Abstract
The development of targeted gene repair is under way and, despite some setbacks, shows promise as an alternative form of gene therapy. This approach uses synthetic DNA molecules to activate and direct the cell’s inherent DNA repair systems to correct inborn errors. The progress of this technique and its therapeutic potential are discussed in relation to the treatment of genetic diseases.
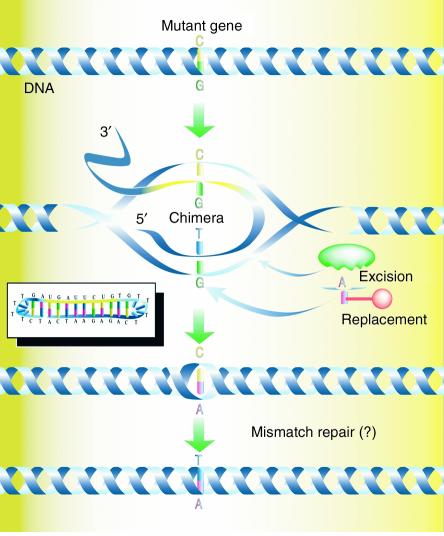
In 1996, we introduced the concept of targeted gene repair, a process in which a synthetic molecule directs the exchange of a single DNA base at a specific site in an episome or chromosome (refs. 1, 2; see ref. 3 for review). This approach is based on seminal work conducted by several groups (4–6) in which gene targeting was used to correct the human βs mutation and develop animal models for testing of novel therapies. In addition, Sherman and colleagues (7–9) provided the first clear evidence that DNA oligonucleotides could be used to alter single bases in yeast. The latter work employed a simpler type of targeting molecule compared with the more traditional vector used for gene targeting protocols. The convergence of these ideas led us to begin testing oligonucleotides in a variety of eukaryotic cells for nucleotide-alteration activity. The general pathway is envisioned as a nucleotide-exchange reaction directed by a double-stranded molecule known as the chimeric RNA/DNA oligonucleotide (chimera). The chimera (Figure 1) is a single-stranded molecule, usually 70–80 bases in length, with sequence complementarity so that it folds into a double-hairpin configuration. This configuration avoids nuclease digestion and concatenation of double-stranded molecules, two adverse molecular events that are common when these types of DNA structures are transfected into mammalian (and other eukaryotic) cells. The base composition includes both RNA and DNA nucleotides with three or four thymidine residues in the “cap turns.” The RNA bases stabilize the intermediate joint molecule on one strand, while the DNA portion of the molecule starts the single-base exchange event. Finally, a unique phosphodiester bond is left unligated so as to facilitate topological intertwining with the target site in the helix.
Figure 1.
Proposed mechanism of targeted gene repair directed by chimeric RNA/DNA oligonucleotides. The RNA/DNA chimera (inset) is a single-stranded oligonucleotide consisting of RNA and DNA residues. The molecule acts by annealing at the site in the target DNA, with the RNA section (yellow) hybridizing with perfect complementarity to one strand and the DNA stretch (blue) hybridizing to the other strand. A T/G base pair mismatch is formed and is acted upon by the cell’s DNA repair systems. In this case, the G residue is excised and replaced by an A to base pair with the T base provided by the chimera. The chimera dissociates, leaving behind a C/A mismatch, which is presumably corrected by the mismatch-repair system to generate a T•A base pair. Here, the action of the chimera directs the exchange of a C•G base pair with a T•A base pair. Reproduced with permission from Science’s STKE (46).
