Abstract
We are developing a system to control G protein signaling in vivo to regulate a broad range of physiologic responses. Our system utilizes G protein-coupled peptide receptors engineered to respond exclusively to synthetic small molecule ligands and not to their natural ligand(s). These engineered receptors are designated RASSLs (receptor activated solely by a synthetic ligand). We have made two prototype RASSLs that are based on the human κ opioid receptor. Small molecule drugs that activate the κ receptor are nonaddictive and safe to administer in vivo. Binding and signaling assays reveal 200–2000-fold reductions in the ability of our RASSLs to bind or be activated by dynorphin, an endogenous peptide ligand of the κ opioid receptor. In a high-throughput signaling assay, these prototype RASSLs expressed in Chinese hamster ovary K1 cells showed little or no response to a panel of 21 opioid peptides but still signaled normally in response to small molecule drugs such as spiradoline. Activation of a RASSL by spiradoline also caused proliferation of rat-1a tissue culture cells. These data provide evidence that G protein-coupled receptors can be made into RASSLs. The potential in vivo applications for RASSLs include the positive enrichment of transfected cells and the development of new animal models of disease.
The family of G protein-coupled receptors is the largest known class of cell surface receptors (1). These receptors respond to odorants, photons, biogenic amines, lipids, peptide hormones, and a variety of other ligands. Activation of G protein-coupled receptors affects a broad range of physiologic events, including heart rate, proliferation, chemotaxis, neurotransmission, and hormone secretion (2). G protein-coupled receptors have a seven-transmembrane domain structure with an extracellular amino terminus and a cytoplasmic carboxyl terminus (3). Ligands outside the cell interact with the receptor to induce a conformational change, which in turn activates a heterotrimeric (αβγ) G protein on the inner surface of the plasma membrane (4, 5).
There are four major classes of G proteins, defined by their α subunits (Gαi, Gαs, Gαq, Gα12). Each couples to a distinct class of receptors and signals through a specific biochemical pathway. In this study, we focused on the Gi-coupled (i.e., activates Gαi) human κ opioid receptor (6–8). The Gi-signaling pathway decreases intracellular cAMP levels by inhibiting adenylyl cyclase. Specific physiologic effects depend on the cell type in which the signaling is activated. For example, signaling via Gi slows the contraction rate of cardiac myocytes (9), increases proliferation of rat-1a cells (10, 11), modulates neurotransmission in the brain (9, 12), and promotes chemotaxis in immune cells (2). Different receptors that signal through the same pathway (e.g., Gi) cause identical responses when activated in tissue culture cells, but it is unclear whether the same holds true in vivo. For example, it is not known whether a Gi-coupled neuronal receptor (such as the κ receptor) can substitute for a Gi-coupled cardiac receptor to slow heart rate.
Studying the effects of G protein-coupled receptor signaling in vivo is complicated by the presence of endogenous ligands, which prevent complete control of receptor activation by the investigator. Receptors engineered to respond to synthetic ligands and not to their natural ligand(s) would eliminate this problem and would be useful reagents for studying the many G protein-coupled signaling pathways. In this report, we describe our efforts to create and characterize such a receptor, which we call a RASSL (receptor activated solely by a synthetic ligand). Our strategy for developing a RASSL centers on mutating amino acid residues involved in binding the natural ligand without affecting the residues involved in binding synthetic small molecule drugs. Peptide receptors are an ideal starting point for RASSL development because peptide ligands typically interact with the extracellular loops of the receptor, whereas small molecules interact with regions closer to the transmembrane domains. Our prototype RASSLs are based on the human κ opioid receptor (8), which signals in response to peptide hormones such as dynorphin and certain enkephalins. The κ receptor has been the focus of intense research by the pharmaceutical industry because κ agonists have the potential to induce analgesia without causing the respiratory depression or addiction associated with μ opioid receptor agonists like morphine (13, 14). As a result, there are many nonaddictive synthetic small molecule κ agonists, some of which are orally available and well tolerated in humans (15, 16). Opioid receptors have also been subjected to extensive mutagenesis, allowing us to choose mutations of the κ receptor that favor the construction of prototype RASSLs.
We have constructed two prototype RASSLs based on the κ opioid receptor. The first, called Ro1 (RASSL based on opioid receptor, no. 1), is a chimeric receptor containing the second extracellular loop (EL2) of the δ opioid receptor. The second, Ro2, contains, in addition to the δ residues, an amino acid substitution at the top of the sixth transmembrane region. We show that both receptors have decreased binding affinity and signaling to a variety of endogenous peptide ligands while maintaining wild-type responses to synthetic small molecule agonists. In addition, activation of Ro2 in rat-1a fibroblasts caused an agonist-dependent growth response. RASSLs may be useful for a variety of in vivo applications, such as creating animal models of disease and allowing positive enrichment of transfected cells.
MATERIALS AND METHODS
Construction of Engineered κ Opioid Receptors.
