Abstract
β-Catenin can function as an oncogene when it is translocated to the nucleus, binds to T cell factor or lymphoid enhancer factor family members, and transactivates its target genes. In this study, we demonstrate that cyclin D1 is one of the targets of β-catenin in breast cancer cells. Transactivation of β-catenin correlated significantly with cyclin D1 expression both in eight breast cell lines in vitro and in 123 patient samples. More importantly, we found that high β-catenin activity significantly correlated with poor prognosis of the patients and was a strong and independent prognostic factor in breast cancer. Our studies, therefore, indicated that β-catenin can be involved in breast cancer formation and/or progression and may serve as a target for breast cancer therapy.
Cyclin D1 overexpression has been found in ≈50% of patients with breast cancer (1, 2), whereas gene amplification accounted for only 15–20% of these cases (3). Therefore, other mechanisms such as up-regulation of gene transcription must have played a substantial role in the overexpression of cyclin D1. By analyzing the promoter region of cyclin D1, we identified a perfect T cell factor 4 (Tcf4)-binding site (CTTTGATC) located between nucleotides −80 and −73, suggesting the potential involvement of the β-catenin/Tcf4 pathway in the regulation of cyclin D1 expression. β-Catenin was first found to be a cell–cell adhesion molecule. However, recent studies have indicated that β-catenin also could be translocated to the nucleus, where it binds to Tcf/lymphoid enhancer factor (Lef) architecture factor family members and activates genes whose promoters contain the binding sites for Tcf/Lef (4–6).
Several mechanisms have been reported to cause this deregulation, including deletion of the adenomatous polyposis coli (APC) gene, mutation of β-catenin, and activation of the Wnt pathway (7). Although deletion of APC and mutation of β-catenin have been found in many types of cancers (7), so far no such defects have been reported in breast cancer. However, many studies have indicated a possible role for the Wnt pathway in breast cancer. For example, mouse Wnt1, Wnt3, and Wnt10b have been found to be among the oncogenes activated by the insertion of mouse mammary tumor virus (MMTV) (8, 9). Mammary hyperplasias also have occurred in Wnt1 transgenic mice (10). In addition, several members of the Wnt family have been shown to induce cell proliferation (11, 12). Moreover, the expression of different Wnt members has been reported to correlate with abnormal cell proliferation in human breast tissue, suggesting the possible involvement of Wnt and the β-catenin pathway in breast cancer (13–15).
Materials and Methods
Cell Lines and Transfections.
All cell lines were obtained from the American Type Culture Collection and maintained in DMEM/F-12 (HyClone) with 10% (vol/vol) fetal bovine serum. Transient transfections were performed by using DC-Chol liposome provided by Leaf Huang, University of Pittsburgh. In brief, exponentially growing 293 cells and MCF7 cells were cultured in six-well plates and transfected with 0.4 μg of reporter, 0.2 μg of pCMVβGal control, and 1 μg of effector constructs or different amounts of β-catenin expression vectors in the dose-dependent experiment or transfected with the control vector pcDNA3 (Invitrogen). The β-catenin, GSK-3β (16), and dnTcf4 effector plasmids have been described (4). Luciferase assays were performed 40 h after transfection and normalized through β-galactosidase activity. Each assay was performed triplicate. The β-catenin stable cell lines were generated by transfecting the 293 cells with the β-catenin phosphorylation mutant (S45Yβ-catenin). Individual clones were selected for resistance to 500 μg/ml G418 (Geneticin, GIBCO/BRL).
Western Blot Analysis.
Cell lysates were separated by SDS/PAGE and transferred onto the nitrocellulose membrane. Protein levels were determined by using antibodies that recognized myc-tagged β-catenin, cyclin D1 (purchased from NeoMarkers, Union City, CA), and α-actin (purchased from Oncogene Science).
Gel Mobility Shift Assays.