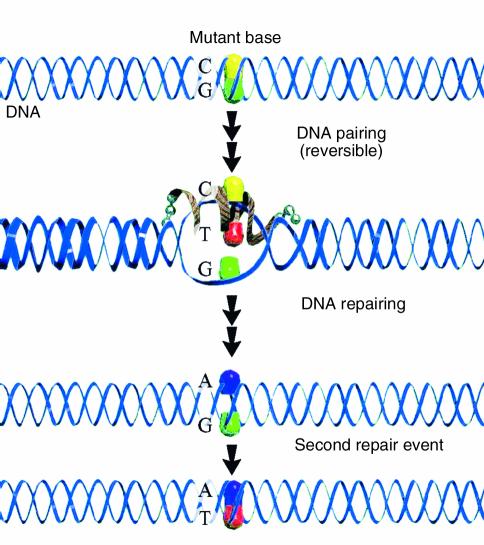
The mechanism of this reaction is currently being elucidated by biochemical (10–12) and genetic studies in yeast (13, 14) (see “Clinical use of gene repair: are we there yet?”). We now believe that the chimera directs nucleotide exchange in two distinct phases: DNA pairing and DNA repairing. The first phase consists of protein-promoted DNA hybridization between the vector (chimera) and its complementary target sequence in the target gene. Once aligned in homologous register, stable base pairs form, except for one predesigned mismatch located in the center of the joint complex. The mismatched base pair illustrated in Figure 1 is G•G. This structural perturbation, in the otherwise perfectly hybridized twin duplex, is recognized by the cell’s endogenous mismatch-repair system, which then catalyzes nucleotide exchange in the second phase, the process of DNA repairing. Once one mismatch is corrected, the chimera dissociates, leaving behind a single mismatched base pair in the original targeted helix. This structure is similar to the one recognized when mismatched base pairs are created during DNA replication. The normal repair pathway is then activated and generates an intact DNA helix. Figure 2 presents a model in which the same sequence of events takes place, except that the process is directed by a single-stranded oligonucleotide with specific terminal linkage modifications. DNA pairing is catalyzed by the same group of proteins, and DNA repair is again carried out by the process of mismatch repair.
Figure 2.
Targeted gene repair with modified single-stranded oligonucleotides. The mutant base is recognized by the single-stranded oligonucleotide, which is designed to hybridize to a region immediately surrounding the targeted base. At one specific site, a mismatch is created (C/T here) with the displacement of the complementary strand, which contains the G residue (G). DNA repair activity catalyzed by the cellular machinery inserts an A on the hybridized strand to pair with the T residue of the template. A second repair event completes the cycle by removing the G residue and replacing it with a T. In this case, a conversion of the G•C base pair to an A•T base pair results from the gene repair reaction. Reproduced with permission from Current Opinion in Molecular Therapeutics (47).
The first reports of successful application of chimera-directed gene repair came from several laboratories, including our own (2). Steer and colleagues pioneered the technique in liver cells, initially in an HUH7 cell line (15), followed by spectacular results in primary hepatocytes (16). Kren et al. then corrected the single base mutation responsible for Crigler-Najjar disease in the prototype Gunn rat model (17). These initial findings have been followed by a steady stream of independent confirmation in mammalian cells and animal models (Table 1). The versatility of the technique has been demonstrated by successful experiments in plant-cell and whole-plant models (Table 1).
Table 1.
The wide scope of gene repair applications.
The reaction to this technique from the gene therapy and gene targeting community was generally disbelief; many scientists believed that the results were most likely artifactual (18). Some went as far as to predict that no one would be able to use the technique independently. But others published impressive data, and independent confirmation followed shortly (19–23). Several misconceptions were then propagated, not the least of which was that no one had shown a phenotypic change. Kren et al. (16), Alexeev et al. (24, 25), Bartlett et al. (20), and Lai et al. (26), to name only a few, had, in fact, done so and had published their results in peer-reviewed journals. The question of reproducibility has plagued this technique, especially when the issue of the high frequencies of gene repair was discussed. Recent evidence published by Tagalakis et al. (27) showed even higher frequencies of gene repair using the same lymphoblastoid cell type used by Cole-Strauss et al. (2) in one of the original reports of this technique. Finally, some claimed that no clonal isolates of permanently altered cells had been produced. As early as 1998, Alexeev and colleagues (24, 25) had clearly demonstrated clonal inheritance, as had the 1998 and 1999 reports of gene repair in plants (28–30).
Clinical use of gene repair
We’ve come a long way in the clinical use of gene repair. Some valid questions do, however, remain unanswered and continue to perplex investigators. The most important one centers on the variability among the data of different research groups. Frequencies of correction differ widely, often ranging from 0.5% to 20% even within the same laboratory. Many current efforts aim primarily to explain this variability.