A human κ receptor cDNA kindly provided by L.-Y. Liu-Chen (Temple University, Philadelphia, PA) was used as a template for further manipulation and mutagenesis. The ORF was first cloned into pcDNA3 mammalian expression vector (Invitrogen). A sequence encoding the prolactin signal peptide followed by sequences encoding a FLAG epitope (DYKDDDD) was then added to the 5′ end (17). The signal peptide was removed by proteolysis during receptor processing, generating a mature receptor with an amino terminus recognized by commercially available anti-FLAG antibodies (IBI). The carboxyl terminus was also altered to encode a hemagglutinin 12CA5 epitope (YPYDVPDYA). Neither of these epitopes affects receptor function as tested by biochemical signaling studies when compared with the untagged receptor (unpublished observations). The Ro1 mutation substituting the EL2 loop of the δ receptor for the EL2 loop of the κ receptor was constructed by subcloning the EcoRI–BglII fragment from rat κ/δ-D (18) into the same sites in κ-WT. E/Q was constructed by PCR mutagenesis using the sense oligonucleotide 5′-CATCCTAGTTCAGGCTCTGGGGAGCACCTCC-3′, which results in an E297Q substitution. The Ro2 construct was made by cloning the BsmBI–EcoNI fragment from E/Q into Ro1. All clones constructed by PCR were confirmed by DNA sequencing.
Expression of Receptors and Binding Assay.
Plasmid DNA (10 μg) was transfected into COS-7 cells growing in 150-mm dishes by adenovirus-mediated transfection (19). After 48–72 h, cells were harvested, and membrane fractions were prepared as described (6). Competition binding assays were carried out in the presence of 1.5 nM [3H]ethylketocyclazocine (EKC) (18.3 Ci/mmol, DuPont/NEN) and 10–16 μg of protein in 50 mM Tris, pH 7.4, in 250 μl for 2 h at room temperature using a Skatron cell harvester. Drugs were tested at 10 or 11 different concentrations in duplicate, and a drug was tested against all receptors in each assay. Ki values were determined with Prism from GraphPad (San Diego). Saturation binding experiments were performed at seven concentrations (0.05–3 nM) of [3H]EKC, with or without 1 μM unlabeled EKC, to determine nonspecific binding.
Cyclase Inhibition Assays.
Chinese hamster ovary (CHO)-K1 cells (106) were transfected with 2 μg of D1 dopamine receptor plasmid and 4 μg of κ-WT, E/Q, Ro1, or Ro2. Cells were labeled, dosed with drug, and processed as described (20). The amount of adenylyl cyclase activity [measured as cAMP/(cAMP + ATP)] in the presence of 1 μM dopamine only was assigned a value of 100%, with an average 10-fold induction of cAMP over basal.
[3H]Thymidine Incorporation Assays.
Rat-1a cells were transfected with either κ-WT or Ro2 plasmid. From a stable pool of transfectants grown in 400 μg/ml G418 (Geneticin, Life Technologies, Gaithersburg, MD), individual high-expressing clones were isolated by fluorescence-activated cell sorting with a phycoerythrin-conjugated FLAG antibody. Cells were seeded at 5 × 105 cells per well in 24-well plates in triplicate. On the next day, the cells were placed in serum-free medium for 24 h. Dynorphin or spiradoline was then added at concentrations ranging from 0.05 pM to 1 μM for 16 h, followed by addition of [3H]thymidine (70–90 Ci/mmol, DuPont/NEN) for 8 h. DNA was harvested as described (21).
Fluorometric Imaging Plate Reader Assay.
CHO-K1 cells were transiently transfected using Lipofectamine (Life Technologies) with the indicated receptor construct and the cDNA encoding the chimeric G protein, Gαqo5 (22). Gαqo5 is a chimeric G protein α subunit that has the Gαq wild-type sequence except for the carboxyl-terminal 5 residues, which were changed to the Gαqo sequence: GCGLY (22). Full information regarding the use of this chimera is available at http://gladstone.ucsf.edu/conklin.html. Twenty-four hours after the start of transfection, the cells were replated into 96-well, black-wall microplates (Corning) at 50,000 cells per well. Approximately 24 h later, the cells were loaded with Fluo-3 fluorescent calcium indicator dye (Molecular Probes) as follows. Culture medium was removed by aspiration and replaced by 0.1 ml of F12 complete growth medium containing 20 mM Hepes, 2.5 mM probenecid [Sigma, from a 250 mM stock solution in 50% Hanks’ balanced salt solution (HBSS) and 50% 1 N NaOH], and 4 μM Fluo-3 (Molecular Probes, from a 4 mM stock solution in dimethyl sulfoxide containing 10% pluronic acid). Probenecid was added to inhibit the extrusion of dye by the multiple drug-resistance pump. After incubation for 1 h in a CO2 incubator, the cells were washed four times with HBSS containing 20 mM Hepes and 2.5 mM probenecid in a Denley cell washer. After the final wash, the solution was aspirated to a residual volume of 90 μl. Ligands were diluted with HBSS containing 20 mM Hepes and 2.5 mM probenecid and aliquoted into a microplate. The fluorometric imaging plate reader (FLIPR, Molecular Devices) transfers 90 μl from the ligand microplate to the cells and makes fluorescence readings for 2 min (every second for the first 60 s and every 2 s for the next 60 s). Total fluorescence counts from the 18-s to 37-s time points are used to determine agonist activity. The instrument software normalizes the fluorescent reading to give equivalent initial readings at time zero.