The gel-shift assays for β-catenin/Tcf4 were performed as described (4). Extracts were prepared from intact nuclei of different breast cancer cell lines. The probe was a double-stranded 15-nt oligomer, CCCTTTGATCTTACC; the control oligomer was CCCTTTGGCCTTACC. The binding reaction contained 5 μg of nuclear protein, 10 ng of radiolabeled probe, and 1 μg of poly(dIdC) in 25 μl of binding buffer (60 mM KCl/1 mM EDTA/1 mM DTT/10% glycerol). Samples were incubated on ice for 30 min, and the probes were added and incubated further at room temperature for 30 min. The β-catenin/Tcf4 bands were confirmed by the competition assays with the excess of cold wild-type or control oligomers and by comparing the complexes derived from the nuclear extract of 293 cells and its β-catenin transfectants.
Immunohistochemical Staining.
Immunohistochemical staining was done by using a modification of the avidin–biotin complex technique described previously (17). The results were analyzed and confirmed by two individuals.
Results
Up-Regulation of Cyclin D1-Promoter Activity and the Protein Expression by β-Catenin.
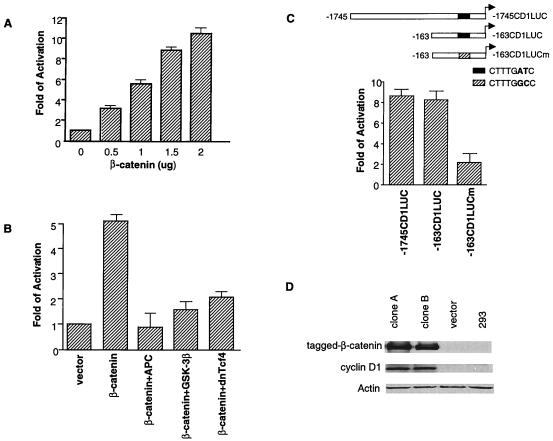
We first sought to determine whether cyclin D1 could be transcriptionally regulated by β-catenin. We found that transient transfection of exogenous human β-catenin in human embryonic kidney 293 cells could activate a cyclin D1 reporter, containing 1,745 bp of the cyclin D1 promoter, up to 11-fold in a dose-dependent manner (Fig. 1A). This activation was blocked when various inhibitors of the β-catenin/Tcf4 pathway were coexpressed such as APC, GSK-3β, and a dominant negative mutant of human Tcf4 (dnTcf4) (Fig. 1B) (18). We chose the 293 cell line to perform our studies because of its low background of β-catenin activity and its previous use for studying the response to β-catenin/Tcf4-mediated transcription (19).
Figure 1.
Up-regulation of cyclin D1 promoter activity and the protein expression by β-catenin. (A) The 293 cells were transfected with 0.4 μg of cyclin D1 reporter (−1745CD1LUC) along with increasing amounts of the wild-type human β-catenin expression plasmid. (B) Cells were transfected with 0.4 μg of cyclin D1 reporter (−1745CD1LUC) with 1.0 μg of β-catenin expression plasmid and 1.0 μg of different negative β-catenin/Tcf regulators or with control pcDNA3 plasmid. (C) Cyclin D1 reporters (0.4 μg) (−1745CD1LUC, −163CD1LUC, and its mutant, 163CD1LUCm, which had an AT to GC change at nucleotides −75 and −74) and 1.5 μg of β-catenin expression plasmid were transfected into 293 cells. (D) The whole cell lysate of 293 cells, 293 cells transfected with empty vector, and two constitutively-activated β-catenin stable transfectants were separated by SDS/PAGE and analyzed by Western blotting with antibodies that recognized myc-tagged β-catenin, cyclin D1 (purchased from NeoMarkers), and α-actin (purchased from Oncogene).