Over the past three years, our laboratory took a decidedly reductionist approach to uncover the sources of variability. Presently, we believe that the efficiency of transfection (or vector uptake into cells) is a key parameter (31, 32). The phase of the cell cycle in which the vector enters the cell also affects gene repair levels, and the cellular level of DNA recombination/repair proteins can also influence the success of gene repair in a positive (enhancing) or negative (suppressive) fashion. Finally, the capacity to produce the full-length, double-stranded chimera at a level of purity at which breakdown products and/or incompletely synthesized products do not interfere with the reaction is critical for obtaining positive results.
The last of these rules prompted us to develop a new vector that could direct gene repair as precisely and efficiently as the chimera, but that had a simplified synthesis protocol. Studies by Gamper et al. (11, 12, 33) indicated that single-stranded DNA with a specific number of linkage modifications appeared to catalyze gene repair efficiently and was easier to synthesize and purify. Both the chimera and the new single-stranded design have been tested in the genetically tractable organism Saccharomyces cerevisiae (34, 35). While this may seem to be regressive, repair proteins in yeast function similarly to their human counterparts. However, the influence of individual protein components on the gene repair process can be assessed using genetic-screening techniques in yeast, but not human cells. Thus, using this simplified vector and the power of yeast genetics, we have now established several tenets of gene repair. (a) There is a clear dose response to gene repair that corresponds to the amount of vector added to the reaction, but there is also a maximum dose above which the repair efficiency falls off precipitously. (b) An optimal length of the vector exists between 70 and 80 bases on the DNA strand; extension of this length to 100 nucleotides reduces repair frequency. (c) Gene repair initiated on the nontranscribed strand is five- to tenfold better than similar events initiated on the transcribed strand. This difference has been estimated to be 1,000-fold (21, 22), but our data suggest a more conservative advantage. (d) Members of the Rad52 epistasis group, a group of genes involved in the repair of double-stranded chromosomal breaks, regulate the gene repair process and probably control the rate-limiting step. Overexpression of some of these genes increases gene repair activity ten- to 20-fold. (e) Finally, a number of DNA repair proteins act as suppressors of the reaction, disrupting the gene repair process and reducing the genetic readout. Studies in yeast have helped define clear roles for proteins such as Rad51, Rad52, Rad54, and Mre11 (36, 37). Mechanistic analyses of the gene repair reaction have demonstrated the importance of DNA pairing in the overall success of the reaction. The choice of which strand to target is an important one, since both transcription and replication processes can modulate the stability of the oligonucleotide bound at the specific site (refs. 38 and 39; E.E. Brachman and E.B. Kmiec, unpublished observations).
There is really only one way to “cure” a genetic disease, and that is to correct the gene at the site of the mutation in the chromosome or at the level of mRNA. Great progress is still being made using eukaryotic viruses as gene therapy vectors to reverse the phenotype of certain disease states. There is little doubt that some of these protocols will result in significant clinical benefit. But the death of Jesse Gelsinger at the University of Pennsylvania in 1999 (40) should serve as a wake-up call to this field. Mr. Gelsinger was a participant in a gene therapy clinical trial that used an adenoviral vector to deliver a corrective gene for a genetic disorder. The operational idea was that the addition of copies of the normal gene transported by a modified virus would alleviate or reverse the disorder. But the amount of virus led to a massive immunological and toxic response, which led to Mr. Gelsinger’s death shortly after it was administered. The serious risks of viral gene therapy must be first openly addressed and then overcome. We hope that novel approaches, such as DNA or RNA gene repair, will be afforded an equal chance of success and that the reasonable levels of financial support provided to viral approaches will be made available to gene repair applications.
In relation to bringing gene repair into the scientific mainstream, however, a number of good things are happening. First, a consortium of scientists, primarily from Europe, is meeting regularly in an open forum to discuss gene repair applications. Second, the subject of gene repair is appearing in a few scientific programs and meetings that primarily focus on the biological use of oligonucleotides. Unfortunately, the key word here is “few.” Third, reports of new approaches to or variations of gene repair (21, 41–45) are beginning to be published in major journals. Doubt still remains within the scientific arena regarding the reality of gene repair emerging as a true gene therapy, but the skepticism is reduced incrementally as the number of successful reports of its application increase. I am reminded of some advice a famous cancer researcher once gave me regarding my work in this area: “Keep pioneering this idea, just in case it works.” I hope we all do, just in case.