Reagents.
Peptide hormones were purchased from Peninsula Laboratories, and small molecule drugs were purchased from Research Biochemicals (Natick MA). Additional supplies of spiradoline were a gift of Pharmacia. All other reagents were from Sigma or Fisher Scientific unless otherwise noted.
RESULTS
Construction of Prototype RASSLs Ro1 and Ro2.
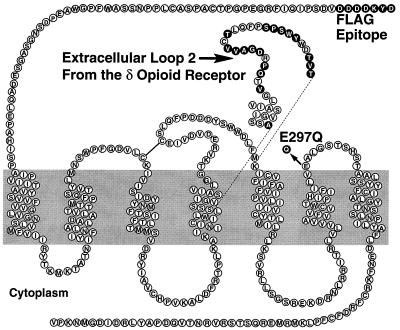
We and others (23–25) have made chimeric receptors in which portions of the κ opioid receptor were replaced by portions of the μ and δ opioid receptors. Results from these experiments showed that extracellular loop 2 (EL2) contains a major determinant for the binding of dynorphin (a κ-selective peptide), whereas EL3 contains the major binding determinants for the δ and μ receptors (23, 24). Consequently, we constructed our first prototype RASSL by substituting the human κ receptor EL2 (residues 198–226) with the δ receptor EL2. We anticipated that this chimera would have decreased binding affinity to κ-specific peptides but would retain activation by κ-specific small molecule drugs because their binding sites are not in the extracellular loops. Two lines of evidence indicated that this strategy would be successful. First, a larger substitution (residues 185–262) in this region in the rat κ receptor decreased dynorphin binding (23, 25). Second, an identical substitution, also in the rat κ receptor, retained a low affinity for certain δ receptor-specific ligands (18), suggesting that our chimera would be unresponsive to δ ligands. We named this first chimera Ro1 (Fig. 1).
Figure 1.
Modifying the human κ receptor to make the Ro1, E/Q, and Ro2 receptors. The wild-type sequence is shown as ○; residues that have been changed are shown as •. The EL2 loop exchange is included in Ro1 and Ro2 constructs. The E297Q mutation is included in the E/Q and Ro2 constructs. The FLAG epitope is included in all the constructs in this study.
To form our second RASSL, we changed the glutamic acid at position 297 in Ro1 to glutamine. Glutamic acid 297, located at the junction of transmembrane domain 6 (TM6) and EL3 in the κ receptor, is thought to contribute to specific opioid peptide binding (26). The cognate residue in the δ receptor is a tryptophan (27). This single amino acid difference could contribute to the binding site specificity of each receptor, as tryptophan is highly polar whereas glutamic acid is negatively charged. We hypothesized that a conservative mutation such as E297Q would remove the charge but minimize other changes to the small molecule binding pocket. We wanted to make this substitution as conservative as possible because a more dramatic change at this position (E297A) decreased the binding of the κ-selective small molecule antagonist nor-BNI (26). This second RASSL, Ro2, thus contains both the δ EL2 sequences and the E297Q substitution (Fig. 1). When the E297Q substitution is present without the δ EL2, the receptor is called E/Q.
Binding and Signaling via the Inhibition of Adenylyl Cyclase.
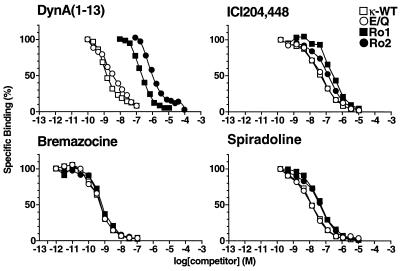
Competitive radioligand-binding assays were used to test the binding affinities of the receptors for various opioid agonists, including the peptide dynorphin A(1–13) and the synthetic drugs bremazocine, spiradoline, and ICI204,448 hydrochloride. Dynorphin is an opioid peptide hormone, bremazocine a nonselective opioid agonist (13), spiradoline a highly selective κ agonist (15, 28), and ICI204,448 a κ agonist that does not cross the blood–brain barrier (29). The most dramatic differences between the receptors were seen in their binding affinities for dynorphin. All the tested receptors have decreased dynorphin binding as compared with the wild-type receptor (κ-WT) (Fig. 2, Table 1). Compared with that of κ-WT, the Ki of dynorphin for Ro1 showed a 200-fold decrease, E/Q a 6-fold decrease, and Ro2 a nearly 2,000-fold decrease in binding activity. All maintained affinities for bremazocine and ICI204,448 that were unchanged from those of κ-WT. Binding of spiradoline was reduced about 5-fold in receptors containing the E297Q substitution (E/Q and Ro2) but was not significantly affected in Ro1. These data demonstrate that the binding domain of dynorphin can be separated from those of small molecule agonists, thus allowing the development of κ receptor-based RASSLs.
Figure 2.
Competition binding assay. Membranes prepared from transfected COS-7 cells were incubated in the presence of 1.5 nM [3H]EKC and various concentrations of dynorphin A(1–13), bremazocine, spiradoline, or ICI204,448. The specific transfected receptor cDNAs are indicated in the figure. Ki values are listed in Table 1. The amount of radioligand bound in the absence of competing ligand was assigned a value of 100%.