To confirm that the Tcf4 site on cyclin D1 promoter was responsible for the activation by β-catenin, we used a deletion construct containing 163 bp of the cyclin D1 promoter as the reporter. Many known transcription factor binding sites had been eliminated from this construct, but it still contained the putative Tcf4 site (−163CD1LUC). As shown in Fig. 1C, expression of β-catenin activated this reporter to a similar extent, suggesting that the responsive element remained within this deletion construct. When the Tcf4 site was mutated so that the AT was changed to GC at nucleotides −75 and −73 (163CD1LUCm), β-catenin no longer sufficiently activated the cyclin D1 gene promoter. These data indicated that the putative Tcf4 site located at −80 to −73 was responsible for the β-catenin-mediated transactivation of the cyclin D1 promoter. In addition to transient transfection, we also generated a stable cell line by transfecting the 293 cells with the β-catenin phosphorylation mutant (S45Yβ-catenin). This mutant has been shown to resist degradation and to increase its activity to transactivate β-catenin/Tcf4-dependent transcription (19). As shown in Fig. 1D, cyclin D1 protein expression in both individual stable transfectants was substantially increased (lanes 1 and 2) compared with the vector control cells (lane 3) and the parental cells (lane 4).
Correlation Between Cyclin D1 Expression and β-Catenin/Tcf4 Activity in Breast Cancer Cell Lines.
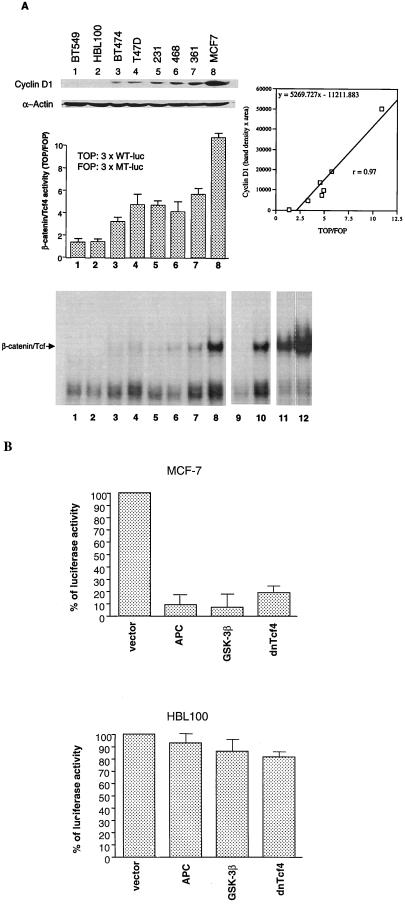
After identifying cyclin D1 as the target gene for β-catenin, we next asked whether β-catenin played an important role in up-regulating the expression of cyclin D1 in breast cancer. We first tested this possibility in breast cancer cell lines in vitro. Eight breast cancer cell lines were chosen to compare their cyclin D1 expression level and their β-catenin/Tcf4 activity. We used reporter constructs that contained three repeats of wild-type (TOP) or mutant (FOP) Tcf4-binding sites (4) to determine the transactivational activity of endogenous β-catenin/Tcf4. Higher ratios of these two reporter activities (TOP/FOP) indicated a higher β-catenin/Tcf4 activity. As shown in Fig. 2A (Top and Middle), cyclin D1 expression in breast cancer cells highly correlated with the β-catenin/Tcf4 activity. The eight cell lines tested could be roughly divided into three groups. BT549 and HBL100 cell lines, which expressed almost no detectable cyclin D1, had the background transactivating activity of β-catenin/Tcf4 (TOP/FOP = 1). In contrast, MCF-7, which expressed the highest level of cyclin D1 protein, had the most significant β-catenin/Tcf4 activity (TOP/FOP = 10). In the other five cell lines, cyclin D1 expression was consistently moderate, as were β-catenin/Tcf4 activities. By linear regression, we demonstrated that cyclin D1 expression indeed was proportionally correlated with β-catenin/Tcf4 activity (r = 0.97). In addition to the reporter assay, we also confirmed the β-catenin/Tcf4 activity by gel-shift assay. Consistent with reporter activity and cyclin D1 expression levels, β-catenin/Tcf4-binding activity was not detectable for either BT549 or HBL100 cells and was detected most strongly in MCF-7 cells as shown in Fig. 2A Bottom.
Figure 2.