Footnotes
Conflict of interest: The author has declared that no conflict of interest exists.
References
- 1.Yoon K, Cole-Strauss A, Kmiec EB. Targeted gene correction of episomal DNA in mammalian cells mediated by a chimeric RNA.DNA oligonucleotide. Proc. Natl. Acad. Sci. U. S. A. 1996;93:2071–2076. doi: 10.1073/pnas.93.5.2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cole-Strauss A, et al. Correction of the mutation responsible for sickle cell anemia by an RNA-DNA oligonucleotide. Science. 1996;273:1386–1389. doi: 10.1126/science.273.5280.1386. [DOI] [PubMed] [Google Scholar]
- 3.Kmiec EB. Targeted gene repair. Gene Ther. 1999;6:1–3. doi: 10.1038/sj.gt.3300789. [DOI] [PubMed] [Google Scholar]
- 4.Shesely EG, et al. Correction of a human beta S-globin gene by gene targeting. Proc. Natl. Acad. Sci. U. S. A. 1991;88:4294–4298. doi: 10.1073/pnas.88.10.4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Snouwaert JN. An animal model for cystic fibrosis made by gene targeting. Science. 1992;257:1083–1088. doi: 10.1126/science.257.5073.1083. [DOI] [PubMed] [Google Scholar]
- 6.Smithies O, Maeda N. Gene targeting approaches to complex genetic diseases: atherosclerosis and essential hypertension. Proc. Natl. Acad. Sci. U. S. A. 1995;92:5266–5272. doi: 10.1073/pnas.92.12.5266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moerschell RP, Tsunasawa S, Sherman F. Transformation of yeast with synthetic oligonucleotides. Proc. Natl. Acad. Sci. U. S. A. 1988;85:524–528. doi: 10.1073/pnas.85.2.524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yamamoto T, Moerschell RP, Wakem LP, Komar-Panicucci S, Sherman F. Strand-specificity in the transformation of yeast with synthetic oligonucleotides. Genetics. 1992;131:811–819. doi: 10.1093/genetics/131.4.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yamamoto T, Moerschell RP, Wakem LP, Ferguson D, Sherman F. Parameters affecting the frequencies of transformation and co-transformation with synthetic oligonucleotides in yeast. Yeast. 1992;8:935–948. doi: 10.1002/yea.320081104. [DOI] [PubMed] [Google Scholar]
- 10.Cole-Strauss A, et al. Targeted gene repair directed by the chimeric RNA/DNA oligonucleotide in a mammalian cell-free extract. Nucleic Acids Res. 1999;27:1323–1330. doi: 10.1093/nar/27.5.1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gamper HB, et al. A plausible mechanism for gene correction by chimeric oligonucleotides. Biochemistry. 2000;39:5808–5816. doi: 10.1021/bi9921891. [DOI] [PubMed] [Google Scholar]
- 12.Gamper HB, et al. The DNA strand of chimeric RNA/DNA oligonucleotides can direct gene repair/conversion activity in mammalian and plant cell-free extracts. Nucleic Acids Res. 2000;28:4332–4339. doi: 10.1093/nar/28.21.4332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu H, Fisher RP, Bailey P, Levine AJ. The CDK7-cycH-p36 complex of transcription factor IIH phosphorylates p53, enhancing its sequence-specific DNA binding activity in vitro. Mol. Cell. Biol. 1997;17:5923–5934. doi: 10.1128/mcb.17.10.5923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rice MC, Czymmek K, Kmiec EB. The potential of nucleic acid repair in functional genomics. Nat. Biotechnol. 2001;19:321–326. doi: 10.1038/86701. [DOI] [PubMed] [Google Scholar]