Table 1.
Binding and signaling characteristics of κ-WT and prototype RASSLs
| Assay | Ligand | Receptor
|
|||
|---|---|---|---|---|---|
| κ-WT | Ro1 | E/Q | Ro2 | ||
| Binding (competition), Ki | DynA(1–13) | 0.06 ± 0.04 | 14.64 ± 2.99 (229) | 0.36 ± 0.11 (5.6) | 124.52 ± 19.38 (1946) |
| Bremazocine | 0.04 ± 0.01 | 0.06 ± 0.03 (1.7) | 0.04 ± 0.02 (0.92) | 0.05 ± 0.02 (1.2) | |
| Spiradoline | 1.32 ± 0.38 | 6.29 ± 1.92 (4.8) | 1.09 ± 0.23 (0.83) | 5.65 ± 1.19 (4.3) | |
| ICI 204,448 | 4.02 ± 2.01 | 30.70 ± 6.76 (7.6) | 3.51 ± 1.10 (0.87) | 12.51 ± 4.90 (3.1) | |
| Binding (saturation), Kd | EKC | 0.17 ± 0.09 | 0.25 ± 0.11 (0.88) | 0.15 ± 0.05 (1.5) | 0.20 ± 0.044 (1.2) |
| Adenylyl cyclase, EC50 | DynA(1–13) | 0.66 ± 0.62 | 61.03 ± 20.56 (92.8) | 1.35 ± 1.4 (2.1) | 1083.73 ± 367 (1649) |
| Bremazocine | 0.07 ± 0.05 | 0.07 ± 0.039 (1.0) | 0.05 ± 0.02 (0.74) | 0.34 ± 0.21 (5.2) | |
| [3H]Thymidine, EC50 | DynA(1–13) | 8.53 ± 4.85 | >1000 (>117) | ||
| Spiradoline | 4.39 ± 3.0 | 5.49 ± 2.81 (1.3) | |||
Data are expressed in nanomolars as mean ± SD. Fold differences from κ-WT are shown in parentheses. For the ligand-binding experiments, each Ki value was derived from four independent experiments, and the Kd values were derived from two independent experiments performed in duplicate. For the adenylyl cyclase inhibition assay, the EC50 values were determined from five (dynorphin) or two (bremazocine) experiments performed in triplicate. Maximal inhibition of adenylyl cyclase was approximately 60% for each receptor. For the [3H]thymidine incorporation assay, EC50 values were determined from three separate experiments performed in duplicate.
We next tested the abilities of these engineered receptors to signal in a physiologic assay by using CHO-K1 tissue culture cells. Because the κ receptor couples to Gαi, receptor activation can be measured biochemically by inhibition of adenylyl cyclase activity. All the receptors inhibited adenylyl cyclase when stimulated with dynorphin, and the IC50 values reflected the differences in their Ki values for dynorphin binding (Table 1). For example, dynorphin activates κ-WT to inhibit adenylyl cyclase with an EC50 value of 0.66 nM, about 92-fold greater than Ro1 and 1,600-fold more than Ro2. All engineered receptors are similar to κ-WT with respect to signaling in response to bremazocine.
Screening Opioid Peptides for Activation of Prototype RASSLs.
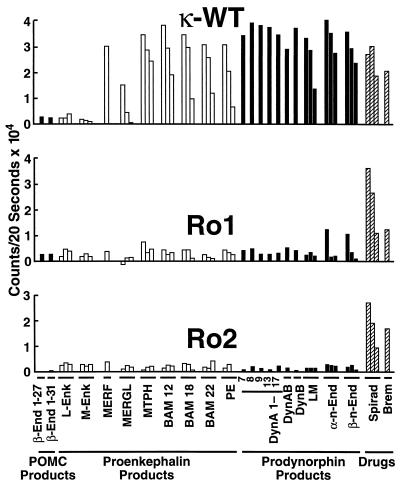
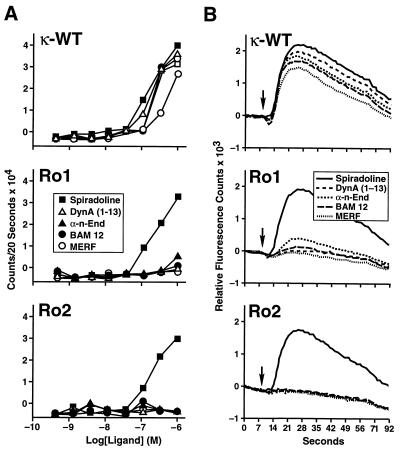
Dynorphin is the classic κ receptor agonist, but there are at least 21 opioid peptides that could theoretically activate the κ receptor, Ro1, or Ro2 (30). To screen all these peptides rapidly and rigorously, we modified a high-throughput screening assay used by the pharmaceutical industry to detect calcium mobilization mediated by G protein-coupled receptors. Although κ receptor stimulation does not normally cause calcium mobilization in CHO cells, it will mobilize calcium when coexpressed with the G protein chimera, Gαqo5 (22). Gαqo5 was designed to allow Gi-coupled receptors (such as the κ receptor) to signal via the Gq pathway and induce calcium mobilization. Although transient, the calcium mobilization can be easily monitored in real time with 96-well microplates and a FLIPR. Because the FLIPR assay provides a more rapid and robust signal than the adenylyl cyclase inhibition assay, we were able to assess the relative agonist activity of the panel of 21 endogenous opioid peptides (Table 2) on κ-WT, Ro1, and Ro2. The results from screening each peptide at concentrations of 0.01, 0.1, and 1.0 μM show that, remarkably, the κ receptor responded to 17 of 21 peptides tested; the only peptides that did not elicit a response were the β-endorphins (1–27 and 1–31), Leu-enkephalin, and Met-enkephalin (Fig. 3). The amino acid changes in Ro1 seemed to decrease signaling by all of the agonist peptides. In fact, at the highest dose tested (1 μM), Ro1 responded significantly to α-neo-endorphin and β-neo-endorphin only. Addition of the E297Q substitution in Ro2 reduced responsiveness to α-neo-endorphin and β-neo-endorphin to basal levels, thereby improving Ro2 as a RASSL. In all cases, the response to spiradoline was relatively unchanged. These results were confirmed by a more thorough dose-response experiment comparing the signaling of κ-WT, Ro1, and Ro2 to spiradoline, dynorphin A(1–13), α-neo-endorphin, MERF, and BAM 12 (Fig. 4).