Correlation between cyclin D1 expression and β-catenin/Tcf4 activity in breast cancer cell lines. (A) Cell lysates from different breast cell lines were separated by SDS/PAGE and analyzed by Western blotting with antibodies, which recognized cyclin D1 and α-actin (Top). The relative β-catenin/Tcf4 activity in different cell lines were determined by the TOP/FOP luciferase activities in each cells (Middle). The density of cyclin D1 bands were quantitated (by NIH image, an analyzing software) and plotted with β-catenin/Tcf4 activity (TOP/FOP) with the r = 0.967 by linear regression. Also, the DNA-binding activities of β-catenin/Tcf4 were determined by gel-shift assay as described previously (Bottom) (4). Lanes 1–8 show the β-catenin/Tcf4-binding activity in indicated cell lines. Lanes 9–12 are the controls to demonstrate the specific binding. Lane 9 and 10, the same as lane 8 except 60-fold excess of wild-type (lane 9) or the mutant (lane 10) cold oligonucleotide was added; lane 11, nuclear extract was from the 293 vector control line; lane 12, nuclear extract was from 293 β-catenin stable line. (B) MCF-7 cells (Top) or HBL100 cells (Bottom) were cotransfected with cyclin D1 reporter (−1745CD1LUC) with different negative β-catenin/Tcf regulators or with the control pcDNA3 plasmid. The absolute luciferase activity of cyclin D1 reporter alone in MCF-7 cells was ≈7-fold higher than that in HBL100 cells.
To further address whether cyclin D1 promoter activity is indeed regulated by β-catenin in these breast cancer cell lines, we cotransfected the cyclin D1 reporter with different negative regulators of the β-catenin/Tcf4 pathway in MCF-7 cells. As shown in Fig. 2B Top, the reporter activity of cyclin D1 promoter was significantly reduced. This reduction of activity could be reversed when β-catenin was coexpressed (data not shown). In contrast, cyclin D1 reporter activity was not affected by the expression of APC, GSK-3β, or dnTcf4 in HBL100 cells in which both β-catenin activity and cyclin D1 expression were low (Fig. 2B Bottom). Our data, therefore, support a substantial role for β-catenin in activating cyclin D1 expression in breast cancer cells.
Correlation Between Activated β-Catenin and Cyclin D1 Overexpression and Their Association with Poor Patient Survival Rate.
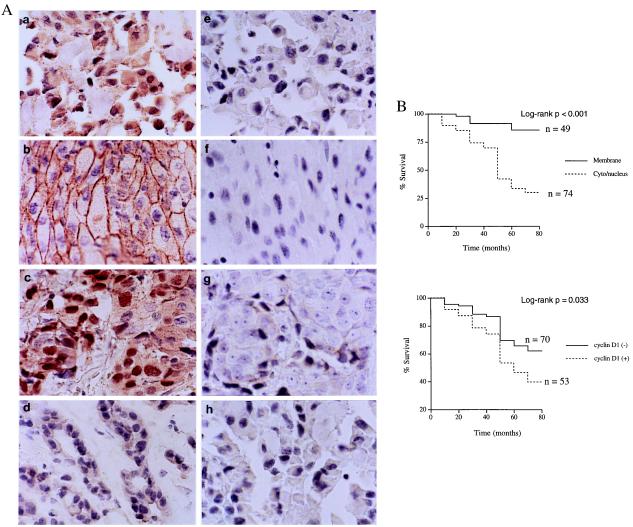
Because cyclin D1 overexpression has been well-documented in patients with breast cancer, we next sought to clinically verify whether β-catenin activity truly contributed to the cyclin D1 overexpression in breast cancer tissues. We determined both cyclin D1 expression and β-catenin activity in 123 primary human breast cancer tissues (age: 26–87 years old; medium: 48 years old) by immunohistochemical staining (Fig. 3A). We determined β-catenin activity by its subcellular localization (20, 21). It has been well documented that accumulated β-catenin in cytoplasm and/or the nucleus increased when cells had stabilized β-catenin and, consequently, the activated β-catenin/Tcf4 activity. In contrast, β-catenin was localized solely at the plasma membrane of cells when its transactivation activity was low. We also have confirmed the correlations between the β-catenin localization and its transactivation activity in various breast cell lines listed in Fig. 2 (data not shown).
