sep;84(1):188–198. doi: 10.1099/00221287-84-1-188. [DOI] [PubMed] [Google Scholar]
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
- Booth I. R. Regulation of cytoplasmic pH in bacteria. Microbiol Rev. 1985 Dec;49(4):359–378. doi: 10.1128/mr.49.4.359-378.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bown A. W. An investigation into the roles of photosynthesis and respiration in h efflux from aerated suspensions of asparagus mesophyll cells. Plant Physiol. 1982 Sep;70(3):803–810. doi: 10.1104/pp.70.3.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Gartner E., Rolfe B. G. Involvement of Genes on a Megaplasmid in the Acid-Tolerant Phenotype of Rhizobium leguminosarum Biovar Trifolii. Appl Environ Microbiol. 1993 Apr;59(4):1058–1064. doi: 10.1128/aem.59.4.1058-1064.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Richardson A. E., Gartner E., Djordjevic M. A., Roughley R. J., Rolfe B. G. Construction of an Acid-Tolerant Rhizobium leguminosarum Biovar Trifolii Strain with Enhanced Capacity for Nitrogen Fixation. Appl Environ Microbiol. 1991 Jul;57(7):2005–2011. doi: 10.1128/aem.57.7.2005-2011.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditta G., Stanfield S., Corbin D., Helinski D. R. Broad host range DNA cloning system for gram-negative bacteria: construction of a gene bank of Rhizobium meliloti. Proc Natl Acad Sci U S A. 1980 Dec;77(12):7347–7351. doi: 10.1073/pnas.77.12.7347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djordjevic M. A., Zurkowski W., Rolfe B. G. Plasmids and stability of symbiotic properties of Rhizobium trifolii. J Bacteriol. 1982 Aug;151(2):560–568. doi: 10.1128/jb.151.2.560-568.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gough J. A., Murray N. E. Sequence diversity among related genes for recognition of specific targets in DNA molecules. J Mol Biol. 1983 May 5;166(1):1–19. doi: 10.1016/s0022-2836(83)80047-3. [DOI] [PubMed] [Google Scholar]
- Jorgensen R. A., Rothstein S. J., Reznikoff W. S. A restriction enzyme cleavage map of Tn5 and location of a region encoding neomycin resistance. Mol Gen Genet. 1979;177(1):65–72. doi: 10.1007/BF00267254. [DOI] [PubMed] [Google Scholar]
- Kobayashi H. A proton-translocating ATPase regulates pH of the bacterial cytoplasm. J Biol Chem. 1985 Jan 10;260(1):72–76. [PubMed] [Google Scholar]
- Kobayashi H., Murakami N., Unemoto T. Regulation of the cytoplasmic pH in Streptococcus faecalis. J Biol Chem. 1982 Nov 25;257(22):13246–13252. [PubMed] [Google Scholar]
- Kobayashi H., Unemoto T. Streptococcus faecalis mutants defective in regulation of cytoplasmic pH. J Bacteriol. 1980 Sep;143(3):1187–1193. doi: 10.1128/jb.143.3.1187-1193.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krulwich T. A., Guffanti A. A. Physiology of acidophilic and alkalophilic bacteria. Adv Microb Physiol. 1983;24:173–214. doi: 10.1016/s0065-2911(08)60386-0. [DOI] [PubMed] [Google Scholar]
- Lowendorf H. S., Alexander M. Identification of Rhizobium phaseoli Strains That Are Tolerant or Sensitive to Soil Acidity. Appl Environ Microbiol. 1983 Mar;45(3):737–742. doi: 10.1128/aem.45.3.737-742.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'hara Graham W., Goss Thomas J., Dilworth Michael J., Glenn Andrew R. Maintenance of Intracellular pH and Acid Tolerance in Rhizobium meliloti. Appl Environ Microbiol. 1989 Aug;55(8):1870–1876. doi: 10.1128/aem.55.8.1870-1876.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padan E., Zilberstein D., Schuldiner S. pH homeostasis in bacteria. Biochim Biophys Acta. 1981 Dec;650(2-3):151–166. doi: 10.1016/0304-4157(81)90004-6. [DOI] [PubMed] [Google Scholar]
- Plazinski J., Cen Y. H., Rolfe B. G. General method for the identification of plasmid species in fast-growing soil microorganisms. Appl Environ Microbiol. 1985 Apr;49(4):1001–1003. doi: 10.1128/aem.49.4.1001-1003.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson A. E., Simpson R. J., Djordjevic M. A., Rolfe B. G. Expression of Nodulation Genes in Rhizobium leguminosarum biovar trifolii Is Affected by Low pH and by Ca and Al Ions. Appl Environ Microbiol. 1988 Oct;54(10):2541–2548. doi: 10.1128/aem.54.10.2541-2548.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott K. F., Hughes J. E., Gresshoff P. M., Beringer J. E., Rolfe B. G., Shine J. Molecular cloning of Rhizobium trifolii genes involved in symbiotic nitrogen fixation. J Mol Appl Genet. 1982;1(4):315–326. [PubMed] [Google Scholar]
- Scott K. F., Rolfe B. G., Shine J. Biological nitrogen fixation: primary structure of the Klebsiella pneumoniae nifH and nifD genes. J Mol Appl Genet. 1981;1(1):71–81. [PubMed] [Google Scholar]
- Slavík J. Intracellular pH of yeast cells measured with fluorescent probes. FEBS Lett. 1982 Apr 5;140(1):22–26. doi: 10.1016/0014-5793(82)80512-7. [DOI] [PubMed] [Google Scholar]
- Staskawicz B., Dahlbeck D., Keen N., Napoli C. Molecular characterization of cloned avirulence genes from race 0 and race 1 of Pseudomonas syringae pv. glycinea. J Bacteriol. 1987 Dec;169(12):5789–5794. doi: 10.1128/jb.169.12.5789-5794.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]