Table 2.
Comparison of opioid peptide sequences
| Peptide | Sequence |
|---|---|
| Proopiomelanocortin products | |
| β-Endorphin(1–27) | YGGFMTSEKSQTPLVL (10) |
| β-Endorphin(1–31) | YGGFMTSEKSQTPLVL (14) |
| Proenkephalin products | |
| Leu-enkephalin | YGGFL |
| Met-enkephalin | YGGFM |
| Met-enkephalin-Arg-Phe | YGGFMRF |
| Met-enkephalin-Arg-Gly-Leu | YGGFMRGL |
| Metorphamide | YGGFMRRV-NH2 |
| BAM 12 | YGGFMRRVGRPE |
| BAM 18 | YGGFMRRVGRPEWW (4) |
| BAM 22 | YGGFMRRVGRPEWW (7) |
| Peptide E | YGGFMRRVGRPEWW (10) |
| Prodynorphin products | |
| Dynorphin A1–7 | YGGFLRR |
| Dynorphin A1–8 | YGGFLRRI |
| Dynorphin A1–9 | YGGFLRRIR |
| Dynorphin A1–13 | YGGFLRRIRPKLK |
| Dynorphin A1–17 | YGGFLRRIRPKLKWDNQ |
| Dynorphin AB1–32 | YGGFLRRIRPKLKWD (17) |
| Dynorphin B1–13 | YGGFLRRQFKVVT |
| Leumorphin | YGGFLRRQFKVVTR (15) |
| α-Neo-endorphin | YGGFLRKYPK |
| β-Neo-endorphin | YGGFLRKYP |
Amino acid sequences of peptides derived from the opioid precursor proteins proopiomelanocortin, proenkephalin, and prodynorphin, using the single letter amino acid notation. Where only a partial sequence is given, the number of additional amino acids is indicated in parentheses.
Figure 3.
Agonist activities of 21 natural opioid peptides on κ-WT, Ro1, and Ro2. Agonist-mediated changes in intracellular calcium were measured by the FLIPR as described in Materials and Methods. For dose responses (indicated by three bars/ligand), agonist concentrations are 1.0, 0.1, and 0.01 μM. For single doses (indicated by a single bar/ligand), the agonist concentration is 1.0 μM. Spirad, spiradoline; Brem, bremazocine. See Table 2 for peptide ligand amino acid sequences. Although abbreviations are used here, the order of the peptide ligands is the same as in Table 2. Data in Figs. 3 and Fig. 4A are expressed as mean of duplicate determinations in a single experiment; two additional experiments gave similar results.
Figure 4.
Agonist activities of selected ligands on κ-WT, Ro1, and Ro2. Agonist-mediated changes in intracellular calcium were measured by the FLIPR as described in Materials and Methods. (A) Dose responses on κ-WT, Ro1, and Ro2. (B) Sample tracings of the actual calcium fluorescence curves from the 1 μM dose. The arrow indicates the addition of agonist.
Proliferation Induced by a Prototype RASSL.
Control of proliferation by G protein-coupled receptors may be valuable for therapeutic applications. For example, one limitation of current gene therapy protocols is the inability to transfect enough cells to generate a therapeutic response. G protein-coupled receptors that cause proliferation could be used, in the presence of exogenously added agonist, to select for transfected cells in vivo.