Figure 3.
Correlation between activated β-catenin and cyclin D1 overexpression and their association with poor patient survival rate. (A) Breast cancer tissue stained with β-catenin antibody (a, cytoplasm/nucleus; b, membrane) and cyclin D1 antibody (c, overexpression; d, negative). The right panels (e–h) showed the respective negative controls for a-d using PBS instead of primary antibodies. (B) Kaplan-Meier analysis for survival correlated with the subcellular localization of β-catenin (Top) and cyclin D1 expression (Bottom). The medium of follow-up of patients was 48 months.
As shown in Table 1, the subcellular localization of β-catenin and cyclin D1 was significantly correlated based on the analysis by Spearman rank correlation (r = 0.6, P < 0.001). The samples stained as either high β-catenin activity with high cyclin D1 expression (40%) or low β-catenin activity with negative cyclin D1 staining (37%). It is worthwhile to mention that, among the 53 cases staining positive for cyclin D1, 49 cases (92%) were positive for β-catenin activity (stained in cytoplasm/nucleus). Thus, the correlation between these two molecules in the primary tumor samples was consistent with our in vitro data from the breast cancer cell lines (Fig. 2A). Therefore, we believe that high β-catenin activity may significantly contribute to cyclin D1 overexpression in breast cancer. These results not only supported our molecular data described above but also further strengthened their clinical biological significance.
Table 1.
Immunoreactivity of β-catenin and cyclin D1 in surgical specimens of breast cancer
| β-Catenin staining | Cyclin D1 staining
|
||
|---|---|---|---|
| Negative (n = 70) | Positive (n = 53) | Total | |
| Cytoplasm/nucleus | 25 (20%) | 49 (40%) | 74 (60%) |
| Membrane only | 45 (37%) | 4 (3%) | 49 (40%) |
| Total | 70 (57%) | 53 (43%) | 123 (100%) |
The 123 surgical specimens of breast cancer were stained with either β-catenin antibodies (purchased from Transduction or Santa Cruz Biotechnology) or cyclin D1 antibody (NeoMarkers) as shown in Fig. 3A. The expression patterns of these two molecules in the samples from each patient were determined and summarized. Correlation of subcellular localization of β-catenin and cyclin D1 expression was analyzed by Spearman rank correlation (r = 0.6, P < 0.001).
More importantly, when the prognostic significance was assessed by Kaplan-Meier analysis and log-rank test, we found that both cyclin D1 overexpression and activated β-catenin were associated with a poorer prognosis and were negatively correlated with patient survival rates (P = 0.033 and P < 0.001, respectively) (Fig. 3B).
To determine whether activated β-catenin was independent of other known prognostic factors in prognosis, multivariate analyses for survival rate were also performed. We found that activated β-catenin was a strong prognostic factor that provided additional and independent predictive information on the patient's survival rate even when other prognostic factors (lymph node metastasis, estrogen receptor and progesterone receptor status, and tumor size) were taken into account (P = 0.001). Cyclin D1 overexpression was also an independent prognostic factor. However, when multivariate analysis was performed including only cyclin D1 expression and β-catenin activity, cyclin D1 was no longer an independent prognostic factor (the P values for cyclin D1 and β-catenin activity were P = 0.457 and P < 0.001, respectively). These results were consistent with the model that cyclin D1 overexpression could be caused by activated β-catenin in breast cancer and consequently correlated to the prognosis.
Discussion
Our studies demonstrated that β-catenin was a poor prognostic marker in human cancer and was implicated in human breast cancer. How β-catenin activity is up-regulated in breast cancer is not clear at this moment. It is possible that activated Wnt pathway may contribute to this up-regulation (13–15). It requires further studies to elucidate the detail mechanisms.