- 15.Kren BT, Cole-Strauss A, Kmiec EB, Steer CJ. Targeted nucleotide exchange in the alkaline phosphatase gene of HuH-7 cells mediated by a chimeric RNA/DNA oligonucleotide. Hepatology. 1997;25:1462–1468. doi: 10.1002/hep.510250626. [DOI] [PubMed] [Google Scholar]
- 16.Kren BT, Bandyopadhyay P, Steer CJ. In vivo site-directed mutagenesis of the factor IX gene by chimeric RNA/DNA oligonucleotides. Nat. Med. 1998;4:285–290. doi: 10.1038/nm0398-285. [DOI] [PubMed] [Google Scholar]
- 17.Kren BT, et al. Correction of the UDP-glucuronosyltransferase gene defect in the gunn rat model of crigler-najjar syndrome type I with a chimeric oligonucleotide. Proc. Natl. Acad. Sci. U. S. A. 1999;96:10349–10354. doi: 10.1073/pnas.96.18.10349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thomas KR, Capecchi MR. Recombinant DNA technique and sickle cell anemia research. Science. 1997;275:1404–1405. doi: 10.1126/science.275.5305.1401e. [DOI] [PubMed] [Google Scholar]
- 19.Bartlett RJ. Long-lasting gene repair. Nat. Biotechnol. 1998;16:1312–1313. doi: 10.1038/4274. [DOI] [PubMed] [Google Scholar]
- 20.Bartlett, R.J., et al. 1998. Targeted reversion of the mdx point mutation. Miami Nature Biotechnology Winter Symposium Short Reports. Volume 9. Advances in gene technology: molecular biology in the conquest of disease. Oxford University Press. Oxford, United Kingdom. 316. (Abstr.)
- 21.Igoucheva O, Peritz AE, Levy D, Yoon K. A sequence-specific gene correction by an RNA-DNA oligonucleotide in mammalian cells characterized by transfection and nuclear extract using a lacZ shuttle system. Gene Ther. 1999;6:1960–1971. doi: 10.1038/sj.gt.3301042. [DOI] [PubMed] [Google Scholar]
- 22.Igoucheva O, Alexeev V, Yoon K. Targeted gene correction by small single-stranded oligonucleotides in mammalian cells. Gene Ther. 2001;8:391–399. doi: 10.1038/sj.gt.3301414. [DOI] [PubMed] [Google Scholar]
- 23.Lai L-W, Lien YH. Homologous recombination based gene therapy. Exp. Nephrol. 1999;7:11–14. doi: 10.1159/000020578. [DOI] [PubMed] [Google Scholar]
- 24.Alexeev V, Yoon K. Stable and inheritable changes in genotype and phenotype of albino melanocytes induced by an RNA-DNA oligonucleotide. Nat. Biotechnol. 1998;16:1343–1346. doi: 10.1038/4322. [DOI] [PubMed] [Google Scholar]
- 25.Alexeev V, Igoucheva O, Domashenko A, Cotsarelis G, Yoon K. Localized in vivo genotypic and phenotypic correction of the albino mutation in skin by RNA-DNA oligonucleotide. Nat. Biotechnol. 2000;18:43–47. doi: 10.1038/71901. [DOI] [PubMed] [Google Scholar]
- 26.Lai, L.-W., Doty, D., Khan, R., Omeara, M., and Lien, Y.-H. Conversion of a single base pair mutation in newborn carbonic anhydrase II deficient mice by chimeric RNA/DNA oligonucleotides. Abstract presented at: 4th Annual Meeting of the American Society of Gene Therapy, May 28–31, 2001, Seattle, Washington, USA.