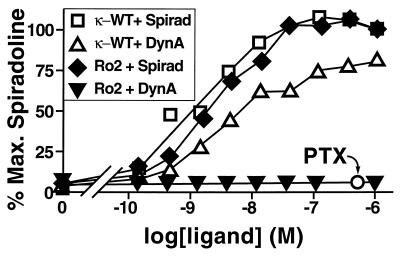
DNA synthesis can be used as an index of cell proliferation and can be measured experimentally by incorporation of [3H]thymidine in response to growth signals (10). Because rat-1a fibroblasts proliferate in response to Gi signaling, we generated clonal rat-1a cell lines expressing either κ-WT or Ro2 to determine if these receptors could induce proliferation. Both receptors caused a robust increase in DNA synthesis in response to spiradoline, with EC50 values of 4.4 nM for κ-WT and 5.5 nM for Ro2 (Fig. 5). Signaling by Ro2 in response to the peptide dynorphin A(1–13) was drastically reduced to the point that it was undetectable at 1 μM. For comparison, spiradoline was maximally effective at 37 nM in this assay and detectable at 5 nM.
Figure 5.
[3H]Thymidine incorporation assay. Amount of [3H]thymidine incorporation in rat-1a cell clones expressing κ-WT or Ro2 in the presence of dynorphin A(1–13) (DynA) or spiradoline (Spirad) as indicated. Ro2 plus spiradoline, plus pertussis toxin (PTX) 200 ng/ml is indicated by ○. The value for 100% is the value for the highest dose of spiradoline for each receptor. The basal point is untreated cells. Data in are expressed as mean ± SD of duplicate determinations in a single experiment; three additional experiments gave similar results.
To demonstrate that signaling by Ro2 was indeed mediated by Gi, cells were pretreated with pertussis toxin, which inactivates Gαi protein subunits (4). Pertussis toxin completely blocked signaling by Ro2 in response to spiradoline, indicating that the proliferative effect is caused by Gi signaling (Fig. 5). Because the basal activity in the treated cells was the same as, and not lower than, that in untreated cells, this result also indicates that this receptor is not constitutively active when overexpressed in tissue culture cells.
DISCUSSION
We have shown that our prototype RASSLs based on the κ receptor have decreased binding affinity and signaling in response to dynorphin A(1–13) and 20 other opioid peptides while maintaining near wild-type affinity and signaling in response to synthetic small molecule agonists. These results demonstrate the feasibility of RASSL development.
The greatest decrease in opioid peptide binding and signaling was seen in Ro1, in which the κ EL2 loop was replaced by δ sequences. Both the enkephalin and the dynorphin peptides were affected by this substitution, which was somewhat surprising because of the diverse nature of these peptide families (see Table 2). The additional E297Q substitution in Ro2 further reduced the dynorphin and enkephalin signaling and more dramatically reduced the residual signaling by α- and β-neo-endorphin. However, both Ro1 and Ro2 have near wild-type binding affinity to, and signaling by, small molecule drugs like spiradoline and bremazocine. It is possible, therefore, that the binding sites for the peptides and small molecules are totally distinct, and one can be eliminated without affecting the other. Alternatively, it may be that the peptides and small molecules have distinct but overlapping binding sites, and the changes made in Ro1 and Ro2 simply affect regions that are peptide-specific. The latter hypothesis seems more likely and is consistent with the view that although peptides may initially bind to the extracellular portion of the receptor, they activate it through contacts deeper within the transmembrane region.
The demonstration that the κ receptor can be made into a RASSL raises the possibility of engineering RASSLs from receptors that signal through different or multiple G protein pathways. The challenge, of course, is to find receptors that have synthetic agonists whose binding sites can be separated from that of the natural ligand(s). Other peptide receptors that respond to small molecule drugs could be used, but relatively few small molecule drugs act on peptide receptors (31). RASSLs could also be made from receptors that have nonpeptide hormones, such as biogenic amines (1, 32). Of the biogenic amine receptors, the serotonin receptors may provide the best opportunity for RASSL development because several synthetic serotonin receptor agonists differ greatly from serotonin in their structure and may bind to the receptor at distinct sites. It seems less likely that a RASSL could be made from an adrenergic receptor because the available synthetic agonists all resemble the natural hormone epinephrine (13). In an elegant study. Strader et al. (40) made a custom ligand specifically designed to complement a mutant adrenergic receptor that had impaired binding to epinephrine. Unfortunately, this custom ligand is unlikely to be useful for most in vivo studies because it is not commercially available and is 1,000-fold less potent (EC50 = 40 μM) than other adrenergic agonists. By comparison, the EC50 for spridoline-induced proliferation in Ro2-transfected rat-1a cells is 5.49 nM (Table 1). Alternatively, although the κ receptor is Gi-coupled, it is possible that the G protein-binding surface could be altered to allow signaling via other G proteins.
The binding experiments indicated that Ro2 has 2,000-fold lower affinity for dynorphin than κ-WT. Is this enough for in vivo RASSL applications? It is difficult to define the level of unresponsiveness to natural hormones that is required. Most peptides are found in the body at concentrations that are 100-fold lower than the 1-μM level we have used for testing. However, as higher local concentrations may be found in certain body subcompartments, this question can be answered only by observing the consequences of expressing RASSLs in vivo.