In the past, β-catenin pathway has been studied mainly in colon carcinoma. Almost 100% of colon cancers have either mutated β-catenin or deleted APC, which is expectedly to activate the β-catenin pathway. In fact, during the time period of our studies, two groups identified cyclin D1 as the β-catenin target in colon carcinoma (22, 23). However, it is worthwhile to mention that cyclin D1 overexpression has been found in only ≈30% of colon cancer (24, 25), which might not be consistent with almost 100% deregulation of the β-catenin pathway, suggesting that the overexpression of cyclin D1 in colon cancer may be more complicated than purely up-regulation by β-catenin. Here, we showed that cyclin D1 was one of the targets for β-catenin in breast cancer. More importantly, we demonstrated the significant role of activated β-catenin in breast cancer both by molecular studies in cell culture and by clinical studies on breast tumor samples. Consistent with these findings, our studies provide strong evidence supporting the biological significance and clinical relevance of this pathway in human breast cancer. In contrast to colon carcinoma, the strong correlation between β-catenin activity and cyclin D1 expression was found in both breast cancer cell lines and breast patient tissue samples. Thus, the data presented in this study may open a new direction in the research of breast cancer involving both cancer formation and progression and provide an opportunity for development of potential therapy by blocking the β-catenin/Tcf4 pathway in breast cancer cells.
Acknowledgments
We thank Dr. Paul Polakis for β-catenin expression constructs; Dr. Hans Clevers for dnTcf4, TOP-, and FOP-luciferase constructs; Dr. Li-Kuo Su for APC expression vector; and Dr. Geoffrey M. Cooper for GSK-3β constructs. We also thank Dr. Pierre McCrea and Dr. Li-Kuo Su of our institution for their critical comments on the manuscript. The work is partially supported by National Cancer Institute Grants R01 CA58880 and R01 CA77858, the M.D. Anderson Faculty Achievement Award (to M.-C.H.), and a predoctoral fellowship from Department of Defense Breast Cancer Research Program DAMD17–98-1–8242 (to S.-Y. L). This work was also supported in part by R29CA70897 and R01CA75503 (to R.G.P.). R.G.P. is a recipient of the Irma T. Hirschl award and an award from the Susan G. Komen Breast Cancer Foundation.
Abbreviations
- Tcf
T cell factor
- Lef
lymphoid enhancer factor
- APC
adenomatous polyposis coli
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.060025397.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.060025397
References
- 1.Gillet C, Fantl V, Fisher C, Bartek J, Dickson C, Barnes D, Peters G. Cancer Res. 1994;54:1812–1817. [PubMed] [Google Scholar]
- 2.Bartkova J, Lukas J, Muller H, Lutzhoft D, Strauss M, Bartek J. Int J Cancer. 1994;11:353–361. doi: 10.1002/ijc.2910570311. [DOI] [PubMed] [Google Scholar]
- 3.Fantl V, Smith R M, Brookes S, Dickson C, Peters G. Cancer Surv. 1993;18:77–94. [PubMed] [Google Scholar]
- 4.Korinek V, Barker N, Morin P J, van Wichen D, de Weger R, Kinzler K W, Vogelstein B, Clevers H. Science. 1997;275:1784–1787. doi: 10.1126/science.275.5307.1784. [DOI] [PubMed] [Google Scholar]