- 27.Tagalakis AD, Graham IR, Riddell DR, Dickson JG, Owen JS. Gene correction of the apolipoprotein (Apo) E2 phenotype to wild-type ApoE3 by in situ chimeraplasty. J. Biol. Chem. 2001;276:13226–13230. doi: 10.1074/jbc.C000883200. [DOI] [PubMed] [Google Scholar]
- 28.Beetham PR, Kipp PB, Sawycky XL, Arntzen CJ, May GD. A tool for functional plant genomics: chimeric RNA/DNA oligonucleotides cause in vivo gene-specific mutations. Proc. Natl. Acad. Sci. U. S. A. 1999;96:8774–8778. doi: 10.1073/pnas.96.15.8774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhu T, et al. Targeted manipulation of maize genes in vivo using chimeric RNA/DNA oligonucleotides. Proc. Natl. Acad. Sci. U. S. A. 1999;96:8768–8773. doi: 10.1073/pnas.96.15.8768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhu T, Mettenburg K, Peterson DJ, Tagliani L, Baszczynski CL. Engineering herbicide-resistant maize using chimeric RNA/DNA oligonucleotides. Nat. Biotechnol. 2000;18:555–558. doi: 10.1038/75435. [DOI] [PubMed] [Google Scholar]
- 31.Bandyopadhyay P, Kren BT, Ma X, Steer CJ. Enhanced gene transfer into HuH-7 cells and primary rat hepatocytes using targeted liposomes and polyethylenimine. Biotechniques. 1998;25:282–292. doi: 10.2144/98252gt03. [DOI] [PubMed] [Google Scholar]
- 32.Bandyopadhyay P, Ma X, Linehan-Stieers C, Kren BT, Steer CJ. Nucleotide exchange in genomic DNA of rat hepatocytes using RNA/DNA oligonucleotides. Targeted delivery of liposomes and polyethyleneimine to the asialoglycoprotein receptor. J. Biol. Chem. 1999;274:10163–10172. doi: 10.1074/jbc.274.15.10163. [DOI] [PubMed] [Google Scholar]
- 33.Gamper HB, Hou YM, Kmiec EB. Evidence for a four-strand exchange catalyzed by the RecA protein. Biochemistry. 2000;39:15272–15281. doi: 10.1021/bi001704o. [DOI] [PubMed] [Google Scholar]
- 34.Liu L, Rice MC, Kmiec EB. In vivo gene repair of point and frameshift mutations directed by chimeric RNA/DNA oligonucleotides and modified single-stranded oligonucleotides. Nucleic Acids Res. 2001;29:4238–4250. doi: 10.1093/nar/29.20.4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rice MC, Bruner M, Czymmek K, Kmiec EB. In vitro and in vivo nucleotide exchange directed by chimeric RNA/DNA oligonucleotides in Saccharomyces cerevisiae. Mol. Microbiol. 2001;40:857–868. doi: 10.1046/j.1365-2958.2001.02407.x. [DOI] [PubMed] [Google Scholar]
- 36.Liu L, Cheng S, van Brabant AJ, Kmiec EB. Rad51p and Rad54p, but not Rad52p, elevate gene repair in Saccharomyces cerevisiae directed by modified single-stranded oligonucleotide vectors. Nucleic Acids Res. 2002;31:2742–2750. doi: 10.1093/nar/gkf397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu, L., Hu, Y., Usher, M., and Kmiec, E.B. 2003. The nuclease activity of Saccharomyces cerevisiae Mre11 functions in targeted nucleotide alteration. Appl. Environ. Microbiol. In press. [DOI] [PMC free article] [PubMed]
- 38.Liu L, et al. Strand bias in targeted gene repair is influenced by transcriptional activity. Mol. Cell. Biol. 2002;22:3852–3863. doi: 10.1128/MCB.22.11.3852-3863.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ellis HM, Yu D, DiTizio T, Court DL. High efficiency mutagenesis, repair, and engineering of chromosomal DNA using single-stranded oligonucleotides. Proc. Natl. Acad. Sci. U. S. A. 2001;98:6742–6746. doi: 10.1073/pnas.121164898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Friedman T. Changing roles for academia and industry in genetics and gene therapy. Mol. Ther. 2000;1:9–12. doi: 10.1006/mthe.1999.0015. [DOI] [PubMed] [Google Scholar]
- 41.Culver KW, et al. Correction of chromosomal point mutations in human cells with bifunctional oligonucleotides. Nat. Biotechnol. 1999;17:989–993. doi: 10.1038/13684. [DOI] [PubMed] [Google Scholar]