Recently, two other types of regulatory systems have been developed to control physiologic responses in vivo. The first type is based on chimeric transcription factors that can be regulated by tetracycline (33, 34), mifepristone (RU486), or ecdysone (35, 36). The second type uses drugs such as FK1012 (37), rapamycin, or coumermycin (38) to initiate protein–protein dimerization to reconstitute transcription factor activity or to activate intracellular signaling pathways. These systems allow precise temporal and spatial regulation of gene expression in transgenic animals and hold great promise as tools for tissue engineering or gene therapy (35). We have now demonstrated a third type of regulatory system that is based on G protein signaling. Activation of G protein-mediated signaling pathways to mimic the effects of natural hormones could allow fine control of complex physiologic processes such as heart rate, chemotaxis, and proliferation.
Ectopic expression and activation of RASSLs in certain tissues may identify pathologies caused by abnormal signaling. In fact, RASSLs may be particularly useful for developing animal models of diseases in which abnormal G protein signaling has been implicated. For instance, increased Gi signaling has been implicated as a potential cause of dilated cardiomyopathy in humans (39). It will be interesting to determine if RASSL-induced Gi signaling will cause a dilated cardiomyopathy in transgenic mice. Similarly, RASSLs may provide a means of testing the hypothetical role of G protein signaling disorders of the thyroid, bone, testis, pituitary, and central nervous system (2). Other applications could include agonist-dependent contraction of muscle implants or secretion of bioactive factors. Finally, transfection of RASSLs into cells where activation causes a proliferative response would enable positive selection of transfected cells and might be useful for gene transfer applications. Current studies in transgenic mice have begun to test some of these potential applications.
Acknowledgments
We thank Henry R. Bourne, Robert W. Mahley, Charles H. Redfern, and Michael Y. Degtyarev for valuable discussions and advice; we also thank Ellen Berman for manuscript preparation and Gary Howard and Stephen Ordway for editorial assistance. This work was supported by the J. David Gladstone Institutes and the National Institutes of Health Grant HL02555 (B.R.C.).
ABBREVIATIONS
- RASSL
receptor activated solely by a synthetic ligand
- FLIPR
fluorometric imaging plate reader
- CHO
Chinese hamster ovary
- EKC
[3H]ethylketocyclazocine
References
- 1.Strader C D, Fong T M, Tota M R, Underwood D. Annu Rev Biochem. 1994;63:101–132. doi: 10.1146/annurev.bi.63.070194.000533. [DOI] [PubMed] [Google Scholar]
- 2.Spiegel A M, Shenker A, Weinstein L S. Endocr Rev. 1992;13:536–565. doi: 10.1210/edrv-13-3-536. [DOI] [PubMed] [Google Scholar]
- 3.Baldwin J M. Curr Opin Cell Biol. 1994;6:180–190. doi: 10.1016/0955-0674(94)90134-1. [DOI] [PubMed] [Google Scholar]
- 4.Conklin B R, Bourne H R. Cell. 1993;73:631–641. doi: 10.1016/0092-8674(93)90245-l. [DOI] [PubMed] [Google Scholar]
- 5.Bourne H R. Curr Opin Cell Biol. 1997;9:134–142. doi: 10.1016/s0955-0674(97)80054-3. [DOI] [PubMed] [Google Scholar]
- 6.Meng F, Hoversten M T, Thompson R C, Taylor L, Watson S J, Akil H. J Biol Chem. 1995;270:12730–12736. doi: 10.1074/jbc.270.21.12730. [DOI] [PubMed] [Google Scholar]
- 7.Yasuda K, Raynor K, Kong H, Breder C D, Takeda J, Reisine T, Bell G I. Proc Natl Acad Sci USA. 1993;90:6736–6740. doi: 10.1073/pnas.90.14.6736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhu J, Chen C, Xue J-C, Kunapuli S, DeRiel J K, Liu-Chen L-Y. Life Sci. 1995;56:PL201–PL207. doi: 10.1016/0024-3205(94)00507-o. [DOI] [PubMed] [Google Scholar]
- 9.Wickman K, Clapham D E. Physiol Rev. 1995;75:865–885. doi: 10.1152/physrev.1995.75.4.865. [DOI] [PubMed] [Google Scholar]
- 10.Pace A M, Wong Y H, Bourne H R. Proc Natl Acad Sci USA. 1991;88:7031–7035. doi: 10.1073/pnas.88.16.7031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Biesen T, Luttrell L M, Hawes B E, Lefkowitz R J. Endocr Rev. 1996;17:698–714. doi: 10.1210/edrv-17-6-698. [DOI] [PubMed] [Google Scholar]