- 5.Morin P, Sparks A B, Korinek V, Barker N, Clevers H, Vogelstein B, Kinzler K W. Science. 1997;275:1787–1790. doi: 10.1126/science.275.5307.1787. [DOI] [PubMed] [Google Scholar]
- 6.Rubinfeld B, Robbins P, El-Gamil M, Albert I, Porfiri E, Polakis P. Science. 1997;275:1790–1792. doi: 10.1126/science.275.5307.1790. [DOI] [PubMed] [Google Scholar]
- 7.Polakis P. Curr Opin Gene Dev. 1999;9:15–21. doi: 10.1016/s0959-437x(99)80003-3. [DOI] [PubMed] [Google Scholar]
- 8.Nusse R, van Ooyen A, Cox D, Fung Y K, Varmus H. Nature (London) 1984;307:131–136. doi: 10.1038/307131a0. [DOI] [PubMed] [Google Scholar]
- 9.Roelink H, Wagenaar E, Lopes da Silva S, Nusse R. Proc Natl Acad Sci USA. 1990;87:4519–4523. doi: 10.1073/pnas.87.12.4519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tsukamoto A S, Grosschedl R, Guzman R C, Parslow T, Varmus H E. Cell. 1988;55:619–625. doi: 10.1016/0092-8674(88)90220-6. [DOI] [PubMed] [Google Scholar]
- 11.Blasband A, Schryver B, Papkoff J. Oncogene. 1992;7:153–161. [PubMed] [Google Scholar]
- 12.Wong G T, Gavin B J, McMahon A P. Mol Cell Biol. 1994;14:6278–6286. doi: 10.1128/mcb.14.9.6278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dale T C, Weber-Hall S J, Smith K, Huguet E L, Jayatilake H, Gusterson B A, Shuttleworth G, O'Hare M, Harris A L. Cancer Res. 1996;56:4320–4323. [PubMed] [Google Scholar]
- 14.Lejeune S, Huguet E L, Hamby A, Poulsom R, Harris A L. Clin Cancer Res. 1995;1:215–222. [PubMed] [Google Scholar]
- 15.Bui T D, Rankin J, Smith K, Huguet E L, Ruben S, Strachan T, Harris A L, Lindsay S. Oncogene. 1997;14:1249–1253. doi: 10.1038/sj.onc.1200936. [DOI] [PubMed] [Google Scholar]
- 16.Pap M, Cooper G M. J Biol Chem. 1998;273:19929–19932. doi: 10.1074/jbc.273.32.19929. [DOI] [PubMed] [Google Scholar]
- 17.Hsu S, Raine L, Fanger H. J Histochem Cytochem. 1981;29:577–580. doi: 10.1177/29.4.6166661. [DOI] [PubMed] [Google Scholar]
- 18.He T, Sparks A B, Rago C, Hermeking H, Zawel L, da Costa L T, Morin P J, Vogelstein B, Kinzler K W. Science. 1998;281:1509–1512. doi: 10.1126/science.281.5382.1509. [DOI] [PubMed] [Google Scholar]
- 19.Porfiri E, Rubinfeld B, Albert I, Hovanes K, Waterman M, Polakis P. Oncogene. 1997;15:2833–2839. doi: 10.1038/sj.onc.1201462. [DOI] [PubMed] [Google Scholar]
- 20.Rimm D L, Caca K, Hu G, Harrison F B, Fearon E R. Am J Pathol. 1999;154:325–329. doi: 10.1016/s0002-9440(10)65278-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fukuchi T, Sakamoto M, Tsuda H, Maruyama K, Nozawa S, Hirohashi S. Cancer Res. 1998;58:3526–3528. [PubMed] [Google Scholar]
- 22.Tetsu O, McCormick F. Nature (London) 1999;398:422–426. doi: 10.1038/18884. [DOI] [PubMed] [Google Scholar]
- 23.Shtutman M, Zhurinsky J, Simcha I, Albanese C, D'Amico M, Pestell R, Ben-Ze'ev A. Proc Natl Acad Sci USA. 1999;96:5522–5527. doi: 10.1073/pnas.96.10.5522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bartkova J, Lukas J, Strauss M, Bartek J. Int J Cancer. 1994;58:568–573. doi: 10.1002/ijc.2910580420. [DOI] [PubMed] [Google Scholar]
- 25.Arber N, Hibshoosh H, Moss S F, Sutter T, Zhang Y, Begg M, Wang S, Weinstein I B, Holt P R. Gastroenterology. 1996;110:669–674. doi: 10.1053/gast.1996.v110.pm8608874. [DOI] [PubMed] [Google Scholar]