- 42.Guo H, et al. Group II introns designed to insert into therapeutically relevant DNA target sites in human cells. Science. 2000;289:452–457. doi: 10.1126/science.289.5478.452. [DOI] [PubMed] [Google Scholar]
- 43.Wilton SD, et al. Specific removal of the nonsense mutation from the mdx dystrophin mRNA using antisense oligonucleotides. Neuromuscul. Disord. 1999;9:330–338. doi: 10.1016/s0960-8966(99)00010-3. [DOI] [PubMed] [Google Scholar]
- 44.Smulevitch SV, Simmons CG, Norton JC, Wise TW, Corey DR. Enhancement of strand invasion by oligonucleotides through manipulation of backbone charge. Nat. Biotechnol. 1996;14:1700–1704. doi: 10.1038/nbt1296-1700. [DOI] [PubMed] [Google Scholar]
- 45.Lan N, Howrey RP, Lee SW, Smith CA, Sullenger BA. Ribozyme-mediated repair of sickle beta-globin mRNAs in erythrocyte precursors. Science. 1998;280:1593–1596. doi: 10.1126/science.280.5369.1593. [DOI] [PubMed] [Google Scholar]
- 46.Parekh-Olmedo H, Czymmek K, Kmiec Eric B. Targeted gene repair in mammalian cells using chimeric RNA/DNA oligonucleotides and modified single-stranded vectors. Sci. STKE. 2001;73:2–3. doi: 10.1126/stke.2001.73.pl1. [DOI] [PubMed] [Google Scholar]
- 47.Brachman EE, Kmiec EB. The ‘biased’ evolution of targeted gene repair. Curr. Opin. Mol. Ther. 2002;4:171–176. [PubMed] [Google Scholar]
- 48.Bartlett RJ, et al. In vivo targeted repair of a point mutation in the canine dystrophin gene by a chimeric RNA/DNA oligonucleotide. Nat. Biotechnol. 2000;18:615–622. doi: 10.1038/76448. [DOI] [PubMed] [Google Scholar]
- 49.Bertoni C, Rando TA. Dystrophin gene repair in mdx muscle precursor cells in vitro and in vivo mediated by RNA-DNA chimeric oligonucleotides. Hum. Gene Ther. 2002;13:707–718. doi: 10.1089/104303402317322276. [DOI] [PubMed] [Google Scholar]
- 50.Rando TA, Disatnik MH, Zhou LZ. Rescue of dystrophin expression in mdx mouse muscle by RNA/DNA oligonucleotides. Proc. Natl. Acad. Sci. U. S. A. 2000;97:5363–5368. doi: 10.1073/pnas.97.10.5363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lai, L.-W., O'Connor, Y.-H., and Lien, Y.-H. Correction of carbonic anhydrase II mutation in renal tubular cells by chimeric RNA/DNA oligonucleotide. Abstract presented at: 1st Annual Meeting of the American Society of Gene Therapy, May 28-31,1998, Seattle, Washington, USA.
- 52.Lai, L.-W., Chua, B., and Lien, Y.-H. In vivo gene targeting in carbonic anhydrase II deficient mice by chimeric RNA/DNA oligonucleotides. 2nd Annual Meeting of the American Society of Gene Therapy, June 6–9, 1999, Washington, DC, USA. p. 236a (Abstr.)
- 53.Li ZH, Liu DP, Yin WX, Guo ZC, Liang CC. Targeted correction of the point mutations of beta-thalassemia and targeted mutagenesis of the nucleotide associated with HPFH by RNA/DNA oligonucleotides: potential for beta-thalassemia gene therapy. Blood Cells Mol. Dis. 2001;27:530–538. doi: 10.1006/bcmd.2001.0416. [DOI] [PubMed] [Google Scholar]
- 54.Tran ND, et al. Efficiency of chimeraplasts gene targeting by direct nuclear injection using a GFP recovery assay. Mol. Ther. 2003;7:248–253. doi: 10.1016/s1525-0016(02)00039-4. [DOI] [PubMed] [Google Scholar]
- 55.Rice MC, May GD, Kipp PB, Parekh H, Kmiec EB. Genetic repair of mutations in plant cell-free extracts directed by specific chimeric oligonucleotides. Plant Physiol. 2000;123:427–438. doi: 10.1104/pp.123.2.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brown BD, Fraser JL, Davey S, Lillicrap D. Targeting a single nucleotide exchange reaction by use of a small oligonucleotide in mammalian and yeast cell-free extracts. Mol. Ther. 2001;3:S402. (Abstr.) [Google Scholar]