- 12.Jan L Y, Jan Y N. Curr Opin Cell Biol. 1997;9:155–160. doi: 10.1016/s0955-0674(97)80057-9. [DOI] [PubMed] [Google Scholar]
- 13.Reisine T, Pasternak G. In: Goodman & Gilman’s The Pharmacological Basis of Therapeutics. Hardman J G, Limbird L E, editors. New York: McGraw-Hill; 1996. pp. 521–555. [Google Scholar]
- 14.Millan M J. Trends Pharmacol Sci. 1990;11:70–76. doi: 10.1016/0165-6147(90)90321-x. [DOI] [PubMed] [Google Scholar]
- 15.Rimoy G H, Wright D M, Bhaskar N K, Rubin P C. Eur J Clin Pharmacol. 1994;46:203–207. doi: 10.1007/BF00192549. [DOI] [PubMed] [Google Scholar]
- 16.Reece P A, Sedman A J, Rose S, Wright D S, Dawkins R, Rajagopalan R. J Clin Pharmacol. 1994;34:1126–1132. doi: 10.1002/j.1552-4604.1994.tb01991.x. [DOI] [PubMed] [Google Scholar]
- 17.Ishii K, Hein L, Kobilka B, Coughlin S R. J Biol Chem. 1993;268:9780–9786. [PubMed] [Google Scholar]
- 18.Meng F, Ueda Y, Hoversten M T, Thompson R C, Taylor L, Watson S J, Akil H. Eur J Pharmacol. 1996;311:285–292. doi: 10.1016/0014-2999(96)00431-1. [DOI] [PubMed] [Google Scholar]
- 19.Forsayeth J R, Garcia P D. BioTechniques. 1994;17:354–359. [PubMed] [Google Scholar]
- 20.Wong Y H. Methods Enzymol. 1994;238:81–94. doi: 10.1016/0076-6879(94)38008-2. [DOI] [PubMed] [Google Scholar]
- 21.Voyno-Yasenetskaya T A, Pace A M, Bourne H R. Oncogene. 1994;9:2559–2565. [PubMed] [Google Scholar]
- 22.Conklin B R, Farfel Z, Lustig K D, Julius D, Bourne H R. Nature (London) 1993;363:274–276. doi: 10.1038/363274a0. [DOI] [PubMed] [Google Scholar]
- 23.Xue J-C, Chen C, Zhu J, Kunapuli S P, de Riel J K, Yu L, Liu-Chen L-Y. J Biol Chem. 1995;270:12977–12979. [PubMed] [Google Scholar]
- 24.Varga E V, Li X, Stropova D, Zalewska T, Landsman R S, Knapp R J, Malatynska E, Kawai K, Mizusura A, Nagase H, Calderon S N, Rice K, Hruby V J, Roeske W R, Yamamura H I. Mol Pharmacol. 1996;50:1619–1624. [PubMed] [Google Scholar]
- 25.Kong H, Raynor K, Yano H, Takeda J, Bell G I, Reisine T. Proc Natl Acad Sci USA. 1994;91:8042–8046. doi: 10.1073/pnas.91.17.8042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hjorth S A, Thirstrup K, Grandy D K, Schwartz T W. Mol Pharmacol. 1995;47:1089–1094. [PubMed] [Google Scholar]
- 27.Valiquette M, Vu H K, Yue S Y, Wahlestedt C, Walker P. J Biol Chem. 1996;271:18789–18796. doi: 10.1074/jbc.271.31.18789. [DOI] [PubMed] [Google Scholar]
- 28.Peters G R, Ward N J, Antal E G, Lai P Y, deMaar E W. J Pharmacol Exp Ther. 1987;240:128–131. [PubMed] [Google Scholar]
- 29.Shaw J S, Carroll J A, Alcock P, Main B G. Br J Pharmacol. 1989;96:986–992. doi: 10.1111/j.1476-5381.1989.tb11911.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mansour A, Hoversten M T, Taylor L P, Watson S J, Akil H. Brain Res. 1995;700:89–98. doi: 10.1016/0006-8993(95)00928-j. [DOI] [PubMed] [Google Scholar]
- 31.Freidinger R M. Prog Drug Res. 1993;40:33–98. doi: 10.1007/978-3-0348-7147-1_4. [DOI] [PubMed] [Google Scholar]
- 32.Schwartz T W. Curr Opin Biotechnol. 1994;5:434–444. doi: 10.1016/0958-1669(94)90054-x. [DOI] [PubMed] [Google Scholar]
- 33.Gossen M, Freundlieb S, Bender G, Müller G, Hillen W, Bujard H. Science. 1995;268:1766–1769. doi: 10.1126/science.7792603. [DOI] [PubMed] [Google Scholar]
- 34.Gossen M, Bujard H. Proc Natl Acad Sci USA. 1992;89:5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Spencer D M. Trends Genet. 1996;12:181–187. doi: 10.1016/0168-9525(96)10013-5. [DOI] [PubMed] [Google Scholar]
- 36.Wang Y, DeMayo F J, Tsai S Y, O’Malley B W. Nat Biotechnol. 1997;15:239–243. doi: 10.1038/nbt0397-239. [DOI] [PubMed] [Google Scholar]
- 37.Spencer D M, Wandless T J, Schreiber S L, Crabtree G R. Science. 1993;262:1019–1024. doi: 10.1126/science.7694365. [DOI] [PubMed] [Google Scholar]
- 38.Farrar M A, Alberola-Ila J, Perlmutter R M. Nature (London) 1996;383:178–181. doi: 10.1038/383178a0. [DOI] [PubMed] [Google Scholar]
- 39.Fu L-X, Magnusson Y, Bergh C-H, Liljeqvist J Å, Waagstein F, Hjalmarson Å, Hoebeke J. J Clin Invest. 1993;91:1964–1968. doi: 10.1172/JCI116416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Strader C D, Gaffney T, Sugg E E, Candelore M R, Keys R, Patchett A A, Dixon R A. J Biol Chem. 1991;266:5–8. [PubMed] [Google Scholar]